Abstract
To investigate the effect of the autoinducer AI-2 on protein expression in Neisseria meningitidis, a luxS mutant of strain MC58 was grown in the presence and absence of in vitro-produced AI-2, and differential protein expression was assessed by two-dimensional differential gel electrophoresis. N. meningitidis did not show a global response to AI-2 signaling activity.
Many bacteria can regulate gene expression in response to their population density, thereby relying on the synthesis, release, and detection of small signaling molecules called autoinducers (quorum sensing) (for recent reviews, see references 3, 14, and 16). While autoinducers usually differ from species to species and work in a species-specific manner, the signaling molecule AI-2 is synthesized by a wide variety of different bacteria (11). Its synthesis requires the luxS gene product, which has been shown to convert the metabolite S-ribosylhomocysteine into the AI-2 precursor 4,5-dihydroxy-2,3-pentanedione (DPD) and l-homocysteine (7, 12, 15). DPD is a highly reactive molecule that is not expected to be stable in solution (12). In Vibrio harveyi, DPD clearly undergoes further reactions as the structure of AI-2 has been determined to be a furanosyl borate diester (4).
The role of AI-2-mediated quorum sensing in bacterial interactions is being studied intensively, and LuxS and AI-2 have been shown to be implicated in the regulation of virulence factors in different bacterial species (22). The presence of a functional luxS gene was shown to be necessary for full meningococcal virulence in an infant rat model (21). In the work presented here, we attempted to identify the cellular targets in Neisseria meningitidis that are controlled by AI-2 in order to elucidate the possible role of quorum sensing in the regulation of meningococcal virulence factors. The response of a non-AI-2-producing N. meningitidis MC58 luxS mutant to in vitro-produced AI-2 was analyzed by proteomics, using two-dimensional differential gel electrophoresis (2D DIGE). The DIGE approach is based on the differential fluorescent dye labeling of proteins derived from different cultures. Equal amounts of labeled protein are mixed and separated by a single 2D gel electrophoresis (19). The resulting 2D gel is submitted to fluorescence emission analysis using the wavelength of each dye, which permits the quantification of the abundance of each protein. Since differentially labeled proteins of the same type will comigrate, their abundances (spot volumes) can be easily compared and their differential protein expression levels can be quantified (spot volume ratios).
While the experiments for this study were ongoing, results of a DNA array analysis demonstrating that AI-2 had only a marginal impact on gene expression in an N. meningitidis serogroup A luxS mutant were published (6).
Mutant strain MC58 luxS does not produce AI-2.
The luxS mutant construction and AI-2 target screening were carried out with the serogroup B strain MC58. It has been demonstrated that this strain produces AI-2 in a luxS-dependent manner (21). Its genome has been entirely sequenced, and 2,158 coding regions were predicted (17). The meningococcal gene luxS (NMB1981) was inactivated by insertion of the ermAM cassette from vector pMGC10 (10) into the natural AgeI site of luxS, previously cloned into vector pUC19. The resulting vector pM2110 was linearized and transformed into N. meningitidis MC58, and the resistance marker was introduced into the chromosomal luxS locus by homologous recombination, which was demonstrated by analytical PCR (data not shown).
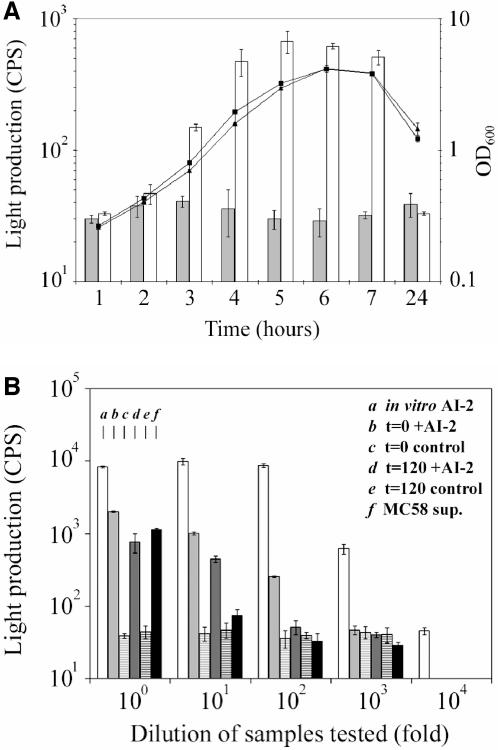
Strains MC58 and MC58 luxS were grown in liquid Catlin MC.2 minimal medium (8), and their growth kinetics were similar (Fig. 1A). AI-2 activities in filtered supernatants were measured during the growth of both strains by determining the bioluminescence response of the V. harveyi reporter strain BB170 to AI-2 (2). AI-2 activity was observed in MC58 supernatants stemming from exponential and stationary growth phases, and no AI-2 activity was detected for MC58 luxS (Fig. 1A).
FIG. 1.
(A) Growth of N. meningitidis MC58 (squares) and MC58 luxS (triangles). The kinetics of AI-2 production of MC58 (white bars) and MC58 luxS (grey bars) was measured through the bioluminescence response of V. harveyi BB170 to AI-2 activity present in cell-free supernatants from the N. meningitidis cultures (light production expressed in counts per second). The response to supernatants from MC58 luxS remained at background level. (B) AI-2 activities present in serial dilutions of in vitro-produced AI-2 (bars a) and cell-free supernatants of a MC58 luxS stationary-phase culture after addition of in vitro-produced AI-2 or control buffer (time [t] = 0; bars b and c) and further growth (t = 120 min; bars d and e) were measured. Finally, AI-2 activity measured in the supernatant of a stationary-phase culture (5 h) of wild-type MC58 is shown (bars f). Bars for each dilution are a, b, c, d, e, and f (left to right).
Preparation of in vitro-produced AI-2.
AI-2 was produced in vitro from S-adenosylhomocysteine as described previously (12). For these purposes, the meningococcal genes pfs (NMB0767) and luxS (NMB1981) were cloned into the pET-28a(+) expression vector (Novagen). The recombinant N-terminal His tag fusion proteins were overexpressed and purified using HiTrap chelating high-performance columns (Amersham). Pfs and LuxS were then used to completely convert 2 mM S-adenosylhomocysteine into the reaction products adenine, l-homocysteine, and DPD. The filtered reaction products were tested for AI-2 activity in the V. harveyi BB170 bioassay (2); serial dilutions of the filtrates were analyzed to take into account the maximization of the assay response at high concentrations of AI-2. The concentration of AI-2 activity present in the in vitro sample (Fig. 1B, bars a) was found to be more than 100-fold higher than the AI-2 activity measured in the supernatants of stationary-phase cultures of the MC58 wild-type strain (Fig. 1B, bars f).
Determination of AI-2 signaling conditions for the MC58 luxS mutant after growth with in vitro-produced AI-2.
Since the incubation time in the presence or absence of AI-2 that would result in the highest number of AI-2-regulated targets was not known, a time course experiment was carried out. An MC58 luxS culture was grown to an optical density at 600 nm of ∼2.5 (beginning of stationary phase) and subsequently split in two: to one-half, in vitro-produced AI-2 was added to a concentration of 10% (vol/vol), and, likewise, in vitro-produced AI-2 reaction buffer (10 mM sodium phosphate buffer, pH 7.5) was added to the other half, which is referred to as the control. Samples were taken from both cultures after 0, 30, 60, and 120 min of further growth for the determination of AI-2 activities and/or preparation of protein extracts. Serial dilutions of the filtered supernatants were tested for AI-2 activity in the V. harveyi BB170 bioassay (Fig. 1B, bars b to e). The activity of the added in vitro-produced AI-2 degraded over time, but after 120 min of growth (Fig. 1B, bars d), it was still greater than the AI-2 activity measured in the supernatants of stationary-phase cultures of the wild-type strain in the same growth medium (Fig. 1B, bars f), demonstrating a good simulation of AI-2 signaling conditions in the experiment.
2D DIGE analysis to identify AI-2-regulated targets in MC58 luxS.
The preparation of bacterial protein extracts, dye labeling, and sample preparation for 2D separation were carried out as described previously (1, 18, 23). Fifty-microgram protein samples collected after the different incubation times from MC58 luxS cultured in the presence or absence of in vitro-produced AI-2 were labeled with the cyanine dyes Cy5 and Cy2 (Amersham), respectively, and two 2D gels per time point were prepared (1, 18). The DIA (differential in-gel analysis) module of the differential analysis software DeCyder (Amersham) was used to calculate protein spot volumes and the normalized volume ratio for each differentially labeled comigrated protein. The values for 2 standard deviations (SD) of the mean volume ratios were between 1.82 and 1.93, and only spots with greater-than-twofold changes in volume in both gels per time point were defined as altered. After incubation of MC58 luxS with or without in vitro-produced AI-2 for 30, 60, and 120 min, only 1, 6, and 10 proteins, respectively, were identified as differentially regulated (Table 1). As the number of identified potential targets of AI-2 was relatively low, we had to consider the possibility that not only the AI-2 signaling activity but also the other two compounds produced in the in vitro AI-2 synthesis reaction (l-homocysteine and adenine) might have contributed to the observed result, since they were not present in the control.
TABLE 1.
Differentially expressed proteins (≥2-fold difference) in N. meningitidis MC58 luxS after various incubation times with or without in vitro-produced AI-2
| Identified protein | Cy5/Cy2 mean ratioa
|
||
|---|---|---|---|
| 30 min | 60 min | 120 min | |
| P1 | +2.55 | +4.03 | +4.08 |
| P2 | +2.39 | +2.47 | |
| P3 | +2.14 | ||
| P4 | −2.77 | −15.77 | |
| P5 | −2.82 | −3.06 | |
| P6 | −10.48 | −9.08 | |
| P7 | +2.66 | ||
| P8 | +2.20 | ||
| P9 | −3.17 | ||
| P10 | −3.25 | ||
| P11 | −4.71 | ||
Cy5 staining used with protein sample from MC58 luxS incubated with in vitro-produced AI-2; Cy2 staining used with protein sample from MC58 luxS incubated with control buffer (phosphate buffer).
2D DIGE analysis to identify AI-2-regulated targets in MC58 luxS under optimized conditions.
The above-described growth experiment was repeated with an optimized control buffer containing 2 mM adenine and 2 mM dl-homocysteine. We chose the incubation time of 120 min to identify a maximum number of AI-2-regulated targets. The experimental design for the subsequent 2D DIGE analysis was also optimized: four analytical 2D gels were prepared as described above and included Cy3-labeled, combined protein samples (25 μg from MC58 luxS with in vitro-produced AI-2 and 25 μg from MC58 luxS with the optimized control buffer) as internal standards for intergel comparison. For two additional 2D gels, 50-μg amounts of combined protein samples were labeled with Cy2, Cy3, or Cy5 in order to determine the degree of fluctuation in the spot volume ratios of image pairs from identical protein samples (control gels). Finally, SYPRO Ruby-stained 2D gels were prepared using 500 μg of protein samples to provide sufficient material for subsequent spot picking and protein identification by mass spectrometry.
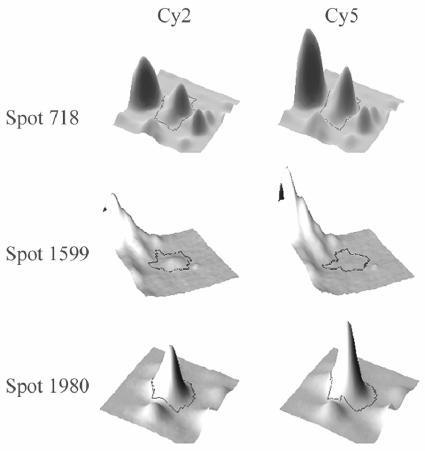
Cy3-Cy2 and Cy3-Cy5 image pairs of the two control gels were created with the DIA mode of DeCyder. An average of 1,300 included spots per image pair resulted in histograms of normally distributed volume ratios (spot frequency/spot volume ratios) with an average 2 SD corresponding to a volume ratio of ±1.3. This ratio was taken as a threshold value above and below which spot volume changes observed in the analytical gels were considered significant. Standardized abundances of all matched protein spots were compared across the four analytical gels, and Student's t test was performed with the DeCyder BVA (biological variation analysis) module to validate the significance of the detected differences between spot volumes from MC58 luxS with in vitro-produced AI-2 and those from MC58 luxS with control buffer (t test P values set at ≤0.01). Surprisingly, only three spots could be proposed to correspond to differentially expressed proteins (Fig. 2). Two proteins were upregulated in MC58 luxS in the presence of AI-2, namely, those corresponding to spots 718 (+1.42-fold, P = 2.2e-006) and 1980 (+1.4-fold, P = 2.3e-006), while one protein was downregulated (that corresponding to spot 1599; −2.23-fold, P = 9.9e-007). It must be noted that spots 718, 1599, and 1980 are identical to three spots that had been identified in the initial DIGE experiment described above (P8, P6, and P1, respectively [Table 1]), where higher differences in their volume ratios had been demonstrated. Since the two experiments differed only in the compositions of the control buffers, the decrease in identified targets from 10 to 3, as well as the smaller volume ratios of the three proteins, appears to be a consequence of the addition of adenine and homocysteine to the control sample in the second experiment. In any case, the very limited number of proteins with altered expression and the very low magnitude of change demonstrate that AI-2 has only a marginal effect, if any, on protein expression in N. meningitidis. Further experiments were carried out to identify the three affected proteins.
FIG. 2.
Three-dimensional simulation of the identified, differentially expressed proteins (by use of DeCyder). Spot 718 and spot 1980 were 1.4-fold upregulated in N. meningitidis MC58 luxS incubated with in vitro-produced AI-2 (Cy5 labeled), whereas spot 1599 showed 2.2-fold-higher expression in MC58 luxS incubated with optimized control buffer (Cy2 labeled).
Analysis of the three differentially regulated proteins by matrix-assisted laser desorption ionization-time of flight (mass spectrometry).
The three selected proteins were excised from the gels and analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry). The mass spectra obtained from spots 718 and 1980 were of good quality and allowed the identification of the corresponding polypeptides, while the spectra obtained from spot 1599 contained very few peaks of weak intensity, which did not allow protein identification. The complete MSDB database was searched using Mascot (Matrix Science). The proteins corresponding to spots 718 and 1980 were identified as 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (MetE) of Neisseria meningitidis MC58 (NMB0944), and 5,10-methylenetetrahydrofolate reductase (MetF) of N. meningitidis MC58 (NMB0943), respectively. The probability-based Mowse scores for the two proteins from Neisseria were ≥139, in contrast to values of ≤63 for other hits (threshold of relevance, 73). Sequence coverage with NMB0944 and NMB0943 was 32 and 39%, respectively, and the theoretical molecular weights and pIs of the proteins corresponded well with the locations of the identified spots on the gels.
The two identified enzymes act closely together in methionine biosynthesis and are located upstream of LuxS in the metabolic pathway that leads to AI-2 synthesis and the release of l-homocysteine (9, 12). MetF catalyzes the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, and MetE transfers a methyl group from 5-methyltetrahydrofolate to l-homocysteine, generating l-methionine. It has been shown that homocysteine is required for full stimulation of metE expression in Escherichia coli and Salmonella enterica serovar Typhimurium (20). The identity of the regulated targets confirms the hypothesis stated above, and it seems very likely that the observed overexpression of MetE and MetF in the AI-2-treated cells is due to the action of l-homocysteine and not to AI-2. It is important to note that 2 mM l-homocysteine was present in the AI-2 sample as a by-product whereas the control sample contained 2 mM of the dl-racemate of homocysteine. It is thus plausible that the effects observed in the second experiment can be attributed entirely to this difference rather than to the action of AI-2. The optimized control might also be the reason why the expression of the other proteins identified in the first experiment was not altered in the second experiment.
As stated above, ∼1,300 protein spots were obtained on the gel images after Cy dye labeling. However, more than 2,100 coding regions have been predicted for the genome of N. meningitidis MC58 (17). Therefore, the possibility that AI-2 regulates targets that have not been visualized by 2D electrophoresis cannot be excluded. Our data suggest that a global cellular response to AI-2 signaling such as that found in E. coli (5, 13) does not take place in N. meningitidis, which is in agreement with the study using a DNA array by Dove et al. that indicates the lack of a concerted response of N. meningitidis to AI-2 on the transcriptional level (6).
Acknowledgments
We thank B. Bassler (Princeton University) for V. harveyi BB170 and L. Aujame, M. Mignon, Y. Berard, T. Krell (Aventis Pasteur, Marcy l'Etoile), S. Pellerin, M. Courcol, and J. F. Leonard (Aventis Pharma, Vitry-sur-Seine, France) for helpful discussions and technical assistance. We thank T. Krell for his contribution to the manuscript.
This work was supported by Aventis Pasteur and a Marie Curie Industry Host Fellowship.
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camara, M., P. Williams, and A. Hardman. 2002. Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect. Dis. 2:667-676. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 5.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 149:1859-1869. [DOI] [PubMed] [Google Scholar]
- 7.Duerre, J. A., and R. D. Walker. 1977. Metabolism of adenosylhomocysteine, p. 43-57. In F. Salvatore, E. Borek, V. Zappia, H. G. Williams-Ashman, and F. Schenk (ed.), The biochemistry of adenosylmethionine. Columbia University Press, New York, N.Y.
- 8.Fu, J., F. J. Bailey, J. J. King, C. B. Parker, R. S. Robinett, D. G. Kolodin, H. A. George, and W. K. Herber. 1995. Recent advances in the large scale fermentation of Neisseria meningitidis group B for the production of an outer membrane protein complex. Bio/Technology 13:170-174. [DOI] [PubMed] [Google Scholar]
- 9.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 10.Nassif, X., D. Puaoi, and M. So. 1991. Transposition of Tn1545-Δ3 in the pathogenic neisseriae: a genetic tool for mutagenesis. J. Bacteriol. 173:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 12.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 13.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturme, M. H., M. Kleerebezem, J. Nakayama, A. D. Akkermans, E. E. Vaugha, and W. M. de Vos. 2002. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Leeuwenhoek 81:233-243. [DOI] [PubMed] [Google Scholar]
- 15.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 18.Tonge, R., J. Shaw, B. Middleton, R. Rowlinson, S. Rayner, J. Young, F. Pognan, E. Hawkins, I. Currie, and M. Davison. 2001. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1:377-396. [DOI] [PubMed] [Google Scholar]
- 19.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071-2077. [DOI] [PubMed] [Google Scholar]
- 20.Urbanowski, M. L., and G. V. Stauffer. 1989. Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winzer, K., Y. Y. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 23.Yan, J. X., A. T. Devenish, R. Wait, T. Stone, S. Lewis, and S. Fowler. 2002. Fluorescence two-dimensional difference gel electrophoresis and mass spectrometry based proteomic analysis of Escherichia coli. Proteomics 2:1682-1698. [DOI] [PubMed] [Google Scholar]