Abstract
A two-dimensional electrophoretic analysis of protein distribution followed by identification of selected proteins by mass spectrometry was performed on fresh bdellovibrio cultures containing attack phase cells of the predatory bacterium Bdellovibrio bacteriovorus strain 109J-1 and the remains of an Escherichia coli or a Pseudomonas syringae pv. tomato prey. Cleavage of the peptidoglycan-associated outer membrane proteins (OMPs) OmpA in E. coli and OprF in P. syringae occurred in both prey. The tryptic peptides obtained from the cleavage products of OmpA and OprF were all located within the 19-kDa pronase-resistant N-terminal parts of the corresponding proteins. The predator cell fraction was separated from the prey ghosts in fresh bdellovibrio cultures by centrifugation on a Percoll-sucrose cushion. Proteins from each fraction were separated by two-dimensional electrophoresis and identified by mass spectrometric analysis. As no prey OMP could be detected in the predator cell fraction, it was concluded that prey OMPs are not transferred to the predator, as had been suggested previously. However, a protein from the predator was found bound to ghost cell envelopes. This protein may correspond to a protein earlier suggested to be associated with the prey outer or cytoplasmic membranes. Along with recently described polypeptides from B. bacteriovorus strains 100 and 114, it forms a new family of putative outer membrane proteins.
The peculiar life cycle of most strains of predatory bacteria belonging to the genus Bdellovibrio and Bdellovibrio-like organisms is composed of an extracellular attack phase and an intraperiplasmic stage. After successful attachment to the outer membrane and penetration of the gram-negative prey's periplasmic space, the invading predatory cell elongates into a filament. The nutrients for growth of the filament and chromosome replication are provided by progressive decomposition of the prey's cytoplasm. Septation brings about fragmentation of the intraperiplasmic filament into progeny, and each synthesizes a flagellum. The cycle culminates with the release of progeny attack cells from the remains of the host (12).
The data present in the recently released annotated genome of B. bacteriovorus type strain 100 mirror the exceptional phenotype of this organism and reflect the adaptation of the bacterium to its predatory life style: of the 3,584 deduced proteins, 369 code for putative lytic activities, 26 code for flagellar and type IV pilus biosynthesis, and 244 code for membrane-inserted transporters, including 147 ABC-type transporters. Nine and 10 pathways for the anabolism and catabolism of amino acids, respectively, are apparently missing from the metabolic arsenal (20).
Previous research has determined that exploitation of prey components include direct use of some fatty acid molecules and uptake of nucleoside monophosphates and ATP (13, 16, 18, 21, 24, 25, 26, 27). However, these data have come under scrutiny. Schwudke et al. (31), showed that Bdellovibrio bacteriovorus synthesizes a unique type of lipid A and that no prey-derived lipooligosaccharide is incorporated into the predator. The annotated genome does not reveal the presence of ATP uptake systems, although one (or more) of the numerous ABC transporters with no defined function may be responsible for nucleoside triphosphate transport.
Previous publications have reported that Bdellovibrio and like organisms also have the capacity to insert various prey outer membrane proteins (OMPs) into their own envelope (6, 7, 8, 32). However, the nature of the imported proteins and even the existence of such a capacity has been a matter of contention (2, 7, 19).
In this work, we show that related, peptidoglycan-associated proteins from different prey cells are similarly degraded by Bdellovibrio cells, that prey OMPs are not detected in the predator, providing strong support that such proteins are not transferred from the prey to the predator, and that a putative outer membrane protein from the predator is bound to the remains of the prey envelope after predatory progeny cells are released.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
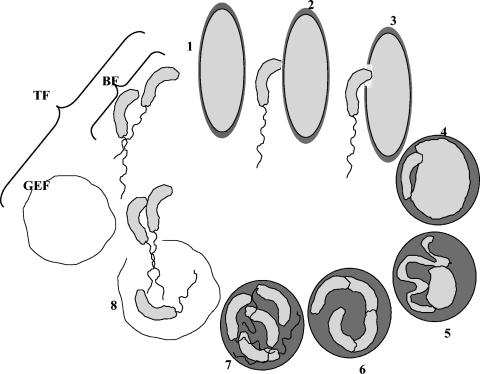
B. bacteriovorus strain 109J-1 (1) was routinely grown in 100 ml in 250-ml Erlemeyer flasks containing a 1:10 dilution of nutrient broth (amended with 2 mM CaCl2 and 3 mM MgCl2 · 7H2O) or in HEPES buffer (25 mM HEPES, 2 mM CaCl2 · 2H2O, 3 mM MgCl2 · 7H2O, pH 7.8) in two-membered cultures with Escherichia coli ML35 as the prey. Alternatively, Pseudomonas syringae pv. tomato from our laboratory collection was used as the prey. Bdellovibrio cultures obtained from two-membered suspensions contained 5 × 1010 to 5 × 1011 predatory cells ml−1. These cultures were either washed three times by centrifugation at 12,000 × g for 10 min at 4°C and resuspended in HEPES buffer (total fraction) or washed and centrifuged on a Percoll-sucrose cushion (see below), yielding a Bdellovibrio attack phase cell fraction, and a ghost envelope fraction (Fig. 1).
FIG. 1.
Life cycle of Bdellovibrio bacteriovorus, and the different fractions analyzed by two-dimensional electrophoresis. 1. Prey search by attack-phase cells. 2. Attachment. 3. Penetration. 4. Establishment. 5. Cell elongation and DNA replication. 6. Cell fragmentation. 7. Differentiation. 8. Progeny release. BF, Percoll-separated attack phase cells. GEF, Percoll-separated envelopes of prey cells. TF, total fraction of a washed fresh bdellovibrio culture.
Percoll separation.
Fresh bdellovibrio cultures were washed three times with HEPES buffer at 4°C by centrifugation for 10 min at 12,000 × g, finally resuspended in 4 ml of phosphate buffer (pH 7.2), and layered onto the surface of 66 ml of a Percoll (Sigma, Rehovot, Israel) solution in a 70-ml ultracentrifuge tube. The solution was prepared by mixing five parts of a Percoll stock solution with four parts of 0.25 M sucrose (11). Centrifugation was performed at 50,000 × g for 30 min at 4°C in a Sorvall Centricon T-1170 with an A-641 rotor (Asheville, N.C.). Attack phase cells were harvested from an opaque band located 1.5 cm from the bottom of the tube. The fraction harvested from a band at the top of the tube constituted the ghost envelope fraction. Percoll was removed from the fractions by dilution with 5 volumes of 14.5 mM NaCl and centrifugation at 20,000 x g for 10 min at 4°C.
Protein extraction.
Cells were frozen in liquid nitrogen, and the total protein fraction was extracted in a buffer containing 0.1 M Tris-HCl, 5% (wt/vol) sucrose, 2% sodium dodecyl sulfate, 5% β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride, pH 8, on ice. An equal volume of saturated phenol was added, and the mixture was shaken for 10 min and centrifuged. The phenolic fraction was again mixed with extraction buffer and separated as above. Proteins were precipitated by the addition of 5 volumes of 0.1 M ammonium acetate in methanol. The protein pellet was washed three times with the same mixture and finally washed with 80% acetone. The pellet was dissolved in rehydration solution (8 M urea, 2% cholamidopropyl-dimethylammonio-propanesulfonate [CHAPS], 0.5% isopropylthiogalactopyranoside, 0.3% dithiothreitol). Protein concentration was determined according to Lowry (14).
Two-dimensional gel electrophoresis.
We loaded 500 μg of protein in 250 μl of rehydration buffer onto a 13-cm nonlinear pH gradient (3 to 10) strip and separated it for 6 h at 30 V, 6 h at 60 V, 1 h at 500 V, 1 h at 2,000 V, and 4 h at 8,000 V in an Ettan IPGphor isoelectric focusing system (Amersham Bioscience, Uppsala, Sweden). The strip was then equilibrated in buffer (1.5 M Tris-HCl, 6 M urea, 30% [vol/vol] glycerol, 2% sodium dodecyl sulfate, 1% dithiothreitol, bromophenol blue, pH 8). Electrophoresis in the second dimension was performed in an 8 to 15% polyacrylamide gradient gel at 35 mA per gel in a Protean II XI cell (Bio-Rad). Gels were stained with Coomassie brilliant blue G-250 (29). Images of the gels were acquired with an AlphaImager System (Alpha Innotech Corporation) and stored as TIFF files. Total, attack phase, and ghost envelope fractions were obtained from at least three independent cultures. Proteins were extracted and separated in independent trials.
Peptide analysis.
Reduction, alkylation, and tryptic digestion steps were carried out in gel as described by Rosenfeld et al. (23). After extraction of the peptides from the gel with 60% CH3CN and 1% CHOOH and evaporation to dryness, the peptide mixtures were solid phase extracted with a C18 resin-filled tip (ZipTip Millipore, Billerica, Mass.) and nanosprayed in a 50% CH3CN and 1% CHOOH solution into a quadrupole orthogonal acceleration time of flight (QTOF) system (Qtof2, Micromass) (37). Mass spectrometry data analysis was performed with the Biolynx package (Micromass), and database searches were performed with the Mascot package (Matrix Science). Similarity searches of sequences, determined via manual analysis, were carried out with the Wisconsin package (version 10.3; Accelrys Inc., San Diego, Calif.), and BLAST searches at the NCBI server (http://www.ncbi.nlm.nih.gov).
Electron microscopy.
A drop of cell suspension was placed on a copper grid, and excess liquid was blotted. The sample was counterstained with a 1% (wt/vol) solution of uranyl acetate for 30 s and examined in a JEOL100-CX transmission electron microscope.
Confocal microscopy.
We used 5 μM Syto13 and 10 μM Fm4-40 (Molecular Probes, Eugene, Oreg.) as nucleic acid and membrane stains, respectively. After staining, the cell suspension was centrifuged, and 10 μl was placed on a gelatin-coated Teflon slide, air-dried, mounted in Citifluor (Citifluor Ltd.), and examined under oil immersion by confocal microscopy (Sp2, Leica) with 488-nm and 514-nm laser light rays for illumination at 100x magnification. A composite picture of both channels was obtained with the platform software.
PCR and sequence analysis.
Amplification of a gene sequence coding for a putative OMP in strain 109J-1was performed with 1 μM each of primers P14s- (5′-ATGAAAAAAMTTTTRGTA-3′ [M = A/C, R = A/G]) and P14e (5′-GAAAGTGTAAGTGTAAGC-3′) in 3 mM MgCl2, 20 μM each of the four deoxyribo-nucleoside triphosphates, 1x reaction buffer (Promega, Madison, Wis.), and 1.25 U of Taq polymerase (Promega) in a total volume of 50 μl. PCR was performed in a Mastercycler Gradient (Eppendorff, Hamburg, Germany) with a denaturation step of 4 min at 94°C, annealing at 50°C for 1 min, and elongation at 72°C for 1 min, followed by 34 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and a final elongation step of 72°C for 5 min. The products were separated in a 1% agarose gel in TAE and purified with the High Pure PCR purification kit (Roche Molecular Biochemicals, Mannheim, Germany). Sequencing reactions were performed with the Big Dye Terminator kit (Perkin-Elmer Inc., Branchburg, N.J.) at the Center for Genomic Technologies at Hebrew University (Jerusalem, Israel). Internal primers were designed and used when needed. Alignments were performed with ClustalW (www.ebi.ac.uk/clustalw) and processed with GenDoc (www.psc.edu/biomed/genedoc). Sequence similarities were obtained with BLAST 2 sequences of the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession number.
The gene sequence coding for the OMP-like protein determined in the present study is available at GenBank under accession no. AY669509 (B. bacteriovorus 109J-1).
RESULTS
Comparative protein analysis of bdellovibrio cultures grown on different hosts.
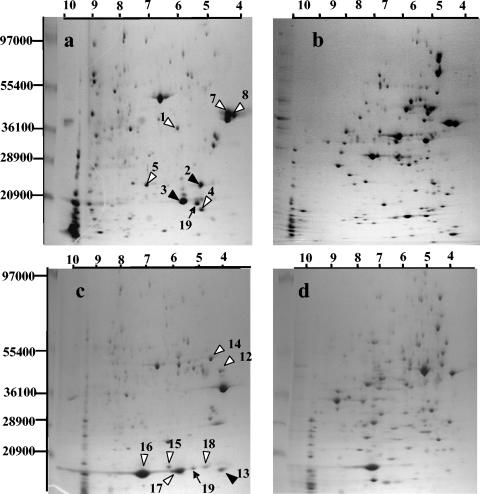
Five abundant proteins from the total fraction that migrated identically to proteins observed in E. coli prey gels were detected (spots 1, 4, 5, 7, and 8, Fig. 2a and b). They were extracted from the gels and analyzed by QTOF, revealing that they were all derived from the prey and corresponded to the major OMPs OmpA, OmpX, YciD, OmpF, and OmpC (numbers 1, 4, 5, 7, and 8, respectively, Table 1). All were unambiguously identified as E. coli-derived polypeptides: two to nine tryptic peptide sequences per spot exactly matched those of E. coli OMPs (Table 1). Two other spots (spots 2 and 3, Fig. 2a) were detected in the total fraction by comparing this fraction with the E. coli prey fraction and with the total fraction from a Bdellovibrio predator grown with P. syringae as the prey. They were identified as E. coli OmpA-derived polypeptides but were of lower molecular weights and different isoelectric points. They probably correspond to cleaved products of the original protein. The identified tryptic fragments from both spots were all situated within the 19-kDa pronase-resistant N-terminal part of OmpA (30).
FIG. 2.
Two-dimensional gel electrophoresis of proteins from (a) a washed, fresh bdellovibrio culture of a two membered culture of Escherichia coli and Bdellovibrio bacteriovorus 109J-1, (b) Escherichia coli, (c) a washed, fresh bdellovibrio culture of a two-membered culture of Pseudomonas syringae pv. tomato and B. bacteriovorus 109J-1, and (d) Pseudomonas syringae pv. tomato. White arrowheads, prey proteins; black arrowheads, degradation products of prey proteins; black arrows, Bdellovibrio proteins. Numbers refer to the entries in Table 1.
TABLE 1.
Identification of proteins after two-dimensional gel electrophoresis of isolated spots from washed fresh bdellovibrio cultures of Escherichia coli and Pseudomonas syringae pv. tomato, attack-phase cells of Bdellovibrio bacteriovorus, and Escherichia coli ghost envelope fraction separated by centrifugation on a Percoll-sucrose cushiona
| Spot no./fractionb | Closest match, protein/organism (accession no.) | E value | No. of peptides identified (no. of identical residues/total) | pI | Mass (Da) |
|---|---|---|---|---|---|
| 1/TF | OmpA/E. coli (gi|15800816) | 3e-93 | 6 (214/346) | 5.99 | 37178 |
| 2/TF | OmpA/E. coli | 1e-37 | 2 (72/346) | 5.2 | 22150c |
| 3/TF | OmpA/E. coli | 2e-31 | 3 (79/346) | 5.8 | 20150c |
| 4/TF | OmpX/E. coli (gi|13360351) | 3e-10 | 4 (95/171) | 5.04 | 16350 |
| 5/TF | Probable outer membrane protein YciD/E. coli (gi|25355362) | 3e-34 | 7 (82/212) | 6.50 | 22908 |
| 6/TF, GEF, BF | Omp-like protein /B. bacteriovorus 109J-1 | 3e-25 | 4 (50/353) | 4.79 | 36851 |
| 7/TF | OmpF/E. coli (gi|15800790) | 1e-34 | 9 (153/362) | 4.64 | 37062 |
| 8/TF | OmpC/E. coli (gi|16130152) | 1e-104 | 9 (253/367) | 4.58 | 40343 |
| 9/TF | EF-Tu/B. bacteriovorus 100 (gi|39576599) | 7e-19 | 6 (93/396) | 5.89 | 43265 |
| 10/TF | Flagellin/B. bacteriovorus 100 (gi|39576653) | 1e-21 | 2 (37/277) | 5.09 | 29212 |
| 11/TF, BF | Alkyl hydroperoxide reductase, C22 subunit/B. bacteriovorus 100 (gi|39576151) | 2e-38 | 11 (91/188) | 6.30 | 21049 |
| 12/TF | OprF-like putative outer membrane protein/P. fluorescens-PfO-1 (gi|23057601) | 1e-26 | 5 (108/344) | 4.83 | 36662 |
| 13/TF | OprF-like putative outer membrane protein/P. fluorescens PfO-1 | 3e-28 | 3 (81/344) | 4.1 | 19150c |
| 14/TF | OmpD-like putative outer membrane protein/P. fluorescens PfO-1 (gi|23058499|) | 7e-30 | 6 (57/448) | 5.13 | 48241 |
| 15/TF | Outer membrane protein W/P. fluorescens PfO-1 (gi|23059868) | 5e-20 | 5 (57/232) | 6.18 | 24738 |
| 16/TF | OmpH1-like putative outer membrane protein/P. fluorescens PfO-1 (gi|48730642) | 1e-50 | 3 (46) | 7.88 | 18100 |
| 17/TF | OmpH1-like putative outer membrane protein/P. fluorescens PfO-1 | 7e-08 | 6 (133/346) | 5.8 | 18100c |
| 18/TF | OmpH1-like putative outer membrane protein/P. fluorescens PfO-1 | 1e-50 | 6 (132/346) | 4.8 | 18160c |
| 19/TF | Hypothetical protein/B. bacteriovorus 100 (gi|39576348) | 1e-21 | 5 (77/384) | 5.72 | 41382 |
| 20/BF | Putative RNA-binding protein/B. bacteriovorus 100 (gi|42522915) | 3e-36 | 9 (64/116) | 8.04 | 11963 |
Values for isoelectric point and mass were deduced from protein sequences obtained from NCBI or from this study unless marked otherwise
TF, total fraction; GEF, ghost envelope fraction; BF, attack-phase cell fraction.
Observed in-gel.
The same procedure was applied with P. syringae pv. tomato as the prey. A comparison of protein distribution between total fraction gels obtained with P. syringae and E. coli as the prey enabled the detection of P. syringae-derived proteins, such as OmpD and OmpW (Fig. 2c and d, Table 1). Spot number 12 was isolated and identified as a Pseudomonas fluorescens OprF-like protein (35). A second spot (number 13) of lower molecular weight, which was not present in gels of the P. syringae prey, was also identified as OprF and most probably represents a cleavage product (Table 1). The tryptic peptides obtained from this spot were situated within the circa 19-kDa pronase-resistant fragment of the related mature polypeptide in Pseudomonas aeruginosa (28). Both this P. syringae OprF and E. coli OmpA have been reported as peptidoglycan-associated OMPs (28, 38).
Spot number 16, which was also detected in the total fraction gels of P. syringae with the same procedure, was an OprH1-like protein. Two additional, more acidic isoforms of this polypeptide were detected in the total fraction (spots 17 and 18) but not in the prey alone (Fig. 2c and d).
Separation of prey ghost cells from predatory attack cells.
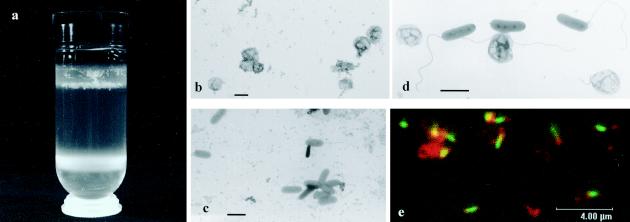
To clarify the origin of the prey proteins within the total fraction, which is a washed suspension of a two-membered culture, fresh bdellovibrio cultures were centrifuged on a Percoll-sucrose cushion, yielding two bands (Fig. 3a; also see Fig. 1). The upper ring was composed of separated ghost envelopes, virtually clear of bdellovibrio cells (Fig. 3b), and of floating fluffy, sticky white material. This material was easily withdrawn from the ring by gentle suction. It was composed mainly of bdelloplasts (infected prey cells), some predator attack cells, and some ghost envelopes. Two-dimensional gel analysis of this fluffy material yielded a picture similar to the one obtained with the total fraction (not shown). The attack bdellovibrio cells formed a concentrated layer at the bottom of the centrifugation tube (the attack-phase fraction) and did not contain prey ghosts, as assessed by microscopic observation (Fig. 3c). In comparison, electron microscopy and dual staining with a nucleic acid stain and a lipid stain followed by confocal microscopy clearly showed that in the washed bdellovibrio cultures (total fraction) that had not been centrifuged on a Percoll-sucrose cushion, ghost envelopes were present along with attack bdellovibrio cells (Fig. 3d and 3e). Centrifugation of bdellovibrio cultures on a Ficoll gradient, as done previously (6), did not yield satisfactory separation of the fractions (not shown).
FIG. 3.
(a) Separation of ghost envelopes (GEF, upper ring) from Bdellovibrio bacteriovorus attack cells (BF, lower ring) on a Percoll cushion. Transmission electron microscopy of (b) ghost envelope fraction; (c) attack-phase fraction, and (d) a washed, fresh bdellovibrio culture. (e) Confocal microscopy of a washed fresh bdellovibrio culture stained with a lipid stain (red) and a nucleic acid stain (green). Bars: 1 μm (b, c, d); 4 μm (e).
Two-dimensional gel electrophoresis analysis of the attack-phase fraction and of the ghost envelope fraction.
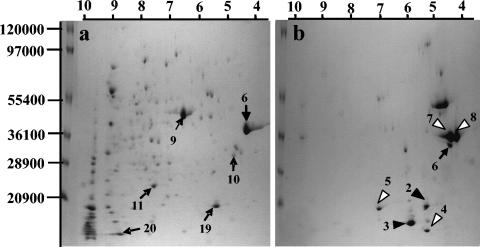
Two-dimensional gels of the bdellovibrio attack phase cell fraction containing purified predatory cells and no ghost envelopes revealed that prey-derived proteins were not detected in this fraction (Fig. 4a), supporting the claim that prey outer membrane proteins are not imported into the predator (2). Furthermore, two-dimensional gels of the ghost envelope fraction contained proteins which migrated like the prey OMPs identified in the total fraction (Fig. 4b). A few other spots which also appeared in the total fraction were apparent in the ghost envelope fraction and were putatively identified as LamB and FadL by comparison with protein gels from E. coli alone and with other published gels of E. coli proteins (17).
FIG. 4.
Two-dimensional gel electrophoresis of proteins from (a) Percoll-separated Bdellovibrio bacteriovorus attack cells and (b) Escherichia coli Percoll-separated ghost envelope fraction. White arrowheads, E. coli proteins; black arrowheads, cleavage products of E. coli proteins; black arrows, B. bacteriovorus proteins. Numbers refer to the entries in Table 1.
One spot which was not present in extracts of prey suspensions was detected in gels originating from ghost envelopes (spot number 6, Fig. 4b). This spot migrated similarly to a protein found in separated attack prey cells (Fig. 4a), suggesting that it originated from the bdellovibrio predator. Extraction and mass spectrometric analysis of this polypeptide revealed that it was derived from the predator (Table 1) and was related to a recently described OMP from B. bacteriovorus type strain 100 (2).
Polypeptides from B. bacteriovorus such as elongation factor Tu, a putative RNA-binding protein, a nondefined protein, a hydroxyperoxidase, and flagellin (Table 1) were isolated and identified in the attack-phase fraction from total protein extracts. None of these proteins, which focused in the basic or acidic parts of the gels, were of lower or higher molecular weights, formed dominant or faint spots, and may have originated from different subcellular fractions, could be detected in the ghost envelope fraction. It is therefore unlikely that the predator-derived OMP-like protein identified in the ghost envelope fraction stemmed from contamination by few remaining entrapped bdellovibrio attack cells within the ghost fraction.
PCR and sequence analysis.
The gene coding for the OMP-like protein identified in ghost envelopes was amplified from B. bacteriovorus cells with primers based on the published sequences from type strain 100 and strain 114 (2). The genes of strains 109J-1 and 100 and of strains 109J-1 and 114 exhibited 86 and 69% identity, respectively, which translated into 82.4% identity (291 out 353 residues) and 86% similarity (307 out 353 residues) and 55% identity (210 out of 376 residues) and 66% similarity (253 out 376 residues), respectively, at the amino acid level.
DISCUSSION
The direct utilization of various “building blocks” has been described as a feature of bdellovibrios and has been used as an explanation for their extreme energy efficiency (22). Previous data have been presented to substantiate the claim that Bdellovibrio predators are able to import prey outer membrane proteins (OMPs) and insert them into their own envelope in a way that retains their functionality (6, 7, 8, 32). In these previous studies, it was also shown that, following host shift, the OMPs appearing in the predator matched those of the new prey.
The great resolution of proteomics extends our view of the processes associated with the growth of bdellovibrio predators at the expense of their prey. Moreover, centrifugation on a Percoll cushion enabled the separation of ghost envelopes and bdellovibrio attack-phase cells. Prey OMPs were identified in fresh bdellovibrio cultures which had been centrifuged and washed a number of times (the total fraction) as well as in the Percoll-sucrose-separated ghost envelope fraction. However, their presence, even as minor components of the Percoll-separated fraction of bdellovibrio attack-phase cells, was not detected, strongly suggesting that prey OMPs are not relocated to the predator, as also shown by Beck et al. in a recent report (2).
Some workers argued that the ability to acquire prey OMPs was greatly dependent upon the prey itself and that the history of the bdellovibrio strain employed also played a role in this ability (32). Rayner et al. (19) were the first to claim that OMPs were not transferred from prey to predator but that B. bacteriovorus synthesized an OmpF-like protein whose apparent molecular weight was very close to that of E. coli OmpF. Also, according to Tudor and Karp (34), an OMP was found inserted into the prey's cytoplasmic membrane. However, the resolution of proteins was not sufficient to determine whether this molecule was transferred OmpF from the E. coli prey's outer membrane or if it was an OmpF-like protein derived from invading predatory cells.
We detected a polypeptide of mass and isoelectric point close to those of OmpF in bdellovibrio attack-phase cells. It was also the only polypeptide of bdellovibrio origin found in protein extracts of Percoll-separated ghost envelope fractions, suggesting that this protein and those reported by Tudor and Karp (34) and by Rayner et al. (19) are the same. As other proteins of the predator, including the strongly expressed cytoplasm-located elongation factor Tu (17), flagellin, and other molecules focusing in different parts of the gels were not detected, we are confident in ruling out contamination by a low level of remaining bdellovibrio attack-phase cells in the ghost envelope fraction. The polypeptide identified in this study is highly homologous to a new, recently described OMP from B. bacteriovorus type strain 100 (2).
Based on 16S rRNA gene analysis, strains 100 and 109J-1 are phylogenetically very close, but strain 114 diverges by about 6.5% from these two strains (5). A comparative analysis of about 105 base pairs from strain 109J-1 with the genome of strain 100 reveals greater than 98% homology at the nucleotide level (not shown). However, although the proteins belong to the same family, the deduced proteins homologous to the new OMP from strain 100 exhibited large differences in strains 109J-1 and 114: the protein sequences of strain 114 versus 109J-1, of 100 versus 109J-1, and of 100 versus 114 were 55, 82.4, and 54% identical, respectively. This suggests high strain specificity for the unknown function filled by this molecule. An extensive search for motifs did not reveal definite functions, and no paralogous genes could be detected in the genome of type strain 100. Clearly, more research is necessary to assign a definite function to these proteins.
In a recent publication (2), the sequence coding for the OMP-like protein in type strain 100 was shown to differ widely from that of a host-independent (HI) strain derived from strain 100 and obtained from a culture collection. However, the OMP-like protein in strain 109J-1 is identical to the one found in the host-independent variant of strain 100 (not shown), suggesting that this mutant is not derived from strain 100 but possibly from strain 109J.
Cover et al. (4) and Beck et al. (2) pointed to a putative degradation product of host OmpA in infected prey cells. Our analysis shows that OmpA is cleaved into at least two peptides that remain in the ghost envelope fraction (Fig. 3b). In a similar manner, an OprF-like protein from P. syringae pv. tomato, an alternative prey of strain 109J-1, was also found to be cleaved as a result of predation. OprF and OmpA are abundant porins (3, 10, 36) in fluorescent pseudomonads and E. coli, respectively, and are involved in maintenance of the cell shape and in transport of nonspecific hydrophilic monomers by passive diffusion (28, 36, 38). The C-terminal domain in both polypeptides is sensitive to pronase digestion and is associated with the peptidoglycan (28). Only fragments from the N-terminal, pronase-resistant part of the proteins (28, 30) were detected in the identified cleavage products of OmpA and OprF after predation of E. coli and P. syringae cells, respectively, by bdellovibrio predators. This suggests that the C-terminal part of these proteins is degraded, loosening the association of the peptidoglycan with the outer membrane layer. Although peptidase activity is detected from the penetration stage onwards during the intraperiplasmic stage (33), OmpA cleavage occurs during the intraperiplasmic growth phase to progeny release (4). Whether the cleavage of peptidoglycan-associated OMPs is essential for progeny release remains to be shown.
OprH1 is an Mg2+- and PhoP-PhoQ-regulated polypeptide in P. aeruginosa (15). Acidic isoforms of an OprH1-like protein were apparent in bdellovibrio cultures of P. syringae pv. tomato and were not detected in suspensions of the prey alone. Such isoforms were also detected in P. aeruginosa grown at low Mg2+ concentrations (9). Whether the expression of isoforms in the prey results from an increased demand for the cation during development of the bdellovibrio cultures or from modifications affecting the protein during infection also remains to be shown.
In conclusion, we identified a number of prey proteins that are selectively and partially degraded or modified during the predator-prey interaction. While these changes may be indispensable for the successful life cycle of the bdellovibrio cell, total degradation may be detrimental, as these polypeptides are essential for the stability of the prey cell envelope. We confirmed that prey OMPs are not transferred to the predator. Our results also suggest that a predator-derived OMP-like protein is associated with the ghost envelope.
Acknowledgments
We give thanks to Ariel Gaaton (Interdepartmental Equipment Unit of Hebrew University of Jerusalem) for excellent technical assistance in protein analysis, to Zach Adam for help in separation in the first dimension, to Philippe Glaser for “snapshot” sequencing of strain 109J-1, and to Christel Talbot for help with confocal microscopy.
This research was funded by the Israel Science Foundation (grants 486/03-16.2 and 132/99).
REFERENCES
- 1.Barel, G., and E. Jurkevitch. 2001. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch. Microbiol. 176:211-216. [DOI] [PubMed] [Google Scholar]
- 2.Beck, S., D. Schwudke, E. Strauch, B. Apple, and M. Linscheid. 2004. Bdellovibrio bacteriovorus strains produce major outer membrane protein during predacious growth in the periplasm of prey bacteria. J. Bacteriol. 186:2766-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond, P. J., and M. S. P. Sansom. 2003. Membrane protein dynamics versus environment: simulations of OmpA in a micelle and in a bilayer. J. Mol. Biol. 329:1035-1056. [DOI] [PubMed] [Google Scholar]
- 4.Cover, W., R. J. Martinez, and S. C. Rittenberg. 1984. Permeability of the boundary layers of Bdellovibrio bacteriovorus 109J and its bdelloplast to small hydrophilic molecules. J. Bacteriol. 157:385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidov, Y., and E. Jurkevitch. 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1439-1452. [DOI] [PubMed] [Google Scholar]
- 6.Diedrich, D. L., C. A. Portnoy, and S. F Conti. 1983. Bdellovibrio possesses a prey-derived OmpF protein in its outer membrane. Curr. Microbiol. 8:51-56. [Google Scholar]
- 7.Diedrich, D. L., C. P. Duran, and S. F. Conti. 1984. Acquisition of Escherichia coli outer membrane proteins by Bdellovibrio sp. strain 109J. J. Bacteriol. 159:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrini, F., V. Romano, M. Valenzi, and M. Di Giulio. 1982. Molecular parasitism in the Escherichia coli-Bdellovibrio bacteriovorus system: translocation of the matrix protein from the host to the parasite outer membrane. EMBO J. 1:1439-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guina, T., M. Wu, S. I. Miller, S. O. Purvine, E. C. Yi, J. Eng, D. R. Goodlett, R. Aebersold, R. K. Ernst, and K. A. Lee. 2003. Proteomic analysis of Pseudomonas aeruginosa grown under magnesium limitation. J. Am. Soc. Mass Spectrom. 14:742-775. [DOI] [PubMed] [Google Scholar]
- 10.Hancock, R. E. W., G. M. Decad, and H. Nikaido. 1979. Identification of the protein production transmembrane diffusion pores in the outer membrane of P. aeruginosa PAO1. Biochim. Biophys. Acta 554:323-329. [DOI] [PubMed] [Google Scholar]
- 11.Higashi, S., S. Katahira, and M. Abe. 1984. Isolation of pea nudole bacteroids utilizing gradient centrifugation. Plant Soil 81:93-99. [Google Scholar]
- 12.Jurkevitch, E. 2000. The genus Bdellovibrio. In M. Dworkin, S. Flakow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. Springer-Verlag, New York, N.Y. [Online.] http://141.150.157.117:8080/prokPUB/index.htm.
- 13.Kuenen, J. G., and S. C. Rittenberg. 1975. Incorporation of long-chain fatty acids of the substrate organism by Bdellovibrio bacteriovorus during intraperiplasmic growth. J. Bacteriol. 121:1145-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry, O. H., N. J. Rosbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 15.Macfarlane, E. L. A., A. Kwasnicka, and R. E. W. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 16.Matin, A., and S. C. Rittenberg. 1972. Kinetics of the deoxyribonucleic acid destruction and synthesis during growth of Bdellovibrio bacteriovorus strain 109D on Pseudomonas putida and Escherichia coli. J. Bacteriol. 111:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molloy, M. P., B. R. Herbert, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:267-2881. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, D. R., and S. C. Rittenberg. 1981. Incorporation of substrate cell lipid A components into the lipopolysaccharide of intraperiplasmically grown Bdellovibrio bacteriovorus. J. Bacteriol. 147:860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner, J. R., W. H. Cover, R. J. Martinez, and S. C. Rittenberg. 1985. Bdellovibrio bacteriovorus synthesizes an OmpF-like outer membrane protein during both axenic and intraperiplasmic growth. J. Bacteriol. 163:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rendulic, S., P. Jagtap, A. Rosinus, M. Eppinger, C. Baar, C. Lanz, H. Keller, C. Lambert, K. J. Evans, A. Goesmann, F. Meyer, R. E. Sockett, and S. C. Schuster. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689-692. [DOI] [PubMed] [Google Scholar]
- 21.Rittenberg, S. C., and D. Langley. 1975. Utilization of nucleotide monophosphates per se for intraperiplasmic growth of Bdellovibrio bacteriovorus. J. Bacteriol. 121:1137-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rittenberg, S. C., and R. B. Hespell. 1975. Energy efficiency of intraperiplasmic growth of Bdellovibrio bacteriovorus. J. Bacteriol. 121:1158-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenfeld, J., J. Capdevieille, J. C. Guillemot, and P. Ferrera. 1992. In-gel digestion of proteins for internal sequence analysis after one-or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 24.Rosson, R. A., and S. C. Rittenberg. 1979. Regulated breakdown of Escherichia coli deoxyribonucleic acid during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J. Bacteriol. 140:620-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosson, R. A., and S. C. Rittenberg. 1981. Pyrimidine metabolism of Bdellovibrio bacteriovorus growth intraperiplasmically and axenically. J. Bacteriol. 146:108-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby, E. G., and J. B. McCabe. 1986. An ATP transport system in the intracellular bacterium Bdellovibrio bacteriovorus 109J. J. Bacteriol. 167:1066-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruby, E. G., J. B. McCabe, and J. I. Barke. 1985. Uptake of intake nucleoside monophosphates by Bdellovibrio bacteriovorus 109J. J. Bacteriol. 163:1087-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saint, N., C. E. Hamel, E. De, and G. Molle. 2000. Ion channel formation by N-terminal domain: a common feature of Oprfs of Pseudomonas and OmpA of Escherichia coli. FEMS. Microbiol. Lett. 190:261-265. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y
- 30.Schweizer, M., I. Hindennach, W. Garten, and U. Henning. 1978. Major proteins of the Escherichia coli outer cell envelope membrane. Interactions of protein II with lipopolysaccharide. Eur. J. Biochem. 82:211-217. [DOI] [PubMed] [Google Scholar]
- 31.Schwudke, D., M. Linscheid, E. Strauch, B. Appel, U. Zähringer, H. Moll, M. Müller, L. Brecker, S. Gronow, and B. Lindner. 2003. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid a containing α-d-mannose that replace phosphate residues. J. Biol. Chem. 278:27502-27512. [DOI] [PubMed] [Google Scholar]
- 32.Talley, B. G., R. L. Mcdada, and D. L. Diedrich. 1987. Verification of the protein outer membrane of Bdellovibrio bacteriovorus as the OmpF protein of its Escherichia coli prey. J. Bacteriol. 169:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomashow, M. F., and S. C. Rittenberg. 1978. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: Solubilization of Escherichia coli peptidoglycan. J. Bacteriol. 135:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tudor, J. J., and M. A. Karp. 1994. Translocation of an outer membrane protein into prey cytoplasmic membranes by Bdellovibrio. J. Bacteriol. 176:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullstrom, C. A., R. Siehnel, W. Woodruff, S. Steinbach, and R. E. Hancock. 1991. Conservation of the gene for outer membrane protein OprF in the family Pseudomonadaceae: sequence of the Pseudomonas syringae oprF gene. J. Bacteriol. 173:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396-401. [DOI] [PubMed] [Google Scholar]
- 37.Wilm, M., and M. Mann. 1996. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68:1-8. [DOI] [PubMed] [Google Scholar]
- 38.Woodruff, W. A., and R. E. W. Hancock. 1989. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J. Bacteriol. 171:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]