ABSTRACT
Ubiquitin (Ub) is a small protein (8 kDa) found in all eukaryotic cells, which is conjugated covalently to numerous proteins, tagging them for recognition by a downstream effector. One of the best characterized functions of Ub is targeting proteins for either selective degradation by the proteasome, or for bulk degradation by the autophagy-lysosome system. The executing arm of the UPS is the 26S proteasome, a large multicatalytic complex. While much is known about the synthesis and assembly of the proteasome's subunits, the mechanism(s) underlying its removal has remained obscure, similar to that of many other components of the ubiquitin-proteasome system. Our recent study identified autophagy as the degrading mechanism for the mammalian proteasome, mostly under stress conditions. Amino acid starvation induces specific ubiquitination of certain 19S proteasomal subunits that is essential for its binding to SQSTM1/p62, the protein that shuttles the ubiquitinated proteasome to the autophagic machinery. SQSTM1 delivers ubiquitinated substrates for proteasomal degradation via interaction of its PB1 domain with the 19S proteasomal subunit PSMD4/Rpn10, in situations where the proteasome serves as a “predator." In contrast, we found that the UBA domain of SQSTM1 is essential for its interaction with the ubiquitinated proteasome and its delivery to the autophagosome, rendering the proteasome a “prey.”
KEYWORDS: autophagy, mTOR, p62, proteasome, protein degradation, ubiquitin
Covalent attachment of Ub to target proteins generating chains of different types and lengths, is catalyzed by 3 enzymes that act in concert: a Ub-activating (E1) enzyme, a Ub-conjugating (E2) enzyme, and a substrate-specific Ub ligase (E3). Ubiquitinated substrates can be recognized directly by the proteasome through its ubiquitin receptors: PSMD2/Rpn1, PSMD4/Rpn10, and ADRM1/Rpn13, or by shuttle proteins that deliver them either to the proteasome or to the phagophore.
The catalytic arm of the UPS is the proteasome, a large 2.5 MDa multisubunit complex, consisting typically of the 20S core particle (CP) and 1 or 2 19S regulatory particles (RPs). The proteasome is responsible for the degradation of the majority of short-lived cellular proteins, and it is tightly involved in the regulation of various vital cellular processes, including maintenance of cellular homeostasis and protein quality control.
The proteasome is long-lived (∼1–2 wk), and under various pathophysiological conditions undergoes different post-translational modifications regulating its function. The elimination of nonfunctional or unnecessary proteasomes, which could be toxic to cells, is a pivotal process. The finding that entire organelles, such as mitochondria and peroxisomes are selectively engulfed by the phagophore, the precursor to the autophagosome, led us to hypothesize that the proteasome can also be selectively degraded by autophagy.
Amino acid deprivation increases significantly the polyubiquitination of specific sites on selective proteasomal subunits—interestingly, on all ubiquitin receptors (PSMD2/Rpn1, PSMD4/Rpn10, and ADRM1/Rpn13), and also on PSMD1/Rpn2. Silencing of E1 or overexpression of nonpolymerizable (no lysine) Ub abrogates proteasomal autophagy, strongly suggesting that polyubiquitination is essential for the process. The Ub ligase(s) involved in this modification is yet to be identified. The known finding that these subunits are located on the most distal part of the proteasome renders them spatially more accessible for Ub modification and autophagic engulfment (‘proteaphagy’). Further, it has been reported that inhibition of the proteasome also induces extensive polyubiquitination of its Ub receptors, which interferes with substrate binding and degradation. The similarity in the ubiquitination pattern demonstrated following proteasomal inhibition and starvation suggests that similar and conserved signals are involved in targeting the proteasome as a “prey” for degradation.
A recent study in A. thaliana demonstrated that chemical or genetic inhibition of the proteasome induces its ubiquitination, resulting in its autophagic degradation. The autophagic engulfment of the ubiquitinated proteasome in A. thaliana is mediated by the AT4G38630/RPN10 proteasomal subunit in its free state. In plants, but not in mammalian and yeast cells, RPN10 can act as a shuttle protein to the phagophore due to its ability to bind both ubiquitin and lipidated ATG8. Another study in yeast demonstrated that autophagic degradation of the ubiquitinated proteasome occurs both under conditions of nitrogen starvation and proteasomal inhibition, and is mediated by the Cue5 autophagy receptor and Hsp42. In our study we discovered that in mammalian cells, the shuttle protein SQSTM1 delivers the ubiquitinated proteasome to the phagophore, as silencing of SQSTM1 inhibits starvation-induced proteaphagy, as well as degradation of the proteasome.
SQSTM1 binds ubiquitinated substrates thought its Ub-associated (UBA) domain and delivers them either to the phagophore by interacting with lipidated LC3 via its LC3-interacting region (LIR), or to the proteasome by interacting with the 19S proteasomal subunit PSMD4/Rpn10 via its Phox and Bem1 (PB1) domain. Serial deletions of SQSTM1 domains revealed a pivotal role for its UBA domain in the interaction with the ubiquitinated proteasome following amino acid starvation (Fig. 1). This finding positions SQSTM1 in a cross road, where dependent on the domain through which it binds to the proteasome, it also determines the fate of the proteasome to be either a “predator” or a “prey.”
Figure 1.
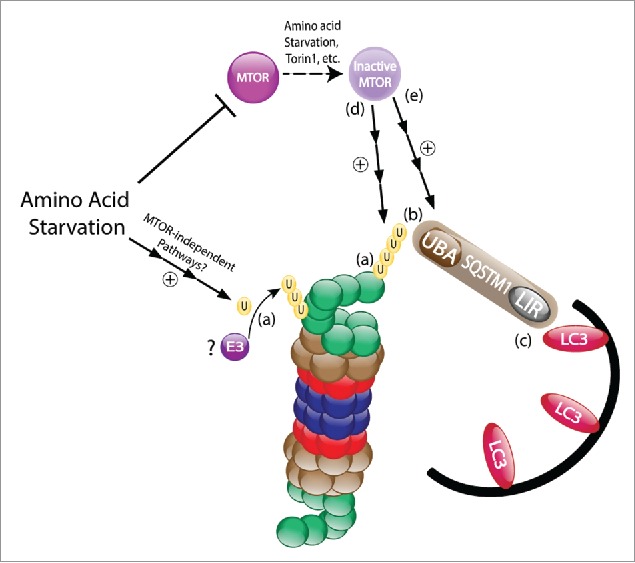
Amino acid starvation induces polyubiquitination and autophagic degradation of the proteasome. (a) Following amino acid deprivation, the ubiquitination of 19S proteasome subunits is upregulated. (b) The ubiquitinated proteasomal subunits are then recognized by the UBA domain of SQSTM1. (c) SQSTM1 targets the proteasome to the evolving phagophore through the binding of its LIR domain to the autophagic protein LC3. (d) This process is mediated, at least in part, via the inhibition of mTORC1. (e) Notably, mTORC1 inactivation was previously shown to also upregulate SQSTM1, as well as other proteins involved in autophagy and lysosome biogenesis. U, ubiquitin; mTOR, mechanistic target of rapamycin; UBA, Ub-associated; LIR, LC3-interacting region.
mTORC1 (mechanistic target of rapamycin complex 1) is a central signaling hub regulating numerous cellular processes including cell metabolism, growth, proliferation, survival and energy homeostasis. The activity of mTORC1 is tightly regulated by different stimuli, such as amino acids, growth factors and glucose levels, as well as genotoxic and ER stress. Inhibition of mTORC1 results in activation of the ULK1 complex, leading to the initiation of autophagy. We further found that specific inhibition of mTORC1 results in increased interaction of the proteasome with SQSTM1 and its subsequent interaction with lipidated LC3, suggesting a transducing role for mTORC1 in phagophore-dependent uptake of the proteasome (Fig. 1).
During the past decade proteasomal inhibition has emerged as an effective strategy for anti-cancer therapy. The findings that inhibition of the proteasome results in its increased ubiquitination, and that inhibition of mTORC1 promotes autophagic engulfment of the ubiquitinated proteasome, can be used for a novel and more effective clinical approach in treatment of multiple myeloma and other diseases by co-administration of proteasome and mTOR inhibitors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research in the laboratory of A.C. is supported by grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Israel Science Foundation (ISF), the I-CORE Program of the Planning and Budgeting Committee and the ISF (Grant 1775/12), the Deutsch-Israelische Projektkooperation, and a special fund for research in the Technion established by Albert Sweet. I.L. is supported by the Foulkes Fellowship. A.C. is an Israel Cancer Research Fund USA Professor.


