ABSTRACT
Accumulating evidence has demonstrated that macroautophagy/autophagy plays an essential role in self-renewal and differentiation in embryonic hematopoiesis. Here, according to the RNA sequencing data sets of 5 population cells related to hematopoietic stem cell (HSC) formation during mouse embryogenesis (endothelial cells, PTPRC/CD45− and PTPRC/CD45+ pre-HSCs in the E11 aorta-gonad-mesonephros (AGM) region, mature HSCs in E12 and E14 fetal liver), we explored the dynamic expression of mouse autophagy-related genes in this course at the single-cell level. Our results revealed that the transcription activity of autophagy-related genes had a substantial increase when endothelial cells (ECs) specified into pre-HSCs, and the upregulation of autophagy-essential genes correlated with reduced NOTCH signaling in pre-HSCs, suggesting the autophagy activity may be greatly enhanced during pre-HSC specification from endothelial precursors. In summary, our results presented strong evidence that autophagy plays a critical role in HSC emergence during mouse midgestation.
KEYWORDS: autophagy, hematopoiesis, hematopoietic stem cell, mouse embryos, single cell
Hematopoietic stem cells (HSCs) are the stem cells that exist in both early embryonic and adult hematopoietic organs, with the capacity to undergo long-term self-renewal and multilineage differentiation into entire mature blood lineages. During mouse embryogenesis, standard HSC potential is initially detectable in the E10.5 AGM region and slightly later in the head, placenta and yolk sac. At E11.5, mature HSCs from these hemogenic sites begin to migrate to fetal liver for further expansion. According to current opinions, HSCs are specified early from embryonic hemogenic endothelial cells and/or pre-HSCs; however, the molecular characteristic of these cells remains elusive because of their rareness and difficulty to achieve efficient isolation.
In recent years, the single-cell RNA-seq technique was widely applied to examine the comprehensive transcriptomes in an individual cell. The development and application of high-throughput single-cell RNA sequencing has already provided an unprecedented opportunity for profound new discoveries in important areas of cell biology. In particular, this approach greatly facilitated the dissection of comprehensive gene expression profiles in rare types of cells, such as in the hemogenic endothelium and pre-HSCs in early embryos, and also heterogeneity within temporally and spatially complex populations of cells. Hence, based on potent surface markers and the single-cell RNA-seq technique, in previous work, we were able for the first time to successfully and efficiently capture the single functional pre-HSCs in the E11 AGM region and HSCs in E12 and E14 fetal liver.
More importantly, recent developments have indicated that autophagy is crucial for maintaining cellular homeostasis and remodeling during embryonic development, and it is also involved in the regulation of various types of stem cells, in particular HSCs, allowing their relatively long life in organs. In a word, autophagy is required for keeping the balance between quiescence, self-renewal and differentiation in embryonic HSCs. However, the underlying mechanism and regulation of autophagy is largely unknown during embryonic hematopoiesis. Hence, we have systematically examined the dynamic expression of the autophagy-related genes in the HSC-competent populations through mouse embryonic development at the single-cell level, providing a critical insight into the mechanistic understanding of the physiological connections between autophagy and mouse embryonic HSC formation.
Dynamic autophagy-related gene expression in mouse embryonic HSCs
According to the 5 population cells related to mouse embryonic HSC formation (ECs, PTPRC/CD45− and PTPRC/CD45+ pre-HSCs in the E11 AGM region, mature HSCs in E12 and E14 fetal liver), we examined dynamic gene expression of 678 autophagy-related genes retrieved from the autophagy database during mouse embryonic HSC formation at the single-cell level. Our results showed that the expression of 82 autophagy-related genes show significant changes between neighboring cell types (on the basis of multiple t-test with p<0.05, false-discovery rate<0.05, and fold change of log2 converted fragments per kilobase per million/FPKM >2 or <0.5). As shown in Figure 1A, 6 clusters were observed among these 82 autophagy-related genes using hierarchical clustering. Consistent with the previous findings that a sharp increase of transcription activity was observed when ECs differentiated into pre-HSCs, over half of the autophagy-related genes significantly increase their expression between ECs and T1 pre-HSCs. Compared with ECs, we observed 13 downregulated autophagy-related genes (such as Bcl2) and 44 upregulated autophagy-related genes (Such as Bad, Pik3cg and Pik3c2b) in T1 pre-HSCs, suggesting the autophagy activity may be greatly enhanced during pre-HSC specification from putative endothelial precursors.
Figure 1.
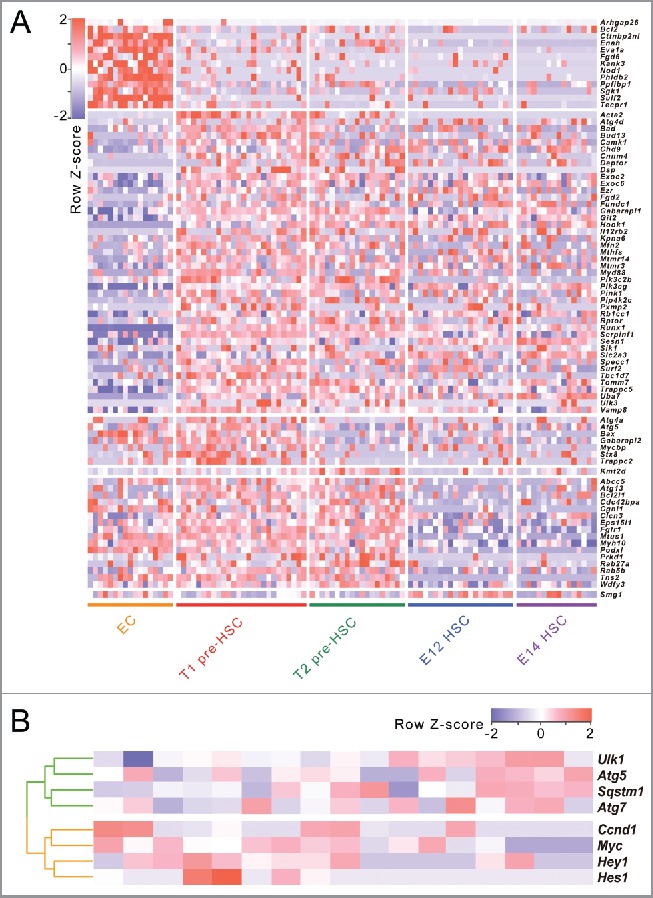
Autophagy-related gene expression dynamics during the process of HSC formation in mouse embryo at the single-cell level. (A) Heat map of differentially expressed autophagy-related genes between each of the 2 consecutive stages during the formation of HSCs in mouse embryo. (B) Hierarchical cluster of gene expression of autophagy- essential genes and NOTCH target genes in pre-HSCs (including both T1 and T2 pre-HSCs) at the single-cell level.
Upregulation of autophagy-essential genes correlates with reduced NOTCH signaling in pre-HSCs
Increasing lines of evidence have indicated that the NOTCH signaling pathway, an important regulator of many fundamental cellular processes such as stem cell maintenance and differentiation during embryonic development, plays a pivotal role in lineage commitment of HSCs. However, it is unclear what the relationship is between autophagy and NOTCH signaling in pre-HSCs (including both T1 and T2 pre-HSCs). To explore the potential relationship between autophagy and the NOTCH signaling pathway in pre-HSCs, we detected the expression of several autophagy-essential genes (Atg5, Atg7, Sqstm1/p62 and Ulk1) and several Notch target genes (Myc/c-Myc, Ccnd1 [cyclin D1], Hes1 and Hey1). Then, we conducted a correlation analysis by means of hierarchical clustering using expression profiles from 17 pre-HSCs. As shown in Figure 1B, the expression of autophagy-essential genes is significantly negatively correlated with the expression of NOTCH target genes (Pearson's correlation coefficient, r = −0.58, on the basis of permutation tests with P = 0.0039), indicating that autophagy activity in pre-HSCs may be correlated with the downregulation of NOCTH signaling during mouse HSC emergence.
Conclusions
In summary, our previous study is the first to isolate 5 populations related to mouse embryonic HSC formation, revealing complex molecular mechanisms regulating step-wise development of HSCs in vivo. Then, our data revealed that the transcription activity of autophagy-related genes has a sharp increase when ECs specify into pre-HSCs, suggesting the autophagy activity may be greatly enhanced during pre-HSC specification from their endothelial precursor cells.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful to Dr. Daniel J. Klionsky for help in editing the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFA0100601), the National Key Basic Research Program of China (2015CB943001), the National Natural Science Foundation of China (31371322, 81530007), Natural Science Foundation of Heilongjiang Province of China (C2015027); WeihanYu Youth Science Fund Project of Harbin Medical University.


