Abstract
Polymeric β-1,6-N-acetyl-d-glucosamine (poly-β-1,6-GlcNAc) has been implicated as an Escherichia coli and Staphylococcus epidermidis biofilm adhesin, the formation of which requires the pgaABCD and icaABCD loci, respectively. Enzymatic hydrolysis of poly-β-1,6-GlcNAc, demonstrated for the first time by chromatography and mass spectrometry, disrupts biofilm formation by these species and by Yersinia pestis and Pseudomonas fluorescens, which possess pgaABCD homologues.
Sessile bacterial growth on a surface or interface is generally referred to as a biofilm. Intense interest in biofilms over the past several years stems largely from the recognition of their roles in protecting microbes from the immune system, predation, and stresses; their association with failed antimicrobial therapies; and their potential to facilitate genetic exchange (2, 3, 6, 7, 9, 12, 24, 31).
Adherence of cells to surfaces and to each other is critical for biofilm formation. A variety of surface organelles have been reported to assist in adherence (4, 20, 29, 32, 36). While capsular polysaccharides affect biofilm architecture, mounting evidence indicates that many of these, including colanic acid of Escherichia coli, Vi antigen of Salmonella enterica, and alginate of Pseudomonas aeruginosa, do not play necessary roles in biofilm formation (5, 13, 30, 38). In contrast, genetic studies and polysaccharide analyses indicate that a cell-bound polysaccharide of E. coli and staphylococci, polymeric β-1,6-N-acetyl-d-glucosamine (poly-β-1,6-GlcNAc), is required for biofilm formation (11, 37). E. coli biofilm is dispersed by meta-periodate, but not by protease, consistent with a role of polysaccharide(s) in maintaining its structural stability but short of demonstrating that poly-β-1,6-GlcNAc performs this role (37).
The four genes of the pgaABCD operon of E. coli are required for biofilm formation and are predicted to encode proteins required for the synthesis and translocation of poly-β-1,6-GlcNAc, also referred to as PGA, through the gram-negative cell envelope (37). Production of staphylococcal β-1,6-GlcNAc polysaccharides, alternatively known as PIA, PS/A, SAE, and PNAG (14, 17, 22, 23, 25, 26), depends upon the icaABCD locus. Two gene products of the pga and ica operons are apparently homologous (37). The UDP-GlcNAc-specific glycosyltransferase, IcaA, polymerizes β-1,6-GlcNAc, and its homologue PgaC is predicted to do the same. IcaB and PgaB are related to polysaccharide N-deacetylases but have not been shown to possess this enzymatic activity. Evidence that genetic loci related to pgaABCD have undergone broad horizontal transfer suggests that poly-β-1,6-GlcNAc may participate in adhesion processes of diverse species (37). These include mammalian and plant pathogens, e.g., Yersinia pestis, Yersinia pseudotuberculosis, Yersinia enterocolitica, Actinobacillus actinomycetemcomitans, Actinobacillus pleuropneumoniae, Bordetella pertussis, Bordetella parapertussis, Bordetella bronchiseptica, Xanthomonas axonopodis, the biocontrol agent Pseudomonas fluorescens, and others. The hmsHFRS operon of Y. pestis is homologous to pgaABCD and has been previously implicated in biofilm formation (6). Species closely related to these do not invariably contain homologous loci, e.g., S. enterica and P. aeruginosa (37).
In the present study, a previously identified β-hexosaminidase and biofilm-dispersing enzyme of A. actinomycetemcomitans, DspB or dispersin B (18, 19), is shown for the first time to specifically hydrolyze the glycosidic linkages of poly-β-1,6-GlcNAc. Growth of cultures in the presence of DspB or treatment of preformed biofilms with this enzyme implicate poly-β-1,6-GlcNAc in the formation and stabilization of diverse bacterial biofilms.
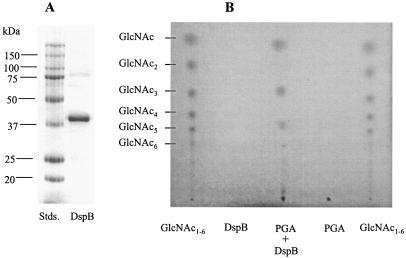
Recombinant DspB containing an N-terminal His tag was produced using a strain containing a dspB plasmid clone [pHis-DspB1]. This clone was prepared by first amplifying the coding region of the dspB gene of A. actinomycetemcomitans HK1651 by PCR using the primers 5′-GGGAATTCCATGCATCATCATCATCATCATAATTGTTGCGTAAAAGGCAATTCCAT (the EcoRI site is underlined) and 5′-TTCTGCAGCTCACTCATCCCCATTCGTCTTATG (the PstI site is underlined). Chromosomal DNA for the PCR was a gift from David Dyer and Allison Gillaspy (University of Oklahoma Health Sciences Center) and was isolated using a PUREGENE DNA isolation kit (Gentra, Minneapolis, Minn.) under the conditions recommended for gram-negative organisms by the manufacturer, with the addition of a phenol-chloroform extraction step prior to ethanol precipitation. The resulting PCR product was cloned into the HincII site of pUC19, sequenced (AnaGen Technologies, Inc., Atlanta, Ga.), and found to be free of PCR-generated mutations. The dspB-containing EcoRI-PstI fragment was excised and subcloned into the corresponding restriction sites of pKK223-3 (1) to produce pHis-DspB1. Strain JM101[pHis-DspB1]) was inoculated (20 ml/liter) and grown with aeration at 37°C in Luria-Bertani (LB) medium (27) containing ampicillin (100 μg/ml). After 4 h of growth, isopropyl-β-d-thiogalactopyranoside (2 mM final concentration) was added to induce dspB expression, and growth was continued for 3 h. The cells were harvested by centrifugation and disrupted by sonication, and DspB protein was purified by affinity chromatography using a His-Trap column (QIAexpressionist), as recommended by the manufacturer (QIAGEN, Valencia, Calif.), and dialyzed against 10 mM sodium phosphate (pH 5.9). This single chromatographic step provided an almost homogeneous preparation of DspB (≥95% purity), as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie blue staining (Fig. 1A) and densitometry. This enzyme was active against 4-nitrophenyl-N-acetyl-β-d-glucosaminide (18). The yield of DspB was ∼6 mg/liter of culture, as determined with a BCA protein assay kit (Pierce, Rockford, Ill.).
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of DspB and TLC separation of products generated by treatment of PGA with DspB. (A) Protein molecular weight standards (Stds.) and 10 μg of recombinant DspB were separated on a 12% polyacrylamide gel and stained with Coomassie blue R-250. (B) Reaction products generated by digestion of PGA for 16 h at 37°C are shown, along with untreated PGA and DspB controls. The GlcNAc standards are β-1,4-linked oligosaccharides derived from chitin.
To investigate the catalytic activity of DspB, a substrate of homopolymeric β-1,6-GlcNAc or PGA was prepared from E. coli K-12 DJ4[pPGA372], as described previously (37), except that the strain was grown in LB medium (27). The genotype of this strain (MG1655 csrA::kan cpsE::Tn10) and plasmid pPGA372 (pUC19 containing pgaABCD) allow PGA to be overproduced (37). Enzymatic treatment of PGA with DspB was carried out for 16 h at 37°C. Thin-layer chromatography (TLC) was used to detect low-molecular-weight oligosaccharides (GlcNAc1-6), as described previously (35). The reaction mixture (75 μl) contained 0.6 mg of PGA in 50 mM ammonium acetate buffer (pH 5.9) along with 7.5 μg of DspB. Apparent reaction products included GlcNAc and GlcNAc oligomers that remain unassigned (Fig. 1B). In contrast, identical enzymatic treatments of two β-1,4-GlcNAc polymers, water-soluble chitin or glycol chitin, produced no degradation products (data not shown). Analysis of the primary amine content of PGA using the ninhydrin reaction (21), with d-glucosamine as a standard, indicated that it contains 3.2% ± 0.2% unacetylated glucosamine, in agreement with the results of previous nuclear magnetic resonance analyses (37). Enzymatic treatment did not significantly increase this value, indicating that DspB does not possess N-deacetylase activity.
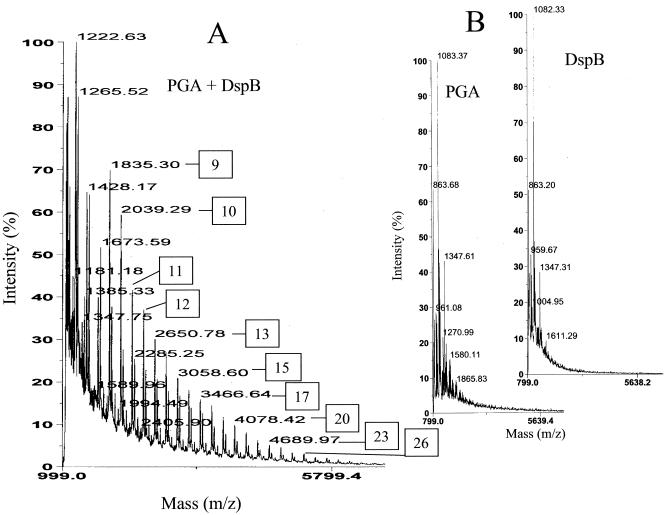
Matrix-assisted laser desorption-ionization time of flight (MALDI-TOF) mass spectrometry allowed detection of individual high-molecular-weight reaction products. Spectra were acquired in a linear mode by using a Voyager STR (Applied Biosystems) employing a nitrogen laser (wavelength, 337 nm). One hundred to 600 laser pulses were used to obtain the spectra. Detection employed delayed extraction and a low-mass gate, which deflected ions lower in mass than the scan range detected. The matrix was a saturated solution of 1,4-hydroxyphenylazobenzoic acid in 50% acetonitrile and 1% trifluoroacetic acid and was mixed in a 10:1 ratio with a sample. One microliter of the mixture was transferred to the sample plate and dried. A reaction identical to the one described above for TLC analysis yielded a series of oligosaccharides, each major product of which differed from the previous one by a mass of 204 Da, equivalent to a single anhydro GlcNAc residue (Fig. 2A). These products were confirmed to be reaction products by their absence in the control spectra of DspB and untreated PGA (Fig. 2B). The experimental masses of the reaction products were indicative of oligomers ranging from GlcNAc9 to >GlcNAc28. Minor products were also observed, many of which differed from the major ions by the mass of a single acetyl moiety. These minor products apparently represent the release of oligosaccharides with preexisting modifications. The low-molecular-weight products that were detected by TLC (Fig. 1) were masked by matrix ions of the mass spectrum. Spectra were acquired up to a mass of 25,000 Da, but no additional products were detected beyond the scale shown in Fig. 2. The PGA substrate for the reaction was not observed because its molecular weight is greater than the range of detection, ≥400,000 Da (37). Together, these findings imply that DspB endolytically hydrolyzes the glycosidic linkages of PGA, ultimately processing the products to GlcNAc and low-molecular-weight oligosaccharides.
FIG. 2.
MALDI-TOF mass spectra of PGA after digestion with DspB for 16 h at 37°C (A) and untreated PGA and DspB (B). The degree of polymerization of a major reaction product is indicated by a boxed number that refers to either the centroid mass (m/z) or ion peak of the spectrum.
Having established that DspB specifically cleaves poly-β-1,6-GlcNAc, we examined the ability of this enzyme to inhibit bacterial biofilm formation. We tested several species that have pgaABCD or icaABCD loci, namely, E. coli K-12 MG1655; its biofilm-forming csrA mutant, TRMG1655; a clinical strain of E. coli that was isolated from a colonized urinary catheter, P18 (16, 33, 37); S. epidermidis 1457 (23); a nonpathogenic Y. pestis strain lacking the 72-kb virulence plasmid, KIM6+, and KIM6, which additionally lacks the 102-kb Pgm locus containing hmsHFRS and the high pathogenicity island (8, 15); and P. fluorescens WCS365 (28). S. enterica serovar Typhimurium ATCC 14028 and P. aeruginosa PAO1 (provided by M. E. Hart) were also tested as examples of related species that lack pga loci (37). As indicated in Fig. 3, these bacteria were grown in microtiter wells containing LB medium (27), tryptic soy broth (TSB) medium (23), colony-forming antigen medium (16), M63 medium (27) supplemented with citric acid (0.4%) and MgSO4 (1 mM), or YPG medium (10). The antibiotics ampicillin (100 μg/ml) and kanamycin (100 μg/ml) were included in growth media as needed.
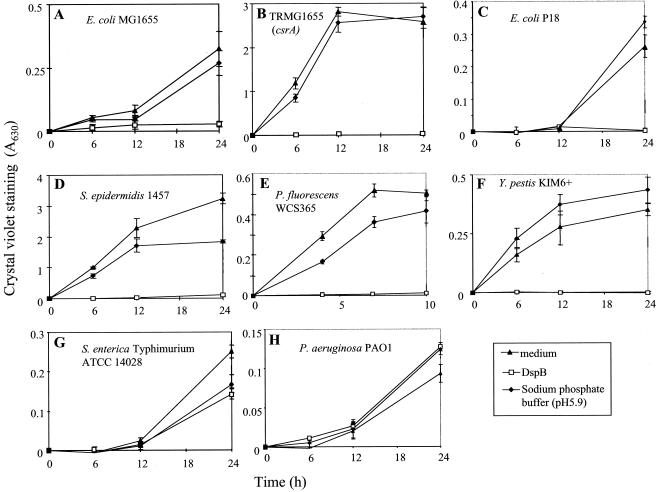
FIG. 3.
Effect of DspB on biofilm formation by the following strains with the listed growth conditions: E. coli MG1655, LB medium, 26°C (A); TRMG1655 (csrA::kan), LB medium, 26°C (B); E. coli P18, TSB, 26°C (C); S. epidermidis 1457, TSB, 26°C (D); P. fluorescens WCS365, M63 supplement, 26°C (E); Y. pestis KIM6+, TSB, 26°C (F); S. enterica serovar Typhimurium, colony-forming antigen medium, 26°C (G); and P. aeruginosa PAO1, YPG, 26°C (H). Biofilm formation in the indicated media, in presence or absence of recombinant DspB or an enzyme buffer control, was determined at the indicated times by crystal violet staining (A630). Error bars represent standard deviations (six replicates) of a representative experiment.
Biofilm formation was measured by crystal violet staining (16). Y. pestis biofilm was somewhat unstable and was examined using a modified procedure. Planktonic cells and spent media were discarded, and adherent cells were gently rinsed twice with deionized water, treated with methanol (100%) for 1 min to fix the cells, and allowed to air dry prior to being stained (http://www.engscientific.com/micro.html). All biofilm experiments were conducted at least twice, with at least six replicas per sample.
Growth in the presence of DspB (50 μg/ml) caused essentially complete inhibition of biofilm formation in species with pgaABCD or icaABCD loci: E. coli MG1655, its csrA mutant, the catheter isolate E. coli P18, S. epidermidis, P. fluorescens, and Y. pestis KIM6+ (Fig. 3). In contrast, DspB did not exhibit a discernible effect on biofilm formation by S. enterica or P. aeruginosa, which lack pga/ica loci. Growth curves in the presence or absence of DspB showed that this enzyme does not inhibit the planktonic growth of these six species (data not shown). Y. pestis KIM6, which lacks hmsHFRS, failed to form biofilm (data not shown).
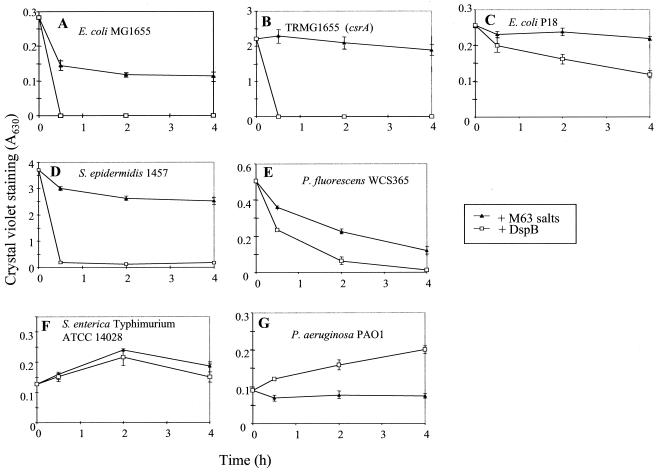
The ability of recombinant DspB to disperse biofilms of these species was examined after 24 h of growth or 10 h for P. fluorescens (28). The media and planktonic cells were removed, M63 minimal salts solution (16) was added with and without DspB (50 μg/ml), and biofilm was measured at various times thereafter. The addition of DspB to E. coli K-12 strains and S. epidermidis led to complete dispersal of the biofilm within 30 min (Fig. 4). DspB exhibited weaker dispersal effects on E. coli P18 and P. fluorescens biofilms, which nevertheless were reproducible. Biofilm of Y. pestis was not dispersed by DspB (data not shown), suggesting that, while growth in the presence of DspB prevents biofilm formation, poly-β-1,6-GlcNAc is inaccessible to the enzyme or other factors are involved in stabilizing the biofilm of Y. pestis. As expected from the results of the biofilm-blocking experiments, biofilms of S. enterica and P. aeruginosa were not dispersed by DspB treatment. The apparent modest stimulation of P. aeruginosa biofilm growth in the presence of DspB has not been further examined but may involve the proteolysis of DspB by secreted proteases (reference 34 and references therein) and uptake of the resulting amino acids and peptides, which might also have affected DspB activity in biofilm inhibition experiments (Fig. 3).
FIG. 4.
Treatment of biofilms of E. coli MG1655 (A), TRMG1655 (csrA::kan) (B), E. coli P18 (C), S. epidermidis 1457 (D), P. fluorescens WCS365 (E), S. enterica serovar Typhimurium (F), and P. aeruginosa PAO1 (G) with DspB or a buffer control (M63 salts). Biofilms were grown under the conditions indicated in the legend to Fig. 3. Error bars represent standard deviations (six replicates) of a representative experiment.
Conclusions.
Studies have recently shown that DspB or dispersin is able to release biofilms of two species, A. actinomycetemcomitans and S. epidermidis (18, 19). However, the biochemical reaction responsible for this activity had not been previously established. Analyses of E. coli PGA depolymerization now reveal that DspB specifically hydrolyzes the glycosidic linkages of poly-β-1,6-GlcNAc. It does not cleave the related β-1,4 linkages of some chitin derivatives or catalyze N deacetylation of PGA. In conjunction with the results of BLAST analyses for pgaABCD loci of other species (37), this information permitted a systematic assessment of the role of this type of polysaccharide in biofilm formation by several eubacteria. The results of this assessment demonstrate that poly-β-1,6-GlcNAc serves as a biofilm adhesin in phylogenetically diverse species, which exploit diverse hosts and environmental niches. Thus, depolymerization of poly-β-1,6-GlcNAc by DspB or perhaps other enzymes that cleave this polymer offers a possibility for inhibiting biofilm formation by several important pathogens which colonize host tissues and/or surfaces of catheters and prosthetic devices. While it is reasonable to expect that species that produce poly-β-1,6-GlcNAc may likewise produce enzymes that degrade this polysaccharide, BLAST analyses of the NCBI microbial-genome database (http://www.ncbi.nlm.nih.gov/BLAST/) revealed the presence of DspB homologues only in Actinobacillus species (data not shown). The possibility that distinct depolymerases have convergently evolved to facilitate biofilm dispersal by other species that utilize this polysaccharide adhesin should be investigated.
Acknowledgments
We thank D. Dyer and A. F. Gillaspy in the Department of Microbiology and Immunology, Health Sciences Center, University of Oklahoma, for providing chromosomal DNA of A. actinomycetemcomitans and T. Watanabe in the Department of Applied Biological Chemistry, Faculty of Agriculture, Niigata University, Niigata, Japan, for providing glycol chitin and water-soluble chitin. We thank D. Mack, M. E. Hart, and G. A. O'Toole for providing strains. We are also grateful to F. H. Strobel of the Emory University Mass Spectrometry Center for the use of the MALDI-TOF mass spectrometer, funded by NIH (1S10 RR 14645-01).
This study was funded in part by the National Institutes of Health (grant GM066794) and by CRIS Project MCS-03703 of the University of Florida Agriculture Experiment Station. Kane Biotech, Inc., may develop applications related to the findings herein. T. Romeo serves as chief scientific advisor for, owns equity in, and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Footnotes
This paper is Journal Series no. R-10438 of the University of Florida Agriculture Experiment Station.
REFERENCES
- 1.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 3.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 4.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 5.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 7.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 9.Ghigo, J. M. 2001. Conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, C., S. Restrepo, J. Tohme, and V. Verdier. 2002. Characterization of pathogenic and nonpathogenic strains of Xanthomonas axonopodis pv. manihotis by PCR-based DNA fingerprinting techniques. FEMS Microbiol. Lett. 215:23-31. [DOI] [PubMed] [Google Scholar]
- 11.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 13.Hanna, A., M. Berg, V. Stout, and A. Razatos. 2002. Role of capsular colanic acid in adhesion of uropathogenic Escherichia coli. Appl. Environ. Microbiol. 69:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 15.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce, J. G., C. Abeygunawardana, Q. Xu, J. C. Cook, R. Hepler, C. T. Przysiecki, K. M. Grimm, K. Roper, C. C. Ip, L. Cope, D. Montgomery, M. Chang, S. Campie, M. Brown, T. B. McNeely, J. Zorman, T. Maira-Litran, G. B. Pier, P. M. Keller, K. U. Jansen, and G. E. Mark. 2003. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr. Res. 338:903-922. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed]
- 20.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 21.Leane, M. M., R. Nankervis, A. Smith, and L. Illum. 2004. Use of the ninhydrin assay to measure the release of chitosan from oral solid dosage forms. Int. J. Pharm. 271:241-249. [DOI] [PubMed] [Google Scholar]
- 22.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 25.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 29.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 30.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 32.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 33.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, T., S. Fujiwara, S. Nishikori, T. Fukui, M. Takagi, and T. Imanaka. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microbiol. 65:5338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD Locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]