Abstract
The characterization of an oxyR insertion mutant provides evidences that katA, which encodes the unique H2O2-inducible HPII catalase, is regulated by OxyR not only in free-living Sinorhizobium meliloti but also in symbiotic S. meliloti. Moreover, oxyR is expressed independently of exogenous H2O2 and downregulates its own expression in S. meliloti.
Sinorhizobium meliloti is a ubiquitous soil α-proteobacterium able to establish symbiosis with alfalfa (Medicago sativa) and related legumes, characterized by the formation of root nodules. The exchange of sophisticated recognition signals between the plant and the bacteria leads to the entering of the bacteria into the root hairs and to the development of primordial cells in the cortex, where the bacteria are released (21, 37). Inside the nodule, the bacteria differentiate into their symbiotic form, the bacteroids, which are able to reduce nitrogen to ammonia; the ammonia is then assimilated by the plant. The key enzyme of nitrogen fixation, the nitrogenase, is subjected to a fragile equilibrium. To avoid its rapid and irreversible inactivation by oxygen, a diffusion barrier in the cortex of nodules limits permeation by oxygen (40), and the plant oxygen carrier, leghemoglobin, delivers the necessary oxygen to the bacteroids (9). Nevertheless, a high respiration rate is required to support the nitrogen fixation process, and this leads to the generation of large amounts of reactive oxygen species (ROS) such as superoxide radicals (O2−) and hydrogen peroxide (H2O2), which can also inactivate the nitrogenase (31). ROS have also been detected in nodules (34); H2O2 accumulation all around bacteria was observed in some infection threads but never inside bacteria or bacteroids, indicating that they contain an efficient H2O2-scavenging system.
To cope with H2O2, S. meliloti possesses three catalases encoded by three different genes: two monofunctional catalases (HPII), KatA (12) and KatC (36), and one bifunctional catalase-hydroperoxidase (HPI), KatB (12). The catalase genes are differentially expressed during free-living growth, oxidative stress, and nodule establishment (13). katA expression has been detected during the exponential growth phase of free-living bacteria only, and katA is the unique catalase gene inducible by exogenous H2O2. In Escherichia coli, inducibility by H2O2 and expression in exponential phase have been observed for the katG catalase gene encoding the catalase-hydroperoxidase HPI (38). The H2O2 induction of katG requires the positive activator OxyR (26), which directly senses oxidative stress (18, 41).
Analysis of the oxyR-katA genetic region.
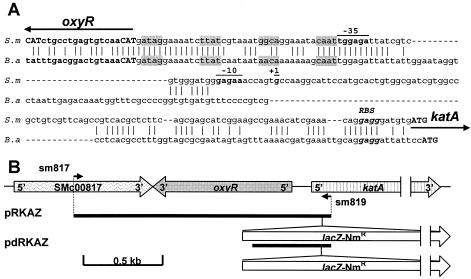
The complete genome sequence of S. meliloti (10) revealed a putative oxyR gene (SMc00818) in front of katA. The oxyR homologous gene in S. meliloti is located 193 bp upstream of and in the strand opposite to katA. The regulation of an HPII-like catalase by OxyR has been described for Brucella abortus only (17). The alignment of the oxyR-katA intergenic regions from S. meliloti and B. abortus (Fig. 1A) revealed highly conserved regions, especially in the DNA-binding site described for B. abortus (16). The S. meliloti sequence (ATAG-N7-TTAT-N7-GGCA-N7-CAAT) is identical to the B. abortus sequence, except that GGCA is replaced by AACA. Moreover, the predicted OxyR amino acid sequences (317 amino acids) for S. meliloti and B. abortus showed 53% identity, and their alignment indicated the location of the OxyR ATG initiation codon of S. meliloti to be 18 bp upstream of the annotated translational start codon. Six amino acids (MLTLRQ) were added to the N-terminal region, four of them being identical in both bacteria (MXTXRQ). Thus, the newly annotated oxyR gene was determined to be located 175 bp upstream of katA and would encode a 317-amino-acid protein in S. meliloti. The two critical cysteines, C200 and C209, which were implicated in the activation of OxyR, are conserved in the OxyR of S. meliloti.
FIG. 1.
(A) Alignment of the oxyR-katA intergenic sequences of S. meliloti (S.m) and B. abortus (B.a). The OxyR DNA-binding site from B. abortus is shaded grey. Initial codons from katA and oxyR are indicated in bold capitals. KatA ribosome binding sites (RBS) are indicated in bold italics. The positions of the katA promoter from S. meliloti are underlined and in bold (+1, −10, and −35 regions). (B) Genetic map of the oxyR-katA region and the pRKAZ and pdRKAZ plasmids used in this study. Positions of the primers used for their construction are indicated. Large arrows indicate the locations and directions of transcription of the identified genes. Small arrows indicate the directions and positions of primers.
Construction of an oxyR insertion mutant and its role in the adaptation to H2O2.
To determine the role of oxyR in the H2O2 response in S. meliloti, an oxyR mutant, named RmoxyR, was constructed using the parental strain Rm1021 by use of insertional inactivation with a uidA transcriptional fusion. Genomic DNA from strain Rm1021 was amplified by PCR with the oligonucleotides sm818a (5′ CTC GCG GAT GTC GGC AGA TTG G) and sm818b (5′ AAA ACA GCG CCC GGG TAA CGA T) (Fig. 1A). The PCR fragment (271 bp) was cloned into the pCR-TOPO vector (Invitrogen), which was followed by a double digestion with BamHI and XbaI in order to clone this fragment upstream of the present uidA gene into the pVO155 suicide vector (29). The mutation was introduced by a single reciprocal recombination into the chromosome of S. meliloti Rm1021 by triparental conjugation as described previously (11). The disruption of the oxyR gene was confirmed by Southern blotting using vector- and gene-specific probes (data not shown). The bacterial strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacteria | ||
| Sinorhizobium meliloti | ||
| RCR2011 | SU47, wild type; Nod+ Fix+ | 33 |
| Rm1021 | Derivative of RCR2011; Smr | 23 |
| RmoxyR | Same as Rm1021 but with oxyR::uidA fusion (oxyR); Smr Nmr | This study |
| 1021-pRKAZ | Same as Rm1021 but with plasmid pRKAZ | This study |
| 1021-pdRKAZ | Same as Rm1021 but with plasmid pdRKAZ | This study |
| oxyR-pRKAZ | Same as RmoxyR but with plasmid pRKAZ | This study |
| oxyR-pdRKAZ | Same as RmoxyR but with plasmid pdRKAZ | This study |
| Escherichia coli | ||
| DH5α | F−supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17(rK−mK+) recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| MT607 | pro-82 thi-1 hsdR17 supE44 recA56 | 8 |
| MT616 | MT607(pRK600) | 8 |
| Plasmids | ||
| pRK600 | ColE1 replicon with RK2 transfer region; Cmr | 8 |
| pBluescriptKS(+) | Derivative of pUC19 with f1(+)oriR; Apr | Stratagene |
| pCR-TOPO | Derivative of pUC19 with f1(+)ori; Apr Kmr | Invitrogen |
| pGEM-T | T vector with f1 oriR; Apr | Promega Corp. |
| pVO155 | Derivative of pUC119 | 29 |
| pKOK5 | Derivative of pSUP202, source of lacZ-Kmr; Apr Kmr | 19 |
| pBBR1-MCS-3 | Derivative of pBBR1-MCS; Tcr | 20 |
| pCR-TOPO-oxyR01 | pCR-TOPO vector with 271-bp PCR-amplified fragment of the oxyR gene | This study |
| pCR-TOPO-oxyR02 | pCR-TOPO vector with 1,780-bp PCR-amplified fragment with the oxyR gene and partial katA gene | This study |
| pGEMoxyR | pGEM-T, with 920-bp PCR-amplified fragment | This study |
| pVO-oxyR | pVO155, with BamHI-XbaI fragment of pCR-TOPO-oxyR01 | This study |
| pBBR-oxyR | pBBR IMCS-3, with SpeI-Apal fragment of pCR-TOPO-oxyR02 | This study |
| pBBR-doxyR | pBBR-oxyR, Sacl-Sacl fragment deleted | This study |
| pRKAZ | pBBR-oxyR, with fusion pkatA-lacZ | This study |
| pdRKAZ | pBBR-doxyR, with fusion pkatA-lacZ | This study |
| pBSKAI-1 | pBluescript, 1.4-kb EcoRI-ApaI fragment with partial katA | 12 |
Abbreviations: Tc, tetracycline; Sm, streptomycin; Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol; Sp, spectinomycin.
Growth inhibition of strain RmoxyR by H2O2 was tested by the halo assay method, which gives a long-term test of resistance to H2O2 exposure. Aliquots (200 μl each) of S. meliloti cultures with optical densities at 600 nm (OD600) of 0.4 to 0.5 were plated onto Luria-Bertani-MC agar plates (12). Paper disks (6-mm diameter) were impregnated with 5 μl of H2O2 (1 M) and placed in the center of the S. meliloti plates. After 2 to 3 days of incubation at 30°C, the diameters of the complete growth inhibition zone averaged 26 and 49 mm for the wild-type Rm1021 strain and the RmoxyR mutant, respectively. The RmoxyR strain complemented with the pBBR-oxyR plasmid carrying the complete oxyR gene showed a growth inhibition zone of 25 mm. To construct the pBBR-oxyR plasmid, a PCR fragment carrying the oxyR gene, the complete intergenic oxyR-katA region, and the beginning of the katA gene was amplified using the oligonucleotides sm819 (5′GCGCTCGCGGTTCTGGTG) and sm817 (5′CTTGCGCCCGATTTCCTGTC), subcloned into the pCR-TOPO vector, and cloned in the vector pBBRMCS-3 by use of the SpeI and ApaI sites. In this assay, strain RmoxyR appears to be more sensitive to H2O2 than its parental strain, indicating that oxyR plays an important role in the protection of S. meliloti against the activated species.
To further investigate the role of OxyR in the adaptation of S. meliloti to H2O2, strains Rm1021 and RmoxyR in exponential phase (OD600, 0.4 to 0.5) were pretreated for 1 h or not with a sublethal dose of H2O2 (1 mM), which is known to activate OxyR in Escherichia coli (41). Cultures were then treated with a lethal dose of H2O2 (20 mM) for various times, and survival was assessed by plating dilutions onto Luria-Bertani-MC agar. With the pretreatment, strain Rm1021 was more resistant to H2O2 than RmoxyR, indicating that oxyR is required for adaptation to H2O2. In contrast, the pretreatment had no effect on the survival of RmoxyR to H2O2 treatment. Unexpectedly, without H2O2 pretreatment, the RmoxyR mutant was more resistant to H2O2 than the parental strain. A similar pattern has been observed for an oxyR mutant of B. abortus, which was more resistant to H2O2 than a wild-type strain when bacteria were not pretreated with H2O2 (16).
oxyR regulates katA expression in free-living conditions.
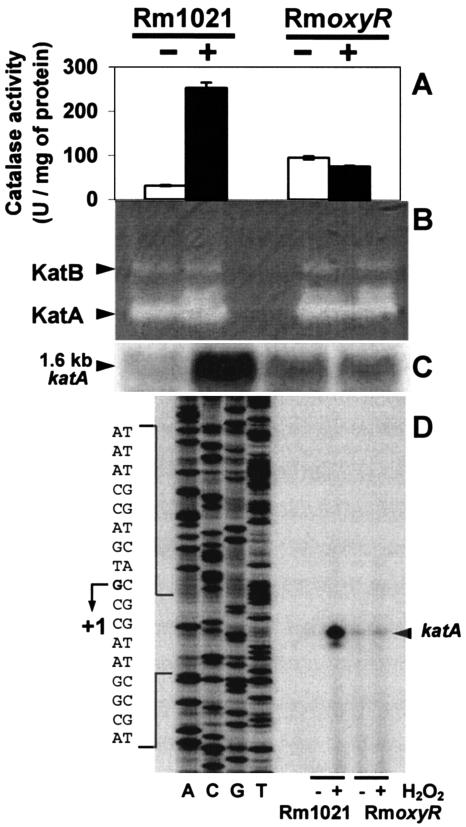
KatA is the major catalase component of an adaptive response to H2O2 (12). To confirm that the results described above were due to KatA deregulation, the RmoxyR mutant and its parental strain were analyzed for total catalase activity spectrophotometrically (Fig. 2A) by monitoring the decomposition of H2O2 at 240 nm (15) and on a native polyacrylamide gel by use of negative diaminobenzidine staining (Fig. 2B) as previously described (5, 12) with or without H2O2 pretreatment. It clearly appeared that the increases in total catalase activity under the different conditions (Fig. 2A) were due solely to KatA, not to KatB or KatC (Fig. 2B). Upon exposure to H2O2, no increase in KatA level was observed in the RmoxyR mutant strain, confirming the hypothesis that OxyR acts as an activator of KatA upon H2O2 exposure. Moreover, the basal KatA level in RmoxyR was higher than that in the parental strain Rm1021 without H2O2 treatment, which is consistent with the higher resistance observed in strain RmoxyR. No difference was observed in catalase activity patterns and in total catalase activities of Rm1021 and RmoxyR with or without H2O2 pretreatment in late stationary phase (data not shown). To verify that KatA deregulations were due to modifications of katA transcription, the accumulation of katA transcripts was quantified in Rm1021 and RmoxyR by Northern blotting (Fig. 2C). RNAs were isolated and separated by electrophoresis as previously described (1, 3), blotted on a nylon membrane, and probed with a 32P-labeled katA probe corresponding to a 450-bp EcoRI-PstI fragment from pBSKA1-1 (12). With or without H2O2 treatment, a single 1.6-kb hybridization band was detected in each strain (Fig. 2C). Variations in katA RNA levels, measured with a phosphorus imager, and total catalase activities (Fig. 2A) were perfectly correlated, indicating that the observed deregulation is essentially at the transcriptional level. To determine the katA transcription start site under oxidative or nonoxidative conditions, primer extension experiments were performed using RNAs from Rm1021 and RmoxyR treated or not with H2O2 (Fig. 2D) as described at the Long laboratory website (http://cmgm.stanford.edu/biology/long/protocols.htm#primer), by using primer pextkatA (5′ GGTGGTGGTGATCGTCGGACGATCTGTCAT), which is specific to katA labeled with [γ-32P]dATP. DNA sequencing was performed using a CycleReader DNA sequencing kit (MBI Fermentas) with [α-33P]dCTP. The transcription start site is located at G (Fig. 2D), corresponding to a position 103 bp upstream of the katA translational start codon, ATG, which is consistent with the hypothetical −35 (TGGAGA) and −10 (GAGAA) boxes and the OxyR binding site (Fig. 1A). The different intensities of the signal for primer extension analysis were also in agreement with the Northern analysis, and no change in the +1 position was observed under the different conditions tested.
FIG. 2.
Effects of H2O2 on catalase activity and katA gene expression in the parental Rm1021 and the mutant RmoxyR strains. Bacteria were treated (+) or not (−) with 1 mM H2O2 for 1 h. Cell extracts were prepared and analyzed for catalase activities spectrophotometrically (A) and on a native polyacrylamide gel (B), using 30 μg of protein per lane and determined with a protein assay kit (Bio-Rad Laboratories GmbH). The positions of KatA and KatB are indicated as described by Sigaud et al. (36). Catalase activities were obtained with triplicate samples from two independent experiments and are given in units per milligram of protein. Data are presented as the means ± standard deviations of results. The expression of the katA gene was monitored using Northern blot (C) and primer extension (D) analyses.
An oxyR gene has been identified in 31 bacterial genomes (6, 27). The position of oxyR is very variable in the different bacterial genomes, but it is often next to a gene involved in oxidative stress protection and regulated by OxyR: for example, ahpC in Mycobacterium tuberculosis (7) and Xanthomonas campestris pv. phaseoli (25), dps in Bacteroides fragilis (32), oxyS in E. coli (4), recG in Pseudomonas aeruginosa (28), kat in B. abortus (17). Similar genomic relationships between oxyR and catalase genes are observed for most members of the Rhizobiales studied so far, such as S. meliloti, Agrobacterium tumefaciens, B. abortus, Mesorhizobium loti, Rhizobium etli, and Rhizobium leguminosarum bv. phaseoli. Nevertheless, OxyR potentially regulates a kat gene encoding a monofunctional catalase (HPII) in S. meliloti and B. abortus only, whereas in other members of the Rhizobiales, oxyR is localized in front of a catalase gene encoding a bifunctional catalase-hydroperoxidase (HPI).
oxyR expression under free-living conditions.
To analyze the expression of the oxyR gene and its regulation by H2O2, we monitored the level of β-glucuronidase activity in the RmoxyR strain, which carried an oxyR::uidA fusion, using p-nitrophenyl β-d-glucuronide as the substrate, according to methods previously described (14). With or without H2O2 treatment, similar glucuronidase (GUS) activities were observed in RmoxyR (5.9 and 6 U per μg of protein, respectively). In RmoxyR complemented by plasmid pBBR-oxyR as well, H2O2 treatment did not modify the GUS activity (3.5 and 3.3 U per μg of protein with and without treatment, respectively). This noninduction of oxyR expression after H2O2 exposure indicates that oxyR is constitutively expressed, independently of exogenous H2O2. Nevertheless, a reduction in β-glucuronidase activity was observed in RmoxyR complemented by plasmid pBBR-oxyR compared to that in the RmoxyR mutant, indicating that oxyR downregulates its own expression as in all bacterial species studied so far (27).
OxyR regulates katA expression in planta.
The effects of oxyR disruption on the nodulation and fixation capacities of the bacteria were analyzed by infection of Medicago sativa and Medicago truncatula plantlets with the RmoxyR mutant and the Rm1021 strain as a control. No significant reductions in nodulation and nitrogen fixation capacities were observed in the RmoxyR mutant compared to those in Rm1021 (data not shown).
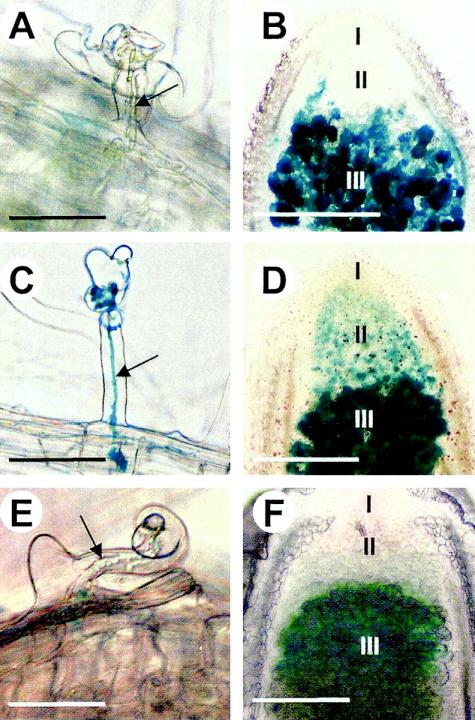
To analyze the role of the OxyR regulator on katA regulation in planta, plants were infected with bacterial strains carrying katA-lacZ plasmid fusions. By use of the two SacI sites present in the oxyR gene and in the polylinker of the vector of the pBBR-oxyR plasmid, the oxyR gene was disrupted by a deletion, resulting in the pBBR-doxyR plasmid. A 3.7-kb PstI-PstI fragment from the pKOK5 vector containing the promoterless lacZ-Kmr cartridge was inserted into the PstI sites of the pBBR-oxyR and pBBR-doxyR plasmids. The recombinant plasmids harboring the correctly orientated katA-lacZ fusions were selected and designated, respectively, pRKAZ and pdRKAZ (Fig. 1B); these plasmids were introduced into the parental strain Rm1021 (resulting in strains 1021-pRKAZ and 1021-pdRKAZ, respectively) and in the RmoxyR mutant (resulting in strains oxyR-pRKAZ and oxyR-pdRKAZ, respectively). To validate the H2O2 inducibility of these katA-lacZ fusions in the different genetic backgrounds, the β-galactosidase activities were measured for the different strains treated or not with 1 mM H2O2 for 1 h (Table 2) using o-nitrophenyl-β-d-galactoside as previously described (24). The results clearly indicated that strains 1021-pRKAZ, 1021-pdRKAZ, and oxyR-pRKAZ showed similar H2O2 induction patterns. In contrast, the oxyR mutant harboring the oxyR-truncated katA-lacZ fusion (oxyR-pdRKAZ) was deregulated, a result that is consistent with the results presented in Fig. 2. Moreover, the provision of a full-length oxyR in trans in the RmoxyR mutant (oxyR-pRKAZ) restores an essentially wild-type katA level when H2O2 is not present. In order to keep a ratio of one oxyR gene to every katA promoter, results obtained with strains 1021-pRKAZ and oxyR-pdRKAZ were taken into account only for in planta experiments. Despite the absence of antibiotic selection pressure during the symbiotic process, the stabilities of recombinant plasmids pRKAZ and pdRKAZ in bacteria were determined to be good by testing the levels of antibiotic resistance of bacteria reisolated from 5-week-old root nodules (93 and 95% of earlier resistance levels, respectively). Analysis of total β-galactosidase activity in 5-week-old nodule extracts showed that the expression of the katA-lacZ fusion was higher in 1021-pRKAZ (1,095 ± 106 Miller units per μg of protein) than in oxyR-pdRKAZ (469 ± 12 Miller units per μg of protein), indicating that OxyR also acts as an activator for katA in symbiotic bacteria. It was noted that equivalent levels of lacZ fusion expression were observed in nodule extracts obtained from both strains 1021-pdRKAZ (1,062 Miller units per μg of protein) and oxyR-pRKAZ (1,014 Miller units per μg of protein), indicating that strains 1021-pRKAZ, 1021-pdRKAZ, and oxyR-pRKAZ showed similar behaviors in planta and in free-living bacteria (Table 2). The histochemical detection of β-galactosidase activity was performed as previously described (2), using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as a substrate (Fig. 3A to D). Nodule sections (150 μm thick) were obtained with a Vibratome 1000 Plus (Labonord, Lille, France) and mounted on slides for observation and photography with an Olympus microscope. Four days after infection, analysis of root hairs revealed no staining inside most infection threads obtained with 1021-pRKAZ (Fig. 3A), whereas a blue staining in all the infection threads was observed with oxyR-pdRKAZ (Fig. 3C). Analysis of 5-week-old nodules showed that the expression of the katA-lacZ fusion is strongly detected in nitrogen-fixing bacteroids (zone III) for both the 1021-pRKAZ and oxyR-pdRKAZ strains (Fig. 3B and D). In contrast, a significant detection of β-galactosidase was observed in infection zone II with the oxyR-pdRKAZ strain only (Fig. 3D). These results are consistent with those observed under free-living conditions: katA is constitutively expressed in the RmoxyR mutant. Thus, OxyR is clearly implicated in the regulation of katA both in free-living bacteria and in planta.
TABLE 2.
Induction of katA-lacZ fusion by H2O2 in strains 1021-pRKAZ, 1021-pKAZ, oxyR-pRKAZ, and oxyR-pdRKAZ
| Strain | β-Galactosidase activitya (Miller units) (mean ± SD)
|
|
|---|---|---|
| −H2O2 | +H2O2 | |
| 1021-pRKAZ | 444 ± 97 | 2,222 ± 265 |
| 1021-pdRKAZ | 509 ± 130 | 1,960 ± 320 |
| oxyR-pRKAZ | 390 ± 157 | 1,825 ± 405 |
| oxyR-pdRKAZ | 1,338 ± 156 | 1,292 ± 198 |
Activity was measured after a 1-h treatment (+) or not (−) with 1 mM H2O2.
FIG. 3.
Histochemical detection of katA (A, B, C, and D) and oxyR (E and F) expression during S. meliloti-M. sativa nodule development. katA expression was monitored in the 1021-pRKAZ (A and B) and oxyR-pdRKAZ (C and D) strains. β-Galactosidase (A, B, C, and D) and β-glucuronidase (E and F) activities were detected using X-Gal and X-Gluc, respectively. katA-lacZ and oxyR-uidA fusions were detected in roots hairs 4 days after infection (A, C, and E) and in 5-week-old nodules (B, D, and F). Arrows indicate infection threads. Spatial development zones (I, II, and III) are indicated on nodule cross sections. Scale bars, 50 (A, C, and E) and 200 (B, D, and F) μm.
oxyR expression in planta.
To analyze the expression of the oxyR gene during the development of the root nodule, Medicago sativa plantlets were infected with the RmoxyR mutant strain harboring the oxyR::uidA fusion. The histochemical detection of β-glucuronidase activity was performed using X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) as the substrate, as previously described (14). Four days after infection, analysis of root hairs revealed no staining (Fig. 3E), suggesting that the oxyR expression level was below the threshold detection level at this step of infection. In contrast, analysis of 5-week-old nodules showed that oxyR::uidA is detectable solely in nitrogen-fixing bacteroids (zone III) (Fig. 3F) and that β-glucuronidase activity from purified RmoxyR bacteroids was exacerbated (36 U per μg of protein) compared to the activity observed in free-living bacteria (5 U per μg of protein). This result indicates an overexpression of oxyR gene transcription in bacteroids. Despite the absence of β-glucuronidase activity inside infection threads, analysis of katA expression with the 1021-pRKAZ and oxyR-pdRKAZ strains showed that OxyR could also repress katA expression in most infection threads and in zone II of mature nodules. An expression level of the oxyR-uidA fusion below the detection threshold could explain these contradictory results. On the other hand, contrary to our previous observations with the GKAZ01 strain (13), expression of the reporter gene in the 1021-pRKAZ strain has also been detected in some infection threads, which could be correlated with the H2O2 accumulation pattern observed during the symbiotic process (34). Recent results suggest that H2O2 could be required for the progression of infection threads (30). It may be proposed that these necessary H2O2 pulses that occur in infection threads could activate OxyR and periodically derepress katA expression.
Despite the fact that the production of a truncated OxyR in the oxyR mutant could disturb the regulation of the katA gene, our results suggest that OxyR could potentially repress katA gene expression in the absence of exogenous H2O2 in S. meliloti. Indeed, the possibility that OxyR could act as a repressor of a catalase gene has been recently demonstrated in Neisseria gonorrhoeae (39). Moreover, OxyR dually regulates ahpC expression in X. campestris pv. phaseoli (22). In this bacterial plant pathogen, ahpC expression is activated by oxidized OxyR and repressed by reduced OxyR. Binding of the reduced form of OxyR blocks the −35 region, preventing binding of RNA polymerase and leading to repression of the gene. Recent results with E. coli indicate that alkyl hydroperoxide reductase (Ahp) is also involved in the primary scavenging of H2O2 when the concentration is very low (35). It must be pointed out that a sophisticated and unusual regulation of genes involved in the H2O2-scavenging system by OxyR has been observed in bacteria interacting with hosts (S. meliloti, B. abortus, and X. campestris pv. phaseoli). It may be suggested that this process optimizes, in all cases, the host-bacterium interactions.
Acknowledgments
We are grateful to Pierre Frendo and Danièle Touati for helpful discussions.
This work was supported by the Improving Human Potential European Program (contract HPRN-CT-2000-00094). E. Kiss has been supported by an INRA postdoctoral fellowship as well as by the Marie Curie Fellowship of the European Community program “Improving the Human Research Potential and the Socio-Economic Knowledge Base” (contract HPMF-CT-2001-01487).
REFERENCES
- 1.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin, C., S. Camut, C. A. Malpica, G. Truchet, and C. Rosenberg. 1990. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabanes, D., P. Boistard, and J. Batut. 2000. Symbiotic induction of pyruvate dehydrogenase genes from Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 13:483-493. [DOI] [PubMed] [Google Scholar]
- 4.Christman, M. F., G. Storz, and B. N. Ames. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 86:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clare, D. A., M. N. Duong, D. Darr, F. Archibald, and I. Fridovich. 1984. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140:532-537. [DOI] [PubMed] [Google Scholar]
- 6.Del Carmen Vargas, M., S. Encarnacion, A. Davalos, A. Reyes-Perez, Y. Mora, A. Garcia-De Los Santos, S. Brom, and J. Mora. 2003. Only one catalase, katG, is detectable in Rhizobium etli, and is encoded along with the regulator OxyR on a plasmid replicon. Microbiology 149:1165-1176. [DOI] [PubMed] [Google Scholar]
- 7.Deretic, V., J. Song, and E. Pagan-Ramos. 1997. Loss of oxyR in Mycobacterium tuberculosis. Trends Microbiol. 5:367-372. [DOI] [PubMed] [Google Scholar]
- 8.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis gene. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, H.-M. 1996. Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol. 4:317-320. [DOI] [PubMed] [Google Scholar]
- 10.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boitard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F.-J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 11.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 12.Hérouart, D., S. Sigaud, S. Moreau, P. Frendo, D. Touati, and A. Puppo. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Hérouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during infection process. Mol. Plant-Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 14.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, D. P. 1982. Intracellular catalase function: analysis of the catalytic activity by product formation in isolated liver cells. Arch. Biochem. Biophys. 214:806-814. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J.-A., and J. E. Mayfield. 2000. Identification of Brucella abortus OxyR and its role in control of catalase expression. J. Bacteriol. 182:5631-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J.-A., Z. Sha, and J. E. Mayfield. 2000. Regulation of Brucella abortus catalase. Infect. Immun. 68:3861-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S. O., K. Merchant, R. Nudelman, W. F. Beyer, Jr., T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109:383-396. [DOI] [PubMed] [Google Scholar]
- 19.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 21.Long, S. R. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 125:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loprasert, S., M. Fuangthong, W. Whangsuk, S. Atichartpongkul, and S. Mongkolsuk. 2000. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 37:1504-1514. [DOI] [PubMed] [Google Scholar]
- 23.Meade, H. M., S. R. Long, G. B. Kuvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutant of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiment in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mongkolsuk, S., S. Loprasert, W. Whangsuk, M. Fuangthong, and S. Atichartpongkun. 1997. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpC-oxyR-orfX from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 179:3950-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, R. W., M. F. Christman, F. S. Jacobson, G. Storz, and B. N. Ames. 1986. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. USA 83:8059-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakjarung, K., S. Mongkolsuk, and P. Vattanaviboon. 2003. The oxyR from Agrobacterium tumefaciens: evaluation of its role in the regulation of catalase and peroxide responses. Biochem. Biophys Res. Commun. 304:41-47. [DOI] [PubMed] [Google Scholar]
- 28.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 30.Rathbun, E. A., M. J. Naldrett, and N. J. Brewin. 2002. Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Mol. Plant-Microbe Interact. 15:350-359. [DOI] [PubMed] [Google Scholar]
- 31.Robson, R. L., and J. R. Postgate. 1980. Oxygen and hydrogen in biological nitrogen fixation. Annu. Rev. Microbiol. 34:183-207. [DOI] [PubMed] [Google Scholar]
- 32.Rocha, E. R., G. Owen, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg, C., P. Boistard, J. Dénarié, and F. Casse-Delbart. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 34.Santos, R., D. Hérouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 35.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigaud, S., V. Becquet, P. Frendo, A. Puppo, and D. Hérouart. 1999. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J. Bacteriol. 181:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stougaard, J. 2000. Regulators and regulation of legume root nodule development. Plant Physiol. 124:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triggs-Raine, B. L., and P. C. Loewen. 1987. Physical characterisation of katG, encoding catalase HPI of Escherichia coli. Gene 52:121-128. [DOI] [PubMed] [Google Scholar]
- 39.Tseng, H.-J., A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witty, J. F., L. Skot, and N. P. Revsbech. 1987. Direct evidence for changes in the resistance of legume root nodules to O2 diffusion. J. Exp. Bot. 38:1129-1140. [Google Scholar]
- 41.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]