ABSTRACT
Macroautophagy is a catabolic process that delivers cytoplasmic components via the autophagosome to lysosomes for degradation. Measuring autophagic activity is critical to dissect molecular mechanisms and functions of autophagy but remains challenging due to the lack of a definitive method. We have recently developed a new fluorescent probe, GFP-LC3-RFP-LC3ΔG, to assess autophagic flux. Upon intracellular expression, the probe is cleaved by ATG4 family proteases into equimolar amounts of GFP-LC3 and RFP-LC3ΔG. The former is degraded by autophagy while the latter persists as an internal control in the cytosol. Autophagic flux can thus be quantified by obtaining the ratio of GFP:RFP signals. Using this method, we have identified several autophagy-modulating drugs by screening an approved drug library. We have also demonstrated that induced and basal autophagic flux can be monitored in zebrafish and mice.
KEYWORDS: autophagic flux assay, autophagy, LC3, mice, zebrafish
Macroautophagy (hereafter, autophagy) is an intracellular degradation system in which a portion of cytoplasm is enclosed by the autophagosome and degraded upon fusion with the lysosome. Autophagy has been implicated in many physiological and pathological settings including human diseases. Accordingly, accurate quantification of the overall process of autophagic degradation, termed ‘autophagic flux,’ is crucial in autophagy studies. However, due to the intricate nature of this pathway, accuracy is often lacking in autophagic flux analyses.
So far, several methods have been used to measure autophagic activity. Autophagosomes are marked by the phosphatidylethanolamine (PE)-conjugated form of MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3; termed LC3-II), a homolog of yeast Atg8 (autophagy-related 8). Measurements can be conducted, for example, by counting the number of autophagosomes visualized by electron or fluorescence microscopy. As the amount of LC3-II reflects the number of autophagosomes, LC3-II levels can also be determined biochemically to ascertain autophagic activity. These assays can reveal accumulation of autophagosomes but are insufficient to measure autophagic flux as an increased number of autophagosomes can result from not only enhanced autophagy induction but also from defects downstream of the process (e.g., in autophagosome-lysosome fusion). To differentiate between the 2 possible causes, it is essential to include a sample treated with lysosomal inhibitors albeit at the risk of additional effects, including an inhibition of MTORC1 (activating autophagy). The complexity of measuring autophagic flux is further compounded when the subject is a whole organism.
We reasoned that the ideal method would allow measurement of cumulative degradation of autophagic substrates without the use of lysosomal inhibitors. It should also be easy, quantitative, and applicable for standard laboratory equipment. To this end, we developed a new fluorescence probe, GFP-LC3-RFP-LC3ΔG, in which the C terminus of GFP-LC3 is fused to RFP-LC3 without the C-terminal glycine (Fig. 1A). When it is expressed in cells, endogenous ATG4 family proteases cleave the peptide bond after the C-terminal glycine of the first LC3, producing equimolar amounts of GFP-LC3 and RFP-LC3ΔG. Upon autophagy induction, GFP-LC3 is conjugated to PE on autophagosomes. Following fusion with the lysosome, GFP-LC3 on the autophagosomal inner membrane is quenched and/or degraded. Incapable of lipidation, RFP-LC3ΔG, by contrast, remains in the cytosol to serve as an internal control. Thus, the GFP:RFP signal ratio inversely correlates with autophagic activity (Fig. 1B).
Figure 1.
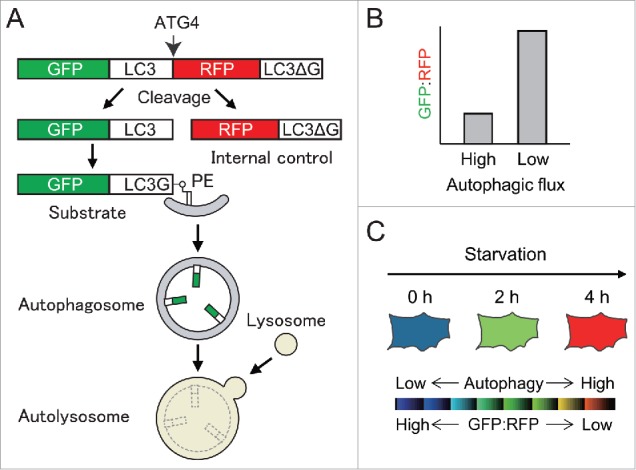
A new method to measure autophagic flux using GFP-LC3-RFP-LC3ΔG probe. (A) A schematic representation to assess autophagic flux using the GFP-LC3-RFP-LC3ΔG probe, which is cleaved by ATG4 into GFP-LC3 (an autophagic substrate) and RFP-LC3ΔG (an internal control). (B) Autophagic flux can be estimated by calculating the GFP:RFP ratio. The ratio decreases when autophagic flux is high, whereas it increases when autophagic flux is low. (C) A schematic example of ratiometric analysis of starvation-induced autophagy in cells expressing GFP-LC3-RFP-LC3ΔG. The blue signal represents high GFP:RFP and low autophagic flux, and the red signal represents low GFP:RFP and high autophagic flux.
As proof of concept, we demonstrated that the GFP:RFP ratio is reduced upon nutrient starvation in wild-type but not in autophagy-deficient cells. Most drugs that were previously reported as autophagy inducers (e.g., Torin1) decreased the GFP:RFP ratio whereas those reported as autophagy inhibitors (e.g., bafilomycin A1 and chloroquine) increased it. This method is applicable for fluorescence microscopy, flow cytometry, and microplate readers (Fig. 1C).
Taking advantage of this high-throughput method, we screened an approved drug library (1,054 drugs) and identified 47 autophagy inducers and 43 autophagy inhibitors. Among them, we identified 13 novel inducers and 18 novel inhibitors. Importantly, 6 out of the above 43 inhibitors were previously reported as “inducers." This may be partly due to inappropriate interpretation of observed autophagosome accumulation in previous studies. This probe is thus particularly useful for large-scale autophagy drug screenings.
This method was also used to monitor autophagic flux in vivo. In zebrafish embryos, the GFP:RFP ratio in the skeletal muscle was reduced upon treatment with Torin1. Furthermore, for the first time, we demonstrated that autophagy is induced 10 min after fertilization in zebrafish, which is interesting as it takes place despite sufficient nutrients stored in the yolk. Similarly, autophagic flux increased after fertilization in mouse early embryos but with much slower kinetics.
This probe is ideal for detection of “basal” autophagic flux because it can quantify autophagic flux as a cumulative index. Indeed, we observed that basal autophagic flux varies among tissues and/or cells. For example, it is higher in the lens compared with the retina in zebrafish. In the skeletal muscle of mice, it differed among muscle fiber types. Thus, the probe is capable of measuring both induced and basal autophagic flux.
What are the differences of the probe with other potential single molecular probes? GFP-LC3-RFP can function similarly to GFP-LC3-RFP-LC3ΔG. However, RFP-LC3ΔG should theoretically serve as a better control than RFP because RFP-LC3ΔG can undergo similar post-translational modifications to GFP-LC3 (except for lipidation). The sensitivity of the GFP-LC3-RFP-LC3ΔG probe is higher than that of tfLC3 (RFP-GFP-LC3) because RFP in tfLC3 is delivered to and degraded in lysosomes. Nevertheless, tfLC3 still maintains a clear advantage in analyzing maturation steps of individual autophagic structures.
What are the limitations of this probe? First, homologous recombination could occur between the 2 LC3s of GFP-LC3-RFP-LC3ΔG, especially during generation of stable transformants. We recommend isolating clones that properly express the probe. Second, time resolution is not high with an apparent reduction of the GFP:RFP ratio requiring 2–4 h. However, this is advantageous for detection of basal autophagy. Last, the GFP:RFP ratio may not change in a uniform manner if expression levels of the probe vary dramatically among cells/tissues. Hence, it is important to compare cells/tissues with similar RFP expression levels. We are currently generating knock-in mice expressing the probe with ubiquitous and higher expression levels to perform comprehensive analyses of autophagic flux in vivo. The plasmids of these probes are available at Addgene.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (Grant Number 25111005) (to N.M.).


