Abstract
Establishment and maintenance of persistent, latent infection by Mycobacterium tuberculosis are dependent on expression of the mprA-mprB regulatory system. Previously, MprA and MprB were shown to participate in phosphotransfer reactions characteristic of two-component signaling systems. To begin identifying downstream effector genes regulated by mprA-mprB during persistent stages of infection, a search for the regulatory sequence(s) recognized by response regulator MprA was carried out. Here, evidence is presented demonstrating that MprA recognizes a 19-bp sequence comprising two loosely conserved 8-bp direct repeat subunits separated by 3 nucleotides. This motif, termed the MprA box, is found upstream of the mprA coding sequence and that of downstream gene pepD (Rv0983). Protein phosphorylation was not required for binding to this DNA sequence by MprA in vitro; however, phosphorylation enhanced DNA binding by MprA and was required for the regulation of mprA and pepD by MprA in vivo. Binding of MprA to the MprA box was dependent on conserved nucleotides contained within repeat subunits and on the spacer length separating these repeats. In addition, recognition of this sequence proceeded via tandem binding of two monomers of MprA. Identification of the genetic determinants regulated by MprA will ultimately enhance our understanding of the mechanisms utilized by M. tuberculosis to undergo latency.
Tuberculosis continues to be one of the most significant infectious diseases in the world despite the availability of a live attenuated vaccine and multiple antitubercular antibiotics (5). The etiological agent, Mycobacterium tuberculosis, is currently predicted to infect more than one-third of the world's population and result in ≈1.8 million human deaths annually (3). Approximately 8 million active infections are documented a year, many of which are likely the result of M. tuberculosis reactivation in latently infected individuals. While fewer than 10% of individuals harboring persistent M. tuberculosis infections undergo reactivation of tuberculosis in their lifetime, natural or induced periods of immune suppression significantly increase this risk to levels approaching 10% per annum (2, 11). Adding to this burden is the increased resistance of M. tuberculosis clinical isolates to commonly used antitubercular antibiotics and the increased incidence of human immunodeficiency virus infection in individuals persistently infected by M. tuberculosis (1, 4, 30). Thus, identification of novel therapeutic targets allowing the elimination of M. tuberculosis during persistent infection or development of therapeutic strategies preventing the reactivation of tubercle bacilli following periods of latency is needed to help diminish the recycling of M. tuberculosis within the human population.
The establishment of persistent infection by M. tuberculosis requires genes from the two-component signal transduction family. Bacterial pathogens frequently use two-component signaling transduction systems to adapt to changing environmental conditions within the host (42). Two-component systems comprise a histidine kinase sensor and a cytoplasmic cognate response regulator. Collectively, these protein pairs sense environmental stimuli and initiate adaptive transcriptional programs in the bacterium by means of phosphotransfer reactions. Of the 11 complete and several orphaned two-component signal transduction systems annotated in the M. tuberculosis genome (8), at least eight are important for aspects of virulence in the tubercle bacillus (32, 33, 35, 36, 44). For example, mutations in a subset of two-component response regulators or sensor kinases either attenuate or slow the growth of M. tuberculosis in macrophages in vitro relative to wild-type M. tuberculosis (35, 36). Interestingly, mutations in other two-component systems enhance the in vivo growth of M. tuberculosis in immunocompetent or immunocompromised SCID mice, suggesting that the genes required for M. tuberculosis virulence are also regulated by a subset of histidine kinase and response regulator proteins that are normally repressed in vivo (32).
The ability of M. tuberculosis to transition from rapid growth during acute periods of infection to the metabolically reduced growth observed during periods of bacterial persistence also appears to be regulated in part by two-component signaling systems. For example, the response regulator Rv3133c (DosR) modulates transcription of a common set of genes in M. tuberculosis in response to hypoxia and nitric oxide exposure, physiological conditions likely to be encountered by the tubercle bacillus during periods of latency (29, 34, 37, 38, 41). In addition, the mprAB two-component system regulates genes required by M. tuberculosis for establishment and maintenance of persistent infection in the host (44).
To begin identifying the genes regulated by this two-component system in M. tuberculosis, a search for the nucleotide motif(s) recognized by the response regulator MprA was carried out. Here we demonstrate that MprA autoregulates its own expression and that of downstream gene Rv0983 (pepD) by recognition of a loosely conserved sequence motif containing two 8-bp direct repeat subunits separated by 3 nucleotides. The sequence requirements for this interaction and the implications for M. tuberculosis pathogenesis are discussed.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Escherichia coli DH5α was used for cloning. E. coli BL21(DE3)/pLysS and Origami B(DE3) (Novagen) were used to overexpress and purify recombinant forms of MprA. E. coli strains were grown at 37°C in Luria-Bertani (LB) broth or LB agar. Ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (50 μg/ml) were added to LB media when appropriate. All antibiotics were purchased from Sigma. The Mycobacterium strains used in this study included Mycobacterium tuberculosis H37Rv (ATCC 27294) and Mycobacterium bovis BCG Pasteur (ATCC 35734). Mycobacterium strains were grown at 37°C in 7H9 broth or 7H10 agar medium (Difco) supplemented with 10% oleic acid-albumin-dextrose-NaCl (Difco). Isopropyl-β-d-thiogalactopyranoside (IPTG; Invitrogen) was added to E. coli cultures to induce protein overexpression at 0.1 mM.
Expression plasmids, primers, and oligonucleotide probes.
The molecular reagents used in this study are described in Table 1. All primers and oligonucleotides were purchased from Qiagen. Expression plasmids encoding wild-type His-tagged MprA [pTZ229] and the phosphorylation-deficient mutant His-MprA (D48A)[pTZ324] have been described previously (45). A thioredoxin (Trx)-tagged derivative of wild-type MprA was also constructed. Briefly, the coding sequence of mprA was amplified from M. tuberculosis H37Rv with mprAstart and mprAstop and cloned into pCR2.1-TOPO (Invitrogen). The resulting plasmid was digested with BamH1 and SalI and separated on an agarose gel, and the DNA fragment containing the mprA coding sequence was recovered and cloned into Trx-tagged pET32b (Novagen) digested with the same restriction enzymes, resulting in pTZ232. Because the N-terminal tag present on pET32b contains six His residues immediately downstream of the Trx tag, recombinant proteins expressed from this vector can be purified with nickel nitrilotriacetic acid column chromatography. DNA probes for electrophoretic mobility shift assays (EMSAs) were generated by PCR or by annealing complementary single-stranded DNA oligonucleotides. PCR products were initially resolved on 1% agarose gels and recovered by gel extraction. Oligonucleotide-derived probes were prepared by mixing complementary single-stranded DNA oligomers in 100 to 150 mM NaCl, heating at 95°C for 5 min, and then cooling to room temperature. All DNA probes were end labeled with [γ32-P]ATP (ICN Biochemical) with T4 polynucleotide kinase (Invitrogen), and unincorporated nucleotides were removed by column elution (Qiagen).
TABLE 1.
Primers, oligonucleotides, and plasmids used in this studya
| Primer, oligonucleotide, or plasmid | Sequence or description | Use, source, or reference |
|---|---|---|
| Primers | ||
| Rv0980 | 5′-GTGGACACTAACTGTGGGGCCACGTT-3′; FP for Rv0980 upstream region | EMSA |
| mprAseq10 | 5′-GTTCTCGTCCGCGCGGGTTTCGCCTG-3′; RP for Rv0980 upstream region | EMSA |
| mprAseq10up | 5′-ACCAACATTGGTGGAACGTGGTGC-3′; FP for mprA upstream region | EMSA, DNase I |
| mprAseq3 | 5′-CACGATCGTCGTCAACGACAAGAATT-3′; RP for mprA upstream region | EMSA, DNase I |
| mprBseq3up | 5′-TCGGATGCCGATTCCGCAGCTT-3′; FP for pepD upstream region | EMSA, DNase I |
| Rv0983NotI | 5′-GCGGCCGCCTAGGTTGCTCTTCCTGTACTAGG-3′; RP for pepD upstream region | EMSA, DNase I |
| Rm60F | 5′-CCCGCATACCACGGCGCG-3′; FP for MprA box upstream of mprA | Competitive EMSA |
| Rm60R | 5′-GGACACCAGTGTCGTCGCAGC-3′; RP for MprA box upstream of mprA | Competitive EMSA |
| mprAfpbs-F | 5′-CCAATGTGGCTGATGTGGCTAAAC-3′; FP for mprA upstream region | DNase I |
| mprAfpbs-R | 5′-AATCATGTCGAGCGCCTCA-3′; RP for mprA upstream region | DNase I |
| TOPOBamH1-fp | 5′-GATCCACTAGTAACGGCCGC-3′; used to generate sequencing ladders | DNase I |
| mprAstart | 5′-GGATCCGTGTCCGTGCGAATTCTTGTC-3′; mprA coding region | MprA expression |
| mprAstop | 5′-GTCGACTCAGGGTGGTGTTTCACGTAG-3′; mprA coding region | MprA expression |
| mprARTsense | 5′-CATTGCTGGAGATGCCTGATCG-3′ | Real-time RT-PCR |
| mprARTantisense | 5′-CTCGGTCTTGCGGCGTAG-3′ | Real-time RT-PCR |
| pepDRTsense | 5′-ACGACGGTAACCTTCTCTGAC-3′ | Real-time RT-PCR |
| pepDRTantisense | 5′-CGGAGACGCCCTGAACAC-3′ | Real-time RT-PCR |
| sigARTsense | 5′-GCCGATGACGACGAGGAG-3′ | Real-time RT-PCR |
| sigARTantisense | 5′-GGCGGATGCGGTGAGTTC-3′ | Real-time RT-PCR |
| Oligonucleotides | ||
| 1 | 5′-CACGGCGCGCCTCTCAGGCCAGTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 2 | 5′-CACGGCGCGCCAAAAAGGCCAGTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 3 | 5′-CACGGCGCGCCTCTCAGGCCAGAAAAAGGCGCTGCGACGACACTG-3′ | EMSA |
| 4 | 5′-CACGGCGCGCCAAAAAGGCCAGAAAAAGGCGCTGCGACGACACTG-3′ | EMSA |
| 5 | 5′-CACGGCGCGCCTCTCAGGCTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 6 | 5′-CACGGCGCGCCTCTCAGGCCTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 7 | 5′-CACGGCGCGCCTCTCAGGCCATCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 8 | 5′-CACGGCGCGCCTCTCAGGCCAGCTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 9 | 5′-CACGGCGCGCCTCTCAGGCCAGCATCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 10 | 5′-CACGGCGCGCCTCTCAGGCCAGCAGTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 11 | 5′-CACGGCGCGCCTCTCAGGCCAGCAGCTCTCAGGCGCTGCGACGACACTG-3′ | EMSA |
| 12 | 5′-CGGCGAACGTTATCTCAGTGGAATCTCAGTCCACGCGCGCAACCT-3′ | EMSA |
| 13 | 5′-CACGCGCGCAACCTAGTTGTGCAGTTACTGTTGAAAGCCACACCC-3′ | EMSA |
| 14 | 5′-CTCGGAGCACGGACATCGAGAACTCTCGGGGTTCGGCGAACGTTA-3′ | EMSA |
| Plasmids | ||
| pTZ229 | His-MprA expression plasmid; mprA coding sequence in pET15b | 45 |
| pTZ232 | Trx-MprA expression plasmid; mprA coding sequence in pET32a | This study |
| pTZ324 | His-MprA(D48A) expression plasmid; mprA(D48A) coding sequence in pET15b | 45 |
FP, forward primer; RP, reverse primer; EMSA, electrophoretic mobility shift assay.
Expression and purification of MprA.
E. coli BL21(DE3)/pLysS or Origami B(DE3) cultures (Novagen) containing overexpression constructs were grown overnight on selective LB agar medium, resuspended in LB broth containing the appropriate antibiotic, grown to mid-exponential phase (optical density at 600 nm of 0.5), and induced for 3 h in the presence of IPTG. Cell extracts were made and nickel-nitrilotriacetic acid-agarose chromatography was performed as previously described (45).
Cellular supernatants and protein eluates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12.5% polyacrylamide gels and stained with Coomassie brilliant blue. For EMSAs, purified recombinant MprA was dialyzed in 10 mM Tris HCl, pH 8.0-150 mM NaCl-1 mM dithiothreitol-20% glycerol at 4°C and stored at −80°C in 40% glycerol. For gel filtration chromatography, Trx-MprA was dialyzed in 50 mM Tris, pH 7.6-50 mM KCl-20 mM MgCl2-10% glycerol overnight at 4°C before loading on the column. The concentrations of recombinant MprA derivatives were determined spectrophotometrically with the BCA kit (Pierce).
When required, acetylphosphate (≥85% purity; Sigma) was used to phosphorylate recombinant MprA in vitro. Briefly, purified wild-type His-MprA or His-MprA (Asp48-Ala) was added to phosphorylation buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl) supplemented with 20 mM MgCl2 and 10 mM acetyl phosphate, and the mixtures were incubated at 37°C for 30 min. Previous studies have indicated that treatment with acetylphosphate under these conditions results in the phosphorylation of ≈25% of the His-MprA in the reaction (45).
Electrophoretic mobility shift assays.
For EMSAs, labeled probe DNA was incubated at room temperature for 10 min with various concentrations of recombinant MprA protein in reaction buffer containing final concentrations of 20 mM KCl, 5% glycerol, 10 ng of salmon sperm DNA per ml, 25 mM Tris-HCl, pH 8.0, 6 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, and 4.0 μg of poly(dI-dC). Ten microliters from each binding reaction was then loaded onto a 4 to 6% nondenaturing polyacrylamide gel and electrophoresed for 2.5 to 3.0 h at 120 V and 4°C. Gels were dried and exposed to Biomax MR (Kodak) X-ray film overnight.
DNase I protection assays.
DNA fragments comprising regions upstream of mprA or pepD were PCR amplified with primers mprAseq10up and mprAseq3 or mprBseq3up and Rv0983NotI and cloned into pCR2.1-TOPO. Inserts were removed from the clones by BamH1 and EcoRV digestion, resolved on a 1% agarose gel, recovered following gel extraction, and labeled at one end with [α-32P]dCTP and Klenow enzyme. Approximately 35 ng of labeled DNA was then incubated with 150 pmol of recombinant His-MprA protein in a reaction buffer containing 25 mM Tris-HCl, pH 8.0, 50 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 10% glycerol, and 0.5 mM dithiothreitol. After 10 min of incubation at room temperature, 0.6 U of DNase I were added to the mixture, and reactions were incubated at room temperature for an additional 2 min. Reactions were stopped by the addition of 2x stop solution (200 mM NaCl, 30 mM EDTA, and 1% sodium dodecyl sulfate). Reaction products were extracted with an equal volume of phenol-chloroform (25:25, vol/vol) and precipitated with 3 volumes of 100% ethanol. DNA was resuspended in 4 μl of loading solution (0.1 M NaOH-formamide [1:2, vol/vol], 0.1% xylene cyanol, 0.1% bromophenol blue) and resolved on a 6% denaturing polyacrylamide gel alongside a DNA sequencing ladder prepared with the Thermo Sequenase radiolabeled terminator cycle sequencing kit (USB). Reactions were separated for 1.5 h at 60 W before the gel was dried and exposed to Biomax MR X-ray film.
Gel filtration chromatography.
Gel filtration chromatography was used to determine the molecular weight of recombinant Trx-MprA purified by nitrilotriacetic acid/Ni2+ affinity chromatography; 5 mg (1 mg/ml) of Trx-MprA and 15 mg (3 mg/ml) of several protein standards (Sigma), including phosphorylase B (92 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa), were subjected to Sephyacryl-200 gel filtration with a 450-ml column equilibrated in running buffer comprising 50 mM Tris, pH 7.6, 50 mM KCl, 20 mM MgCl2 and 10% glycerol. The column was run at 0.3 ml/min, and 5.3-ml fractions were collected. The concentration and molecular mass of all proteins from collected fractions were determined by spectrophotometry and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were stained with Coomassie brilliant blue.
Real-time reverse transcription-PCR.
Total RNA was prepared from wild-type Mycobacterium bovis BCG and its isogenic derivatives as previously described (17). Briefly, 15-ml cultures of each strain were grown in 7H9 to an optical density at 600 nm of ≈1.20. Each culture was split into two, and one sample was treated with sodium dodecyl sulfate at a final concentration of 0.05%. Treated and untreated cultures were then incubated at 37°C for 90 min with shaking at 225 rpm. Cells were transferred to a 15-ml conical tube, concentrated by centrifugation, resuspended in RNAlater (Ambion), and incubated overnight at 4°C. Mycobacteria were mechanically lysed with a bead beater in the presence of RNA-Bee (Tel-Test). Total RNA was chloroform extracted and precipitated by the addition of isopropanol, and the resulting RNA pellet was resuspended in diethylpyrocarbonate-treated water. Turbo DNase I (Ambion) was used to remove contaminating genomic DNA.
cDNA was synthesized with 500 ng of total RNA in a 20-μl reverse transcription reaction containing arbitrary tuberculosis decamers (12) and SuperScript III reverse transcriptase (Invitrogen). Identical reactions lacking reverse transcriptase were also performed to confirm the absence of genomic DNA in all samples. Subsequent PCRs were performed on an iCycler iQ real-time PCR detection system (Bio-Rad) with gene-specific primer sets and iQ SYBR Green Supermix. Transcripts were quantified with a standard curve that was generated by amplifying serial fivefold dilutions of plasmid DNA carrying the mprA coding region with primers mprARTsense and mprARTantisense. The copy number of plasmid DNA used in these reactions ranged from 3 × 106 to 960. PCR efficiency in all reactions was determined to be between 90 and 100%, and single PCR products were confirmed by performing a postamplification melting curve analysis for each reaction. To compensate for potential variations in transcript numbers between treated and untreated cultures, mprA and pepD expression levels between strains were normalized to that of sigA because its expression is constitutive and unresponsive to stresses in M. tuberculosis (20). mprA and pepD induction levels following SDS treatment for each strain were calculated as follows: ([number of mprA or pepD transcripts]SDS induced/[number of sigA transcripts]SDS induced) ÷ ([number of mprA or pepD transcripts]untreated/[number of sigA transcripts]untreated).
Statistical analysis.
All statistical analyses (analysis of variance and Fisher's protected least-significant difference) were performed with ANOVA (version 1.11; Abicus Software).
RESULTS
MprA binds DNA sequences upstream of mprA and pepD.
Two-component signal transduction systems often regulate their own expression or the expression of nearby genes. To determine if the MprAB system behaved in a similar manner, the ability of the response regulator MprA to bind regions upstream of Rv0980c (PE_PGRS18), Rv0981 (mprA)-Rv0982 (mprB), and Rv0983 (pepD) was investigated by electrophoretic mobility shift assays (EMSAs). His-MprA was able to bind DNA sequences upstream of mprAB (Fig. 1A, region 2) and pepD (Fig. 1A, region 3) in a concentration-dependent manner (Fig. 1B, compare lane 1 with lanes 2 to 7). In contrast, His-MprA remained unable to bind the DNA sequence immediately upstream of PE_PGRS18 even at the highest concentration of protein used (data not shown). When binding reactions were repeated in the presence of increasing concentrations of unlabeled competitor DNA from the same upstream region, binding of His-MprA to the mprA and pepD upstream regions was reduced (Fig. 1B, compare lane 8 with 9 to 11), demonstrating that the interaction of His-MprA with DNA from these regions was specific. As expected, His-MprA binding to the mprA and pepD upstream region was not significantly reduced following the addition of unlabeled competitor DNA from the PE_PGRS18 upstream region (Fig. 1B, compare lane 12 with 13 to 15). A small reduction in His-MprA binding was discernible for the pepD upstream region in the presence of a 600-fold excess of unlabeled competitor DNA from the PE_PGRS18 upstream region; however, we attribute this to a nonspecific interaction due to the high concentration of unlabeled DNA used in this reaction. Thus, MprA binds to the mprAB and pepD upstream regions in a specific and concentration-dependent manner.
FIG. 1.
Binding of mprA and pepD upstream regions by His-MprA. (A) Genetic organization of the mprAB operon and adjacent genes. DNA fragments used in electrophoretic mobility shift assays (EMSAs) were PCR amplified from M. tuberculosis H37Rv and correspond to regions immediately upstream of coding sequences for PE_PGRS18 (region 1; −211 to +50), mprAB (region 2; −240 to +37), or pepD (region 3; −164 to +50). (B) His-MprA binding to mprA and pepD upstream regions; 10 ng of DNA from regions upstream of mprA (upper) and pepD (lower) was incubated in the absence of His-MprA (lane 1), increasing amounts of His-MprA (lanes 2 to 7), or fixed amounts of His-MprA and increasing amounts of unlabeled competitor DNA from the same (lanes 8 to 11) or adjacent (lanes 12 to 15) region. The amount of His-MprA used in binding reactions was 6.5, 13, 26, 52, 104, and 144 pmol for lanes 2 to 7, respectively, and 20 pmol for lanes 8 to 15. Unlabeled competitor DNA from the same region was added in 0-, 10-, 100-, or 200-fold molar excess of the labeled DNA used in lanes 8 to 11, respectively, while unlabeled competitor DNA from the adjacent region was added in 0-, 100-, 200-, or 600-fold molar excess of the labeled DNA in lanes 12 to 15, respectively. (C) Phosphorylation is not required for but enhances the binding of mprA and pepD upstream regions by His-MprA; 10 ng of DNA from the mprA or pepD upstream region was incubated in the absence (lanes 1 and 4) orpresence of 55 pmol of His-MprA(D48A) (lanes 2 and 3) or wild-type His-MprA (lanes 5 and 6). His-MprA derivatives were not phosphorylated (lanes 2 and 5) or were phosphorylated in vitro with acetylphosphate (lanes 3 and 6). B, bound; F, free.
Phosphorylation enhances but is not essential for binding of MprA to mprAB and pepD upstream regions in vitro.
Phosphorylation is thought to be essential for the ability of response regulators to bind DNA and activate or repress gene expression in vivo. However, response regulators frequently bind their cognate recognition sequence in vitro in the absence of protein phosphorylation. To determine if protein phosphorylation was required for MprA binding in vitro, binding reactions and EMSAs with the mprAB and pepD upstream regions were repeated with His-MprA(D48A), a mutant derivative that is unable to be phosphorylated in vitro (45). His-MprA(D48A) remained able to bind labeled DNA from both the mprAB and pepD upstream regions (Fig. 1C, compare lanes 1 and 2), suggesting that phosphorylation is not essential for His-MprA binding to these regions in vitro.
Because phosphorylation has also been shown to enhance the interaction of response regulators with DNA in vitro (19, 25), the ability of phosphorylated His-MprA to bind the mprAB and pepD upstream regions was also investigated. His-MprA was phosphorylated in these assays with acetylphosphate, a phosphodonor capable of phosphorylating ≈25% of the His-MprA in these reactions (45). Phosphorylation of wild-type His-MprA in vitro with acetylphosphate enhanced the ability of recombinant MprA to bind the mprAB and pepD upstream regions relative to untreated protein (Fig. 1C, compare lanes 5 and 6). For example, only a small fraction of labeled DNA from each region remained unbound in binding reactions with phosphorylated His-MprA. Interestingly, a diffuse protein-DNA banding pattern was observed when the pepD upstream region was incubated with phosphorylated His-MprA. We attribute this mobility pattern to either enhanced bending of the DNA by the phosphorylated protein or the presence of multiple MprA binding sites in this region. As expected, enhanced binding of the mprA and pepD upstream regions was not observed following incubation of His-MprA(D48A) with acetylphosphate (Fig. 1C, compare lanes 2 and 3). Thus, binding of the mprAB and pepD upstream regions by MprA in vitro is not dependent on but appears to be enhanced by protein phosphorylation.
MprA protects DNA regions containing an 8-bp direct repeat motif.
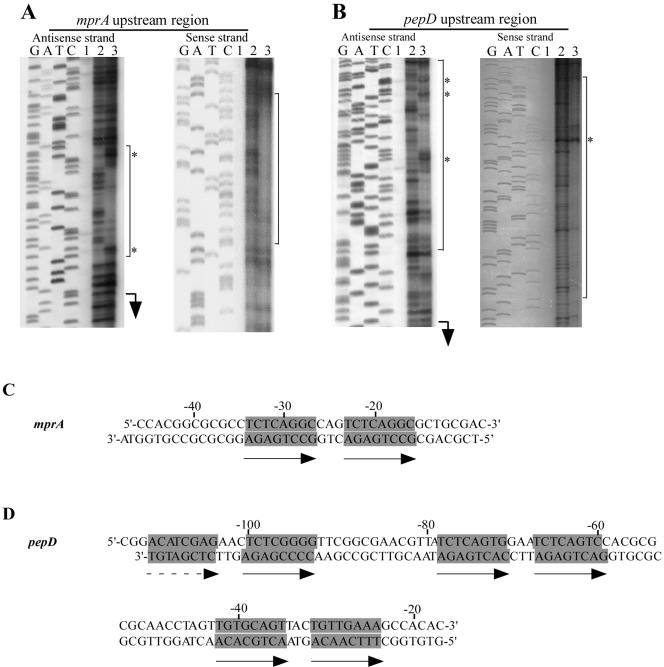
The results of the EMSAs indicated that His-MprA bound the mprAB and pepD upstream regions in a specific manner. To determine whether a specific nucleotide sequence motif was present in these regions, the area of DNA bound by His-MprA in these regions was determined by DNase I footprint mapping. His-MprA protected an area of ≈40 bp in the mprA upstream region, beginning 8 nucleotides upstream of the translational start site (Fig. 2A). Similarly, a region of ≈98 bp was protected by His-MprA in the pepD upstream region, beginning 17 nucleotides upstream of the translational start site (Fig. 2B). Addition of His-MprA also resulted in production of several DNase I-hypersensitive sites in both regions, suggesting that binding of His-MprA may induce conformational changes in DNA structure.
FIG. 2.
DNase I protection assays of mprA and pepD upstream regions. DNA encompassing the mprA (A) and pepD (B) upstream regions were PCR amplified from M. tuberculosis H37Rv; 35 ng of DNA from each region was incubated with His-MprA in the presence or absence of DNase I. G, A, T, and C designate the DNA sequence ladder generated for each strand. Reactions contained no His-MprA and no DNase I (lanes 1), no His-MprA and 0.6 U of DNase I (lane 2), or 150 pmol of His-MprA and 0.6 U of DNase I (lane 3). Translational starts are indicated with arrows, and sites of DNase I hypersensitivity are indicated by *. The protected regions on each strand are indicated by black bars. (C) mprA upstream region protected by His-MprA. (D) pepD upstream region protected by His-MprA. Nucleotides comprising a conserved 8-bp direct repeat found within each protected region are highlighted and indicated with arrows.
When each protected region was examined more closely, a loosely conserved 8-bp direct repeat motif could be observed in each sequence. For example, the mprAB upstream region contained a single repeat motif comprising nucleotides TCTCAGGC-cag-TCTCAGGC (Fig. 2A and C). Similarly, three loosely conserved direct repeat motifs (TCTCAGTG-gaa-TCTCAGTC, TGTGCAGT-tac-TGTTGAAA, and ACATCGAG-aac-TCTCGGGG) were identified in the protected sequence from the pepD upstream region (Fig. 2B and D). Thus, MprA binds DNA sequences containing an 8-bp direct repeat(s).
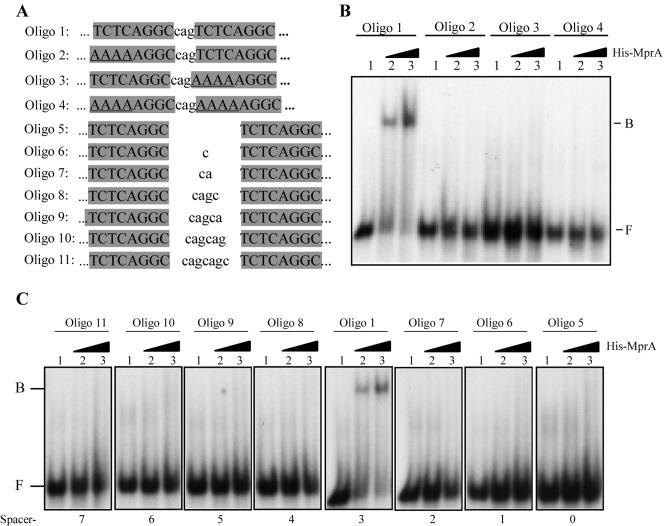
MprA binding is dependent on direct repeat subunits and the intervening spacer region.
Because His-MprA protected a DNA region containing an 8-bp direct repeat upstream of mprA, we next investigated whether this sequence motif was important for DNA binding. We radiolabeled 45-mer oligonucleotides (Fig. 3A) carrying the wild-type direct repeat sequence upstream of mprA (oligonucleotide 1) or sequence variants that were altered in repeat subunits (oligonucleotides 2 to 4) or the intervening spacer region (oligonucleotides 5 to 11) with [γ-32P]ATP, annealed them with their complementary oligonucleotides, and used them in binding reactions with His-MprA. His-MprA strongly bound the DNA probe carrying the wild-type sequence (Fig. 3B and 3C, oligonucleotide 1), suggesting that this motif alone is likely responsible for DNA interaction. In contrast, His-MprA was unable to bind probes in which conserved nucleotides from one or both repeat subunits had been mutated (Fig. 3B, oligonucleotides 2 to 4). Similarly, addition or deletion of nucleotides from the spacer region between subunit repeats also abolished the ability of His-MprA to bind oligonucleotide probes (Fig. 3C, oligonucleotides 5 to 11). Thus, binding of MprA to the mprA upstream region is dependent on direct repeat subunits and is sensitive to the spacer length between repeat subunits.
FIG. 3.
His-MprA binding to the mprA upstream region is dependent on conserved 8-bp direct repeat and the spacer region between repeat subunits. (A) Sequences of 45-mer oligonucleotide-based DNA probes used for EMSAs. Oligonucleotide 1 contains the wild-type sequence derived from the mprA upstream region. Oligonucleotides 2 to 4 carry substitutions in either or both direct repeat subunits. Oligonucleotides 5 to 11 have alterations in the spacer region between direct repeat subunits. (B) EMSA of various oligonucleotide-based DNA probes with His-MprA. Reactions were performed with 2.5 ng of labeled probe DNA in the absence of His-MprA (lanes 1) or with 75 pmol (lane 2) or 265 pmol (lane 3) of His-MprA. Note that nucleotides outside the boxed regions are identical in all DNA probes. B, bound; F, free.
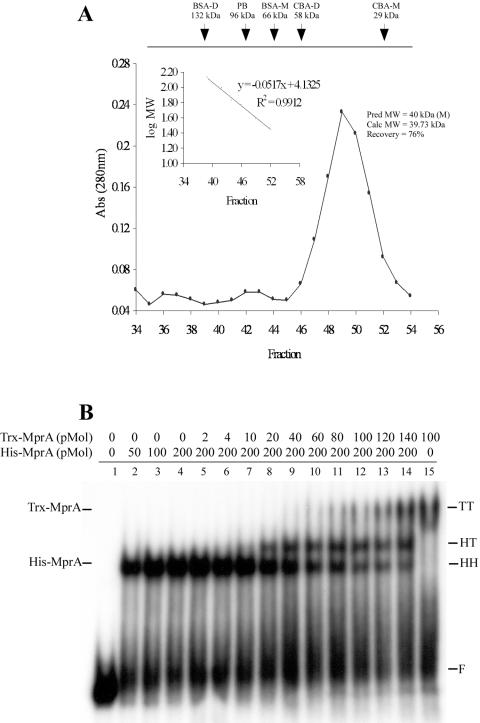
Direct repeat subunits are tandemly bound by two molecules of MprA.
Many two-component response regulators form dimers or other higher-order complexes to bind target DNA and initiate transcriptional programs (21, 25, 28). To determine the number of MprA molecules bound to the repeat sequence upstream of mprA, we next performed gel filtration chromatography on purified MprA and carried out competitive EMSAs with the DNA fragment from this region. Trx-tagged and His-tagged MprA proteins were used for these experiments because the differences in molecular mass between these recombinant proteins could easily be distinguished by EMSA. Purified Trx-MprA eluted predominantly as a single protein peak in gel filtration chromatography experiments, representing ≈76.2% of the total protein loaded on the column (Fig. 4A). Compared to protein standards, including phosphorylase B, bovine serum albumin, and carbonic anhydrase, the estimated molecular mass of Trx-MprA was calculated to be 39.7 kDa. This is in good agreement with the predicted molecular mass of 43.01 kDa for the monomeric form of Trx-MprA. Minor protein peaks with estimated molecular masses of 81.2 and 165.8 kDa were also observed following column chromatography, but these fractions represented less than 5% of the total protein loaded. Thus, MprA appears to be predominantly monomeric in solution when purified from E. coli.
FIG. 4.
The 8-bp direct repeat binds two molecules of MprA tandemly. (A) Sephacryl-200 gel filtration chromatography was performed on 5 mg (1 mg/ml) of Trx-MprA or 15 mg (3 mg/ml) of several protein standards, including bovine serum albumin (BSA), phosphorylase B (PB), and carbonic anhydrase (CBA). A standard curve (inset) of protein standards was generated to determine the approximate mass of Trx-MprA; ≈76% of Trx-MprA eluted from the column at a molecular mass calculated to be 39.73 kDa. (B) Competitive EMSAs with the direct repeat from the mprA upstream region and recombinant derivatives of MprA; 2.8 ng of a 60-bp PCR-derived DNA probe carrying the direct repeat upstream of mprA was incubated in binding reactions with recombinant forms of MprA. Binding reactions contained no protein (lane 1), 50, 100, or 200 pmol of His-MprA alone (lanes 2 to 4), 200 pmol of His-MprA and 2, 4, 10, 20, 40, 60, 80, 100, 120, or 140 pmol of Trx-MprA (lanes 5 to 14), or 100 pmol of Trx-MprA alone (lane 15). F, free DNA; HH, two molecules of His-MprA; TT, two molecules of Trx-MprA; HT, one molecule each of His-MprA and Trx-MprA.
To determine the number of MprA molecules that were bound to each direct repeat, competitive EMSAs with Trx-MprA and His-MprA were performed with a 60-bp PCR-generated probe carrying the wild-type direct repeat sequence upstream of mprA. As observed previously, addition of His-MprA to the binding reaction resulted in production of a single protein-DNA complex (Fig. 4B, lanes 2 to 4). As expected, a single protein-DNA complex with a molecular mass greater than that observed with His-MprA was also observed in binding reactions containing Trx-MprA alone (Fig. 4B, lane 15). Addition of increasing concentrations of Trx-MprA to reactions containing fixed amounts of His-MprA resulted in production of a third protein-DNA complex that migrated at a position between those observed with His-MprA and Trx-MprA (Fig. 4B, lanes 5 to 14). This pattern of mobility (15) is consistent with one MprA molecule being bound to each direct repeat subunit. Together with the results demonstrating that alterations to the direct repeat subunits completely abolish MprA binding (Fig. 3B), we conclude that the interaction of MprA with the 8-bp direct repeat requires tandem binding by two protein monomers.
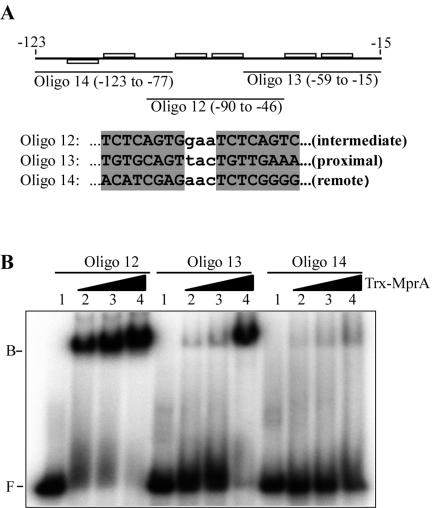
MprA binds three 8-bp repeat sequences upstream of pepD with various affinities.
The results from DNase I footprint experiments with the pepD upstream region suggested that His-MprA protected a large DNA region that contained one well-conserved and two less-conserved 8-bp direct repeat motifs. To determine whether MprA bound to all three sequences, we designed 45-mer oligonucleotides specific to each region and used them as probes in binding assays with Trx-MprA. Trx-MprA bound efficiently to the DNA probe carrying the well-conserved intermediate direct repeat upstream of pepD (Fig. 5B, oligonucleotide 12). Similarly, Trx-MprA also bound the DNA probe carrying the less-conserved proximal repeat, albeit with much lower affinity than that observed with the intermediate repeat (Fig. 5B, oligonucleotide 13). Finally, binding of Trx-MprA to the remote direct repeat was poor, even at the highest concentrations of Trx-MprA used (Fig. 5B, oligonucleotide 14). Thus, MprA recognizes three direct repeats upstream of pepD and binds to each with different affinities.
FIG. 5.
pepD upstream region contains multiple MprA binding sites with different affinities for MprA. (A) Regional location of 8-bp direct repeats upstream of pepD and sequences of oligonucleotide-based DNA probes used for EMSAs. Oligonucleotides 12 to 14 correspond to oligonucleotides containing individual direct repeats that are intermediate, proximal, and distal from the translational start site, respectively. Note that nucleotides outside the boxed regions are identical to the wild-type sequence upstream of pepD. (B) EMSA of individual direct repeats upstream of pepD with Trx-MprA. Reactions were performed with 2.5 ng of probe DNA in the absence of recombinant MprA (lanes 1) or with 108 pmol (lanes 2), 216 pmol (lanes 3), or 450 pmol (lanes 4) of Trx-MprA. B, bound; F, free.
MprA regulates mprA and pepD expression in vivo in a phosphorylation-dependent manner.
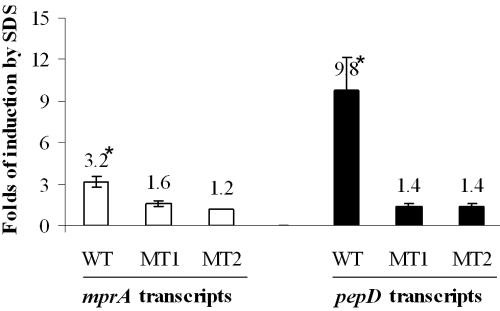
To determine whether MprA regulated expression of mprA and pepD in vivo, real-time reverse transcription-PCR was used to quantify mprA and pepD transcripts from wild-type and mprA mutant strains of M. bovis BCG. As the levels of MprA in M. bovis BCG grown in standard culture medium are normally low (44), mprA expression was induced by a 90-min exposure to low (0.05%) concentrations of SDS, a condition previously demonstrated to induce expression of mprA in M. tuberculosis H37Rv (27). mprA and pepD expression levels in each strain were also normalized to that of sigA, as expression of this transcript has been shown to remain unchanged under growth in various conditions (20). Treatment of wild-type M. bovis BCG with SDS resulted in a statistically significant induction of both mprA (3.2-fold ± 0.4-fold, P < 0.01) and pepD (9.8-fold ± 2.4-fold, P < 0.01) expression relative to that of untreated cultures. In contrast, induction of mprA or pepD was not observed in the mprA::Kmr mutant or the mprA::Kmr mutant carrying the mprA(D48A) allele following exposure to SDS. Thus, MprA regulates the expression of both mprA and pepD in vivo in response to SDS exposure, and this regulation requires phosphorylated protein.
DISCUSSION
Reactivation of persistent bacilli in latently infected individuals continues to be an important health concern and likely contributes to the recycling of tuberculosis in the human population (26). Thus, understanding the processes that regulate M. tuberculosis entry into cells, maintenance, and reactivation from persistent infection will be crucial to worldwide control of tuberculosis. Like other bacterial pathogens, M. tuberculosis uses two-component transduction systems to adapt to changing conditions within the host (10, 16, 18, 34, 35, 43, 44). In the low-dose murine model of tuberculosis, expression of the mprAB two-component system was shown to be required for long-term persistence of M. tuberculosis infection in the lungs and spleen of infected animals (44). Here, we have continued the characterization of this regulatory system and report that MprA recognizes a loosely conserved 8-bp direct repeat motif comprising two repeat subunits separated by 3 nucleotides, a sequence present upstream of its own coding sequence and that of the downstream gene pepD.
While expression of several two-component systems has been shown to be important for M. tuberculosis virulence, the downstream effector determinants regulated by these systems remain largely unknown. The best characterized of the M. tuberculosis response regulators, DosR, has been shown to modulate its own expression and that of a 47-gene regulon in M. tuberculosis in response to hypoxia and nitric oxide exposure (29, 38, 41). DosR recognizes a 20-bp palindromic sequence that is present upstream of nearly all genes regulated in response to these stimuli (13, 34). TrcR, another response regulator implicated in M. tuberculosis pathogenesis, also undergoes autogenous regulation by recognition of a 36-bp AT-rich region upstream of its coding sequence (16), although the stimulus processed by this two-component system remains unknown. In contrast to DosR and TrcR, MprA recognizes a 19-bp nucleotide motif, termed the MprA box, comprising two 8-bp direct repeat subunits separated by a 3-nucleotide spacer region. MprA belongs to the OmpR subfamily (45), a group of structurally conserved response regulators characterized by their conserved C-terminal winged helix-turn-helix DNA binding motif. All members of this subfamily characterized to date appear to recognize direct repeat DNA sequences (28). However, differences in the nucleotide sequence of the cognate recognition site, the spacing between repeat subunits, and the number of repeats present in a given promoter region allow these transcription factors to regulate diverse biological processes.
In M. tuberculosis, there is a single MprA box located upstream of the mprA coding sequence and three MprA boxes upstream of pepD (Fig. 2). While the sequences of these boxes are not identical, they all appear to be capable of binding MprA, albeit with various affinities (Fig. 1 and 5). Sites possessing the highest binding affinity for MprA have the sequence TCTCNGNN (where N is any nucleotide), which appears to be highly conserved in both subunits of the repeat (Fig. 3B, oligonucleotide 1; Fig. 5B, oligonucleotide 12). Alterations of this sequence, as observed with the proximal and distal MprA boxes upstream of pepD, are tolerated by MprA, although they do exhibit reduced binding affinity (Fig. 5B, oligonucleotides 13 and 14). In particular, the first 4 nucleotides of each box appear to be critical for binding, as mutations of this sequence abolish MprA binding (Fig. 3B, oligonucleotides 2 to 4).
Importantly, MprA binding to repeat subunits appears to be exquisitely sensitive to the spacer length separating these elements. For example, increasing or decreasing the distance between repeat subunits by even one nucleotide abolishes the ability of MprA to recognize the sequence (Fig. 3C). The essentiality of 3-bp spacing may be due to the requirement for cooperativity between the two MprA monomers that we postulate bind each repeat. The combined distance between two repeat subunits (11 nucleotides) is approximately equivalent to one full turn of the DNA helix. Thus, this spacing would allow two MprA monomers to bind in a side-by-side manner on the same face of the DNA helix. In addition, mutations within a single repeat subunit that alter the nucleotide composition but do not change the spacer length between repeat subunits prevent interaction of MprA with the DNA (Fig. 3B, oligonucleotides 2 and 3). While we cannot formally rule out the possibility that MprA binds DNA as a dimer or other higher-order complex or that phosphorylation increases the propensity of MprA to form dimers in solution, MprA is predominantly monomeric in solution when prepared in buffer that is similar to the one used in in vitro DNA binding assays. These results are consistent with those observed with OmpR, which has been shown to be monomeric in solution and to bind multiple repeat elements upstream of target genes in a tandem fashion (15, 21, 28).
Some response regulators require phosphorylation to bind to their DNA recognition targets in vitro (6, 23). However, for other response regulators, phosphorylation only enhances the binding affinity of the protein for its target sequence (14, 19). In M. tuberculosis, DosR binds to its palindromic recognition sequence in a phosphorylation-independent manner (37). Similarly, phosphorylation is not required for MprA binding to the mprA and pepD upstream region but rather appears to enhance the interaction of MprA with these regions. Phosphorylation is required by MprA for the in vivo regulation of mprA and pepD, as a M. bovis BCG derivative mutated in the endogenous mprA gene but expressing the mprA(D48A) allele can no longer induce expression of these genes in response to SDS exposure (Fig. 6). This observation is also consistent with previous results demonstrating that M. bovis BCG expressing the mprA(D48A) allele is attenuated for growth in macrophages relative to wild-type M. bovis BCG (45).
FIG. 6.
MprA regulates mprA and pepD expression in vivo in Mycobacterium bovis BCG in response to SDS. Wild-type M. bovis BCG (WT), the mprA::Kmr mutant (MT1), and the mprA::Kmr mutant carrying the mprA(D48A) allele in trans (MT2) were grown to an optical density at 600 nm of ≈1.2 and split into two cultures; one culture was induced for 90 min with 0.05% SDS. Following SDS treatment, bacteria were processed and RNA was extracted for quantitative real-time reverse transcription-PCR. Data are presented as induction of mprA (open bars) or pepD (solid bars) for each BCG strain following SDS treatment. All values have been normalized to that of sigA and are presented as the mean and standard deviation of reactions performed in triplicate.
The observations that both mprA and pepD are inducible by exposure to SDS (27) and that this regulation is dependent on MprA suggest a role for this two-component system as a regulator of stress, possibly in stress manifesting in the cell wall or outer membrane of the organism. pepD encodes a trypsin-like serine protease (39). Serine proteases are thought to aid in the degradation of misfolded proteins that are generated in response to stresses (7, 31). In pathogenic bacteria, many serine proteases are also surface exposed or secreted and are predominant antigens recognized by the host immune system (9, 22, 24, 31, 39). In M. tuberculosis, PepD has been demonstrated to be secreted into the culture medium in vitro (39). However, its role in virulence has not yet been established.
Interestingly, mprAB and pepD lie in a cluster of in vivo-expressed genes that may constitute a 34.1-kb pathogenicity island that is present in the genome of M. tuberculosis and other pathogenic mycobacteria but not in saprophytic strains such as Mycobacterium smegmatis (40). In addition, the majority of genes in this region appear to be involved in cell wall biosynthesis and lipid metabolism. DNA microarray analyses of wild-type M. tuberculosis H37Rv and the isogenic mprA mutant also suggested a role for MprA in the regulation of determinants thought to comprise or be associated with the cell wall, including those from the proline-glutamine (PE)- and proline-proline-glutamine (PPE)-rich families (17) (data not shown). Thus, we speculate that the biological function of MprAB may be to act as a general sensor of stress response at the bacterial cell surface.
In summary, MprA regulates its own expression and that of a putative serine protease through the recognition of a loosely conserved 8-bp direct repeat motif. Further delineation of the core “consensus” sequence required for binding is currently under way and will aid in the identification of additional gene products that are regulated by MprA in M. tuberculosis. Ultimately, this information will help to better define the biological processes that are required by M. tuberculosis for persistent infection in the host.
Acknowledgments
We thank members of the Zahrt and Dara Frank laboratories for useful discussions regarding this work. We also thank Kent Wilcox for technical assistance and Joe Barbieri for careful review of the manuscript.
This work was supported by NIH grant AI51669 to T.C.Z.
REFERENCES
- 1.Anderson, R. M. 1999. The pandemic of antibiotic resistance. Nat. Med. 5:147-149. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. World Health Organization Global Tuberculosis Programme fact sheet no. 13: tuberculosis. World Health Organization, Geneva, Switzerland.
- 3.Anonymous. 2004. World Health Organization Global Tuberculosis Programme fact sheet no. 104. World Health Organization, Geneva, Switzerland.
- 4.Bloom, B. R., and C. J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, B. R., and P. M. Small. 1998. The evolving relation between humans and Mycobacterium tuberculosis. N. Engl. J. Med. 338:677-678. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, P. E., and S. Stibitz. 1995. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J. Bacteriol. 177:6486-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Cortes, G., B. de Astorza, V. J. Benedi, and S. Alberti. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuberc. Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 11.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAM. 282:677-686. [DOI] [PubMed] [Google Scholar]
- 12.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forst, S. A., J. Delgado, and M. Inouye. 1989. DNA-binding properties of the transcription activator (OmpR) for the upstream sequences of ompF in Escherichia coli are altered by envZ mutations and medium osmolarity. J. Bacteriol. 171:2949-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 16.Haydel, S. E., W. H. Benjamin, Jr., N. E. Dunlap, and J. E. Clark-Curtiss. 2002. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J. Bacteriol. 184:2192-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessner, M. J., V. K. Singh, X. Wang, S. Khan, M. R. Tschannen, and T. C. Zahrt. 2004. Utilization of a labeled tracking oligonucleotide for visualization and quality control of spotted 70-mer arrays. BMC Genomics 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himpens, S., C. Locht, and P. Supply. 2000. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146:3091-3098. [DOI] [PubMed] [Google Scholar]
- 19.Holman, T. R., Z. Wu, B. L. Wanner, and C. T. Walsh. 1994. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33:4625-4631. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Y., and A. R. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, K. J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615-628. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 23.Li, J., S. Kustu, and V. Stewart. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 241:150-165. [DOI] [PubMed] [Google Scholar]
- 24.Li, S. R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 29.Ohno, H., G. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol 5:637-648. [DOI] [PubMed] [Google Scholar]
- 30.Pablos-Mendez, A., M. C. Raviglione, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, and P. Nunn. 1998. Global surveillance for antituberculosis-drug resistance, 1994-1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 338:1641-1649. [DOI] [PubMed] [Google Scholar]
- 31.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 32.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423-1435. [DOI] [PubMed] [Google Scholar]
- 34.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez, E., S. Samper, Y. Bordas, C. Guilhot, B. Gicquel, and C. Martin. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179-187. [DOI] [PubMed] [Google Scholar]
- 36.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 314:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skeiky, Y. A., M. J. Lodes, J. A. Guderian, R. Mohamath, T. Bement, M. R. Alderson, and S. G. Reed. 1999. Cloning, expression, and immunological evaluation of two putative secreted serine protease antigens of Mycobacterium tuberculosis. Infect. Immun. 67:3998-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 43.Zahrt, T. C., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zahrt, T. C., C. Wozniak, D. Jones, and A. Trevett. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]