Abstract
AdpA is the key transcriptional activator for a number of genes of various functions in the A-factor regulatory cascade in Streptomyces griseus, forming an AdpA regulon. Trypsin-like activity was detected at a late stage of growth in the wild-type strain but not in an A-factor-deficient mutant. Consistent with these observations, two trypsin genes, sprT and sprU, in S. griseus were found to be members of the AdpA regulon; AdpA activated the transcription of both genes by binding to the operators located at about −50 nucleotide positions with respect to the transcriptional start point. The transcription of sprT and sprU, induced by AdpA, was most active at the onset of sporulation. Most trypsin activity exerted by S. griseus was attributed to SprT, because trypsin activity in an sprT-disrupted mutant was greatly reduced but that in an sprU-disrupted mutant was only slightly reduced. This was consistent with the observation that the amount of the sprT mRNA was much greater than that of the sprU transcript. Disruption of both sprT and sprU (mutant ΔsprTU) reduced trypsin activity to almost zero, indicating that no trypsin genes other than these two were present in S. griseus. Even the double mutant ΔsprTU grew normally and developed aerial hyphae and spores over the same time course as the wild-type strain.
The gram-positive, soil-dwelling, filamentous bacterial genus Streptomyces is characterized by its ability to produce a wide variety of secondary metabolites and by its complex morphological differentiation, culminating in sporulation. A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is a chemical signaling molecule or microbial hormone that triggers aerial mycelium formation and secondary metabolism at an extremely low concentration in Streptomyces griseus (3-8). A-factor switches on the transcription of adpA, encoding a transcriptional activator, by binding to ArpA, the A-factor receptor protein that has bound the promoter of adpA, and dissociating the DNA-bound ArpA from the DNA (27). The target of ArpA appears to be only adpA (12). The AdpA thus induced activates a number of genes required for morphological development and secondary-metabolite formation, forming an AdpA regulon (4, 36).
We previously observed a remarkable difference in protein profiles between the wild-type S. griseus strain IFO13350 and an A-factor-deficient mutant, HH1. When the profile of total mycelial proteins of the A-factor-deficient mutant was compared with that of the wild-type strain by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, more than 10 proteins of a wide range of molecular sizes were found only in the wild-type strain, whereas a few proteins were present only in the mutant. Addition of A-factor to mutant HH1 during growth made its protein profile almost the same as that of the wild-type strain (5). We can say that these observations result from the remarkable pleiotropic effect of A-factor and that some of the proteins produced in response to A-factor are under the direct control of AdpA.
We have so far identified several genes as targets of AdpA. These include strR, which serves as a pathway-specific transcriptional activator for streptomycin biosynthetic genes (27); an open reading frame encoding a probable pathway-specific regulator for a polyketide compound (36); adsA, which encodes an extracytoplasmic function sigma factor of RNA polymerase essential for aerial mycelium formation (33); sgmA, which encodes a metalloendopeptidase probably involved in apoptosis of substrate hyphae during aerial mycelium development (13); ssgA, which encodes a small acidic protein essential for spore septum formation (34); and amfR, essential for aerial hyphae formation (35). Many of these genes were identified by a PCR-gel mobility shift method (9, 33).
In addition to these gene products, we previously observed that the A-factor-deficient S. griseus mutant HH1 shows a much diminished level of extracellular trypsin-like activity and that addition of A-factor to the mutant restored its productivity to the level of the wild-type strain (21). Trypsin is one of the major extracellular proteases of S. griseus. Since we assumed that many other genes are members of the AdpA regulon, these observations prompted us to identify the trypsin gene and determine its dependence on A-factor and AdpA. For these purposes, we devised a convenient method to identify extracellular proteins, followed by cloning of a trypsin gene and determination of its transcriptional dependence on A-factor and AdpA. The nucleotide sequence of the cloned gene showed that it is the orthologue of sprT previously cloned by Kim et al. (20).
This paper deals with the transcriptional activation of sprT, encoding the trypsin-like protease by A-factor, via AdpA, a transcriptional activator in the A-factor regulatory cascade. In other words, sprT is a member of the AdpA regulon. An unexpected finding was that S. griseus produces an additional trypsin-type protease, named SprU, showing end-to-end similarity to SprT, which also turned out to be controlled by AdpA. Gene disruption of either or both of the protease genes gave no apparent phenotypic changes, suggesting that sprT and sprU are not important for morphological differentiation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO13350 was obtained from the Institute of Fermentation, Osaka, Japan. The S. griseus mutants ΔadpA (27) and HH1 (8) were described previously. Streptomyces strains were grown in YMPD medium (yeast extract [Difco], 0.2%; meat extract [Kyokuto], 0.2%; Bacto peptone [Difco], 0.4%; NaCl, 0.5%; MgSO4 · 7H2O, 0.2%; glucose, 1%; pH 7.2). YMPD agar contained 2% agar. R2YE medium (15) was used for the regeneration of protoplasts. Thiostrepton (50 μg/ml) and neomycin (20 μg/ml) were added when necessary. A low-copy-number plasmid, pKU209 (11), containing the ampicillin and thiostrepton resistance genes, with a copy number of one or two per genome was used. Escherichia coli JM109 and vector pUC19 for DNA manipulation were purchased from Takara Shuzo. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA for gene disruption. Histidine-tagged AdpA (AdpA-H) was purified from E. coli BL21(DE3) harboring pET-adpA as described previously (33). The media and growth conditions for E. coli were described by Maniatis et al. (23). Ampicillin (50 μg/ml) and kanamycin (50 μg/ml) were used when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Shuzo. [α-32P]dCTP (110 TBq/mmol) for DNA labeling with a BcaBest DNA labeling system (Takara Shuzo) and [γ-32P]ATP (220 TBq/mmol) for end labeling at the 5′ ends with T4 polynucleotide kinase were purchased from Amersham Pharmacia Biotech. DNA was manipulated in Streptomyces spp. (15) and in E. coli (1, 23) as described earlier. Nucleotide sequences were determined by the dideoxy chain termination method with a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham).
Partial purification of extracellular proteins and identification of SprT.
S. griseus strains IFO13350 and HH1 were grown in 60 ml of R2YE medium in a 250-ml baffled flask at 28°C with vigorous shaking. After 3 days of cultivation, each culture was used to inoculate 6 liters of fresh R2YE medium and further cultured for 3 days under the same conditions. After centrifugation of the culture broth at 15,000 rpm for 30 min at 4°C, the resulting supernatant was concentrated by trichloroacetic acid (TCA) precipitation. The precipitate was resuspended in buffer A (10 mM potassium phosphate, pH 7.0) and dialyzed overnight against the same buffer. The dialyzed sample was loaded onto a Resource-S cation exchange column (Amersham) previously equilibrated with buffer A and eluted at a flow rate of 1 ml/min with an NaCl gradient (0 to 1 M) in buffer A. The fractions containing proteins were concentrated by TCA and analyzed by polyacrylamide gradient gel electrophoresis on sodium dodecyl sulfate-4 to 20% polyacrylamide. Protein concentrations were determined by the method of Bradford with bovine serum albumin as the standard. For determination of the N-terminal amino acid sequence of extracellular proteins by the Edman degradation procedure, the proteins on the gel were electrophoretically transferred to a polyvinylidene difluoride Immobilon membrane (Millipore).
Cloning of the DNA fragments containing sprT and sprU.
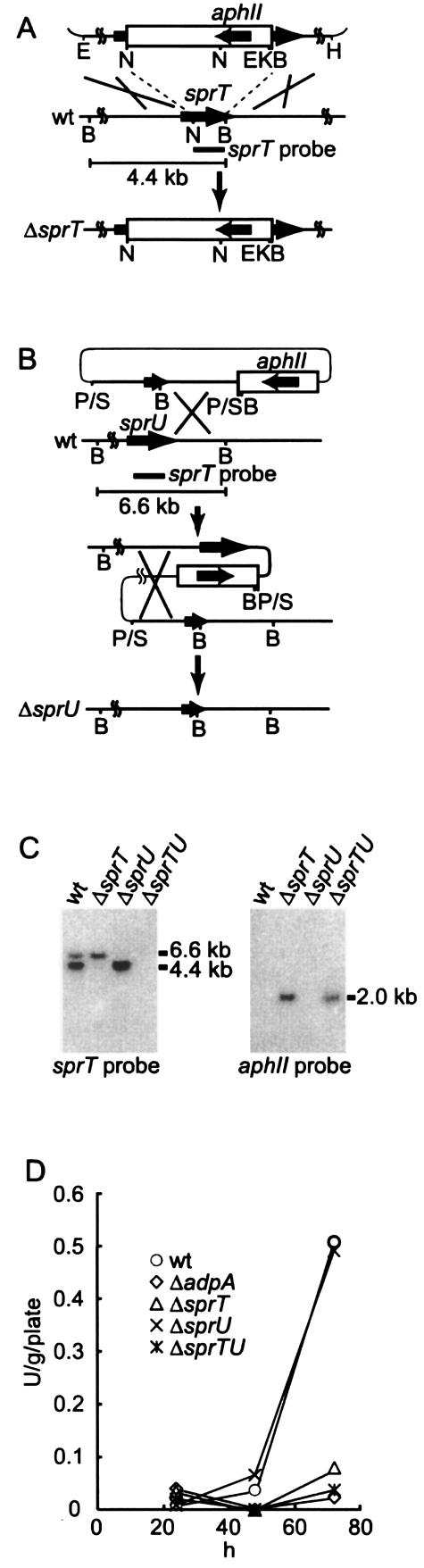
For cloning the DNA fragments containing sprT by PCR, we used the DNA sequence registered in the NCBI sequence databank and cloned a 6.6-kb DNA fragment containing sprT (see Fig. 2A) by standard DNA manipulation, including Southern hybridization and colony hybridization. A 6.6-kb fragment containing sprU, an sprT homologue, which we had happened to find during these cloning experiments was cloned similarly with a 0.5-kb NcoI-BamHI fragment within the sprT coding region as a hybridization probe.
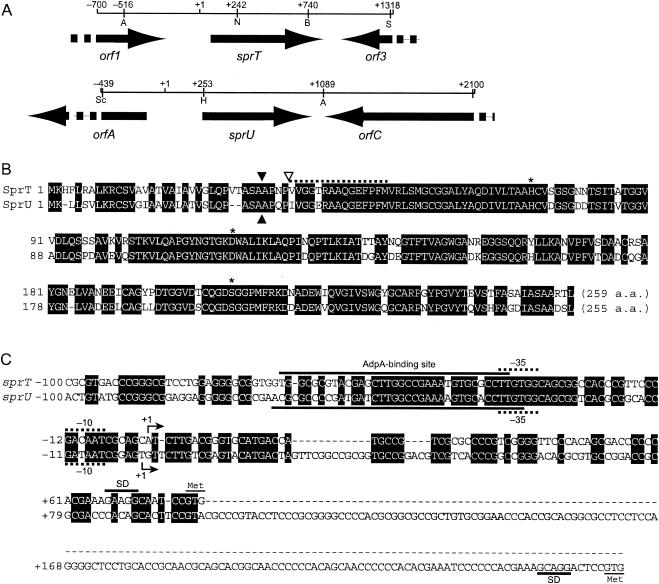
FIG. 2.
Gene organization in the cloned fragments containing sprT and sprU (A), amino acid sequences of SprT and SprU (B), and nucleotide sequences of the promoter and operator regions of sprT and sprU (C). (A) The positions and directions of open reading frames predicted by FramePlot analysis (10) of the nucleotide sequence are indicated by arrows. Restriction sites used in the construction of plasmids are also shown. Abbreviations for restriction enzymes: A, Aor51HI; B, BamHI; H, HindIII; N, NcoI; S, SalI; and Sc, SacI. The numbers on the restriction sites are the nucleotide positions, taking the transcriptional start point as +1. (B) The amino acid sequences of SprT and SprU were deduced from the nucleotide sequences and aligned. The amino acid sequence of SprT, starting at Val-37, determined with the purified protein is shown with a dotted line. The amino acids that form a His-Asp-Ser catalytic triad are indicated by stars. The N terminus of SprT is indicated by an open triangle, and sites in SprT and SprU that are probably cleaved by a signal peptidase are also shown by solid triangles. (C) The promoter and operator sequences of sprT and sprU are shown. The transcriptional start points determined by high-resolution S1 mapping (see Fig. 3B) are taken as +1. The AdpA binding sites determined by DNase I footprinting (see Fig. 5) are indicated. Probable −35 and −10 sequences for the sprT and sprU promoters are indicated with dotted lines (31). Purine-rich sequences, which may serve as ribosome-binding (Shine-Dalgarno) sites, are also shown.
Gel mobility shift assay.
Purification of histidine-tagged AdpA (AdpA-H) from E. coli BL21(DE3) and the gel mobility shift assay were described previously (33). The DNA fragments used for 32P-labeled probes were amplified by PCR and 32P labeled with T4 polynucleotide kinase. For gel mobility shift assays with AdpA-H and various regions in front of sprT, six probes, T1 to T6 (see Fig. 4A), were used. Table 1 lists the forward (F) and reverse (R) primer sequences for preparing these probes: T1-F and T1-R for probe T1; T2-F and T2-R for probe T2; T1-F and T3-R for probe T3; T4-F and T1-R for probe T4; T5-F and T3-R for probe T5; and T1-F and T6-R for probe T6. Similarly, for the gel mobility shift assays with various regions around the transcriptional start point of sprU, seven probes, U1 to U7 (see Fig. 4B), were used: U1-F and U1-R for probe U1; U2-F and U2-R for probe U2; U3-F and U3-R for probe U3; U1-F and U4-R for probe U4; U5-F and U5-R for probe U5; U6-F and U2-R for probe U6; and U6-F and U1-R for probe U7.
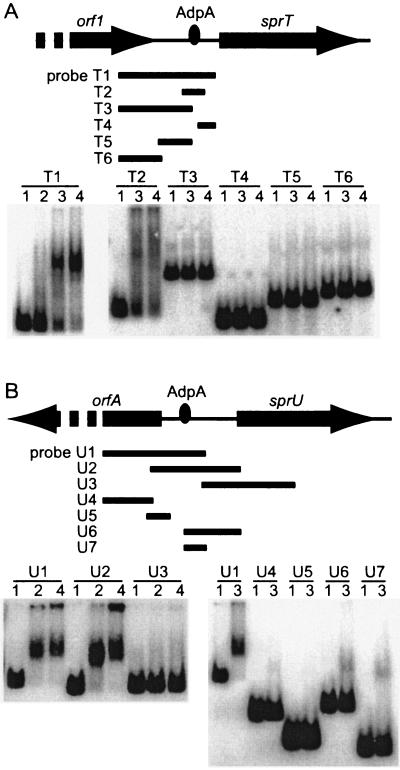
FIG. 4.
Binding of AdpA-H to the operator regions of sprT (A) and sprU (B), as determined by gel mobility shift assays. (A) The amounts of AdpA-H used were 0.04 μg (lane 2), 0.2 μg (lane 3), and 0.4 μg (lane 4). Lane 1 is a control to which no AdpA-H was added. The probes used were T1 (positions −448 to +61), T2 (positions −112 to +7), T3 (positions −448 to −61), T4 (positions −34 to +61), T5 (positions −238 to −61), and T6 (positions −448 to −219). AdpA-H was found to bind to a single site, as shown. (B) The amounts of AdpA-H used were the same as in A. The probes used were U1 (positions −444 to +96), U2 (positions −200 to +277), U3 (positions +77 to +566), U4 (positions −444 to −181), U5 (positions −193 to −67), U6 (positions −24 to +277), and U7 (positions −24 to +96). AdpA-H was also found to bind to a single site, as shown.
TABLE 1.
Primers used in this study
| Gene | Primer | Positions | Sequence (5′ to 3′) |
|---|---|---|---|
| sprT | |||
| T1-F | −448 to −429 | CCCGGGGCATCGCCCGCCGC | |
| T1-R | +61 to +42 | TGGGGGGTCGCTGTGGGAAC | |
| T2-F | −112 to −93 | GCGTCCCCGGTCCGCGTGAC | |
| T2-R | +7 to −13 | TCAAGATGCTGCGATTGTCG | |
| T3-R | −61 to −80 | ACGCGCCACCACCGCCCCTC | |
| T4-F | −34 to −15 | GTGGCAGCGGCCACCCGTTC | |
| T5-F | −238 to −219 | GCCCTCCGGCAGACGGGTAG | |
| T6-R | −219 to −238 | CTACCCGTCTGCCGGAGGGC | |
| TS1H-F | −112 to −93 | GCGTCCCCGGTCCGCGTGAC | |
| TS1L-F | −313 to −294 | AACTGTCCGCGCCCGTCGAG | |
| TS1L-R | +60 to +41 | GGGGGGTCGCTGTGGGAACC | |
| Tfp-R | +7 to −13 | TCAAGATGCTGCGATTGTCG | |
| T-RT | +629 to +610 | CTCGTTGCCGTAGGCGGAGC | |
| Trt-F | +5 to +24 | TGACGGGTGCATGACCATGC | |
| Trt-R | +278 to +259 | GATGTCCTGGGCGTAGAGGG | |
| Tbd-F | −162 to −143 | GGCGTACGTGACACGACCGC | |
| Tbd-R | +77 to +58 | GGATTGCCTTCTTTCGTGGG | |
| sprU | |||
| U1-F | −444 to −425 | GAGCTCGATGAAGTACTCGG | |
| U1-R | +96 to +77 | GGAAGTGCTGTGGGTCGCGC | |
| U2-F | −200 to −181 | GAGCGGACCGTGTCCAGATG | |
| U2-R | +277 to +258 | CACCTTTTCAGCACGCTCAG | |
| U3-F | +77 to +96 | GCGCGACCCACAGCACTTCC | |
| U3-R | +566 to +547 | GGCCTGGAGGACCTTGGTGG | |
| U4-R | −181 to −200 | CATCTGGACACGGTCCGCTC | |
| U5-F | −193 to −174 | CCGTGTCCAGATGCGGAAAG | |
| U5-R | −67 to −86 | CGTTCGCGGCCCCCTCCTCC | |
| U6-F | −24 to −5 | GTCACGCGGCACCGATAATC | |
| Ufp-F | −113 to −94 | GTCGACGGACTCCACTGTAT | |
| Ufp-R | +29 to +10 | CCGAACTAGTCATGTACTCG | |
| U-RT | +392 to +373 | CATGAACGGGAACTCGCCCT | |
| Urt-F | +77 to +96 | GCGCGACCCACAGCACTTCC | |
| Urt-R | +331 to +312 | GAGGCGGGCTGGAGACTGAC | |
| Ubd-F | −164 to −145 | ACAGGTGCGCCCGGAGGGCC | |
| Ubd-R | +76 to +57 | GGTCCGGCACGCGTGTCCCG |
S1 nuclease mapping.
Total RNA was isolated with Isogen (Nippon Gene) from cells grown on cellophane on the surface of YMPD agar medium. S1 nuclease mapping was described by Kelemen et al. (14). Hybridization probes were prepared by PCR with a pair of 32P-labeled and unlabeled primers. The PCR primers used for high-resolution S1 nuclease mapping were TS1H-F and T1-R (Table 1) for sprT and U5-F and U1-R for sprU. The primers for low-resolution S1 mapping were TS1L-F and TS1L-R for sprT and U5-F and U1-R for sprU. For the adpA probe, 5′-CGGTCGGTAATCCGGCCCTG-3′ (positions −274 to −255 with respect to the transcriptional start point of adpA) and 5′-TCTACTGCGTCGCGTGGTCC-3′ (positions +150 to +131) were used. hrdB encoding a principal σ factor of RNA polymerase was used to check the purity and amount of RNA used, as described previously (27). Protected fragments were analyzed on 6% polyacrylamide DNA sequencing gels by the method of Maxam and Gilbert (24).
Reverse transcription-PCR.
Total RNA was isolated from cells grown on YMPD agar medium. The concentration of RNA was determined by measuring the A260 by spectrophotometry. Reverse transcription-PCR was done with 3 μg of RNA, 2 pmol of primer T-RT for sprT (Table 1) or primer U-RT for sprU, SuperScript III reverse transcriptase (Invitrogen), and RNase H (Invitrogen) according to the manufacturer's instructions. Primers Trt-F and Trt-R were used for PCR amplification of the 274-bp fragment of sprT, and primers Urt-F and Urt-R were used for amplification of the 255-bp fragment of sprU (see Fig. 6C). The PCR conditions were 98°C for 20 s, 61°C for 30 s, and 72°C for 45 s for a total of 25 cycles.
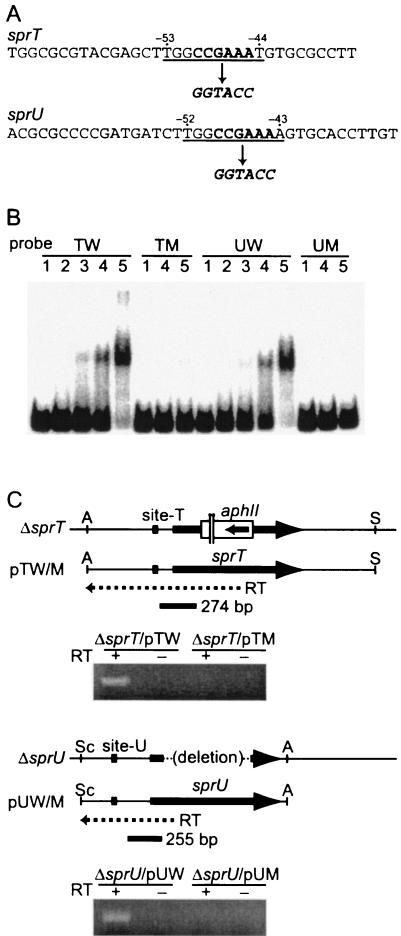
FIG. 6.
Effects of a mutation in the AdpA-binding sites on the promoter activities of sprT and sprU. (A) Mutations were introduced in the AdpA-binding sites for sprT and sprU. The consensus AdpA-binding sequences are underlined. The sequences indicated by bold letters in the consensus sequence were changed to generate a KpnI cleavage sequence. (B) Gel mobility shift assays for determination of AdpA binding to the mutated sequences. Probe TW (positions −162 to +77) contained the intact operator for sprT, and probe TM contained the mutated operator. Probe UW (positions −164 to +76) contained the intact operator for sprU, and probe UM contained the mutated operator. The amounts of AdpA-H used in lanes 1, 2, 3, 4, and 5 were 0, 15, 45, 150, and 300 ng, respectively. (C) Transcriptional analysis of sprT and sprU with mutated AdpA-binding sites by reverse transcription-PCR. RNA was prepared from cells grown at 28°C for 72 h on YMPD agar medium. The primers for reverse transcription were designed within the sequences that were deleted from the chromosome in the ΔsprT and ΔsprU hosts so that only mRNA derived from the plasmids was reverse transcribed into cDNA. After PCR of the cDNA in a total of 25 cycles, the amplified fragments were separated by 2% agarose gel electrophoresis. The lanes for sprT contained 10 μl each of the amplified solution in a total of 100 μl, and the lanes for sprU contained 20 μl each of the same volume of the amplified solution.
DNase I footprinting.
The method for DNase I footprinting was described by Yamazaki et al. (33). For analysis of AdpA-binding sites, a 32P-labeled fragment was prepared by PCR with primers TS1H-F and Tfp-R for sprT and primers Ufp-F and Ufp-R for sprU.
Alterations of the AdpA-binding sequences by PCR.
Mutations were introduced in the AdpA-binding sites of sprT and sprU by PCR. The CCGAAA sequence in the middle of the AdpA-binding site for sprT (see Fig. 6A) was changed to a KpnI recognition sequence, GGTACC, as follows. Two fragments were amplified with primers 5′-GGAATTCCGACGACGATCCCGTCCTCG-3′ (positions −540 to −521; italics indicate an EcoRI site) and 5′-CGGGGTACCCCAAGCTCGTACGCGCCACC-3′ (positions −51 to −70; italics indicate a KpnI site); and 5′-CGGGGTACCTGTGCGCCTTGTGGCAGCGG-3′ (positions −44 to −25; italics indicate a KpnI site) and 5′-CCTGGATCCACTCGTCGGCGTTG-3′ (positions +747 to +725; italics indicate a BamHI site). The amplified fragments were connected via the KpnI site and inserted into pUC19. The absence of PCR errors was checked by nucleotide sequencing. The mutation thus generated was called TM.
Similarly, the CCGAAA sequence in the middle of the AdpA-binding site for sprU (see Fig. 6A) was changed to a KpnI recognition sequence with two pairs of primers, RV (Takara Shuzo) and 5′-CGGGGTACCCCAAGATCATCGGGGCGCGT-3′ (positions −50 to −69; italics indicate a KpnI site), and 5′-CGGGGTACCAGTGCACCTTGTGGCAGCGG-3′ (positions −43 to −24; italics indicate a KpnI site) and M4 (Takara Shuzo). The template used was a SacI-HindIII fragment (positions −439 to +253) cloned on pUC18. The mutation thus generated was called UM. For determination of AdpA binding to the mutated sites by gel mobility shift assays, 32P-labeled probes of 239 bp (positions −162 to +77) for TM and 240 bp (positions −164 to +76) for UM were prepared by PCR with a pair of primers, Tbd-F and Tbd-R, and primers Ubd-F and Ubd-R, respectively (Table 1).
For construction of plasmid pTW, the 3.5-kb Aor51HI (position −516; Fig. 2A)-BamHI fragment containing sprT was placed in pUC19. For construction of pTM with the mutated AdpA-binding site, the 1.3-kb Aor51HI-BamHI (positions −516 to +740) fragment on the pUC plasmid was replaced with the mutated fragment. Digestion of these pUC plasmids with EcoRI (in the multicloning site of pUC19) and SalI (position +1318) yielded a DNA fragment containing the complete sprT coding and promoter sequences and either the intact or the mutated AdpA-binding site. The EcoRI-SalI fragments were cloned separately between the EcoRI and XhoI sites of pKU209, resulting in pTW and pTM.
For construction of plasmid pUW containing sprU, the 1.5-kb SacI-Aor51HI fragment (positions −439 to +1089; Fig. 2A) was placed in pUC19. For construction of pUM with the mutated AdpA-binding site, the 0.7-kb EcoRI (in pUC18)-HindIII (position +253) fragment was replaced with the mutated fragment. These fragments on pUC19 were excised as an EcoRI-SalI fragment by use of the restriction sites in the multicloning site and cloned between the EcoRI and XhoI sites of pKU209, resulting in pUW and pUM.
Gene disruption.
The chromosomal sprT gene of S. griseus IFO13350 was disrupted as follows. An upstream region of 4.6 kb from sprT and a downstream region of 2.0 kb were placed in the BamHI site of pUC19. The 1.7-kb EcoRI-NcoI (position +242; Fig. 2A) fragment containing the 5′ side of sprT and a 2.4-kb NcoI-KpnI fragment carrying the kanamycin resistance gene from Tn5 (2) were inserted by three-fragment-ligation into the EcoRI-KpnI site (multicloning site) of plasmid pUC19 containing a 2.0-kb BamHI (position +740)-BamHI fragment at the BamHI site. In this construction, the kanamycin resistance gene was inserted between the NcoI and BamHI sites (corresponding to Met-56 to Ile-222) of sprT. This plasmid prepared from E. coli JM110 was digested with DraI, alkali denatured with 0.1 M NaOH, and introduced by protoplast transformation into S. griseus IFO13350, as described (26). Correct replacement of the disrupted sprT sequence with the intact chromosomal sprT sequence, as a result of double crossover, was checked by Southern hybridization. Doubly sprT and sprU disrupted mutants were similarly generated by disrupting sprT in an sprU disruptant (see below).
We generated null sprU mutants by in-frame deletion. For deletion of the sequence from +366 to +1004 (corresponding to Ala-40 to Ala-252), the 1.1-kb fragment upstream of the Arg-39 codon and the 1.1-kb fragment downstream of the Asp-253 codon were amplified by PCR. For PCR to amplify the upstream region, 5′-CCGGAATTCGTACGCCGTGTACGCCTGGG-3′ (italics indicate an EcoRI site) and 5′-CGCGGATCCGCGTTCCCCGCCGACGATCG-3′ (italics indicate a BamHI site) were used. For amplifying the downstream region, 5′-CGCGGATCCGACTCGCTCTGACCCCGGTC-3′ (italics indicate a BamHI site) and 5′-CCCAAGCTTCAGGACGAGTCGTTCCAGCC-3′ (italics indicate a HindIII site) were used. After the absence of PCR errors had been confirmed by nucleotide sequencing, these two fragments were ligated via the BamHI site and inserted between the EcoRI and HindIII sites in pUC19. The fragment with a deletion of the sequence encoding from Ala-40 to Ala-252 was excised as a PvuII fragment by use of the PvuII sites outside the multicloning site and inserted in the SmaI site in plasmid pUC19 containing the kanamycin resistance gene. This plasmid was introduced by transformation into S. griseus IFO13350, and kanamycin-resistant transformants were first isolated. Kanamycin-sensitive colonies, derived after a second crossover in one of the transformants, were candidates for sprU-disrupted strains. The correct replacement was checked by Southern hybridization.
Trypsin activity assay.
S. griseus strains were grown for various periods as a lawn on cellophane on the surface of YMPD agar medium in a petri dish. After the cellophane and the cells had been removed, an agar plate (5 by 5 cm) was cut out and homogenized in 10 ml of trypsin assay buffer. The trypsin activity in the agar was measured spectrophotometrically by the release of p-nitroanilides due to the enzymatic hydrolysis of an artificial chromogenic substrate, N-benzoylarginine p-nitroanilide (BAPNA). The reaction mixture, composed of 890 μl of a reaction buffer (50 mM Tris-HCl [pH 8.0] and 20 mM CaCl2) and 10 μl of 50 mM BAPNA in dimethyl sulfoxide was warmed at 37°C for 5 min, rapidly mixed with 100 μl of the enzyme solution, and incubated for 15 min. The reaction was stopped by adding 400 μl of 30% acetic acid in dioxane, and the absorbance at 405 nm was recorded. One unit of hydrolytic activity was defined as the amount of enzyme needed to produce 1 μmol of p-nitroanilide per min (29).
Nucleotide sequence accession numbers.
The nucleotide sequences of sprT and sprU have been deposited in the DDBJ, EMBL, and GenBank databases under accession numbers AB182575 and AB182576, respectively.
RESULTS AND DISCUSSION
Production of SprT in response to A-factor.
To compare the protein profiles of the extracellular fractions of the wild-type strain IFO13350 and the A-factor-deficient mutant HH1, we cultured both strains at 28°C in R2YE medium and prepared the supernatants by centrifugation. Proteins in the supernatants were precipitated with trichloroacetic acid, and the precipitates were resuspended in 10 mM potassium phosphate buffer (pH 7.0) and dialyzed against the same buffer. Because we had learned that SprT flowed through a Resource-S cation exchange column during purification of SprT from Pronase, a commercial protease mixture (22), the dialyzed sample was loaded onto a Resource-S cation exchange column and the flowthrough fraction was collected. The proteins were concentrated with trichloroacetic acid and analyzed by denaturing 4 to 20% gradient polyacrylamide gel electrophoresis (Fig. 1).
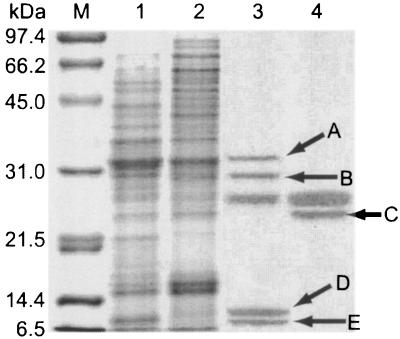
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of extracellular proteins that were obtained by TCA precipitation of the culture broth of S. griseus, followed by Resource-S cation exchange column chromatography. Lane M, molecular size standards; lane 1, TCA precipitate of the wild-type strain S. griseus IFO13350; lane 2, TCA precipitate of the A-factor-deficient S. griseus mutant HH1; lane 3, flowthrough fraction of the TCA precipitate of strain IFO13350 from the Resource-S cation exchange column; and lane 4, flowthrough fraction of the TCA precipitate of mutant HH1 from the Resource-S cation exchange column. Each lane contained appropriate amounts of proteins. Four proteins, A, B, D, and E, are observed only in the wild-type strain, and one protein, C, is observed only in mutant HH1.
Four proteins, marked A, B, D, and E, were produced only by the wild-type strain IFO13350. On the other hand, protein C was found only in the culture broth of the A-factor-deficient mutant HH1. This is consistent with our previous observation on the pleiotropic effect of A-factor on protein biosynthesis, either positive or negative (5). Because SprT purified from Pronase had a molecular mass of about 28 kDa (22), we transferred protein B to a polyvinylidene fluoride membrane, and its N-terminal amino acid sequence was determined by Edman degradation. The N-terminal amino acid sequence was Val-Val-Gly-Gly-Thr-Phe-Ala-Ala-Gln-Gly-Glu-Phe-Pro-Phe-Met, which matched that of the mature form of SprT from S. griseus ATCC 10137 (20), except for the sixth Phe. The sixth amino acid of SprT of strain IFO13350 turned out to be Arg from the nucleotide sequence. These findings suggested that SprT of S. griseus IFO13350 was biosynthesized in response to A-factor. The other four proteins have not been characterized.
Cloning and nucleotide sequencing of sprT and sprU.
We cloned the sprT gene on the basis of the nucleotide sequence of sprT from strain ATCC 10137 and determined the nucleotide sequence of sprT for detailed transcriptional analysis. We also cloned a longer DNA fragment containing sprT in a total of 6.6 kb for gene disruption. The gene organization in the DNA fragment containing sprT is shown in Fig. 2A. The DNA database predicted that orf1 encoded a putative oxidoreductase found in Streptomyces avermitilis and orf3 encoded a hypothetical protein found in Streptomyces coelicolor A3(2).
The deduced amino acid sequence of SprT, containing the amino acid sequence determined with purified SprT, is shown in Fig. 2B. Since the PSORT program (http://psort.nibb.ac.jp/) predicted that a signal peptidase cleaves the Ala-Ala peptide bond indicated by a solid triangle, SprT may be matured by a second cleavage between the Pro-Val bond indicated by an open triangle. The computer program also predicted that the Ala-Ala bond of SprU (see below), indicated by a solid triangle, is cleaved by a signal peptidase. A catalytic His-Asp-Ser triad (Fig. 2B) can be predicted by alignment with other serine proteases. As already reported by Read and James (28) and Kim et al. (20), trypsin-like proteases in Streptomyces spp., including SprT, are similar in amino acid sequence to mammalian trypsin rather than to bacterial serine proteases.
As described later, during Southern hybridization for confirmation of correct gene replacement (see Fig. 7C) we found that an sprT-like gene was also present in S. griseus. Because subsequent study of this sprT-like gene showed that it actually encoded an enzymatically active SprT-like protease, we cloned it and determined its nucleotide sequence. We named this gene sprU. sprU was present between orfA, encoding a hypothetical protein found in S. avermitilis, and orfC, encoding a β-galactosidase (Fig. 2A). SprU showed 80% amino acid identity to SprT and had a conserved His-Asp-Ser catalytic triad as is found for trypsin-type proteases (Fig. 2B).
FIG. 7.
Disruption of the chromosomal sprT and sprU genes and trypsin activity in the disruptants. (A and B) Schematic representationof disruption of sprT and sprU on the chromosome of S. griseus IFO13350. Restriction enzymes: N, NcoI; B, BamHI; K, KpnI; P, PvuII; Sm, SmaI; H, HindIII; and E, EcoRI. (C) Southern hybridization to check the correct disruption of sprT and sprU with the indicated sprT probe against the BamHI-digested chromosome. The sprT probe also hybridized to the sprU sequence. ΔsprTU, doubly disrupted sprT and sprU mutant. (D) Trypsin activities of the ΔsprT, ΔsprU, and ΔsprTU mutants as a function of cultivation time. Strains were grown at 28°C for 72 h on YMPD agar medium.
Dependence of transcription of sprT and sprU on adpA.
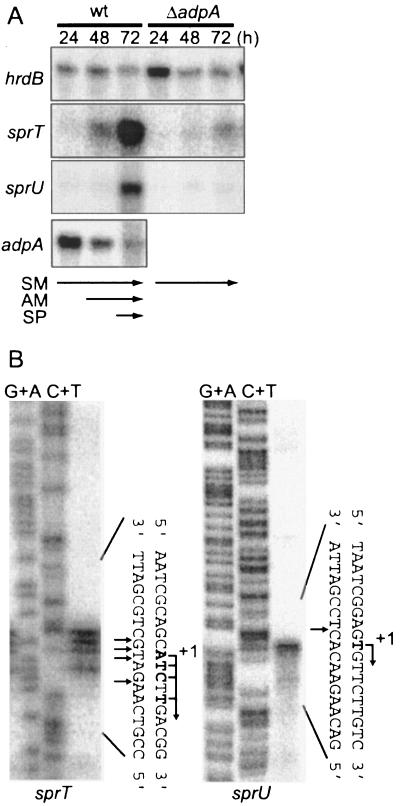
Because SprT was produced in response to A-factor, we examined the transcription of sprT in the presence and absence of adpA. RNA was purified from the wild-type strain IFO13350 and an adpA disruptant (ΔadpA) that were grown on cellophane on the surface of YMPD agar medium. The transcription of sprT was observed at 48 h, when cells began to form aerial hyphae, and greatly enhanced at 72 h, when sporulation began, as determined by low-resolution S1 nuclease mapping (Fig. 3A). On the other hand, sprT was not transcribed in an adpA-disrupted mutant, ΔadpA. These results showed that sprT was transcribed in an adpA-dependent manner. High-resolution S1 mapping identified multiple transcriptional start points about 80 nucleotides upstream of the start codon of sprT (Fig. 3B and 2C).
FIG. 3.
Transcriptional analysis of sprT and sprU. (A) The time courses of sprT and sprU transcription were followed by low-resolution S1 mapping with RNA prepared from cells grown at 28°C for the indicated hours on cellophane on the surface of YMPD medium. As controls, transcription of adpA and hrdB was also determined with the same RNA samples. hrdB, which is transcribed throughout growth, was used to check the purity and amount of mRNA used. The wild-type (wt) strain grew as substrate mycelium (SM) at 24 h, as a mixture of substrate and aerial mycelium (AM) at 48 h, and as a mixture of aerial hyphae and spores (SP) at 72 h. (B) The transcriptional start points of sprT and sprU were determined by high-resolution S1 nuclease mapping. RNA (40 μg each) prepared from wild-type cells grown at 28°C for 72 h on YMPD solid medium was used. The arrows indicate the positions of S1 nuclease-protected fragments.
We also examined the dependence of sprU transcription on adpA (Fig. 3A). sprU was transcribed at 72 h, when sporulation began. The transcription of sprU was also adpA dependent, because no transcription was observed in the adpA disruptant. The amount of the sprU transcript at 72 h was estimated to be much smaller than that of the sprT transcript from the intensities of the S1 signals. High-resolution S1 mapping identified the transcriptional start point to be 248 nucleotides upstream of the start codon (Fig. 3B and 2C).
adpA is transcribed actively at a rather early stage of growth, probably at or near the decision point at the middle of the exponential growth phase (12), as is seen in Fig. 3A. The transcription continues until the late stage, after aerial hyphae have formed, because one of the targets of AdpA is ssgA, whose gene product is essential for septum formation in aerial hyphae (34). We assume that AdpA activates the genes not only for aerial mycelium formation and secondary metabolism but also for rather late events such as septation in aerial hyphae. The examples for the former case are σAdsA, essential for aerial hyphae formation (33), sgmA, important for aerial mycelium formation (13), and strR, a pathway-specific transcriptional activator for streptomycin biosynthesis (27), and the examples for the latter case are ssgA, essential for septation in aerial hyphae, and the two trypsin genes in this study. The low transcription of sprT and sprU at an early stage of growth, where adpA is actively transcribed, suggests the presence of a mechanism by which their transcription is repressed during the early growth stage.
Binding of AdpA to a single site upstream of sprT and sprU.
Since the transcription of both sprT and sprU was dependent on adpA, we determined the ability of AdpA to bind somewhere around their transcriptional start points. AdpA has been shown to activate target genes by binding to various positions with respect to the transcriptional start point, for example, more than 200 bp upstream and 25 bp downstream from the transcriptional start point (36). In addition, some target genes contain multiple AdpA-binding sites, and AdpA activates ssgA, for example, by binding to two sites, from −255 to −216 and from −127 to −95 (34). We therefore designed 32P-labeled probes covering various regions and used them for gel mobility shift assays with AdpA (Fig. 4).
Probe T1, covering the region from −448 to +61 with respect to the transcriptional start point of sprT, and probe T2, covering the region from −112 to +7, gave a single retarded signal (Fig. 4A). On the other hand, probes T3 to T6 gave no retarded signals. We therefore concluded that AdpA bound a single site at positions −112 to +7. A single binding of AdpA to this region was confirmed by DNase I footprinting (see below).
Probe U1, covering the region from −444 to +96 with respect to the transcriptional start point of sprU, and probe U2, covering the region from −200 to +277, gave a single retarded signal, whereas probes U3, U4, U5 (positions −193 to −67), U6 (−24 to +277), and U7 did not. Thus, AdpA was suggested to bind a single site between positions −67 and −24.
Binding sites of AdpA upstream of sprT and sprU.
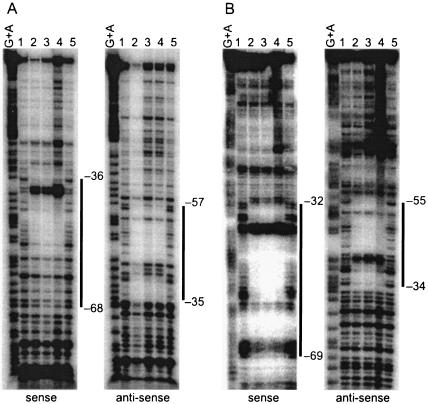
We performed DNase I footprinting to determine the exact location of the AdpA-binding sites in front of sprT and sprU. For the AdpA-binding site in front of sprT, a 32P-labeled probe covering the region from −112 to +7 was used. AdpA protected a sequence from positions −36 to −68 with respect to the transcriptional start point of sprT on the sense strand and from −35 to −57 on the antisense strand (Fig. 5A). Positions −39 and −40, at which enhanced DNase I digestion was observed, in the protected region were probably exposed on the surface of the AdpA-DNA complex. The AdpA-binding site for sprT contains a sequence, 5′-TGGCCGAAAT-3′, which is in agreement with the consensus AdpA-binding sequence, 5′-TGGCSNGWWY-3′ (where S is G or C; W is A or T; Y is T or C; and N is any nucleotide) (36) (Fig. 2C).
FIG. 5.
DNase I footprinting for determining the AdpA-binding sites in front of sprT (A) and sprU (B). DNase I footprinting assays were performed on the sense and antisense strands. The amounts of AdpA-H used in lanes 1, 2, 3, 4, and 5 were 0, 0.2, 0.4, 0.8, and 0 μg, respectively. The DNase I digests were run with the same probes that were chemically cleaved (G+A lanes).
For the AdpA-binding site in front of sprU, a 32P-labeled probe covering the region from −113 to +29 was used. AdpA protected from DNase I digestion a sequence from −32 to −69 on the sense strand and from −34 to −55 on the antisense strand (Fig. 5B). Enhanced DNase I digestion in the protected region was observed at positions −38 and −39. The AdpA-binding site for sprU also contains 5′-TGGCCGAAAA-3′, which is similar to the AdpA consensus sequence. The location of the consensus AdpA-binding sequences with respect to the transcriptional start point of sprU is from −52 to −43, which is almost the same as that (from −53 to −44) for sprT.
Direct transcriptional control of sprT and sprU by AdpA.
To determine whether the transcription of sprT and sprU is directly controlled by AdpA, we introduced a mutation in the consensus AdpA-binding sequences by site-directed mutagenesis and measured the effects of the mutation on the promoter activity by reverse transcription-PCR. As shown in Fig. 6A, the six nucleotides in the consensus AdpA-binding sequences for sprT and sprU were changed to a KpnI recognition sequence of six nucleotides. The 32P-labeled probe TM (positions −162 to +77) containing the mutated AdpA-binding site and the promoter of sprT gave almost no retarded signal in the gel mobility shift assay with AdpA, whereas a similar probe, TW, containing the intact AdpA-binding site gave a distinct retarded signal (Fig. 6B). Probe UM (positions −164 to +76) containing the mutated AdpA-binding site and the sprU promoter gave almost no retarded signal, whereas a similar probe, UW, containing the intact AdpA-binding site gave a retarded signal. These results showed that the mutations almost abolished the ability of AdpA to bind to these sequences. In addition, by comparing the intensities of signals observed with the intact AdpA-binding sites, we roughly estimated that AdpA bound the binding sites for sprT and sprU at the same affinity.
We constructed pTW, containing a 1.8-kb Aor51HI-SalI fragment, in which the sprT promoter, together with its upstream region as far as position −516, and the complete sprT coding sequence were contained, with the low-copy-number plasmid pKU209. We also constructed a similar plasmid, pTM, in which the AdpA-binding site was replaced by the TM mutation. We introduced them into an sprT disruptant (mutant ΔsprT) and measured transcription from the sprT promoter on the plasmid by reverse transcription-PCR. Reverse transcription was performed to amplify mRNA as cDNA from +1 to +629. The RNA sample was prepared from cells grown at 28°C for 72 h on cellophane on the surface of YMPD agar medium. The PCR was performed to amplify the cDNA from +5 to +278 in a total of 25 cycles. Since the chromosomal sprT sequence in mutant ΔsprT lost the primer region for the reverse transcription, the amplified DNA should come from the mRNA starting at the sprT promoter on pTW. As shown in Fig. 6C, a distinct amplified signal was detected for mutant ΔsprT harboring pTW, whereas no such signal was detected for mutant ΔsprT harboring pTM under the conditions used (Fig. 6C). In control lanes to which no reverse transcriptase was added, no amplification from the RNA sample occurred, indicative of no contamination of DNA. These results showed that direct binding of AdpA to the AdpA-binding site was essential for transcriptional activation of sprT.
Similarly, we constructed two plasmids, pUW containing a 1.5-kb SacI-Aor51HI fragment with the intact AdpA-binding site, the sprU promoter, and the sprU coding sequence, and pUM, containing the UM mutation instead of the intact AdpA-binding site. These plasmids were introduced into an sprU disruptant (mutant ΔsprU). Reverse transcription-PCR to amplify mRNA from +1 to +392 and PCR to amplify the cDNA from +77 to +331 were performed. Mutant ΔsprU lost the primer region for the reverse transcription. A distinctly amplified signal for mutant ΔsprU harboring pUW was detected, whereas no signal was detected for ΔsprU harboring pUM (Fig. 6C). Therefore, AdpA activates the sprU promoter by directly binding to the AdpA-binding site at the operator position.
Operators for sprT and sprU.
AdpA is a representative of a large subfamily of the AraC/XylS family (36). Some AdpA-binding sites consist of an inverted repeat of the consensus sequence, 5′-TGGCSNGWWY-3′, that is located at various positions with respect to the transcriptional start point of the target genes. On the other hand, the AdpA-binding sites of other target genes, such as sgmA (13) and orf1 (36), contain the consensus sequence in a single copy. Since the AdpA-binding sites for sprT and sprU contain the consensus sequence in a single copy at about −50 nucleotide positions in the same orientation as that of transcription, the mechanism of transcriptional activation of these genes by AdpA can be included in the latter group. The binding position at −50 and the same orientation of the recognition sequence as of transcription is typical of several AraC/XylS family members that function as dimers, such as RhaS at rhaBAD, AraC at araBAD, and XylS at Pm (30, 32). We assume that AdpA binds sites of this type by anchoring the DNA via the subunit near the −35 and −10 sequences so that this subunit can interact with RNA polymerase for transcriptional activation (36).
No phenotypic alterations in morphological development by mutants ΔsprT, ΔsprU, or ΔsprTU.
Trypsin-like proteases had been suggested to be important for morphological differentiation, particularly in aerial hyphae formation, on the basis of the observations that trypsin-specific inhibitors such as tosyl lysyl chloromethyl ketone and leupeptin caused several Streptomyces species to remain bald or to delay aerial hypha formation, depending on the concentration (16, 18, 25). Kim et al. (17, 19) also assumed that a leupeptin-inactivating enzyme controlled the amount of leupeptin for normal colony development at an appropriate time in leupeptin-producing Streptomyces exfoliatus. However, the phenotypes observed with inhibitors and inhibitor-degrading enzymes might result from indirect effects of these agents. No experiments on disruption of trypsin-like protease genes have been reported. We therefore disrupted the trypsin genes to determine the exact effects of SprT and SprU on morphological differentiation.
The chromosomal sprT gene was disrupted by inserting the kanamycin resistance gene between the NcoI and BamHI sites (i.e., between the codons for Met-56 and Ile-222) in the sprT coding sequence (Fig. 7A), and the sprU gene was disrupted by in-frame deletion of the sequence encoding from Ala-40 to Ala-252 (Fig. 7B). Correct disruption was checked by Southern hybridization with the NcoI-BamHI fragment within the sprT coding sequence (Fig. 7C). An unexpected finding was that the wild-type strain showed a 6.6-kb signal in addition to the expected 4.4-kb signal in the BamHI-digested chromosomal DNA. Subsequent cloning of the 6.6-kb fragment revealed the presence of an additional gene encoding a trypsin-like enzyme, named SprU, as described above. Correct disruption of sprT was confirmed by the absence of the 4.4-kb signal. In addition, correct deletion of sprU was confirmed by the absence of the 6.6-kb signal. We also disrupted the sprT gene in mutant ΔsprU and confirmed its correct disruption by a similar Southern hybridization assay, generating mutant ΔsprTU (Fig. 7C).
The trypsin activities of the three mutants grown on YMPD agar medium were compared to that of the wild-type strain (Fig. 7D). Mutant ΔsprT showed very low trypsin activity, and mutant ΔsprU showed almost the same activity as the wild-type strain, which indicated that most of the trypsin activity of S. griseus was represented by SprT. Because the ΔsprTU mutant showed almost no trypsin activity, the activity exerted by SprU was estimated by the activity shown by the ΔsprT mutant. The almost total lack of trypsin activity shown by the ΔsprTU mutant suggested that no other trypsin genes were present in S. griseus. Under the growth conditions used, aerial hypha formation began at approximately 30 h, and sporulation was observed at approximately 50 h. Both SprT and SprU were produced at 50 h, when sporulation began, and their production was highly active throughout sporulation. The timing of the production of these proteases was in good agreement with the transcriptional patterns of these genes (Fig. 3A), in which transcription of both genes was most active at 72 h.
The ΔsprT, ΔsprU, and ΔsprTU mutants together with the wild-type strain were grown on YMPD and R2YE medium as a lawn by spreading a lump of mycelium and as a colony by inoculating a lump of mycelium or spores with a toothpick. Contrary to our expectation, however, no apparent phenotypic changes were observed in the mutants; the three mutants grew normally and formed aerial hyphae and spores on the same time course as the wild-type strain, and no changes in streptomycin production were observed. We therefore conclude that these trypsin genes do not play an important role in morphological differentiation or secondary metabolism. In S. griseus IFO13350, a zinc-containing metalloendopeptidase has been shown by gene disruption experiments to be important for aerial hypha formation, either in apoptotic lysis of proteins in substrate hyphae or in activation of some other enzymes important for aerial hypha formation by processing them into their mature forms (13).
We examined the possible effects of a trypsin inhibitor on the morphogenesis of S. griseus IFO13350 and the ΔsprTU mutant by placing a paper disk (diameter, 9 mm) containing various amounts of leupeptin, known as a trypsin-specific inhibitor, on the surface of a cell lawn at different times. Even when 2 mg of leupeptin was supplied, cells of both strains in the vicinity of the paper disks developed aerial hyphae at the same time, although sporulation of the wild-type strain appeared to be slightly delayed (data not shown). No effect of leupeptin on aerial hypha formation is in good agreement with the facts that the ΔsprT, ΔsprU, and ΔsprTU mutants developed normal aerial hyphae and spores, that transcription of these genes is most active during spore formation, and that trypsin activity is also detected during sporulation. These observations on S. griseus IFO13350 are different from those on other Streptomyces strains, in which trypsin inhibitors caused a delay or complete inhibition of aerial hypha formation (16).
The difference may result from physiological differences among Streptomyces strains; for example, the number of proteases of the trypsin type and of other types and the amounts of these proteases that are produced at specific times must differ from one strain to another. S. avermitilis contains a gene, SAV6208, encoding a protein showing 81% identity in amino acid sequence to SprT and three other genes (SAV2034, SAV2443, and SAV4044) encoding a protein showing about 35% identity. S. coelicolor A3(2) does not contain a gene encoding a protein showing high similarity, although it contains two genes (SCO5821and SCO7461) encoding a protein showing about 30% identity. In addition, trypsin activity in Streptomyces roseus is already high at the time of aerial hypha formation, and that in S. coelicolor A3(2) is considerably lower (16). These observations suggest that trypsin-type proteases contribute significantly to morphological differentiation in some Streptomyces species, but in other species its contribution is too small to be detectable.
Acknowledgments
J. Kato was supported by the Japan Society for the Promotion of Science (JSPS). This work was supported, in part, by a Joint Research Project under the Japan-Korea Basic Scientific Cooperation Program of JSPS, the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan, and a Grant-in-Aid for Scientific Research on Priority Areas from Monkasho.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 4.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi, S., and T. Beppu. 1990. Autoregulatory factors of secondary metabolism and morphogenesis in actinomycetes. Crit. Rev. Biotechnol. 10:191-204. [DOI] [PubMed] [Google Scholar]
- 6.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377-398. [DOI] [PubMed] [Google Scholar]
- 7.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 8.Horinouchi, S., Y. Kumada, and T. Beppu. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J. Bacteriol. 158:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horinouchi, S., H. Onaka, H. Yamazaki, S. Kameyama, and Y. Ohnishi. 2000. Isolation of DNA fragments bound by transcriptional factors, AdpA and ArpA, in the A-factor regulatory cascade. Actinomycetologica 14:11-16. [Google Scholar]
- 10.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 11.Kakinuma, S., Y. Takada, H. Ikeda, H. Tanaka, S. Omura, and D. A. Hopwood. 1991. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J. Antibiot. (Tokyo) 44:995-1005. [DOI] [PubMed] [Google Scholar]
- 12.Kato, J., I. Miyahisa, M. Mashiko, Y. Ohnishi, and S. Horinouchi. 2004. A single target is sufficient to account for the biological effects of the A-factor receptor protein of Streptomyces griseus. J. Bacteriol. 186:2206-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, J., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser, H., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 16.Kim, I. S., S. G. Kang, and K. J. Lee. 1995. Physiological importance of trypsin-like protease during morphological differentiation of streptomycetes. J. Microbiol. 33:315-321. [Google Scholar]
- 17.Kim, I. S., Y. B. Kim, and K. J. Lee. 1998. Characterization of the leupeptin-inactivating enzyme from Streptomyces exfoliatus SMF13 which produces leupeptin. Biochem. J. 331:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, I. S., and K. J. Lee. 1995. Physiological roles of leupeptin and extracellular proteases in mycelium development of Streptomyces exfoliatus SMF13. Microbiology 141:1017-1025. [DOI] [PubMed] [Google Scholar]
- 19.Kim, I. S., and K. J. Lee. 1996. Trypsin-like protease of Streptomyces exfoliatus SMF13, a potential agent in mycelial differentiation. Microbiology 142:1797-1806. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. C., S. H. Cha, S. T. Jeong, S. K. Oh, and S. M. Byun. 1991. Molecular cloning and nucleotide sequence of Streptomyces griseus trypsin gene. Biochem. Biophys. Res. Commun. 181:707-713. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J.-M., and S.-K. Hong. 2000. Streptomyces griseus HH1, an A-factor deficient mutant, produces diminished level of trypsin and increased level of metalloproteases. J. Microbiol. 38:160-168. [Google Scholar]
- 22.Koo, B.-J., H.-B. Kwang, S.-M. Byun, and S.-K. Hong. 1998. Purification and characterization of Streptomyces griseus trypsin overexpressed in Streptomyces lividans. J. Microbiol. Biotechnol. 8:333-340. [Google Scholar]
- 23.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 25.Nicieza, R. G., J. Huergo, B. A. Connolly, and J. Sanchez. 1999. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. Analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J. Biol. Chem. 274:20366-20375. [DOI] [PubMed] [Google Scholar]
- 26.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 28.Read, R. J., and M. N. James. 1988. Refined crystal structure of Streptomyces griseus trypsin at 1.7 Å resolution. J. Mol. Biol. 200:523-551. [DOI] [PubMed] [Google Scholar]
- 29.Sarath, G., R. S. De La Motte, and F. W. Wagner. 1989. Protease assay methods, p. 25-55. In R. J. Beynone and J. S. Bond (ed.), Proteolytic enzymes: a practical approach. IRL Press, Oxford, England.
- 30.Schleif, R. 2003. AraC protein: a love-hate relationship. Bioessays 25:274-282. [DOI] [PubMed] [Google Scholar]
- 31.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]