Abstract
Regulation of cell-specific cellulase synthesis (expressed in milligrams of cellulase per gram [dry weight] of cells) by Clostridium thermocellum was investigated using an enzyme-linked immunosorbent assay protocol based on antibody raised against a peptide sequence from the scaffoldin protein of the cellulosome (Zhang and Lynd, Anal. Chem. 75:219-227, 2003). The cellulase synthesis in Avicel-grown batch cultures was ninefold greater than that in cellobiose-grown batch cultures. In substrate-limited continuous cultures, however, the cellulase synthesis with Avicel-grown cultures was 1.3- to 2.4-fold greater than that in cellobiose-grown cultures, depending on the dilution rate. The differences between the cellulase yields observed during carbon-limited growth on cellulose and the cellulase yields observed during carbon-limited growth on cellobiose at the same dilution rate suggest that hydrolysis products other than cellobiose affect cellulase synthesis during growth on cellulose and/or that the presence of insoluble cellulose triggers an increase in cellulase synthesis. Continuous cellobiose-grown cultures maintained either at high dilution rates or with a high feed substrate concentration exhibited decreased cellulase synthesis; there was a large (sevenfold) decrease between 0 and 0.2 g of cellobiose per liter, and there was a much more gradual further decrease for cellobiose concentrations >0.2 g/liter. Several factors suggest that cellulase synthesis in C. thermocellum is regulated by catabolite repression. These factors include: (i) substantially higher cellulase yields observed during batch growth on Avicel than during batch growth on cellobiose, (ii) a strong negative correlation between the cellobiose concentration and the cellulase yield in continuous cultures with varied dilution rates at a constant feed substrate concentration and also with varied feed substrate concentrations at a constant dilution rate, and (iii) the presence of sequences corresponding to key elements of catabolite repression systems in the C. thermocellum genome.
Clostridium thermocellum is a thermophilic, anaerobic, cellulolytic bacterium that grows on soluble β-glucans, including cellobiose and cellodextrins, as well as on cellulosic substrates, including Avicel, filter paper, solka floc, and pretreated mixed hardwood. The extracellular multienzyme cellulase complex produced by C. thermocellum, the cellulosome (1, 29), exhibits high activity against crystalline cellulose (26). To date, more than 20 catalytic subunits have been sequenced, and these subunits collectively exhibit a variety of activities, including endoglucanase, exoglucanase, xylanase, mannanase, chitinase, and lichenase (46, 48). The C. thermocellum cellulosome includes a nonhydrolytic subunit (scaffoldin) and three main catalytic cellulolytic subunits (CelA [endoglucanase], CelS [processive exoglucanase], and CelK [cellobiohydrolase]), as well as a xylanolytic subunit (XynC) (46, 48). CelS is the most abundant catalytic subunit, and the hydrolytic properties of CelS closely resemble those of the cellulosome (28, 48).
Regulation of cellulase synthesis by C. thermocellum is an important feature of the physiology of this microorganism, particularly in light of the substantial investment of ATP that cellulase synthesis represents (34, 35). Moreover, this regulation is a central determinant of hydrolysis and growth rates and thus is of interest for understanding cellulose utilization in both natural environments and industrial processes. Johnson et al. (25) reported that synthesis of true cellulase activity (i.e., degradation of crystalline cellulose) was markedly repressed by cellobiose. mRNA corresponding to endoglucanases CelA, CelF, and CelD were found to be regulated at the level of transcription by a mechanism analogous to catabolite repression (40). Recently, the numbers of CelS and CipA transcripts per cell were shown to decrease with increasing growth rate (8, 9), the numbers of CelS transcripts were found to be higher for growth under cellobiose limitation than for growth under nitrogen limitation (8), and control of scaffoldin and CelS transcription was shown to involve the housekeeping sigma-A factor (9). Based on the inverse correlation observed between the growth rate and the synthesis of key cellulosome gene transcripts (celS, cipA, olpB, and orf2), as well as the apparent absence of key components of catabolite repression systems (catabolite responsive element [CRE] sequences), Dror et al. (9) inferred that that the growth rate plays a role in regulation of the cellulosome-related genes that have been studied but the known mechanisms of catabolite repression do not play a role.
Studies of the control of cellulase synthesis by microorganisms have been carried out using a variety of methodological approaches. These approaches include measurement of supernatant protein (11), measurement of activity in culture supernatants (2, 3, 12, 19, 26, 30), measurement of cellular mRNA content (8-10, 16, 17, 20, 23, 31, 37, 40), sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (27, 49), fast liquid chromatography (21), and immunoblotting (39). Activity measurements have the desirable feature of being specific to cellulase synthesis, but they have limited value in a physiological context (e.g., for calculation of parameters such as yields and ATP investment). mRNA measurements are valuable because the expression of many genes can be monitored simultaneously. However, there is little evidence for a proportional relationship between mRNA levels and protein synthesis, there is some strong evidence to the contrary in yeast (18), and there are increasing indications that posttranslational control of gene expression is important in prokaryotes (13, 24, 43). Quantitative measurement of cellulase synthesis is not subject to these limitations and also provides information that can readily be interpreted in a physiological context. Proteomic analysis of microbial synthesis of cellulase, in which synthesis of many cellulase components is evaluated simultaneously, is of interest but has been used to date to a very limited extent and awaits development of appropriate protein arrays and related methodologies.
For anaerobic, cellulolytic microorganisms, including but not limited to C. thermocellum, a large portion of the total amount of cellulase synthesized (and in many cases most of it) is bound to cells and/or substrate (2, 8, 34, 38, 56) and is thus not present in culture supernatants. Moreover, the fraction of bound cellulase can vary substantially as a function of the growth conditions (2, 38, 56). Thus, measurement of the cellulase present in culture supernatants, based either on mass or on activity, gives a decidedly incomplete picture of cellulase expression. Recently, we developed an enzyme-linked immunosorbent assay (ELISA)-based assay using antibodies raised against a peptide sequence from the C. thermocellum scaffoldin protein, and we showed that the signal from this assay can be used to infer the cellulosome concentration (or the total cellulase concentration given an appropriate standard curve) in cell-associated, cellulose-bound, and unbound forms with a coefficient of variation of about 5% (56). The study described here was undertaken in order to investigate control of cellulase synthesis with this assay.
MATERIALS AND METHODS
Chemicals and strains.
All chemicals were reagent grade and were obtained from Sigma (St. Louis, Mo.) unless indicated otherwise. C. thermocellum ATCC 27405 was a gift from Arnold Demain in 1983 and has been maintained in our lab since then as described elsewhere (34). Stock cultures were maintained in MTC medium (see below) containing 10 g of Avicel (PH105; FMC Corp., Philadelphia, Pa.) per liter as the carbon source (56, 57).
Medium composition and preparation.
C. thermocellum was grown under anaerobic conditions at 60°C. Chemically defined MTC medium was prepared by combining six sterile solutions under a nitrogen atmosphere. Sterilization was accomplished by filter sterilizing preparations with a 0.2-μm-pore-size filter (Pall Corp., Ann Arbor, Mich.) for solution E and by autoclaving preparations for solutions A, B, C, D, and F. Solution A contained distilled water, Avicel, and 0.2% resazurin (optional). Solution B, which was concentrated 25-fold relative to the final medium, contained citric acid tripotassium salt, citric acid monohydrate, NaSO4, KH2PO4, and Na2CO3. Solution C, which was concentrated 50-fold, contained NH4Cl and urea. Solution D, which was concentrated 50-fold, contained MgCl2 · 6H2O, CaCl2 · 2H2O, FeCl2 · 4H2O, and l-cysteine hydrochloride monohydrate. Solution E, which was concentrated 50-fold, contained pyridoxamine dihydrochloride, p-aminobenzoic acid, d-biotin, vitamin B12, and thiamine. Solution F, which was concentrated 1,000-fold, contained MnCl2 · 4H2O, CoCl2 · 6H2O, ZnSO4 · 7H2O, CuSO4 · 5H2O, H3BO3, Na2MoO4, NiCl2 · 6H2O, and citric acid monohydrate. The final concentrations have been reported elsewhere (56).
Batch and continuous cultures.
Batch and continuous fermentations were carried out in round-bottom reactors (Applikon Dependable Instruments, Foster City, Calif.) with agitation provided by a marine (six-pitched-blade turbine) impeller at 300 rpm, and the working volumes and feed substrate concentrations as indicated below. Continuous fermentation of cellobiose was also carried out in 60-ml (working volume) jacketed glass fermentors (NDS Glass, Vineland, N.J.) with medium addition, medium removal, and base addition via 2-mm-internal diameter stainless steel tubes penetrating a buytl rubber no. 6 stopper and with agitation provided by a magnetic stir bar turning at approximately 200 rpm. Avicel was delivered to continuous cultures via a peristaltic pump as described previously (33). The temperature was maintained at 60 ± 1°C by means of water circulated through a jacket. The pH was maintained at 6.80 with an Applikon ADI 1020 controller via addition of 8 M sodium hydroxide. For continuous cultures, at least four steady-state samples were harvested at each dilution rate for which data are reported below, and at least 0.5 residence time elapsed between the times that samples were harvested. Cultures were considered to be at a steady state when the variation among samples was less than 5% and exhibited no consistent increasing or decreasing trend over time.
Quantification of protein, dry weight, substrate, and fermentation products.
The protein concentrations in supernatant samples were determined by the Bradford protein assay with bovine serum albumin as the standard (56). The concentrations of protein in culture pellets were determined by the Lowry method after samples were lysed with SDS in the presence of NaOH (56). Dry weights were determined by filtering 10-ml samples through Pall Corp. 0.2-μm-pore-size Metricel membrane filters and drying the samples at 72°C until a constant weight was achieved. Residual cellulose was measured by quantitative saccharification as described elsewhere (34). For determination of cellobiose concentrations below the high-performance liquid chromatography (HPLC) detection limit (∼0.15 g/liter), 0.8 ml of cell-free broth after centrifugation was mixed with 0.2 ml of 10% (wt/wt) sulfuric acid and then autoclaved for 40 min to convert the cellobiose to glucose. After neutralization to pH 4 to 6 with solid CaCO3 and centrifugation, the concentration of glucose in the supernatant was measured using the Sigma Infinity glucose kit reagents at double the concentrations recommended (Sigma G2020), which can detect a concentration of glucose as low as 0.005 g/liter (57). Ethanol, lactate, acetate, cellobiose, and glucose in samples taken from steady-state continuous fermentors were acidified by adding 0.0565 ml of sulfuric acid (10%, wt/wt) to 1-ml samples, which were subsequently analyzed by HPLC by using a Bio-Rad HPX-87H column (Bio-Rad, Hercules, Calif.) operated at 55°C with a 0.01% (vol/vol) H2SO4 running buffer and a refractive index detector.
Cellulase purification.
Cellulase purification for the purpose of visualizing the protein composition and development of standard curves was carried out for cellobiose-grown and Avicel-grown cultures by affinity digestion involving cold adsorption to amorphous cellulose followed by hydrolysis at an increased temperature to release cellulase (56). SDS-PAGE was carried out at 150 V by using a Bio-Rad 7.5% polyacrylamide Ready Gel (Tris-HCl). Protein-containing samples were denatured by mixing equal volumes of 3% (wt/vol) SDS, 10% (vol/vol) glycerol, 5% (vol/vol) mercaptoethanol, and 0.02% (wt/vol) bromophenol blue in 62.5 mM Tris buffer (pH 6.8) and boiling the preparation for 3 min (29).
Cellulase-based ELISA and cell mass determination.
Antibody production was carried out by Genosys Co. (The Woodlands, Tex.). The mass concentrations of cellulase and cells were calculated based on an indirect ELISA by using antibody raised against a sequence from the C. thermocellum scaffoldin protein as described previously (56). For cells grown on cellobiose, cell mass (X) was calculated based on the difference between the dry cell mass and the mass of cell-associated cellulase measured by the ELISA. In the case of Avicel-grown cultures, cell mass was calculated with the equation X = (PP − EP)/fP/X, where PP is the pellet protein concentration, EP is the cellulase concentration, and fP/X is the ratio of noncellulase pellet protein weight to noncellulase dry weight calculated as follows: (pellet protein weight − pellet cellulase weight)/(dry weight − pellet cellulase weight). For Avicel-grown batch cultures, a fP/X value of was 0.52 was used based on end point measurements for a cellobiose-grown batch culture (56). For Avicel-grown continuous cultures, we used fP/X values that were obtained from steady-state continuous cultures grown on cellobiose at the same dilution rate and feed substrate concentration.
Cell-specific cellulase yield (YE/X) (expressed in milligrams of cellulase per gram of cells [dry weight]) was defined as the ratio of total cellulase (including free, cell-associated, and cellulose-bound cellulase) to cell mass exclusive of cell-associated cellulase.
RESULTS
Cellulase composition and ELISA.
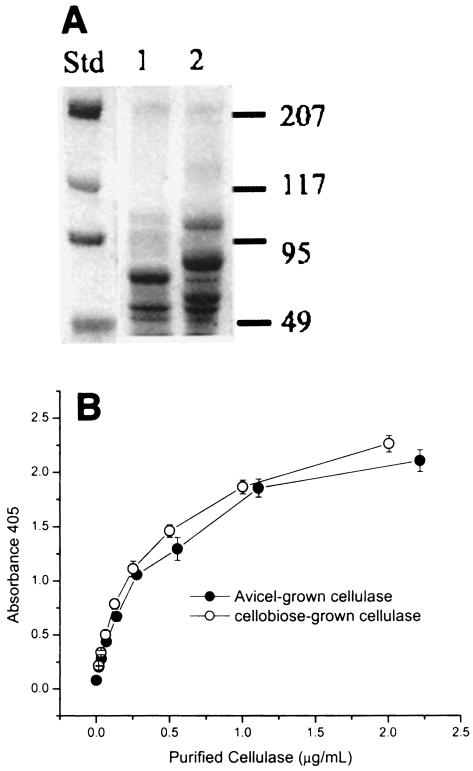
Cellulases from the supernatants of stationary-phase cellobiose-grown and Avicel-grown batch cultures of C. thermocellum ATCC 27405 were purified by affinity digestion, and 80 and 87% of the activity originally present, respectively, were recovered. The compositions of the cellulosomes produced during growth on these two carbon sources were different, as shown by SDS-PAGE (Fig. 1A). The affinity digestion-purified cellulases from cellobiose- and Avicel-grown continuous cultures produced SDS-PAGE patterns similar to the patterns obtained for cellulases obtained from batch cultures and appeared to be similar at various dilution rates (data not shown). The specific activity of cellulase prepared from cellobiose-grown cultures on Avicel PH-105 (1.8 IU/mg of cellulase protein) was 25% lower than the specific activity previously measured for supernatant from Avicel-grown cultures (2.4 IU/mg). Standard curves for ELISA carried out with purified cellulases from Avicel- and cellobiose-grown cultures are presented in Fig. 1B. The ELISA results were similar for the two preparations, although the mean ratio of absorbance to concentration of purified cellulase produced for cellobiose-grown cultures was greater than that for Avicel-grown cultures by a small amount (≤10%). The higher absorbance was consistent with the greater relative amount of scaffoldin observed by SDS-PAGE on a weight basis. The cellulase concentrations for Avicel- and cellobiose-grown cultures reported below were determined with reference to ELISA standard curves for the respective substrates.
FIG. 1.
SDS-PAGE (A) and ELISA results (B) for purified cellulases prepared from Avicel- and cellobiose-grown cultures. (A) Lane 1, cellobiose-grown cellulase; lane 2, Avicel-grown cellulase; lane Std, Bio-Rad prestained high-range protein standards, including myosin (207 kDa), β-galactosidase (117 kDa), bovine serum albumin (95 kDa), and ovalbumin (49 kDa).
Avicel and cellobiose batch cultures.
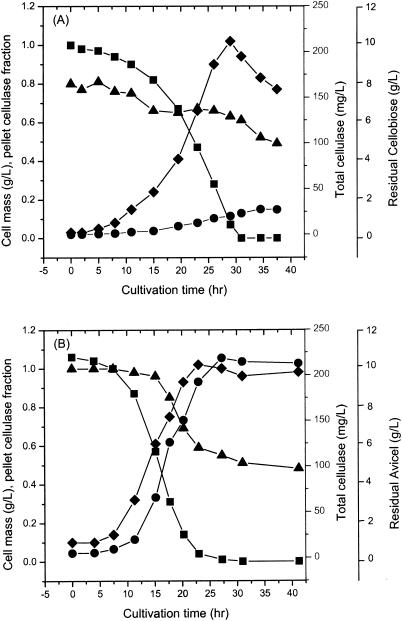
Figure 2 shows the profiles of substrate, cell, and total cellulase concentrations as a function of time for batch cultures of C. thermocellum ATCC 27405 grown on cellobiose and Avicel at an initial concentration of 10 g/liter. The total cellulase included cellulase that was free, cellulase that was cell associated, and (for cellulose-grown cultures) cellulase that was cellulose bound. At the end of fermentation, the total cellulase concentration was 223 mg/liter for the Avicel-grown culture, compared to 24.9 mg/liter for the cellobiose-grown culture. The amount of the pellet cellulase (cellulase bound to cells, to cellulose, or to both cells and cellulose) expressed as a fraction of the total cellulase decreased from 0.8 to 0.52 and from 1 to 0.48 on cellobiose and Avicel, respectively, as the fermentation proceeded. The total cellulase synthesis by stationary-phase cultures represented 21% of the cell mass for growth on cellulose and 3% of the cell mass for cellobiose-grown cells. The concentration of supernatant protein (consisting of the cellulosome released from cells, as well as noncellulosomal proteins) was nearly threefold higher in the Avicel-grown culture than in the cellobiose-grown culture (data not shown).
FIG. 2.
Batch cultures of C. thermocellum grown with ∼10 g of cellobiose per liter (A) and ∼10 g of Avicel per liter (B). Symbols: ♦, cell mass; ▴, pellet cellulase fraction (pellet cellulase/total cellulase); •, total cellulase; ▪, residual cellobiose (A) or residual Avicel (B).
Avicel and cellobiose continuous cultures at low feed concentrations.
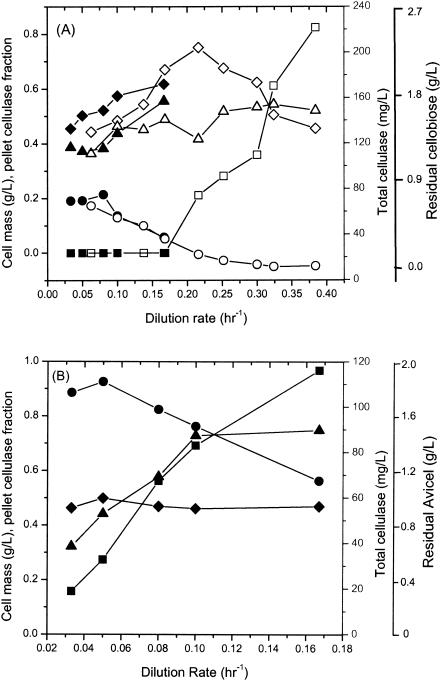
To investigate cellulase synthesis under steady-state conditions and at a variety of growth rates, continuous cultures were grown on both cellobiose and Avicel with a feed concentration of about 5 g/liter. Figure 3A shows steady-state data for fermentation of cellobiose in two reactors. Data for dilution rates from 0.0333 to 0.167 h−1 were obtained in a round-bottom reactor with a 1.5-liter working volume and with agitation by a turbine with six pitched blades rotating at 300 rpm. Data for dilution rates from 0.0625 to 0.384 h−1 were also obtained with a 60-ml (working volume) reactor with agitation by a stir bar. The results for these two reactors obtained at the same dilution rates agree very well (Fig. 3A). The cell mass increased from 0.44 to 0.67 g/liter as the dilution rate increased from 0.0333 to 0.167 h−1, but it decreased at higher dilution rates to 0.454 g/liter at a dilution rate of 0.384 h−1, at which more than one-half of the feed substrate was not consumed. As the dilution rate increased over the range examined, the total cellulase concentration decreased from 80 to 16 mg/liter, and approximately one-half of the cellulase was cell associated.
FIG. 3.
Continuous cultures of C. thermocellum grown with ∼5 g of cellobiose per liter (A) and ∼5 g of Avicel per liter (B) with different dilution rates. Symbols: ♦ and ⋄, cell mass; ▴ and ▵, pellet cellulase fraction; • and ○, total cellulase; ▪ and □, residual cellobiose (A) or residual Avicel (B). In panel A the solid symbols indicate data for a large reactor (1.5 liters) and the open symbols indicate data for a small reaction (60 ml).
Figure 3B shows steady-state data for fermentation of ∼5 g of Avicel per liter in continuous cultures with dilution rates ranging from 0.0333 to 0.167 h−1 in a 1.5-liter (working volume) reactor. As the dilution rate increased over this range, the residual Avicel concentration increased from 0.31 to 1.94 g/liter, the cell mass was nearly constant at about 0.5 g/liter, and the fraction of pellet cellulase increased from 0.32 to 0.78. A dilution rate of 0.2 h−1 resulted in washout. Soluble reducing sugar could not be detected in the liquid phase by HPLC (limit of detection, 0.15 g/liter). The total cellulase concentration decreased somewhat from 111 to 67 mg/liter.
Comparison of cellulase and cell yields on Avicel and cellobiose.
Table 1 shows a comparison of the cellulase and cell yields from batch and continuous cultures on cellobiose and Avicel. The cellulase yield is defined as the ratio of the total cellulase to the cell mass exclusive of cellulase, and the cell yield is defined as the ratio of the cell mass exclusive of cellulase to the mass of substrate consumed on a glucose equivalent basis (see Materials and Methods). The cellulase yield observed for stationary-phase Avicel-grown cultures (223 mg of cellulase per g of cells) was nearly ninefold higher than the cellulase yield observed for stationary-phase cellobiose-grown cultures. The cellulase yields declined with increasing dilution rate in continuous cultures grown on both Avicel and cellobiose, and the cellulase yields at a particular dilution rate were 1.34- to 2.36-fold higher for Avicel cultures. As the dilution rate increased from 0.033 to 0.167 h−1, the cellulase yield for Avicel-grown cultures decreased from 230 to 144 mg of cellulase/g of cells, whereas the cellulase yield for cellobiose-grown cultures decreased from 172 to 61 mg of cellulase/g of cells. Over this range of dilution rates, the fermentor substrate concentration was below the detection limit for cellobiose-grown continuous cultures (Fig. 3A). The cell yields in stationary-phase batch cultures were similar for Avicel and cellobiose (about 0.1 g of cells/g of glucose equivalents). The cell yields increased with increasing dilution rates in continuous cultures grown on both Avicel and cellobiose, and the cell yields at a particular dilution rate were 13.8% ± 5.3% higher for Avicel cultures. As the dilution rate increased from 0.033 to 0.167 h−1, the cell yield for Avicel-grown cultures increased from 0.101 to 0.151 g of cells/g of substrate consumed, whereas the cell yield for cellobiose-grown cultures increased from 0.083 to 0.127 g of cells/g of substrate consumed.
TABLE 1.
Cellulase synthesis in batch and continuous cultures
| Substrate | Type of culture | Dilution rate (h−1) | Initial concn (g/liter) | Final concn (g/liter) | Cell yield (g of cells/g of glucose equivalents) | YE/X (mg of cellulase/g of cells) |
|---|---|---|---|---|---|---|
| Cellobiose | Batch | 9.54 | 0 | 0.099 | 24.9 | |
| Continuous | 0.033 | 4.65 | 0 | 0.083 | 173 | |
| 0.05 | 4.65 | 0 | 0.104 | 139 | ||
| 0.08 | 4.65 | 0 | 0.108 | 144 | ||
| 0.1 | 4.65 | 0 | 0.118 | 98 | ||
| 0.167 | 4.65 | 0 | 0.127 | 61 | ||
| Avicell | Batch | 10.6 | 0 | 0.102 | 223 | |
| Continuous | 0.033 | 4.85 | 0.31 | 0.101 | 230 | |
| 0.05 | 4.85 | 0.59 | 0.114 | 223 | ||
| 0.08 | 4.85 | 1.18 | 0.124 | 211 | ||
| 0.1 | 4.85 | 1.38 | 0.128 | 198 | ||
| 0.167 | 4.85 | 1.93 | 0.151 | 144 |
Cellobiose continuous cultures with increasing feed concentrations.
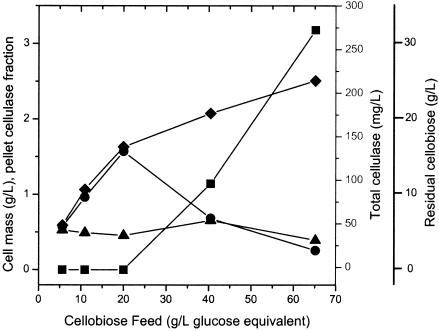
Steady-state cellobiose-grown continuous cultures were incubated in a 400-ml (working volume) reactor with feed concentrations of 5.6 to 65 g/liter at a fixed dilution rate of 0.0556 h−1, with results shown in Fig. 4. For feed concentrations of 5.6 to 20 g/liter, residual substrate could not be detected by HPLC (≤0.15 g/liter), and the cell mass increased from 0.59 to 1.63 g/liter. Increasing the feed cellobiose concentration to more than 20 g/liter resulted in significant and progressively increasing amounts of unutilized cellobiose and a modest increase in the cell mass over that observed for a 20-g/liter cellobiose feed. For cellobiose-limited growth at feed concentrations of ≤20 g of cellobiose per liter, the total cellulase synthesis increased from 46.9 to 133 mg/liter, and YE/X was nearly constant at 78.9 ± 3 mg of cellulase/g of cells. For growth at higher feed concentrations, at which significant concentrations of unutilized cellobiose were present, the total cellulase concentrations fell to 56 and 19 mg/liter, and the cellulase yields fell to 27 and 7.7 mg of cellulase/g of cells for feed concentrations of 40 and 65 g of cellobiose per liter, respectively.
FIG. 4.
Continuous cultures of C. thermocellum with an increase in the cellobiose feed concentration from 5 to 60 g/liter at a dilution rate of 0.0556 h−1. Symbols: ♦, cell mass; ▴, pellet cellulase fraction; •, total cellulase; ▪, residual cellobiose.
Cellulase yield in relation to cellobiose concentration.
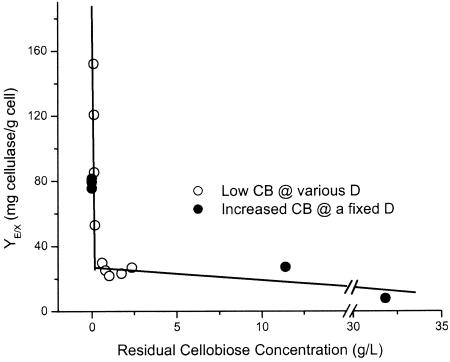
Figure 5 presents the cellulase yield as a function of the fermentor cellobiose concentration for cellobiose-grown continuous cultures maintained either with a 5-g/liter feed and various dilution rates (Fig. 3) or with a fixed dilution rate of 0.0556 h−1 and various feed concentrations. Fermentor samples from cellobiose-grown continuous cultures with substrate concentrations below the HPLC detection limit were reanalyzed by using a more sensitive enzymatic assay (see Materials and Methods), whose detection limit was 0.15 g/liter. It was found that the cellulase yield data correlated well with the fermentor cellobiose concentration. In particular, YE/X decreased by more than sixfold from 173 mg of cellulase/g of cells to about 27 mg of cellulase/g of cells as the cellobiose concentration increased from 0.11 to 0.2 g/liter and then continued to decline but did so much more gradually as the cellobiose concentration increased from 0.2 to 32 g/liter. The relationship between cellulase yield (YE/X) (expressed in milligrams of cellulase per gram of cells) and cellobiose concentration (CB) (expressed in grams per liter) is described well (r2 = 0.966, 0.851) by the following functions, respectively:
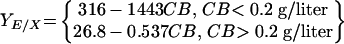 |
FIG. 5.
Relationship between cell-specific cellulase yield and extracellular cellobiose concentration. The data are from steady-state, cellobiose-grown continuous cultures (see Fig. 3A and 4). CB, cellobiose concentration; D, dilution rate.
DISCUSSION
An ELISA-based protocol based on antibody raised against an amino acid sequence from the ninefold-repeated type I cohesin region of the scaffoldin protein was used to quantify the production of C. thermocellum cellulase, which exists in unbound, cellulose-bound, and cell-bound forms (56). The SDS-PAGE banding patterns for affinity digestion-purified cellulosome were different for Avicel- and cellobiose-grown cultures, which is consistent with some (2, 8) but not all (3) previous reports. The ELISA standard curves were very similar but not identical for cultures grown on Avicel and cultures grown on cellobiose. Lamed et al. (29) concluded that the major characteristics of the cellulolytic system of C. thermocellum correspond to those of the purified cellulosome. Consistent with this, we showed previously that both SDS-PAGE banding patterns and cellulosome specific activity were essentially identical for unbound, cell-bound, and cellulose-bound affinity digestion-purified cellulosome preparations from Avicel-grown cultures (56). In light of these observations, we inferred that expression of various cellulosome components is differentially regulated during growth on Avicel and cellobiose but that total cellulase production can be estimated for both substrates by using the ELISA-based protocol that we have developed together with standard curves, obtained under conditions of interest.
The cell yields (in grams [dry weight] of cells exclusive of the amount of cellulase per gram of glucose equivalents consumed) were higher for continuous cultures grown on Avicel than for continuous cultures grown on cellobiose at every dilution rate tested over the range from 0.033 to 0.167 h−1. Similar cell yields were observed for Avicel- and cellobiose-grown stationary-phase batch cultures. However, the cell yields determined for batch cultures may be affected by cell lysis and or endogenous metabolism after the substrate is exhausted and are regarded as less reliable than the values determined for continuous cultures. Higher cell yields for growth on cellulose than for growth on cellobiose have been observed for the mesophilic cellulolytic anaerobe Clostridium cellulolyticum (6, 7), and cell yields have been observed to increase with increasing chain length for cellobiose and soluble cellodextrins for C. thermocellum (52), as well as several other cellulolytic bacteria (32, 47, 53). Phosphorolytic cleavage of β-glucosidic bonds by cellobiose and cellodextrin phosphorylases may contribute to the increasing cell yields with increasing substrate chain length (34). Consistent with this, the rates of phosphorolytic cleavage of both cellobiose and cellopentaose are much higher (e.g., 20-fold higher) than the rates of hydrolytic cleavage in C. thermocellum cell extracts grown on either Avicel or cellobiose (57). Lower ATP expenditure for substrate transport has also been suggested as a factor that contributes to higher cell yields for substrates with greater chain lengths (34). Higher cell yields on Avicel than on cellobiose as a result of either phosphorolytic cleavage of β-glucosidic bonds or a higher transport efficiency appear to require that the mean chain length assimilated during growth on cellulose be greater than 2 during growth on cellulose (34). While the available information suggests that compounds having a higher molecular weight than cellobiose may be important intermediates in cellulose hydrolysis by C. thermomcellum, as well as other cellulolytic bacteria, this has not been definitively proven.
Cellulase yields measured in continuous cultures provide a basis for inference regarding regulation of cellulase synthesis in C. thermocellum. Classical chemostat theory stipulates that the concentration of a single rate-limiting growth substrate is uniquely determined by the dilution rate (44). Consistent with this observation, if cellobiose were the only soluble intermediate of cellulose hydrolysis, then the steady-state bulk-phase cellobiose concentration would be the same at a specified dilution rate whether the fermenter is grown on Avicel or on cellobiose. The fact that the cellulase yields in continuous cultures were substantially higher (1.34- to 2.36-fold higher, depending on the dilution rate) for growth on Avicel than for growth on cellobiose seems to be a response to one or both of the following environmental factors. (i) Cellobiose is in fact not the only soluble hydrolysis product during growth on cellulose; hence, the concentrations of cellobiose (and other soluble hydrolytic products of cellulose) are not the same during growth on Avicel and during growth on cellobiose at a particular dilution rate, and this results in different cellulase yields through repression and/or induction. (ii) The presence of insoluble cellulose is sensed by the cell (e.g., in association with attachment to the substrate surface), triggering synthesis of an intracellular regulatory molecule which acts to increase the cellulase yield. Elucidation of the relative importance of a different profile of hydrolysis products and the presence of the cellulose surface as factors that affect cellulase synthesis awaits further investigation.
Our results suggest that cellulosome synthesis by C. thermocellum is regulated by carbon catabolite repression (CCR). The ninefold-higher YE/X value observed during batch growth on Avicel than during batch growth on cellobiose is consistent with CCR by cellobiose, since the concentrations of cellobiose are manyfold higher during batch growth on this substrate than during batch growth on Avicel. The fact that YE/X values correlate well with cellobiose concentrations (Fig. 5) is also consistent with the hypothesis that CCR is operative. The strongest indication that CCR by cellobiose is an important if not dominant mechanism for control of cellulosome synthesis by C. thermocellum is the fact that data from two quite different experiments, one involving changing the dilution rate at a constant substrate concentration and the other involving changing the substrate concentration at a constant dilution rate, fall on the same curve for YE/X versus cellobiose concentration. Both changing the dilution rate and changing the feed cellobiose concentration result in changes in the fermentor cellobiose concentration, which appears to be sufficient to determine the cellulase yield under the conditions examined.
CCR-mediated control of cellulosome synthesis in C. thermocellum is further supported by the observation that the three key components of a CCR system (a LacI/GalR family regulatory protein, an HPr protein and an HPr kinase, and a 14-bp cis-acting catabolite responsive element binding sequence) are present in the draft C. thermocellum genomic sequence (GenBank ID AABG00000000; assembly date, November 2003; http://genome.ornl.gov/microbial/cthe/). Several putative LacI/GalR family genes are found in C. thermocellum (9). Although the levels of amino acid identity for these C. thermocellum open reading frames are rather low (e.g., in the range from 29 to 50%) compared to those for Bacillus subtilis ccpA, such low levels of identity are typical of LacI/GalR family proteins. For example, the levels of amino acid identity for confirmed LacI/GalR proteins relative to the protein encoded by B. subtilis ccpA are 38% for regA from Clostridium acetobutylicum (4), 33% for malR from Clostridium butyricum (14), and 30% for B. subtilis ccpB (45). Warner and Lolkema (54) identified in the C. thermocellum genome genes corresponding to an HPr kinase-like protein, HPr-like proteins containing a regulatory site serine residue, and an HPr-like protein missing the active site histidine residue. We were able to locate many (>100) putative CRE sequences, including two putative CREs inside the cipA structural gene (positions +953 and +5231) by using the more degenerate CRE consensus sequence (WGWNANCGNTNNCW). Dror et al. (9) identified only two CRE sequences by using the less degenerate sequence (TGWAARCGYTWNCW). Substantial degeneracy of CRE sequences is supported by results for B. subtilis. Whereas Chauvaux (4) found 29 CRE sequences based on a consensus sequence with 7 of the 14 bases degenerate, Moreno et al. (41) later found by using DNA arrays that ∼330 genes are regulated by CCR. Moreover, whole-genome analysis of B. subtilis indicated that the CRE sequence is not strictly conserved and that CRE variation provides a means to alter the affinities of regulatory proteins to CRE sequences, thereby modulating regulation (15, 41). It should be noted that several glycosyl hydrolases have been reported to be regulated by CCR; these enzymes include Bacillus spp. cellulases, xylanases, and mannases (22), C. acetobutylicum amylase (5, 55), Streptomyces chintiase 63 (42), Trichoderma reesei xylanase I (36), and Thermobifida fusca endoglucanase CelE (50, 51). Identification of specific CCR components and associated coeffecters involved in control of cellulase synthesis in C. thermocellum is an important area for further research.
Acknowledgments
This work was supported by grant DE-FG02-02ER15350 from the Department of Energy and by grant 60NANB1D0064 from the National Institute of Standards and Technology.
REFERENCES
- 1.Bayer, E. A., R. Kenig, and R. L. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Setter, and R. L. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat, S., P. W. Goodenough, E. Owen, and M. K. Bhat. 1993. Cellobiose: a true inducer of cellulosome in different strains of Clostridium thermocellum. FEMS Microbiol. Lett. 111:73-78. [Google Scholar]
- 4.Chauvaux, S. 1996. CcpA and HPr(ser-P): mediators of catabolite repression in Bacillus subtilis. Res. Microbiol. 147:518-522. [DOI] [PubMed] [Google Scholar]
- 5.Davison, S. P., J. D. Santangelo, S. J. Reid, and D. R. Woods. 1995. A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology 141:989-996. [DOI] [PubMed] [Google Scholar]
- 6.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Kinetics of metabolism of cellulose degradation at high substrate concentrations in steady-state continuous culture of Clostridium cellulolyticum on a chemically defined medium. Appl. Environ. Microbiol. 67:3827-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Carbon flux distribution and kinetics of cellulose fermentation in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. J. Bacteriol. 183:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal CelS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J. Bacteriol. 185:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Gogary, S., A. Leite, O. Crivellaro, D. E. Eveleigh, and H. El-Dorry. 1989. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc. Natl. Acad. Sci. USA 86:6138-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esterbauer, H., W. Steiner, I. Labudova, A. Hermann, and M. Hayn. 1991. Production of Trichoderma cellulase in laboratory and pilot scale. Biores. Technol. 36:51-65. [Google Scholar]
- 12.Garcia-Martinez, D. V., A. Shinomyo, A. Madia, and A. L. Demain. 1980. Studies on cellulase production by Clostridium thermocellum. Eur. J. Appl. Microbiol. Biotechnol. 9:189-197. [Google Scholar]
- 13.Glanemann, C., A. Loos, N. Gorret, L. B. Willis, X. M. O'Brien, P. A. Lessard, and A. J. Sinskey. 2003. Disparity between changes in mRNA abundance and enzyme activity in Corynebacterium glutamicum: implications for DNA microarray analysis. Appl. Microbiol. Biotechnol. 61:61-68. [DOI] [PubMed] [Google Scholar]
- 14.Goda, S. K., O. Eisa, M. Akhter, and N. P. Minton. 1998. Molecular analysis of the malR gene of Clostridium butyricum NCIMB 7423, a member of the LacI-GalR family of repressor proteins. FEMS Microbiol Lett. 165:193-200. [DOI] [PubMed] [Google Scholar]
- 15.Gosseringer, R., E. Kuster, A. Galinier, J. Deutscher, and W. Hillen. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266:665-676. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, N. M., R. A. J. Warren, D. G. Kilburn, and R. C. Miller, Jr. 1987. Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi. J. Bacteriol. 169:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg, N. M., R. A. J. Warren, D. G. Kilburn, and R. C. Miller Jr. 1987. Regulation and initiation of cenB transcripts of Cellulomonas fimi. J. Bacteriol. 169:4674-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halliwell, G., T. M. Phillips, and N. Halliwell. 1995. Microcrystalline forms of cellulose as substrates for strains of Clostridium thermocellum and cellulase formation. Proc. Biochem. 30:243-250. [Google Scholar]
- 20.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayn, M., and H. Esterbauer. 1985. Separation and partial characterization of Trichoderma reesei cellulase by fast chromatofocusing. J. Chromatogr. 329:379-387. [Google Scholar]
- 22.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 23.Ilmen, M., A. Saloheimo, M.-L. Onnela, and M. E. Penttila. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaacs, F. J., D. J. Dwyer, C. Ding, D. D. Pervouchine, C. R. Cantor, and J. J. Collins. 2004. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22:841-847. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, E. A., F. Bouchot, and A. L. Demain. 1985. Regulation of cellulase formation in Clostridium thermocellum. J. Gen. Microbiol. 131:2303-2308. [Google Scholar]
- 26.Johnson, E. A., M. Sakajoh, G. Halliwell, A. Madia, and A. L. Demain. 1982. Saccharification of complex cellulosic substrates by the cellulase system from Clostridium thermocellum. Appl. Environ. Microbiol. 43:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruus, K., W. K. Wang, J. Ching, and J. H. D. Wu. 1995. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 177:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, E., and D. B. Wilson. 1987. Regulation of β-1,4,-endoglucanase synthesis in Thermomospora fusca. Appl. Environ. Microbiol. 53:1352-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, E., and D. B. Wilson. 1988. Transcription of the celE gene in Thermomonospora fusca. J. Bacteriol. 170:3838-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou, J., K. A. Dawson, and H. J. Strobel. 1997. Cellobiose and cellodextrin metabolism by the ruminal bacterium Ruminococcus albus. Curr. Microbiol. 35:221-227. [DOI] [PubMed] [Google Scholar]
- 33.Lynd, L. R., H. E. Grethlein, and R. H. Wolkin. 1989. Fermentation of cellulosic substrates in batch and continuous cultures by Clostridium thermocellum. Appl. Environ. Microbiol. 55:3131-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynd, L. R., and Y. H. Zhang. 2002. Quantitative determination of cellulase concentration as distinct from cell concentration in studies of microbial cellulose utilization: analytical framework and methodological approach. Biotechnol. Bioeng. 77:467-475. [DOI] [PubMed] [Google Scholar]
- 36.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 37.Margolles-Clark, E., M. Ilmen, and M. Penttila. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 38.Mayer, F., M. P. Coughlan, Y. Mori, and L. G. Ljungdahl. 1987. Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53:2785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messner, R., and C. P. Kubicek. 1991. Carbon source control of cellobiohydrolase I and II formation by Trichoderma reesei. Appl. Environ. Microbiol. 57:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra, S., P. Beguin, and J.-P. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanases genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analysis. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 42.Ni, X., and J. Westpheling. 1997. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc. Natl. Acad. Sci. USA 94:13116-13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogueira, T., and M. Springer. 2000. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 3:154-158. [DOI] [PubMed] [Google Scholar]
- 44.Novick, A., and L. Szilard. 1950. Description of the chemostat. Science 112:715-716. [DOI] [PubMed] [Google Scholar]
- 45.Saier, M. H., Jr., S. Chauvaux, G. M. Cook, J. Deutscher, I. T. Paulsen, J. Reizer, and J.-J. Ye. 1996. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology 142:217-230. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 47.Shafer, M. L., and K. W. King. 1965. Utilization of cellulose oligosaccharides of Cellvibrio gilvus. J. Bacteriol. 89:113-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Biotechnol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 49.Spiridonov, N. A., and D. B. Wilson. 1998. Regulation of biosynthesis of individual cellulases in Thermomonospora fusca. J. Bacteriol. 180:3529-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of celR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 51.Spiridonov, N. A., and D. B. Wilson. 2000. A celR mutation affecting transcription of cellulase genes in Thermobifida fusca. J. Bacteriol. 182:252-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strobel, H. J. 1995. Growth of the thermophilic bacterium Clostridium thermocellum in continuous cultures. Curr. Microbiol. 31:210-214. [Google Scholar]
- 53.Thurston, B., K. A. Dawson, and H. J. Strobel. 1993. Cellobiose versus glucose metabolism by the ruminal bacterium Ruminococcus albus. Appl. Environ. Microbiol. 59:2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner, J. B., and J. S. Lolkema. 2003. Ccp-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woods, D. R., and S. J. Reid. 1995. Regulation of nitrogen metabolism, starch utilization and the β-hbd-adh1 gene cluster in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:299-306. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y. H., and L. R. Lynd. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an ELISA-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 75:219-227. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y.-H. P., and L. R. Lynd. 2004. Kinetics and relative importance of phosphorolytic and hydrolytic cleavage of cellodextrins and cellobiose in cell extracts of Clostridium thermocellum. Appl. Environ. Microbiol. 70:1563-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]