ABSTRACT
Autophagy is a catabolic cellular process required to maintain protein synthesis, energy production and other essential activities in starved cells. While the exact nutrient sensor(s) is yet to be identified, deprivation of amino acids, glucose, growth factor and other nutrients can serve as metabolic stimuli to initiate autophagy in higher eukaryotes. In the early-branching unicellular parasite Trypanosoma brucei, which can proliferate as procyclic form (PCF) in the tsetse fly or as bloodstream form (BSF) in animal hosts, autophagy is robustly triggered by amino acid deficiency but not by glucose depletion. Taking advantage of the clearly defined adenosine triphosphate (ATP) production pathways in T. brucei, we have shown that autophagic activity depends on the levels of cellular ATP production, using either glucose or proline as a carbon source. While autophagosome formation positively correlates with cellular ATP levels; perturbation of ATP production by removing carbon sources or genetic silencing of enzymes involved in ATP generation pathways, also inhibited autophagy. This obligate energy dependence and the lack of glucose starvation-induced autophagy in T. brucei may reflect an adaptation to its specialized, parasitic life style.
KEYWORDS: amino acid starvation, AMPK, autophagy, cell respiration, glycolysis, Trypanosoma brucei
Introduction
Autophagy is a conserved cellular process during which unnecessary or dysfunctional proteins or organelles are engulfed by autophagosomes and targeted for bulk degradation in lysosomes. This ‘self-eating’ pathway can support cell survival under starvation or other stress conditions by maintaining the cell energy level, recycling the amino acids for essential new protein synthesis, and eliminating harmful cellular materials.1
Autophagy can be triggered by a multitude of stress conditions. Among them, low cellular energy charge or deprivation of essential nutrients, including glucose and amino acids have been extensively studied.2 The AMP-activated protein kinase (AMPK) senses low energy levels (high AMP:ATP ratio) in cells, that usually occur upon glucose depletion, and acts as a checkpoint for cell growth, autophagy and metabolism coordination.3 However, the exact role of glucose in autophagy is still controversial (reviewed in4). On the other hand, limitations of nonessential amino acids trigger autophagy by at least 2 distinct mechanisms. First, accumulation of uncharged tRNA species upon amino acid starvation activates EIF2AK4/Gcn2 (eukaryotic translation initiation factor 2 α kinase 4), thus blocking protein synthesis and inducing autophagy.5 Second, depletion of lysosomal amino acids can lead to inactivation of MTORC1 by dissociating the complex from lysosomal surface.6-8 Amino acid deprivation has also been reported in mammalian and nonmammalian models to affect intracellular levels of acetyl-CoA (AcCoA) and α-ketoglutarate and thus trigger autophagy.7,9,10 While autophagy is required to maintain ATP production in starved cells,11 ATP is also required for at least several autophagy steps.12 However, cellular ATP levels under amino acid starvation have been rarely investigated. Increased,7 unchanged13,14 and decreased15-17 cellular ATP have all been reported in various organisms, making it difficult to extrapolate the correlation between ATP production and amino acid starvation-induced autophagy.
Despite it being a highly conserved eukaryotic pathway, most of the autophagy studies have focused on mammalian cells and yeast. Much less is known about autophagy in the early-branching protozoan parasites, which include many important human and animal pathogens. Studies of autophagy in these unicellular parasites however, may provide insights to the diverse functions and the evolution of the autophagic process.18 Until now, approximately 40 AuTophaGy-related (ATG) genes have been identified in yeast or mammalian cells. Among them, only half have been found in the protozoan parasites. While the Atg8–PE conjugation system is highly conserved, proteins involved in other autophagy steps lack conservation or have not been detected in the parasite genomes, suggesting the presence of a divergent autophagic pathway.19 The core autophagy events, from double-membrane autophagosome formation to the final degradation in lysosomes is conserved,20 although differences in autophagy induction and regulation have been noted. As an example, our recent study in Trypanosoma brucei has revealed an essential role of the acidocalcisomes, which are lysosome-related organelles found in many protozoan parasites, in autophagy induction.21 Additionally, taking advantage of the molecular genetic accessibility of T. brucei, a role of autophagy in cell death and glycosome-turnover and differentiation has been proposed.20,22-24 The prodeath role of autophagy is also reported in Toxoplasma gondii and Plasmodium berghei.25,26
Trypanosoma brucei transmits between mammal hosts via tsetse flies. During the transmission, trypanosomes encounter distinct host environments with varying nutrients, temperatures and other conditions. To cope with this, the parasite cells undergo extensive remodeling in their cellular metabolic pathways in different hosts. Most notably, the BSF and PCF trypanosomes generate energy with distinct pathways. The BSF trypanosomes produce ATP via glycolysis using glucose (abundant in blood) as carbon source; and the PCF cells generate ATP mainly via oxidative phosphorylation using proline (abundant in tsetse flies) as carbon source. When both glucose and proline are available, the PCF cells prefer to metabolize glucose as carbon source.27,28 Taking advantage of these simple and well-defined energy metabolism pathways in T. brucei, we systematically analyzed the relationship between energy metabolism and autophagy, and demonstrated a positive correlation between cellular ATP level and autophagic activity in both PCF and BSF cells. Inhibition of ATP production by deprivation of carbon sources or perturbation of ATP synthesis also inhibited autophagic activity. Furthermore, we showed that glucose starvation could not induce autophagy and AMPK was dispensable in amino acid starvation-induced autophagy, suggesting the absence of an autophagy signaling mechanism through sensing of low energy charge in T. brucei. Autophagy was also monitored in the BSF T. brucei in culture, supporting that amino acid starvation and continuous ATP production fueled by glucose are both relevant factors to induce autophagy in this protozoan parasite.
Results
Cellular ATP production is enhanced upon amino acid starvation
To evaluate the relationship between energy level and autophagy induction in starving PCF trypanosomes, we first measured the ATP level and autophagosome formation in cells cultivated in medium or starved in Hank's balanced salt solution supplemented with glucose (gHBSS). Consistent with our previous study, autophagosome formation using YFP-Tb927.7.5910/TbATG8.2 as a marker20 is observed at 30 min upon starvation, and autophagosome number reaches equilibrium after 2 h (Fig. 1A and B).20 During this time course, a significant increase in cellular ATP was observed in starving T. brucei. The cellular ATP level was elevated by ∼20% (from 3.68 ± 0.14 nmol/107 cells to 4.40 ± 0.62 nmol/107 cells) at 30 min poststarvation, and ∼60% at 2 h poststarvation (5.92 ± 0.58 nmol/107 cells) (Fig. 1C). The AMP and ADP level did not change significantly during the course of the experiment (Fig. S1B). Due to the continuous increase in ATP levels (i. e. reduced AMP:ATP ratio, c.f. Fig. S1B) upon starvation, the amino acid starvation-induced autophagy was unlikely to be triggered by a low cellular energy charge (or high AMP:ATP ratio) in T. brucei. The enhanced ATP level upon starvation could be a consequence of reduced ATP consumption due to decreased cellular activities including protein synthesis, and/or increased energy metabolism via glycolysis or cell respiration, with AMP influx through nucleotide interconversion29,30 or other unknown mechanisms.
Figure 1.
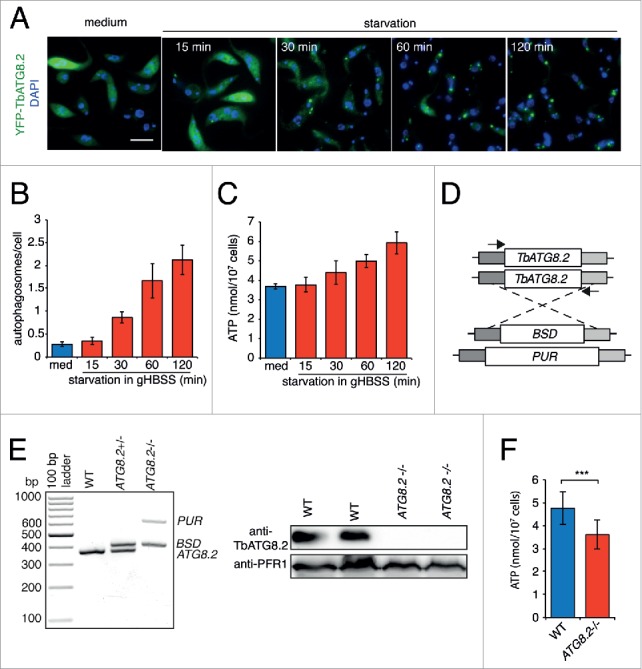
Enhanced cellular ATP level and glucose and/or proline metabolism in amino acid-starved cells. (A to C) Steady increase of autophagosomes and cellular ATP upon starvation. Total cellular autophagosomes and ATP levels were determined in YFP-TbATG8.2 cells cultivated in medium or starved in gHBSS for 15 min, 30 min, 60 min and 120 min. Quantification results are shown as mean ± SD of 3 independent experiments. The cells were costained with DAPI, which labels nuclear (large blue ovals) and mitochondrial DNA (small blue dots) in T. brucei. Scale bar: 5 μm. (D) Schematic representation of the TbATG8.2 loci and the drug cassettes used for generation of TbATG8.2−/− cell line by PCR-based approach. BSD, blasticidin-resistance gene; PUR, puromycin-resistance gene. The 2 arrows present the position of primer pair (TbATG8.2-con-F and TbATG8.2-con-R, Table S1) used for knockout validation. (E) Knockout of TbATG8.2 expression was confirmed by PCR using primers indicated in (D) (Table S1) flanking the TbATG8.2 ORF region, and by immunoblots with anti-TbATG8.2. Anti-PFR1 was used as a loading control. (F) ATP levels in WT and TbATG8.2−/− cells starved in gHBSS for 2 h. Depletion of TbATG8.2 signification inhibited ATP production in starved cells. Quantification results are shown as mean ± SD of 3 independent experiments. ***P < 0.001.
Autophagy is required to maintain ATP production in starved yeast or mammalian cells31,32 and this is also the case in T. brucei. We constructed a TbATG8.2 knockout (TbATG8.2−/−) cell line by replacing the 2 endogenous TbAtg8.2 alleles with the blasticidin-resistant gene (BSD) and the puromycin-resistant gene (PUR), respectively (Fig. 1D and E). The successful knockout was confirmed by PCR with primers flanking the TbATG8.2 open reading frame (ORF) (Fig. 1E, left panel), and by immunoblotting using TbATG8.2 specific antibody (Fig. 1E, right panel). The increase in cellular ATP level as observed in wild-type cells upon starvation were also inhibited in TbATG8.2−/− cells (compare Fig. 1F with Fig. 1C), supporting that the elevated ATP production and ATP level requires autophagy.
Autophagic activity positively correlates with cellular ATP level
In our previous study, we have shown that the presence of glucose in Hank's balanced salt solution (HBSS) is crucial for autophagy induction,20 suggesting that ATP production is required for autophagy. However, PBS only without glucose or other carbon source has also been used to induce autophagy induction in T. brucei in several other studies,24,33 casting doubts on the requirement of ATP production in autophagy induction.
To address this contradiction, we starved the YFP-TbATG8.2 cells in gHBSS or HBSS, with or without 2-DG, a nonmetabolizable glucose analog (Fig. 2A). Compared with cells starved in gHBSS, cells starved in HBSS with or without 2-DG showed only basal level autophagosome formation, comparable to that observed in cells cultured in medium (Fig. 2A and quantified in 2B). This was further confirmed by immunoblotting assays to monitor the presence of form II (phagophore and autophagosome-associated) of TbATG8.2 (Fig. 2D). Strikingly, ATP was not detectable in cells starved with HBSS or HBSS with 2-DG (Fig. 2C). More than 30% HBSS-starved cells were positive for propidium iodide staining (Fig. S2), suggesting necrotic cell death. These results also suggest that cellular ATP production critically depends on extracellular carbon source supplied and may be required for autophagy induction. Consistent with this, the addition of 2-DG (equal amount with glucose) inhibited both autophagy and ATP production in gHBSS (Fig. 2A to D).
Figure 2.
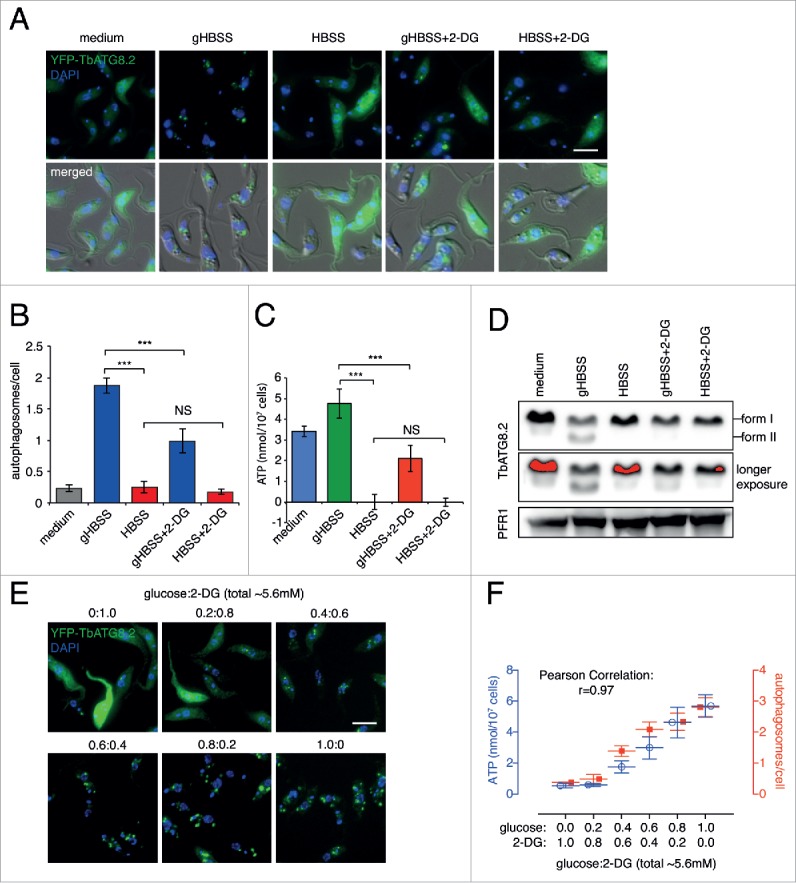
Glucose supports autophagy induction in an ATP-dependent manner. (A) YFP-TbATG8.2 cells were cultivated in medium or starved in gHBSS or HBSS, with or without 2-DG, for 2 h. Scale bar: 5 μm. (B) The average number of autophagosomes per cell was quantified in >200 cells for each condition and shown as mean ± SD of 3 independent experiments. (C) Cellular ATP was measured in the same cells treated as in (A). (D) Immunoblotting analysis of TbATG8.2 in cells treated as in (A). The autophagosome-associated form II and the un-associated form I of TbATG8.2 were separated by 12.5% urea-SDS-PAGE and probed with anti-TbATG8.2. The same blots were probed with anti-PFR1 as loading control. (E, F) YFP-TbATG8.2 cells were washed with and resuspended in HBSS, then glucose and 2-DG were added at varying ratio (∼5.6 mM total) and starved for 2 h. Autophagosome formation and cellular ATP were measured as described above. Scale bar: 5 μm. Quantification results are shown as mean ± SD of 3 independent experiments. *** P < 0.001 and NS as not significant.
To further analyze the relationship between ATP production and autophagy, cellular ATP level and autophagosome formation were measured in cells starved in HBSS supplemented with glucose and 2-DG at varying ratio, to a total concentration of ∼5.6 mM (Fig. 2E). Both ATP level and autophagosome formation increased as glucose concentration increased (and 2-DG concentration decreased), in a dose-dependent manner. Autophagosome formation was thus positively affected by the cellular ATP level, exhibiting a strong correlation with r = 0.97 (Fig. 2F).
In addition to glucose, the PCF trypanosomes have adapted to using proline that is abundantly available in tsetse fly, as a carbon source for ATP production.34 We therefore tested whether HBSS supplemented with proline (pHBSS) can induce autophagy. Robust autophagosome formation (Fig. 3A and B), cellular ATP production (Fig. 3C), and increase in the autophagosome-associated form II of TbATG8.2 (Fig. 3D) were observed in cells starved with HBSS containing glucose, proline or both as carbon source. On the contrary, cells starved in HBSS alone or HBSS containing alanine (a control amino acid that cannot be used for ATP production in T. brucei),28,35 showed little autophagic activity and no detectable cellular ATP (Fig. 3A to D). Similar to glucose, proline fueled ATP production and autophagosome formation in a dose-dependent manner until 6.5 mM (0.75 g/L), at which point both ATP and autophagosome formation reached maximum. Again, a strong positive correlation (r = 0.931) was observed between ATP level and autophagic activity (Fig. 3E and F).
Figure 3.

Proline also supports autophagy induction in an ATP-dependent manner. (A to D) YFP-TbATG8.2 cells were cultivated in medium or starved in pHBSS, HBSS, HBSS supplemented with alanine, which could not be used as carbon source for energy metabolism (negative control), and gHBSS (positive control). Controls using gHBSS supplemented with proline or alanine were also included to exclude possible inhibitory effects proline or alanine on autophagy. Cells were then processed for fluorescence microscopy (A), autophagosome quantification (B), cellular ATP measurements (C) and TbATG8.2 immunoblotting assays (D). Scale bar: 5 μm. ***P < 0.001. (E, F) YFP-TbATG8.2 cells were starved in HBSS containing increasing amount of proline (0 to 17.4 mM) for 2 h. Autophagosome formation and cellular ATP were measured as described above. Scale bar: 5 μm. Quantification results were shown as mean ± SD of 3 independent experiments.
Together, these results strongly suggest that the autophagic activity in amino acid-starved trypanosomes is influenced by the cellular energy level, which corresponds to the availability of glucose or proline in starvation medium. The strong positive correlation between cellular ATP levels and autophagosome formation prompted us to further examine the effects of inhibited ATP production, using both genetic and chemical treatments, on autophagy initiation.
Blocking glucose or proline metabolism inhibits autophagy in an ATP-dependent mode
Glucose metabolism in T. brucei relies on glycolysis.34 Through glycolysis, one mole of glucose is converted to 2 moles of pyruvate, which is then imported into mitochondria and converted to acetyl-CoA (AcCoA) by a pyruvate dehydrogenase (PDH) complex. AcCoA is not further metabolized via the tricarboxylic acid (TCA) cycle, although all TCA cycle related enzymes are expressed in T. brucei.27,36 Instead, AcCoA can be converted into acetate by ASCT (acetate:succinate CoA-transferase, Tb927.11.2690) and ACH (acetyl-CoA thioesterase, Tb927.3.4260) in mitochondria,37,38 and produce ATP through an ASCT-SCoAS (succinyl-CoA synthetase, Tb927.3.2230) cycle (Fig. 4A).38,39
Figure 4.
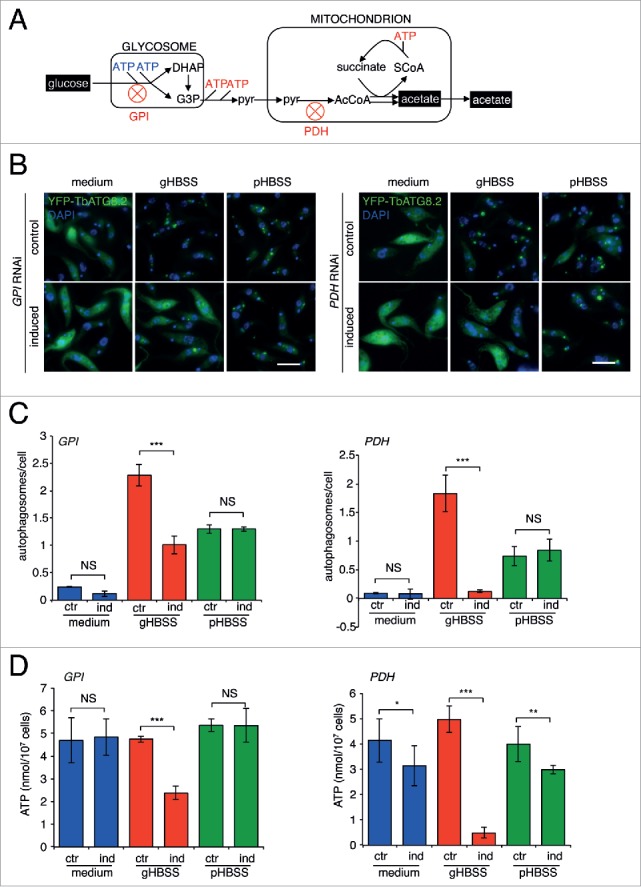
Genetic perturbation of energy metabolism from glucose inhibits autophagosome formation in gHBSS but not in pHBSS. (A) Schematic diagram showing the energy-related glucose metabolism pathways in T. brucei. ((B) to D) YFP-TbATG8.2 control (uninduced) and GPI- or PDH- RNAi (induced) cells were cultivated in medium or starved in gHBSS or pHBSS for 2 h. The cells were processed for fluorescence microscopy, autophagosome quantification and ATP measurement as described above. Scale bar: 5 μm. Quantification results were shown as mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
Tb927.1.3830/GPI (glucose-6-phosphate isomerase, glycosomal) and Tb927.10.12700/PDH (pyruvate dehydrogenase E1 α subunit, putative) were selected for RNAi analyses (Fig. S3). Depletion of either enzyme led to inhibited autophagosome formation (Fig. 4B and C), and decreased ATP (Fig. 4D) in gHBSS-starved cells. On the contrary, autophagosome formation and cellular ATP were not affected in pHBSS-starved cells, confirming that the inhibition on autophagy was specifically due to inhibited ATP production via glucose metabolism, rather than a nonspecific effect on the autophagy machinery. It was of interest to note that depletion of PDH had a strong inhibitory effect on ATP and autophagy in gHBSS, despite 2 ATPs can be produced through glycolysis in the steps before PDH (from glucose to pyruvate; Fig. 4A). This observation supports the view that ASCT-SCoAS cycle plays a major role in ATP generation.37
In the absence of glucose, proline can be used by PCF T. brucei as carbon source. In the mitochondria, proline is converted to glutamate, producing one FADH2 and one NADH. Glutamate is subsequently converted to 2-oxoglutarate, which enters a fragment of the TCA cycle, producing succinate and generating one NADH and one ATP (Fig. 5A). To block proline metabolism, Tb927.7.210/PRODH (proline dehydrogenase, mitochondrial) and Tb927.11.9980/OGDH (2-oxoglutarate dehydrogenase E1 component, putative) were depleted by RNAi. Neither enzyme was essential for cell proliferation in medium (Fig. S3), as the cultured cells prefer glucose over proline as carbon source for ATP generation.28 Consistently, RNAi depletion of PRODH or OGDH did not affect autophagy initiation (Fig. 5B and C) and ATP level (Fig. 5D) in cells starved in gHBSS. Interestingly, when starved in pHBSS, both autophagy initiation and ATP generation were inhibited in PRODH-RNAi cells, but not in OGDH-RNAi cells (Fig. 5B to D). This is likely due to the production of FADH2 and NADH before the OGDH catalytic step (Fig. 5A). Both FADH2 and NADH can be used for ATP generation through cell respiration.40
Figure 5.

Genetic perturbation of proline metabolism or oligomycin treatment inhibits ATP production and autophagy in pHBSS but not in gHBSS. (A) Schematic diagram shown the energy-related proline metabolism by substrate level phosphorylation in T. brucei. (B to D) YFP-TbATG8.2 control (uninduced) and PRODH- or OGDH-RNAi (induced) cells were cultivated in medium or starved in gHBSS or pHBSS for 2 h. Cells were then processed for fluorescence microscopy (B), autophagosome quantification (C) and ATP measurements (D) as described above. Scale bar: 5 μm. Quantification results were shown as mean ± SD of 3 independent experiments. ***P < 0.001; NS, not significant. (E) Schematic diagram showing the energy-related proline metabolism by oxidative phosphorylation in T. brucei. Oligomycin is an ATP synthase inhibitor and CCCP can collapse the membrane potential (ΔΨ) and pH gradient (ΔpH) of mitochondria. (F) MitoTracker Red CMXRos accumulation in mitochondria was inhibited by CCCP treatment. Scale bar: 5 μm. (G) Starvation-induced acidocalcisome acidification was inhibited by CCCP treatment. Scale bar: 5 μm. (H to J) YFP-TbATG8.2 cells were cultivated in medium or starved in gHBSS or pHBSS, in the absence and presence of oligomycin or CCCP. Cells were then processed for fluorescence microscopy (H), autophagosome quantification (I) and ATP measurements (J) as described above. Scale bar: 5 μm. Quantification results were shown as mean ± SD of 3 independent experiments. ***P < 0.001; NS, not significant.
Both oligomycin and CCCP were then used to block ATP production using FADH2 and NADH, metabolites from the proline pathway (Fig. 5E). Oligomycin inhibited autophagosome formation and greatly reduced cellular ATP level in pHBSS but not in gHBSS (Fig. 5H to J), further attesting to the crucial role of efficient ATP production via cell respiration in pHBSS-induced autophagy. Interestingly, the protonophore CCCP inhibited autophagosome formation and ATP production in both pHBSS and gHBSS (Fig. 5H to J). Like in other organisms, CCCP disrupted membrane potential at mitochondria as well as at acidocalcisomes, a lysosome-related acidic organelle in T. brucei (Fig. 5F and G). While mitochondrial membrane potential is critical for cell respiration, acidocalcisome acidification is required for autophagy induction.21 It is not clear how CCCP may affect ATP production via the glucose pathway. One possibility is that disruption of mitochondrial membrane potential affected pyruvate uptake into the mitochondria and ATP production through the ASCT-SCoAS cycle.
Taken together, through specific molecular genetic perturbations and drug treatments to block ATP production, we have confirmed the ATP requirement for autophagy initiation in T. brucei, using glucose or proline as carbon source. Without active ATP production, autophagy cannot be induced by amino acid starvation.
Glucose starvation-induced autophagy is absent in trypanosomes
In higher organisms, energy shortage that usually occurs upon glucose starvation can induce autophagy, where AMPK senses low energy level (i.e. high AMP:ATP ratio) and mediates autophagy initiation.41,42 We have demonstrated thus far that T. brucei autophagic activity is ATP-dependent, suggesting that trypanosomes may lack the energy-sensing mechanism for autophagy induction.
To address this possibility, we first tested if autophagy could be induced in cells starved in sugar-free medium that contained the complete set of amino acids except for proline. The cells were cultivated up to 12 h in this sugar-free medium supplemented with increasing amount of proline, during which the cells remained viable (data not shown). Autophagosome formation was not detected during the course of the treatment despite increasing proline (Fig. 6A). Therefore, the deprivation of sugar from the cultivation medium is not able to induce autophagy in T. brucei.
Figure 6.
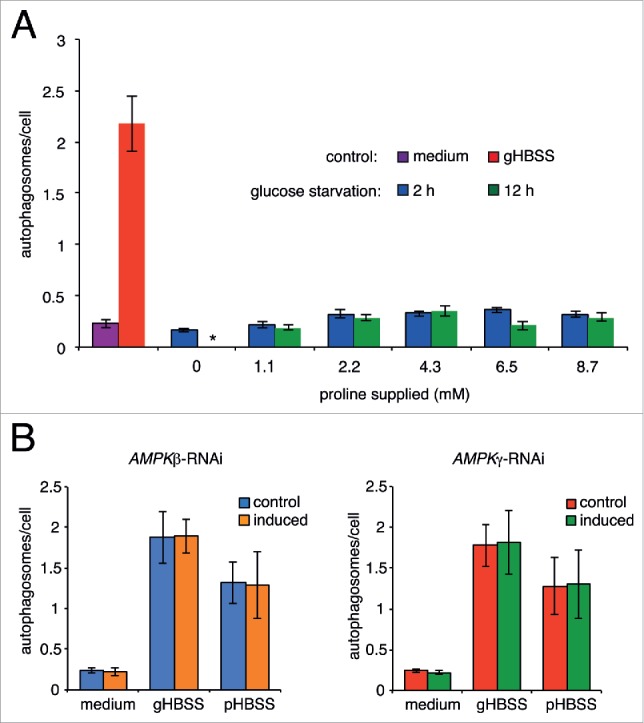
Glucose-deprivation does not induce autophagy in T. brucei. (A) YFP-TbATG8.2 cells were starved in sugar-free medium containing all amino acids except for proline. Proline was then supplemented to the starvation medium at varying doses from 1.0 to 8.7 mM. Cells cultivated in medium or starved in gHBSS were used as negative and positive controls, respectively. Regardless of proline concentration, only basal level autophagosome formation was observed, at 2 h or 12 h in glucose starvation conditions. (B) T. brucei AMPK did not affect amino acid starvation-induced autophagy. YFP-TbATG8.2 control or cells with inducible depletion of AMPK subunit β or subunit γ (induced) were cultivated in medium or starved in gHBSS or pHBSS. Autophagosome formation under either amino acid starvation conditions was not affected by the depletion of either AMPK subunits.
Since glucose starvation did not induce autophagy, the function of AMPK in autophagy was studied in amino acid-starved cells. Tb927.8.2450/AMPKβ (AMPK subunit β; SNF1-related protein kinase regulatory subunit β; 5′-AMP-activated protein kinase subunit β) and Tb927.10.3700/AMPKγ, confirmed AMPK subunits in T. brucei,43 were depleted by RNAi. Depletion of either subunit arrested cell proliferation after 48 h induction (Fig. S3), hence autophagy was analyzed at 48 h postinduction before the proliferation arrest. Depletion of either AMPK subunit had no detectable effects on autophagosome formation in gHBSS or in pHBSS (Fig. 6B and C). These results indicated that trypanosome AMPK is dispensable for autophagy induced by amino-acid deprivation. Unlike the higher eukaryotes, trypanosome may lack the AMPK-dependent energy-sensing pathway for autophagy induction.
Energy-dependence of autophagy in bloodstream form trypanosomes
Compared with PCF cells, BSF trypanosomes only generate ATP by glycolysis using glucose as carbon source. As autophagy in BSF trypanosomes was more efficiently induced in cytomix (Fig. 7A; and see Methods), all subsequent starvation experiments on BSF cells were performed using cytomix containing varying concentrations of glucose (1.1, 2.2, 3.3, 4.4 and 5.5 mM). In cytomix without glucose, all cells swelled up, stopped moving and died after 2 h starvation, with or without proline or 2-DG (Fig. 7B). The presence of glucose in cytomix, even at 1.1 mM (0.2 g/L) glucose, supported cell survival (based on cell motility and morphology) for at least 2 h (data not shown), allowing autophagosome quantification to be performed. Cells starved in cytomix supplemented with 3.3 mM (0.6 g/L) glucose (close to the physiological concentration in blood) exhibited the highest autophagic activity, as well as the highest cellular ATP level (Fig. 7C and D). Overall, the autophagic activity correlated with the ATP level (r = 0.956), just as in PCF cells.
Figure 7.
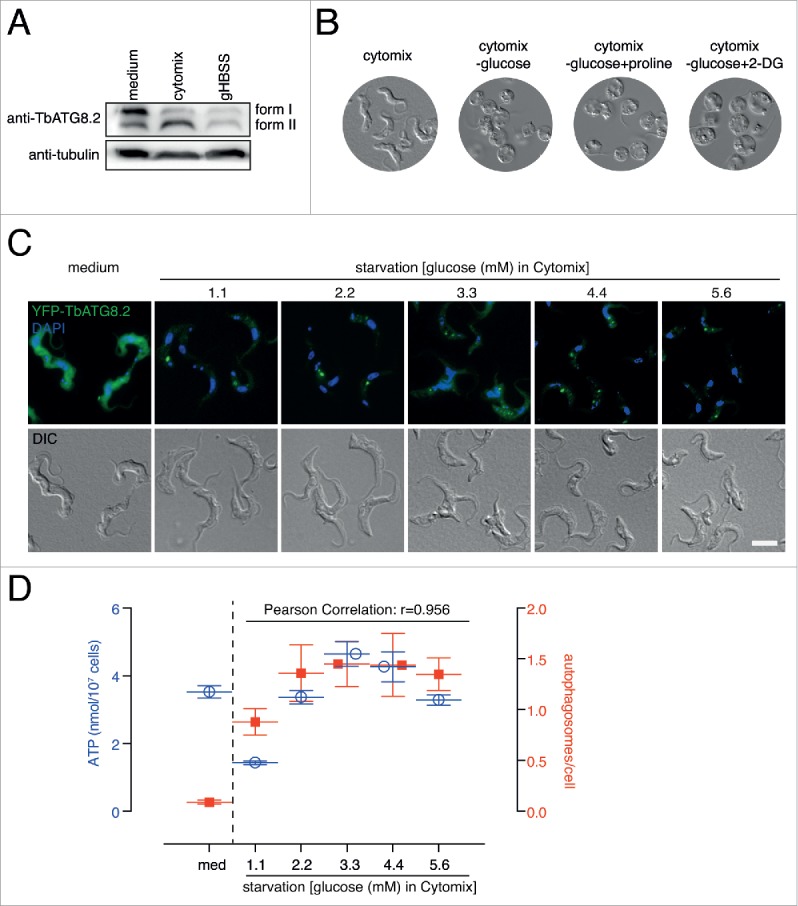
Amino acid starvation-induced autophagy in bloodstream trypanosomes is ATP-dependent. (A) For BSF cells, autophagy was more efficiently induced in cytomix than in gHBSS. The autophagosome associated form II and the un-associated form I of endogenous TbATG8.2 were separated by 12.5% urea-SDS-PAGE and probed with anti-TbATG8.2. The same blots were probed with anti-tubulin as loading control. (B) Cytomix without glucose could not support BSF cell survival. All cells swelled up after 2 h. (C) and (D) YFP-TbATG8.2 BSF cells were cultivated in medium or starved in cytomix supplemented with 1.1, 2.2, 3.3, 4.4 or 5.6 mM glucose for 2 h. The cells were processed for fluorescence microscopy, autophagosome quantification and ATP measurements as described above. Quantification results were shown as mean ± SD of 3 independent experiments.
Discussion
It is generally accepted that in glucose-starved cells, the low energy level and high AMP:ATP ratio is sensed by AMPK, which subsequently initiates autophagy.41,42 However, the relationship between energy level and autophagy in amino acid-starved cells has not been systematically analyzed and existing results are contradictory and confusing.7,13,15-17,44,38 This may be due to the complexity of ATP production pathways and potential metabolic shift under starvation stress in the higher eukaryotes.45-47 In trypanosomes, the energy metabolism by glycolysis, TCA cycle and oxidative phosphorylation using glucose or proline are compartmentalized in glycosomes and mitochondria. While the BSF cells use only glucose as carbon source for ATP production, the PCF cells have a strong preference for glucose as carbon source when it is present, and switch to proline when glucose is absent.28 Although gluconeogenesis occurs in T. brucei, it is thought to contribute mainly to NADPH generation to resist the oxidative stress but not to ATP production.35 The relatively simple and well-defined energy metabolism pathway in T. brucei thus provides a unique opportunity to dissect the relationship between ATP production and autophagy induction.
Our results established a requirement of cellular ATP for autophagy induction (Fig. 2 to 5 and 7). Robust autophagy induction was observed in gHBSS- or pHBSS-starved PCF cells, as cellular ATP level increased (Figs. 1 to 3). On the contrary, cells starved in HBSS alone showed little autophagic activity and undetectable cellular ATP. Genetic perturbations and drug treatments further supported the specific contribution of glucose and proline in ATP production and autophagy induction (Figs. 4 and 5). The ATP-dependence of autophagy induction was also observed in BSF cells (Fig. 7).
These findings indicated a positive correlation between cellular energy level and autophagy, and emphasized the critical role of ATP production in determining autophagy activity. Such correlation is yet to be experimentally demonstrated in other organisms. However, based on these results, we suggest that energy generation should be considered as a part of the autophagic flux (Fig. 8A), which describes the dynamic process of autophagosome formation, fusion with lysosomes and degradations.48 Perturbation of cellular ATP levels can influence the autophagosome-based autophagy assay, but not the core autophagy machinery and subsequent fusion and degradation steps. In cases where a treatment is observed to inhibit autophagosome formation, an assay on cellular ATP levels can help to distinguish whether this treatment inhibits autophagy specifically or indirectly via inhibition of cell bioenergetics.
Figure 8.
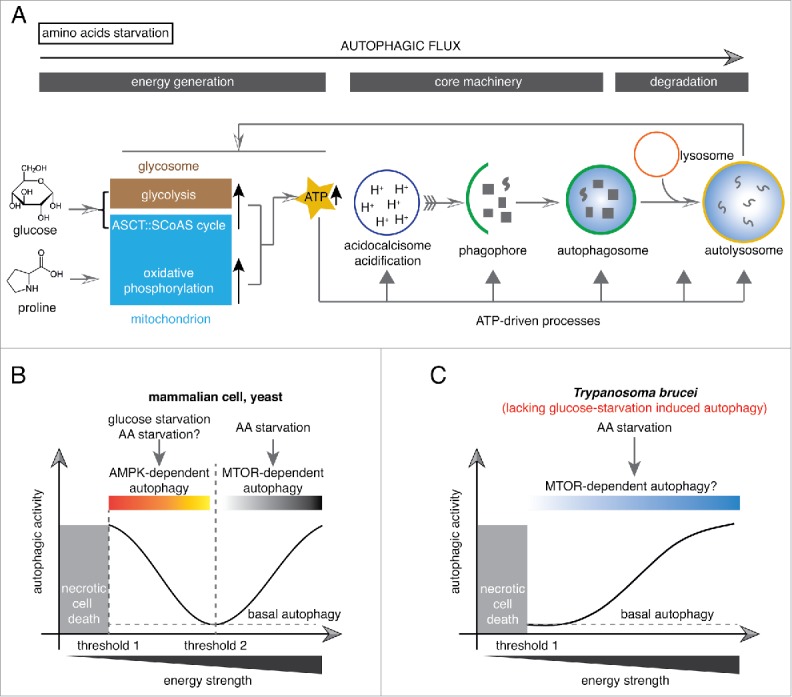
ATP-driven and AMPK-independant autophagy in Trypanosoma brucei. (A) Upon amino acid starvation, the cellular ATP level was enhanced due to increased glycolysis or oxidative phosphorylation activity with supplement of glucose or proline, respectively, as carbon sources. Robust ATP production drives autophagy occurrence and conversely autophagy prompts enhanced energy metabolism. (B) T. brucei lacks an AMPK-mediated energy sensing mechanism. In both mammalian cells and trypanosomes, the cells will undergo necrotic cell death when the cellular energy level dropped harshly (threshold 1). An additional energy threshold (threshold 2) exists in nutrient starved mammalian cells and yeast: below this threshold, AMPK is activated and further leads to autophagy induction; above this threshold, autophagy is triggered by amino acid starvation through an MTOR-mediated pathway with an ATP-dependent mode. In contrast, threshold 2 does not exist in T. brucei.
Additionally, our results caution against the use of starvation buffers without any carbon source, as previously used in some autophagy studies in T. brucei.24,33 In these studies, autophagy has been observed in PCF T. brucei starved in PBS only, using YFP-ATG8.2 (same as in this study) as a marker. It is unlikely the difference was due to different cell lines used. In addtion to PCF 29.13, we have also performed starvation assays on PCF YTat1.1 and BSF Lister 427 cells. All cell lines responded the same, i.e. elevated autophagosome formation in gHBSS or cytomix containing glucose, but little autophagic activity observed in HBSS or cytomix without glucose. One possible explanation lies in the levels of autophagosome formation. While earlier autophagy studies lack quantification so it is not possible to make comparisons, more recent studies quantify autophagic activity by showing the number of autophagosomes per cell, following the recommendations made by Mizushima et al.48 It appears that starvation with gHBSS consistently produces higher autophagic activity20,21,49 compared with starvation with PBS only.33,50 As has been shown in our current work, autophagic activity depends critically on the amount of carbon source and the cellular ATP level, it is possible that the low autophagic activity observed in PBS starvation was at basal level or slightly above in case of insufficient washing before starvation.
The energy-requirement for autophagy initiation has also been reported in other organisms, and several steps in the autophagic cascade consume energy.2 While low-energy level induces autophagy, a rapid reduction of energy level below a critical limit triggers cell death rather than autophagy (Ref.51 and schematically shown in Fig. 8B; Fig. S2). In T. brucei, ATP production was required for autophagy (Figs. 2 to 5 and 7) and the ATP is at least required for acidification of the acidocalcisomes (Fig. 8A and Fig. S4), a lysosome-related organelle that is essential for autophagy induction.21 Furthermore, autophagic activity was positively affected by cellular ATP level (Fig. 2, Figs. 3, 7), which relied on proline or glucose in a dose-dependent manner. Thus, unlike the higher eukaryotes, an energy sensing mechanism that allows autophagy induction in response to low energy level is unlikely to be present in T. brucei (Fig. 8B). Further supporting this conclusion, depletion of AMPK β- and δ-subunits did not affect amino acid starvation-induced autophagy (Fig. 6). In a previous study, increased cellular AMP:ATP ratios and autophagosomes have been found in cells depleted of Tb927.11.1350/TbMCU (calcium uniporter protein, mitochondrial).52 However, no evidence is provided for the dependence or correlation between these 2 observations. It would be of great interest to study the role of AMPK in TbMCU-RNAi-induced autophagy.
The role of glucose in autophagy induction is also controversial in the higher eukaryotes. Glucose deprivation has been reported to induce autophagy via AMPK.53 However, a later study reports that glucose deprivation and the subsequent ATP reduction do not activate autophagy in 4 different mammalian cell lines.54 In the same study, 2-deoxyglucose (2-DG), a nonmetabolic glucose analog, has been shown to induce autophagy.54 However, the induction is due to 2-DG-triggered endoplasmic reticulum stress rather than ATP depletion.55 In our study, neither glucose starvation- nor 2-DG-induced autophagy was observed in PCF T. brucei. On the contrary, 2-DG treatment in starved cells inhibited gHBSS-induced autophagy, accompanied with decreased ATP level (Fig. 2). For BSF cells, depletion of glucose from cytomix resulted in cell swelling and death (Fig. 7). Our results thus suggest that glucose-deprivation does not induce autophagy in T. brucei.
The lack of glucose starvation-induced autophagy and the absence of an energy-sensing mechanism for autophagy induction in T. brucei is likely a specialized adaptation to its parasitic life style. Glucose is stably available in mammalian blood, and proline is abundant in tsetse fly, which are the natural habitats of BSF and PCF T. brucei, respectively. In animals, blood glucose level is tightly regulated by hormones such as glucagon and insulin. Normal blood glucose concentration in humans is ∼4 mM (0.72 g/L) and it is higher after a meal or under diabetic conditions. Low blood glucose <0.389 mM (0.7 g/L) can occur in hypoglycemia and can cause seizure and coma, thus unlikely to be present as a long-lasting condition. In the insect vector tsetse fly, proline is the major source of energy to support flight and reproduction.56 Proline is synthesized mainly from lipid in the fat-bodies and oxidized to alanine in the muscle.57 The alanine is then transported to and reserved in fat-bodies for proline resynthesis.57 Due to this cyclic production and consumption of proline, the level of proline in the tsetse fly is likely kept at a consistent level even during starvation or T. brucei infection. Indeed, T. brucei parasitemia at 4 to 7 d postinfection is not changed in starved tsetse flies compared with fed controls, suggesting a steady supply of proline.58
While glucose and proline supplies are likely not a constraint, the amino acids availability in host environments may vary. Almost 40 y ago, Newport et al. measured the free serum amino acids in field voles infected with T. b. gambiense, a subspecies of T. brucei causing chronic human trypanosomiasis in central and west Africa.59 The infected voles are in a state of hypoaminoacidemia, with reduced levels of threonine, serine, valine, isoleucine, leucine, tyrosine and tryptophan.59 Since T. b. gambiense parasitemia is low in infected animals, the reduction of amino acid level is expected to be markedly higher in T. b. brucei or T. b. rhodesiense infected animals, which often have high parasitemia. More recently, a global urine and plasma metabolic responses to T. b. brucei infection have been systemically studied in mice.60 A significant reduction (by 80% to 90%) in leucine, isoleucine and valine levels is detected in the infected plasma.60 On the other hand, the plasma glucose level in the infected animals is tightly regulated and does not vary as much (with a ∼10% increase 1 d after infection and returning to preinfection level before d 21, and with a ∼10 % decrease on d 28 and 33).60 The constant glucose level during T. brucei infection (until just before host death) is also reported by Balmer et al.61 We therefore propose that trypanosomal autophagy can occur in hypoaminoacidemia hosts, induced by high parasitemia or by changed host metabolism.62 Future animal infection studies that allow simultaneous monitoring of plasma amino acid levels and autophagosome formation in T. brucei would be needed to substantiate this hypothesis. How this might affect parasite infection and pathogenesis also remains to be investigated, using pleomorphic T. brucei strains that are capable of differentiation in animal models.
Materials and methods
Cell lines and reagents
All experiments were performed in procyclic (PCF) T. brucei YTat1.1 and 29.1363,64 or in bloodstream form (BSF) Single-Marker (SM)64 strains. The cells were maintained in Cunningham's medium65 supplemented with 15% fetal bovine serum (FBS; Hyclone SH30071) or tetracyclin-free FBS (Clontech, 631106) for PCF, or in HMI-9 medium (IMDM (Gibco, G-12200–028) supplemented with 1 mM hypothanxine (Sigma, H9377), 1 mM sodium pyruvate (Sigma, P5280), 0.16 mM thymidine (Sigma, T1845), 0.05 mM bathocuprone sulfate (Sigma, B1125) and 1.5 mM L-cysteine (Sigma, C7352) supplemented with 10% FBS for BSF, with appropriate antibiotics. Oligomycin (75351), CCCP (C2759) and 2-deoxy-D-glucose (D3179) were purchased from Sigma. LysoTracker Red (10 mM; L7528) and MitoTracker Red CMXRos (0.1 μg/ml; M7512) were purchased from Invitrogen. All other chemicals were purchased from Sigma. Antibodies used in this study are shown below.
Vector construction and transfection
For inducible RNAi of Tb927.1.3830/GPI, Tb927.10.12700/PDH, Tb927.7.210/PRODH, Tb927.11.9980/OGDH and Tb927.8.2450/AMPKβ and Tb927.10.3700/AMPKγ, suitable fragments were selected using RNAit,66 to avoid off-target effects. The fragments were amplified with the primers listed in Table S1, digested with XbaI and subcloned into p2T7 vectors.67 For stable transfections, DNA constructs were linearized with NotI and electroporated into 29.13 cells stably expressing YFP-TbATG8.2. Stable transformants were selected with 10 μg/ml phleomycin (Sigma, P9564).
For ectopic expression of YFP-TbATG8.2 in BSF cells, YFP and TbATG8.2 cDNA fragments were inserted into pTSA-rib vector sequentially using XhoI/BamHI and BamHI/BamHI sites, respectively. The construct was linearized with BglII or SphI and electroporated into SM cells using Nucleofector™ (Lonza, Switzerland) with Human T cell transfection reagent (Lonza, VPA-1002). Stable transformants were selected with 5 μg/ml phleomycin.
Construction of TbATG8.2 knockout cell line
A PCR based method68 was used to replace the 2 TbATG8.2 alleles with blasticidin (BSD) and puromycin (PUR) resistance markers, respectively. The drug cassettes were amplified with primer pairs ATG8.2-KO-bsd-f (attcaccactggtacatataaaaagaaagcacagaagaattagtgtacataatggccaagcctttgtctcaa) and ATG8.2-KO-bsd-r (atgttttattgcgcatcaaagcaactaatggtttactaacactgaaggaaattagccctcccacacataacca), or ATG8.2-KO-pur-f (attcaccactggtacatataaaaagaaagcacagaagaattagtgtacataatgaccgagtacaagccca) and ATG8.2-KO-pur-r (atgttttattgcgcatcaaagcaactaatggtttactaacactgaaggaaatcaggcaccgggcttgc). Transfection of YTat1.1 cells was performed using the PCR products and stable transformants were selected with 10 μg/ml blasticidin (Invitrogen, R210–01) or 5 μg/ml puromycin (Sigma, P8833). Selected TbATG8.2−/− clones were confirmed by PCR using primers listed in Table S1 and by immuoblots using anti-TbATG8.2.20
Starvation conditions and autophagy measurements
For amino acids starvation of PCF cells, T. brucei cells harvested from mid-LOG phase culture were washed once with HBSS (137 mM NaCl, 5.4 mM KCl, 0.25 mM Na2HPO4, 0.44 mM KH2PO4, 1.3 mM CaCl2, 1 mM MgSO4, 4.2 mM NaHCO3, pH 7.3), gHBSS (HBSS with 5.6 mM (1 g/L) glucose),20 pHBSS (HBSS with 17.4 mM (2 g/L) proline) or aHBSS (HBSS with 22 mM (2 g/L) β-alanine and 12 mM (1.09 g/L) DL-alanine) with or without 2-DG and then resuspended in the same starvation buffer at a concentration of 5 × 106 cells/ml and incubated at 27°C for 2 h unless otherwise indicated.
For amino acids starvation of BSF trypanosomes, cells harvested from mid-LOG phase culture were washed once with cytomix (2 mM EGTA, pH 7.6, 120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, pH 7.6, 25 mM HEPES, pH 7.6, 5 mM MgCl2, 0.5% glucose, 100 μg/ml BSA (Sigma, A7906), 1 mM hypoxanthine (Sigma, H9377), pH 7.6), glucose-depleted cytomix with or without 2-DG or proline, or cytomix with varying concentrations of glucose (1.1 to 5.6 mM (0.2–1.0 g/L)) and then resuspended in the same buffer at a concentration of 5 × 105 cells/ml and incubated at 37°C with 5% CO2 for 2 h.
For glucose starvation, cells were washed once and then resuspended with sugar-free medium with or without proline at a concentration of 5 × 106 cells/ml and incubated at 27°C for up to 12 h. To monitor the effects of oligomycin and CCCP on autophagy, cells stably expressing YFP-TbATG8.2 were pretreated with 5 μg/ml oligomycin or 50 μM CCCP for 30 min, washed once with starvation buffer, followed by starvation in the presence of inhibitors for another 2 h. The formation of autophagosomes was monitored by fluorescence microscopy.
Fluorescence microscopy
For autophagosome visualization and quantification, control or treated cells were washed and resuspended in serum-free medium or starvation buffers with or without drugs, and attached to coverslips. Cells were fixed with 4% paraformaldehyde (Sigma, P6148) in phosphate-buffered saline (PBS; 1st BASE, BUF-2040), pH 7.4 for 20 min at room temperature. Nuclear and mitochondrial DNA was stained with 2 μg/ml DAPI (Invitrogen, D1306) and observed using an Axioplan2 inverted microscope (Carl Zeiss MicroImaging, Jena, Germany) equipped with a CoolSNAP HQ2 camera (Photometrics, Tucson, Arizona, USA) and a Plan-Apochromat 63×/1.40 oil DIC objective. The acidocalcisome staining and documentation using LysoTracker Red was published previously.21
Adenine nucleotides (ATP, ADP and AMP) measurements
The adenine nucleotides were extracted with boiling water method.69 Briefly, a total of 2 × 107 cells from different treatments were pelleted and resuspended in 100 μL ice-cold dH2O, which was immediately heated in a boiling water bath for 10 min. The boiled lysates were centrifuged at 17,000 g for 5 min at 4°C and the supernatants were collected for adenine nucleotides concentration measurements using an ATP, ADP and AMP Assay kit (BMR, A-125), following the manufacturer's protocol.
In this protocol, the AMP and ADP were converted to ATP by adenylate kinase and pyruvate kinase, respectively (supplied in the kit). Three sets of reactions were measured for each sample: SET I (AMP+ADP+ATP), SET II (ADP+ATP) and SET III (ATP). The concentration of each adenine nucleotide was calculated as followed: AMP concentration = [ATP]SET I – [ATP]SET II; ADP concentration = [ATP]SET II – [ATP]SET III; ATP concentration = [ATP]SET III. The measurement was performed by a luciferin–luciferase bioluminescence assay in GloMax® 96 Microplate luminometer (Promega, Madison, Wisconsin, USA). Relative Light Units (RLUs) were converted to concentration (nmol adenine nucleotide per 107 cells) based on the standard curve constructed with 1 to 100 μM ATP standard (Fig. S1A).
Measurement of mitochondrial membrane potential
The measurement was performed following published protocol by Graef and Nunnari16 with slight modifications. Briefly, after 1 h of amino acid starvation (gHBSS or pHBSS), the cells were stained with 0.1 μg/ml MitoTracker Red CMXRos (Molecule Probes, M7512) for 30 min at 27°C. Control or starved cells were washed with serum-free medium, gHBSS or pHBSS once and imaged using an Axioplan2 inverted microscope (Carl Zeiss MicroImaging, Jena, Germany) equipped with a CoolSNAP HQ2 camera (Photometrics, Tucson, Arizona, USA) and a Plan-Apochromat 63×/1.4×0 oil DIC objective.
Immunoblotting analyses
Cells were cultivated in medium or starved in HBSS, gHBSS, pHBSS or aHBSS with or without 2-DG for 2 h. Cell lysates were prepared from 5 × 106 cells, and then separated using 12.5% SDS-PAGE gels containing 6 M urea.20 The proteins were immobilized onto PVDF membrane (Bio-Rad, 162–0177) and probed with anti-Tb927.7.5910/TbATG8.220 or anti-Tb927.3.4290/PFR121 as loading control. Following incubation with goat-anti-rat IgG conjugated to horseradish peroxidase (Sigma, A9037), the blots were processed using the Clarity Western ECL system (Bio-Rad, 170–5060) and documented by ImageQuant LAS 4000 mini Luminescent Image Analyzer (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
RNA extraction and reverse transcription (RT)-PCR
Total RNA was extracted from 2 × 107 control or RNAi cells with TRIzol® Reagent (Life Technologies, 15596–026) following the manufacturer's protocol. The RNA samples were treated with RNase-free DNase I (Roche, 04 716 728 001) and the first-strand cDNA was synthesized with M-MLV Reverse Transcriptase (Invitrogen, 28025–013). PCR was performed using the primers listed in Table S1.
Statistical analyses
For autophagosome quantification and adenine nucleotides quantification, 3 independent experiments were performed and statistical analyses were performed using the 2-tailed, equal variance Student t test. P values <0.05, <0.01 and <0.001 were determined to be statistically significant (*), highly significant (**) and extremely significant (***), respectively. P value > 0.05 was determined as not statistically significant (NS).
Supplementary Material
Abbreviations
- 2-DG
2-deoxy-D-glucose
- AcCoA
acetyl-CoA or acetyl coenzyme A
- AMPK
AMP-activated protein kinase
- ASCT
acetate, succinate CoA-transferase
- ATG
autophagy related
- ATP
adenosine triphosphate
- BSF
bloodstream form of Trypanosoma brucei
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- DMSO
dimethyl sulfoxide
- FADH2
flavin adenine dinucleotide hydroquinone form
- gHBSS
Hank's balanced salt solution supplemented with glucose
- HBSS
Hank's balanced salt solution
- NADH
nicotinamide adenine dinucleotide
- OGDH
2-oxoglutarate dehydrogenase
- PCF
procyclic form of Trypanosoma brucei
- PDH
pyruvate dehydrogenase
- GPI
glucose-6-phosphate isomerase or phosphoglucose isomerase
- pHBSS
Hank's balanced salt solution supplemented with proline
- PRODH
proline dehydrogenase
- SCoAS
succinyl-CoA synthetase
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCA cycle
tricarboxylic acid cycle or citric acid cycle or Krebs cycle
- WT
wild type
- YFP
yellow fluorescent protein
Disclosure of potential conflicts of interest
There were no potential conflicts of interest that needed to be disclosed.
Acknowledgments
We would like to thank Anais E.S.C. Brasseur in our lab for the pTSA-YFP-TbATG8.2 vector.
Funding
This study is supported by a Tier 1 research grant (to C.Y.H.) from the Singapore Ministry of Education and a grant (#31472058 to Z.R.L.) from the National Science Foundation of China.
References
- [1].Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280-93; PMID:20965422; http://dx.doi.org/ 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell 2014; 159:1263-76; PMID:25480292; http://dx.doi.org/ 10.1016/j.cell.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 2011; 13:1016-23; PMID:21892142; http://dx.doi.org/ 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moruno F, Perez-Jimenez E, Knecht E. Regulation of autophagy by glucose in Mammalian cells. Cells 2012; 1:372-95; PMID:24710481; http://dx.doi.org/ 10.3390/cells1030372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 2010; 29:2082-96; PMID:20473272; http://dx.doi.org/ 10.1038/emboj.2010.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/ 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al.. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 2014; 53:710-25; PMID:24560926; http://dx.doi.org/ 10.1016/j.molcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- [8].Duran RV, MacKenzie ED, Boulahbel H, Frezza C, Heiserich L, Tardito S, Bussolati O, Rocha S, Hall MN, Gottlieb E. HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 2013; 32:4549-56; PMID:23085753; http://dx.doi.org/ 10.1038/onc.2012.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Mariño G, Pietrocola F, Harger A, Zimmermann A, et al.. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab 2014; 19:431-44; PMID:24606900; http://dx.doi.org/ 10.1016/j.cmet.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, et al.. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature 2014; 510:397-401; PMID:24828042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 2005; 6:439-48; PMID:15928708; http://dx.doi.org/ 10.1038/nrm1660 [DOI] [PubMed] [Google Scholar]
- [12].Plomp PJ, Wolvetang EJ, Groen AK, Meijer AJ, Gordon PB, Seglen PO. Energy dependence of autophagic protein degradation in isolated rat hepatocytes. Eur J Biochem 1987; 164:197-203; PMID:3830181; http://dx.doi.org/ 10.1111/j.1432-1033.1987.tb11011.x [DOI] [PubMed] [Google Scholar]
- [13].Campbell JM, Weissbach H. The effect of amino acid starvation on nucleoside uptake and RNA synthesis in Tetrahymena. J Biol Chem 1980; 255:4691-7; PMID:6154697 [PubMed] [Google Scholar]
- [14].DeVorkin L, Go NE, Hou YC, Moradian A, Morin GB, Gorski SM. The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB. J Cell Biol 2014; 205:477-92; PMID:24862573; http://dx.doi.org/ 10.1083/jcb.201303144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Grummt I, Grummt F. Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell 1976; 7:447-53; PMID:947552; http://dx.doi.org/ 10.1016/0092-8674(76)90175-6 [DOI] [PubMed] [Google Scholar]
- [16].Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J 2011; 30:2101-14; PMID:21468027; http://dx.doi.org/ 10.1038/emboj.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paquette JC, Guerin PJ, Gauthier ER. Rapid induction of the intrinsic apoptotic pathway by L-glutamine starvation. J Cell Physiol 2005; 202:912-21; PMID:15389638; http://dx.doi.org/ 10.1002/jcp.20194 [DOI] [PubMed] [Google Scholar]
- [18].Klionsky DJ. What can we learn from trypanosomes? Autophagy 2006; 2:63-4; PMID:16874079; http://dx.doi.org/ 10.4161/auto.2.2.2418 [DOI] [PubMed] [Google Scholar]
- [19].Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, et al.. Autophagy in protists. Autophagy 2011; 7:127-58; PMID:20962583; http://dx.doi.org/ 10.4161/auto.7.2.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li FJ, Shen Q, Wang C, Sun Y, Yuan AY, He CY. A role of autophagy in Trypanosoma brucei cell death. Cell Microbiol 2012; 14:1242-56; PMID:22463696; http://dx.doi.org/ 10.1111/j.1462-5822.2012.01795.x [DOI] [PubMed] [Google Scholar]
- [21].Li FJ, He CY. Acidocalcisome is required for autophagy in Trypanosoma brucei. Autophagy 2014; 10:1978-88; PMID:25484093; http://dx.doi.org/ 10.4161/auto.36183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Herman M, Perez-Morga D, Schtickzelle N, Michels PA. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 2008; 4:294-308; PMID:18365344; http://dx.doi.org/ 10.4161/auto.5443 [DOI] [PubMed] [Google Scholar]
- [23].Cull B, Godinho JLP, Rodrigues JCF, Frank B, Schurigt U, Williams RAM, Coombs GH, Mottram JC. Glycosome turnover in Leishmania major is mediated by autophagy. Autophagy 2014; 10:2143-57; PMID:25484087; http://dx.doi.org/ 10.4161/auto.36438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brennand A, Rico E, Rigden DJ, Van Der Smissen P, Courtoy PJ, Michels PA. ATG24 Represses Autophagy and Differentiation and Is Essential for Homeostasy of the Flagellar Pocket in Trypanosoma brucei. PLoS One 2015; 10:e0130365; PMID:26090847; http://dx.doi.org/ 10.1371/journal.pone.0130365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ghosh D, Walton JL, Roepe PD, Sinai AP. Autophagy is a cell death mechanism in Toxoplasma gondii. Cell Microbiol 2012; 14:589-607; PMID:22212386; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01745.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eickel N, Kaiser G, Prado M, Burda PC, Roelli M, Stanway RR, Heussler VT. Features of autophagic cell death in Plasmodium liver-stage parasites. Autophagy 2013; 9:568-80; PMID:23388496; http://dx.doi.org/ 10.4161/auto.23689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].van Weelden SW, Fast B, Vogt A, van der Meer P, Saas J, van Hellemond JJ, Tielens AG, Boshart M. Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J Biol Chem 2003; 278:12854-63; PMID:12562769; http://dx.doi.org/ 10.1074/jbc.M213190200 [DOI] [PubMed] [Google Scholar]
- [28].Lamour N, Riviere L, Coustou V, Coombs GH, Barrett MP, Bringaud F. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J Biol Chem 2005; 280:11902-10; PMID:15665328; http://dx.doi.org/ 10.1074/jbc.M414274200 [DOI] [PubMed] [Google Scholar]
- [29].Fish WR, Looker DL, Marr JJ, Berens RL. Purine metabolism in the bloodstream forms of Trypanosoma gambiense and Trypanosoma rhodesiense. Biochim Biophys Acta 1982; 719:223-31; PMID:6817814; http://dx.doi.org/ 10.1016/0304-4165(82)90092-7 [DOI] [PubMed] [Google Scholar]
- [30].Fish WR, Marr JJ, Berens RL. Purine metabolism in Trypanosoma brucei gambiense. Biochim Biophys Acta 1982; 714:422-8; PMID:6800408; http://dx.doi.org/ 10.1016/0304-4165(82)90149-0 [DOI] [PubMed] [Google Scholar]
- [31].Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 2005; 280:31582-6; PMID:16027116; http://dx.doi.org/ 10.1074/jbc.M506736200 [DOI] [PubMed] [Google Scholar]
- [32].Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab 2011; 13:495-504; PMID:21531332; http://dx.doi.org/ 10.1016/j.cmet.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Proto WR, Jones NG, Coombs GH, Mottram JC. Tracking autophagy during proliferation and differentiation of Trypanosoma brucei. Microbial Cell 2014; 1:9-20; http://dx.doi.org/ 10.15698/mic2014.01.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol 2006; 149:1-9; PMID:16682088; http://dx.doi.org/ 10.1016/j.molbiopara.2006.03.017 [DOI] [PubMed] [Google Scholar]
- [35].Allmann S, Morand P, Ebikeme C, Gales L, Biran M, Hubert J, Brennand A, Mazet M, Franconi JM, Michels PA, et al.. Cytosolic NADPH homeostasis in glucose-starved procyclic Trypanosoma brucei relies on malic enzyme and the pentose phosphate pathway fed by gluconeogenic flux. J Biol Chem 2013; 288:18494-505; PMID:23665470; http://dx.doi.org/ 10.1074/jbc.M113.462978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Durieux PO, Schutz P, Brun R, Kohler P. Alterations in Krebs cycle enzyme activities and carbohydrate catabolism in two strains of Trypanosoma brucei during in vitro differentiation of their bloodstream to procyclic stages. Mol Biochem Parasitol 1991; 45:19-27; PMID:1904988; http://dx.doi.org/ 10.1016/0166-6851(91)90023-Y [DOI] [PubMed] [Google Scholar]
- [37].Riviere L, van Weelden SW, Glass P, Vegh P, Coustou V, Biran M, van Hellemond JJ, Bringaud F, Tielens AG, Boshart M. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem 2004; 279:45337-46; PMID:15326192; http://dx.doi.org/ 10.1074/jbc.M407513200 [DOI] [PubMed] [Google Scholar]
- [38].Millerioux Y, Morand P, Biran M, Mazet M, Moreau P, Wargnies M, Ebikeme C, Deramchia K, Gales L, Portais JC, et al.. ATP synthesis-coupled and -uncoupled acetate production from acetyl-CoA by mitochondrial acetate:succinate CoA-transferase and acetyl-CoA thioesterase in Trypanosoma. J Biol Chem 2012; 287:17186-97; PMID:22474284; http://dx.doi.org/ 10.1074/jbc.M112.355404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Van Hellemond JJ, Opperdoes FR, Tielens AG. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc Natl Acad Sci U S A 1998; 95:3036-41; PMID:9501211; http://dx.doi.org/ 10.1073/pnas.95.6.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Duarte M, Tomas AM. The mitochondrial complex I of trypanosomatids–an overview of current knowledge. J Bioenerg Biomembr 2014; 46:299-311; PMID:24961227; http://dx.doi.org/ 10.1007/s10863-014-9556-x [DOI] [PubMed] [Google Scholar]
- [41].Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology 2011; 13:132-U71; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al.. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; PMID:21205641; http://dx.doi.org/ 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Clemmens CS, Morris MT, Lyda TA, Acosta-Serrano A, Morris JC. Trypanosoma brucei AMP-activated kinase subunit homologs influence surface molecule expression. Exp Parasitol 2009; 123:250-7; PMID:19647733; http://dx.doi.org/ 10.1016/j.exppara.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Johnson MA, Vidoni S, Durigon R, Pearce SF, Rorbach J, He JY, Brea-Calvo G, Minczuk M, Reyes A, Holt IJ, et al.. Amino Acid Starvation has opposite effects on mitochondrial and cytosolic protein synthesis. Plos One 2014; 9:e93597; PMID:24718614; http://dx.doi.org/ 10.1371/journal.pone.0093597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shao ML, Shan B, Liu Y, Deng YP, Yan C, Wu Y, Mao T, Qiu Y, Zhou Y, Jiang S, et al.. Hepatic IRE1 alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPAR alpha axis signalling. Nat Commun 2014; 5:3528; PMID:24670948 [DOI] [PubMed] [Google Scholar]
- [46].O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 2013; 493:346-55; PMID:23325217; http://dx.doi.org/ 10.1038/nature11862 [DOI] [PubMed] [Google Scholar]
- [47].Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell 2005; 16:2503-17; PMID:15758028; http://dx.doi.org/ 10.1091/mbc.E04-11-0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/ 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Schmidt RS, Butikofer P. Autophagy in Trypanosoma brucei: amino acid requirement and regulation during different growth phases. PLoS One 2014; 9:e93875; PMID:24699810; http://dx.doi.org/ 10.1371/journal.pone.0093875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Proto WR. Characterisation of autophagy and a metacaspase in Trypanosoma brucei. 2010; PhD thesis, University of Glasgow. [Google Scholar]
- [51].Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, Alnemri ES, Altucci L, Andrews D, Annicchiarico-Petruzzelli M, et al.. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 2015; 22:58-73; PMID:25236395; http://dx.doi.org/ 10.1038/cdd.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Huang G, Vercesi AE, Docampo R. Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat Commun 2013; 4:2865; PMID:24305511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251-62; PMID:22436748; http://dx.doi.org/ 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ramirez-Peinado S, Leon-Annicchiarico CL, Galindo-Moreno J, Iurlaro R, Caro-Maldonado A, Prehn JH, Ryan KM, Muñoz-Pinedo C. Glucose-starved cells do not engage in prosurvival autophagy. J Biol Chem 2013; 288:30387-98; PMID:24014036; http://dx.doi.org/ 10.1074/jbc.M113.490581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, Savaraj N, Lampidis TJ. 2-Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol 2011; 67:899-910; PMID:20593179; http://dx.doi.org/ 10.1007/s00280-010-1391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bursell E, Slack E. Oxidation of Proline by Sarcosomes of Tsetse-Fly, Glossina-Morsitans. Insect Biochem 1976; 6:159-67; http://dx.doi.org/ 10.1016/0020-1790(76)90026-3 [DOI] [Google Scholar]
- [57].Bursell E. Synthesis of proline by fat-body of Tsetse-Fly (Glossina-Morsitans) - Metabolic Pathways. Insect Biochem 1977; 7:427-34; http://dx.doi.org/ 10.1016/S0020-1790(77)90068-3 [DOI] [Google Scholar]
- [58].Welburn SC, Maudlin I. Control of Trypanosoma brucei brucei infections in tsetse, Glossina morsitans. Med Vet Entomol 1997; 11:286-9; PMID:9330261; http://dx.doi.org/ 10.1111/j.1365-2915.1997.tb00408.x [DOI] [PubMed] [Google Scholar]
- [59].Newport GR, Page CR 3rd, Ashman PU, Stibbs HH, Seed JR. Alteration of free serum amino acids in voles infected with Trypanosoma brucei gambiense. J Parasitol 1977; 63:15-24; PMID:321737; http://dx.doi.org/ 10.2307/3280098 [DOI] [PubMed] [Google Scholar]
- [60].Wang Y, Utzinger J, Saric J, Li JV, Burckhardt J, Dirnhofer S, Nicholson JK, Singer BH, Brun R, Holmes E. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc Natl Acad Sci U S A 2008; 105:6127-32; PMID:18413599; http://dx.doi.org/ 10.1073/pnas.0801777105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Balmer O, Stearns SC, Schotzau A, Brun R. Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology 2009; 90:3367-78; PMID:20120806; http://dx.doi.org/ 10.1890/08-2291.1 [DOI] [PubMed] [Google Scholar]
- [62].Gobert AP, Daulouede S, Lepoivre M, Boucher JL, Bouteille B, Buguet A, Cespuglio R, Veyret B, Vincendeau P. L-Arginine availability modulates local nitric oxide production and parasite killing in experimental trypanosomiasis. Infect Immun 2000; 68:4653-7; PMID:10899869; http://dx.doi.org/ 10.1128/IAI.68.8.4653-4657.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ruben L, Egwuagu C, Patton CL. African Trypanosomes Contain Calmodulin Which Is Distinct from Host Calmodulin. Biochim Biophys Acta 1983; 758:104-13; PMID:6135450; http://dx.doi.org/ 10.1016/0304-4165(83)90290-8 [DOI] [PubMed] [Google Scholar]
- [64].Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 1999; 99:89-101; PMID:10215027; http://dx.doi.org/ 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- [65].Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool 1977; 24:325-9; PMID:881656; http://dx.doi.org/ 10.1111/j.1550-7408.1977.tb00987.x [DOI] [PubMed] [Google Scholar]
- [66].Redmond S, Vadivelu J, Field MC. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol 2003; 128:115-8; PMID:12706807; http://dx.doi.org/ 10.1016/S0166-6851(03)00045-8 [DOI] [PubMed] [Google Scholar]
- [67].Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol 2002; 125:211-6; PMID:12467990; http://dx.doi.org/ 10.1016/S0166-6851(02)00238-4 [DOI] [PubMed] [Google Scholar]
- [68].Melville SE. Parasite genomics protocols Totowa NJ: Humana Press, 2004. [Google Scholar]
- [69].Yang NC, Ho WM, Chen YH, Hu ML. A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal Biochem 2002; 306:323-7; PMID:12123672; http://dx.doi.org/ 10.1006/abio.2002.5698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


