Summary
Many animal tissues/cells are photosensitive, yet only two types of photoreceptors (opsins and cryptochromes) have been discovered in metazoans. The question arises as to whether unknown types of photoreceptors exist in the animal kingdom. LITE-1, a seven-transmembrane gustatory receptor (GR) homolog, mediates UV light-induced avoidance behavior in C. elegans. However, whether LITE-1 functions as a chemoreceptor or photoreceptor has not been determined. Here, we show that LITE-1 directly absorbs both UVA and UVB light with an extinction coefficient 10–100 times that of opsins and cryptochromes, indicating that LITE-1 is highly efficient in capturing photons. Unlike typical photoreceptor proteins employing a prosthetic chromophore to capture photons, LITE-1 strictly depends on its protein conformation for photon absorption. We further identified two tryptophan residues critical for LITE-1 function. Interestingly, unlike GPCRs, LITE-1 adopts a reversed membrane topology. Thus, LITE-1, a taste receptor homolog, represents a distinct type of photoreceptor in the animal kingdom.
Keywords: Photosensation, Chemosensation, Neuron, Photopigment, Photosensory, Chemosensory
Graphical abstract
Introduction
Light sensation is critical for all phyla of life, ranging from bacteria to humans (Wang and Montell, 2007; Yau and Hardie, 2009). Organisms have evolved various types of photoreceptor proteins (hereinafter referred to as photoreceptors) to detect light (Falciatore and Bowler, 2005; Wang and Montell, 2007; Yau and Hardie, 2009). These photoreceptors show different spectral properties with some sensing blue and others detecting green and red, covering a wide spectrum of light (Falciatore and Bowler, 2005; Wang and Montell, 2007; Yau and Hardie, 2009). Photoreceptors are typically composed of two moieties: a host protein and a prosthetic chromophore (e.g. retinal), the latter of which is responsible for light absorption (Wang and Montell, 2007; Yau and Hardie, 2009). In addition to image-forming photoreceptor cells in the retina, a growing list of non-image-forming photosensitive cells/tissues has been identified in a wide range of animal species (Wang and Montell, 2007; Yau and Hardie, 2009). For example, a sub-set of ganglion and horizontal cells in the vertebrate retina are photosensitive (Yau and Hardie, 2009). Photosensitive cells are also found in the skin (e.g. keratinocytes and melanocytes) of mammals, the pupil of most vertebrates, the pineal of non-mammalian vertebrates, the hypothalamus of birds, and the body surface of insects (Bellono et al., 2013; Foster and Soni, 1998; Moore et al., 2013; Xiang et al., 2010; Yau and Hardie, 2009). However, in contrast to microbes and plants which express many types of photoreceptors, only two such groups of proteins have been identified in the animal kingdom: opsins and cryptochromes (Wang and Montell, 2007; Yau and Hardie, 2009). The question thus arises as to whether unknown types of photoreceptors exist in metazoans.
The nematode C. elegans detects and responds to a wide variety of sensory cues such as mechanical forces (e.g. touch and stretch), chemicals (e.g. odorants and tastants), and temperature, representing a popular genetic model organism for the study of sensory perception (de Bono and Maricq, 2005). Despite the lack of eyes, C. elegans also responds to light (Edwards et al., 2008; Ward et al., 2008). Specifically, short wavelengths of light, particularly UV light, induce avoidance behavior (negative phototaxis) in C. elegans, which is mediated by a group of photosensory neurons, providing a protective mechanism for the worm to avoid lethal doses of UV in the sunlight (Liu et al., 2010; Ward et al., 2008). LITE-1, a member of the invertebrate seven-transmembrane (7-TM) gustatory receptor (GR) family, is required for UV light-induced avoidance behavior (Edwards et al., 2008; Liu et al., 2010). Ectopic expression of LITE-1 can confer photo-sensitivity to photo-insensitive cells (Edwards et al., 2008; Liu et al., 2010). Despite such indirect evidence suggesting LITE-1 as a candidate photoreceptor, other possibilities remain. For example, unlike long wavelengths of light, UV illumination produces reactive-oxygen-species (ROS) such as H2O2, which in turn can evoke an avoidance behavioral response similar to that induced by UV light (Bhatla and Horvitz, 2015). Given that LITE-1 is a member of the gustatory receptor (GR) family, it has thus been suggested that LITE-1 may function as a chemoreceptor (Yau and Hardie, 2009). In this case, LITE-1 would sense light-produced chemicals but not light per se.
To address this conundrum, here we purified LITE-1 protein from worm lysate and found that it directly absorbs UVA and UVB light. This property of LITE-1, together with its capacity in producing light-evoked functional outputs in vivo, indicates that LITE-1 is a photoreceptor. LITE-1 bears a number of unique features that distinguish it from other photoreceptors. These include an exceptionally high efficiency in photoabsorption, an ability to sense both UVA and UVB light, a strict dependence on conformation for photoabsorption, a strong resistance to bleaching by UV light, and a reversed membrane topology compared to opsins. These results identify LITE-1, a taste receptor homolog, as a unique photoreceptor with features not seen in any known photoreceptors. Thus, novel types of photoreceptors are present in the animal kingdom. Furthermore, we identified two tryptophan residues in LITE-1 that are critical for photoabsorption. Remarkably, introducing such a tryptophan residue into another GR family member promotes photosensitivity, opening up the intriguing prospect that it might be possible to genetically engineer new photoreceptors.
Results
LITE-1 adopts a membrane topology opposite to conventional 7-TM receptors
As a first step, we considered whether LITE-1 is related to any known photoreceptors. LITE-1 is predicted to contain 7-TM domains (Figure 1A). The only known 7-TM photoreceptors in metazoans are opsins, but LITE-1 has no significant homology with opsins at the sequence level (Edwards et al., 2008; Liu et al., 2010). As both insect OR (olfactory receptors) and GR (gustatory receptors) members were shown to possess a membrane topology opposite to conventional 7-TM receptors (Benton et al., 2006; Zhang et al., 2011), we thus questioned whether LITE-1 and opsins are even related at the membrane topology level. To probe the membrane topology of LITE-1, we raised antibodies against the N- and C-termini of LITE-1 (Figure 1A). Immunostaining with these antibodies did not reveal consistent LITE-1 expression in worm tissues (A.W. and X.Z.S.X. unpub.), suggesting that LITE-1 is expressed at a very low level in vivo. We therefore generated transgenic animals expressing LITE-1 in muscle cells using a muscle-specific promoter, as LITE-1 can be functionally expressed in these cells at a higher level, though it remains possible that recombinant LITE-1 may not fully preserve all the functional properties of native proteins (Edwards et al., 2008; Liu et al., 2010) (also see below). We found that our LITE-1 antibodies can detect LITE proteins in primary cultured muscle cells (Figure 1B). Surprisingly, the C-terminal end of LITE-1 appears to be extracellular, as antibodies against LITE-1’s C-terminus can detect LITE-1 when applied extracellularly under non-permeabilizing conditions (Figure 1B). This staining is specific for LITE-1 since no signal was observed in control muscle cells (Figure S1). By contrast, the same protocol failed to detect LITE-1 with antibodies against its N-terminal end, though the protein was clearly expressed in these cells as shown under permeabilizing conditions (Figure 1B). To provide additional evidence, we fused a Myc tag to the N-terminal end of LITE-1 and obtained the same result (Figure 1B). This suggests that the N-terminal end of LITE-1 is intracellular.
Figure 1. LITE-1 adopts an unusual membrane topology with its C-terminus facing extracellularly and N-terminus located intracellularly.
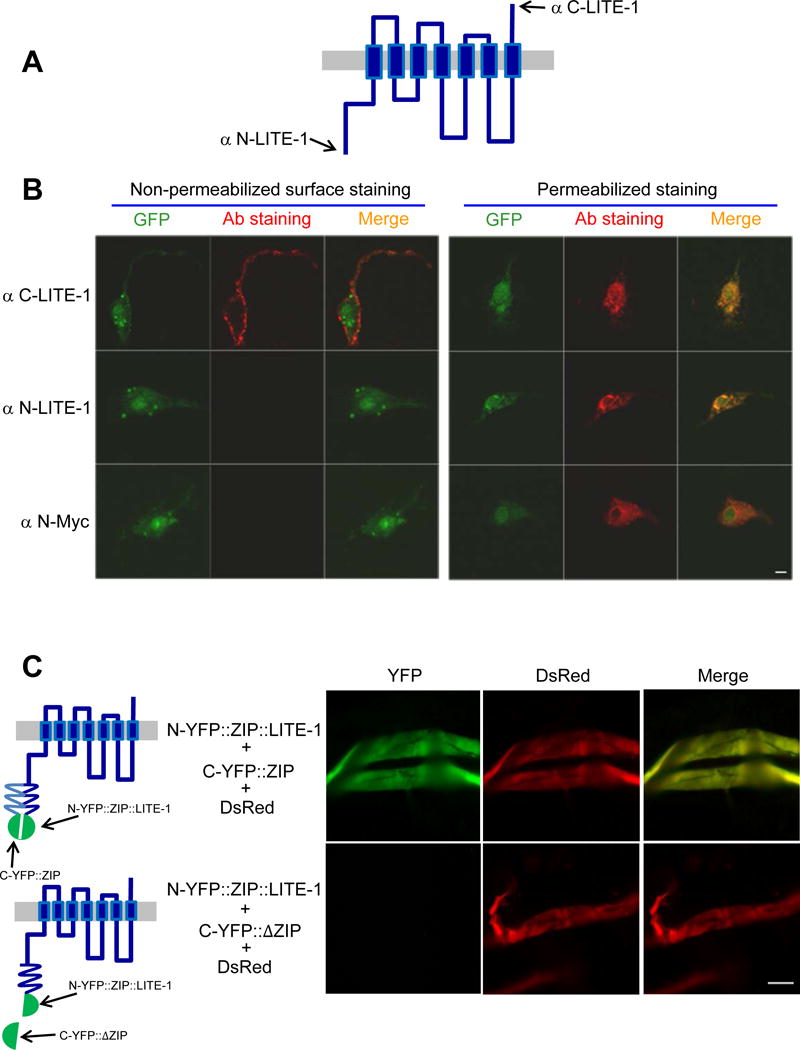
(A) A schematic of LITE-1 membrane topology. Antibodies were raised against the N-terminal (αN-LITE-1) and C-terminal (αC-LITE-1) peptide (15 aa) of the LITE-1 long isoform.
(B) LITE-1 displays a distinct membrane topology with its C-terminus facing extracelluarly and it N-terminus located in the cytoplasm. Shown are confocal images from immunofluorescence staining. LITE-1 (long form) was co-expressed with GFP as a transgene in muscles under the myo-3 promoter. Staining was performed on primary cultured cells under non-permeabilizing conditions for surface staining or under permeablizing conditions to stain the entire cell. αN-LITE-1 and αC-LITE-1 detects the N- and C-terminal end of LITE-1, respectively. αN-Myc stains the Myc tag fused to the N-terminal end of LITE-1. See Figure S1 for controls. Scale bar: 2 μm.
(C) BiFC images showing that the N-terminus of LITE-1 is located in the cytoplasm. Shown on the left are schematics describing the design of the BiFC approach. Shown on the right are fluorescence images. N-YFP∷ZIP∷LITE-1 was expressed as a transgene in muscles using the myo-3 promoter. C-YFP∷ZIP (or C-YFP∷ΔZIP that lacks a zip domain) and DsRed were co-expressed as a separate transgene in muscles using the same promoter. Two transgenes were crossed together to examine reconstitution of YFP fluorescence in muscles. Only if the N-terminus of LITE-1 is located intracellularly would one be able to detect YFP fluorescence. Muscles were acutely dissected out from transgenic worms using a protocol described previously (Liu et al., 2013). Scale bar: 100 μm.
Also see Figure S1
To collect further evidence, we employed the BiFC (Bimolecular Fluorescence Complementation) approach (Hu et al., 2002). In this approach, the N- and C-terminal fragment of YFP is fused to a leucine zipper domain to generate N-YFP∷ZIP and C-YFP∷ZIP, respectively (Figure 1C). The zipper domains then bring the two YFP fragments together to reconstitute a fluorescent YFP protein (Figure 1C). We attached N-YFP∷ZIP to the N-terminus of LITE-1, and found that this N-YFP∷ZIP∷LITE-1 fusion complemented with C-YFP∷ZIP to reconstitute YFP fluorescence in live muscle cells acutely dissected from the animal, but not with C-YFP∷ΔZIP that lacked the zipper domain (Figure 1C). This observation further demonstrates that the N-terminus of LITE-1 is located intracellularly. We conclude that LITE-1 adopts a reversed membrane topology compared to opsins. Thus, LITE-1 does not seem to be closely related to any known photoreceptors at the sequence or structural levels.
Purification of LITE-1 protein from worm lysate
Is LITE-1 a photoreceptor? A lack of clear similarity to known photoreceptors does not necessarily disqualify LITE-1 as a photoreceptor. To address this question, a simple, yet definitive approach is to examine whether purified LITE-1 protein can capture photons by spectrophotometry (Wang and Montell, 2007; Yau and Hardie, 2009). All known photoreceptors were verified by this approach (Figure 3C). To this end, we searched for an expression system that would allow us to purify a sufficient amount of LITE-1 protein for spectrophotometric analysis. Muscle cells thus came to our attention, as they constitute a major mass of worm tissues and have been successfully utilized as a heterologous system to functionally express receptors and channels (Salom et al., 2012; Wang et al., 2012). Importantly, it has been shown that LITE-1 can be functionally expressed in muscles, as its expression can confer photo-sensitivity to these otherwise photo-insensitive cells (Edwards et al., 2008; Liu et al., 2010), though it remains unclear whether such photosensitivity results from light or light-produced chemicals. Indeed, as previously reported (Edwards et al., 2008; Liu et al., 2010), UV light can induce the contraction of body-wall muscles ectopically expressing LITE-1, leading to body paralysis (Figure 2A, Figure S2, and Suppl. Video 1 & 2). To provide more direct and quantitative evidence, we recorded the response of muscle cells to UV light by calcium imaging using the genetically-encoded calcium sensor RCaMP. We found that UV illumination induced robust calcium transients in muscle cells ectopically expressing LITE-1, but not in control muscle cells (Figure 2B–D). These experiments show that LITE-1 was functionally expressed in muscle cells. They also show that LITE-1 can indeed confer photo-sensitivity to photo-insensitive cells, demonstrating that it can be potentially used as an optogenetic tool.
Figure 3. Photoabsorption by LITE-1 relies on its conformation.
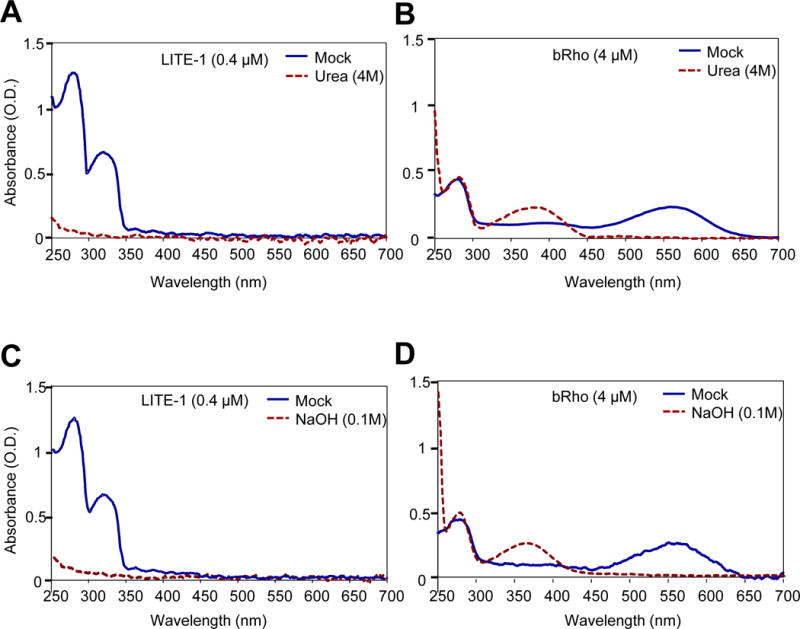
(A) Denaturing LITE-1 with urea completely abolishes its photoabsorption. Shown are spectral data for mock- and urea-treated LITE-1. LITE-1 was treated with urea (4 M) for 5 min at room temperature prior to spectral analysis.
(B) Denaturing bacterial rhodopsin (bRho) with urea does not eliminate its photoabsorption. Urea treatment shifts bRho’s 568 nm absorbance peak to 370 nm. bRho was treated with urea (4 M) for 5 min at room temperature prior to spectral analysis.
(C) Denaturing LITE-1 with NaOH completely abolishes its photoabsorption. LITE-1 was treated with NaOH (0.1 M) for 5 min at room temperature prior to spectral analysis.
(D) Denaturing bacterial rhodopsin (bRho) with NaOH does not eliminate its photoabsorption. NaOH treatment shifts bRho’s 568 nm absorbance peak to 370 nm. bRho was treated with NaOH (0.1 M) for 5 min at room temperature prior to spectral analysis.
LITE-1 concentration: 0.4 μM. bRho concentration: 4 μM.
Also see Figure S4.
Figure 2. LITE-1 absorbs UVA and UVB light, and ectopic expression of LITE-1 confers photo-sensitivity to photo-insensitive cells.
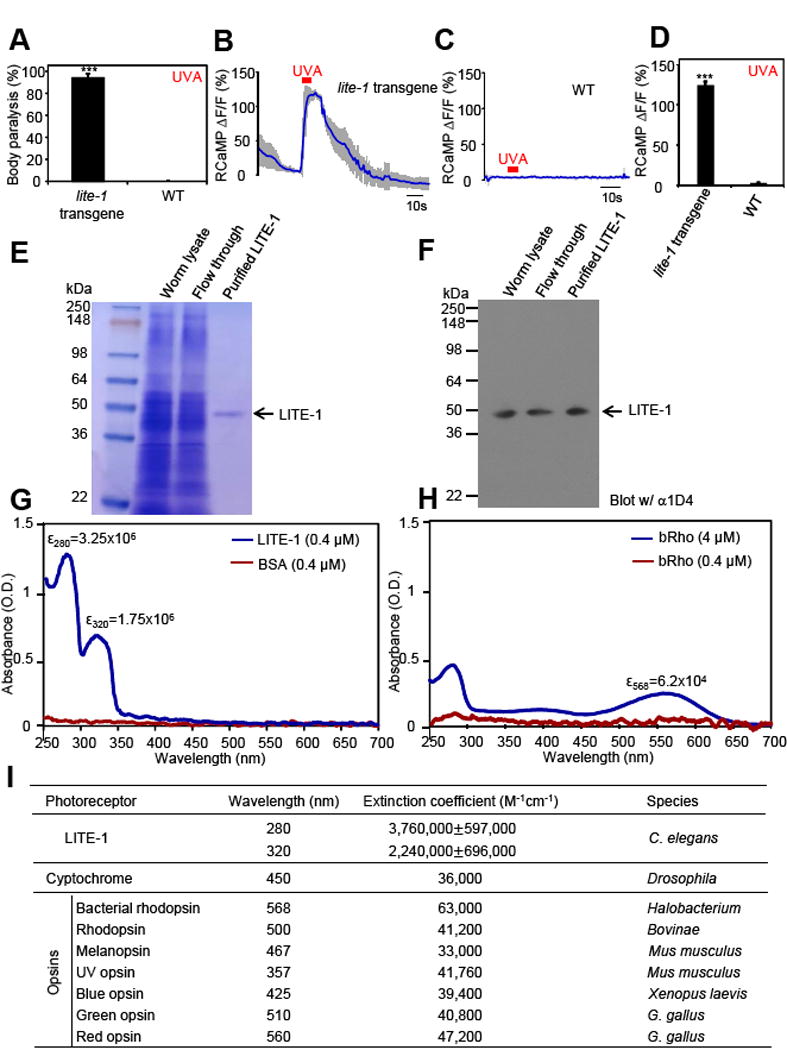
(A) Transgenic expression of LITE-1 in muscle cells confers photosensitivity shown by behavioral assays. LITE-1 was expressed as a transgene in muscle cells under the myo-3 promoter. WT (wild-type) and LITE-1 transgenic worms were exposed to a 20 sec pulse of UVA light (350±20 nm, 0.8 mW/mm2). Animals showing muscle contraction-induced paralysis during light illumination were scored positive. n=20. Error bars: SEM. ***p<0.00001 (ANOVA with Bonferroni test).
(B–D) Transgenic expression of LITE-1 in muscle cells confers photosensitivity shown by calcium imaging. RCaMP was expressed as a transgene in muscle cells under the myo-3 promoter. A 5 sec pulse of UVA light (340±20 nm, 0.7 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (B) and (C) represent SEM. (D) Bar graph. n≥7. *p<0.0001 (t test).
(E–F) Purification of LITE-1. Worm lysate, flow through, and purified LITE-1 were loaded. Shown in (E) is an SDS-PAGE gel stained with coomassie blue. Shown in (F) is a Western blot probed with anti-1D4 that recognizes the 1D4 tag attached to the C-terminal end of LITE-1. The amount of each sample loaded in (F) was 1/10 of that in (E). Samples for SDS-PAGE and Western were prepared at room temperature under non-reducing conditions (free of β-ME and DTT) to avoid aggregation of LITE-1.
(G) LITE-1 shows strong absorption of UVA and UVB light while BSA does not. The same concentration of purified LITE-1 and BSA (0.4 μM) was subjected to UV-visible spectrophotometric analysis. The extinction coefficient (ε) for both peaks of LITE-1 was noted. Unit: M−1cm−1. Note: these numbers only represent the LITE-1 sample shown here and those in Figure 3, as they were from the same batch of purification. See (I) for averaged data for LITE-1 from different batches of purification.
(H) Bacterial rhodopsin (bRho) shows much weaker absorption of light compared to LITE-1. The results from low and high concentrations of bRho were shown. bRho was purchased from Sigma.
(I) LITE-1 is far more efficient in photon absorption than cryptochromes and opsins. The extinction coefficients for LITE-1 were averaged from samples from seven independent purifications. “±” represents SEM. The numbers for cryptochromes and opsins were from published literature: cryptochrome (Thompson and Sancar, 2002), bacterial rhodopsin (Oesterhelt and Hess, 1973), rhodopsin (Okano et al., 1992), melanopsin (Matsuyama et al., 2012), UV opsin (Insinna et al., 2012), blue opsin (Vought et al., 1999), green opsin, and red opsin (Kolesnikov et al., 2014).
Also see Figure S2–3 and Supplemental movie 1–2.
Since our LITE-1 antibodies are not suitable for affinity-purification, we then tested a number of monoclonal antibodies against small affinity tags such as Myc, FLAG, and 1D4, and found that 1D4 antibody worked most efficiently. Using this antibody, we were able to affinity-purify LITE-1, a membrane protein, to homogeneity, as determined by SDS-PAGE followed by coomassie staining (Figure 2E), and by Western blot (Figure 2F). This result was also verified by silver staining (not shown).
Purified LITE-1 protein absorbs photons
By subjecting purified LITE-1 protein to spectrophotometric analysis, we found that it exhibited strong absorption of UV light, with two absorbance peaks at 280 nm and 320 nm (Figure 2G). Thus, LITE-1 can capture both UVB and UVA light (WHO definition of UVB: 280–315 nm; UVA: 315–400 nm). As a comparison, at the same concentration (0.4 μM), BSA showed no such absorption (Figure 2G). In addition, bacterial rhodopsin (bRho), which is a commercial product obtained from Sigma Co., exhibited minimal absorption at its signature peak 568 nm (Figure 2H). Only at 10× concentration (4 μM) were we able to detect modest light absorption in bacterial rhodopsin (bRho), which was still much weaker than that found in LITE-1 (Figure 2H). It should be noted that though bRho exhibited weaker photoabsorption compared to LITE-1, its extinction coefficient (62,000 in Figure 2H vs. 63,000 in Figure 2I), as well as its spectral properties, were both in line with those reported in literature (Figure 2H–I), indicating that the quality of bRho samples was reliable. The extinction coefficient of both absorbance peaks of LITE-1 is >106 M−1cm−1, which is 10–100 times that of all known photoreceptors (Figure 2I). Thus, LITE-1 has a high efficiency in capturing photons.
To make a more direct comparison, we purified bovine rhodopsin (Rho) ectopically expressed in worm muscles (Salom et al., 2012), and did so side-by-side with LITE-1 under the same conditions (Figure S3A–B). Compared to LITE-1, purified bovine rhodopsin (Rho) also showed much weaker photoabsorption at its signature peak (Figure S3C–D), providing additional evidence demonstrating that LITE-1 is highly efficient in capturing photons. The relatively weak photoabsorption by bovine rhodopsin (Rho) was not because our purified Rho samples were of low quality, as the extinction coefficient of the purified Rho was in fact very similar to that reported in literature (Figure S3D vs. Figure 2I). In addition, the signature absorbance peak of the purified Rho was 500 nm, which was identical to that published in literature (Figure S3D vs. Figure 2I). This set of control experiments also validated our experimental system, including protein expression, purification, concentration determination, and spectral analysis.
In another control experiment, we purified mammalian adenosine A2A receptor (A2AR) ectopically expressed in worm muscles (Salom et al., 2012) (Figure S3A–B). Like LITE-1 and opsins, A2AR is also a 7-TM receptor but not expected to be photosensitive. Indeed, we found that as predicted, this receptor did not absorb light when purified and tested side-by-side with LITE-1 and bovine Rho (Figure S3C). Thus, multiple control experiments support that LITE-1 absorbs photons and does so at a high efficiency. This property of LITE-1, together with its capacity in producing various light-induced functional outputs [e.g. light-induced muscle contraction and calcium transients, and avoidance behavior (Figure 2A–D, Figure S2, and Suppl. Video 1 &2)], indicates that LITE-1 is a photoreceptor. LITE-1 is also the only photoreceptor that shows strong absorption of both UVA and UVB light.
LITE-1 strictly depends on its conformation for photoabsorption
We next sought to characterize the photoabsorption of LITE-1. A photoreceptor is usually composed of two moieties: a host protein and a prosthetic chromophore (Falciatore and Bowler, 2005; Wang and Montell, 2007; Yau and Hardie, 2009). The spectral properties of a photoreceptor are certainly affected by the host protein. However, the absolute ability of a photoreceptor to absorb light does not rely on the host protein, as light absorption is mediated by the chromophore (e.g. retinal, flavin, bilin, and p-coumaric acid) (Falciatore and Bowler, 2005; Marti et al., 1991; Radding and Wald, 1956). Consequently, denaturing a photoreceptor usually shifts its absorbance peaks to different wavelengths but does not eliminating them, as they are mediated by the associated chromophore (Dutta et al., 2010; Hagins, 1973; Hubbard, 1969; Maglova et al., 1989). This, surprisingly, does not appear to the case for LITE-1. Denaturing LITE-1 with urea completely abolished the light absorption by LITE-1, eliminating both the 280 and 320 nm peaks (Figure 3A). As a comparison, the same urea treatment failed to abolish the light absorption by bacterial rhodopsin (bRho), but instead shifted its absorbance peak from 568 nm to 370 nm (Figure 3B), the latter of which is the signature peak of free retinal, the chromophore of bRho (Sperling and Rafferty, 1969). A similar phenomenon was observed with our purified bovine rhodopsin (Rho) (Figure S3E–F). It is notable that the 280 nm peak of denatured bRho remained unchanged (Figure 3B), consistent with the notion that this peak was mediated by the intrinsic light absorption by tryptophan residues of the bRho protein. This peak was not that distinct in denatured LITE-1 in Figure 3A since the concentration of LITE-1 used was 1/10 that of bRho. We also treated LITE-1 using other denaturing agents such as NaOH, and observed a similar phenomenon (Figure 3C–D). These observations demonstrate that unlike typical photoreceptors, LITE-1 strictly depends on its conformation for photoabsorption.
We also tested H2O2. Interestingly, H2O2 treatment completely abolished LITE-1’s photoabsorption (Figure S4A). As an oxidizing agent, H2O2 can damage the function of proteins, lipids and nucleic acids (Fridovich, 2013). Oxidization of LITE-1 may affect the conformation of LITE-1, which is required for its absorption of light. Similarly, H2O2 treatment also destroyed the spectral fingerprint of bRho by shifting its absorbance peak from 568 nm to 370 nm (Figure S4B). Thus, H2O2 appears to inhibit the photoabsorption of both LITE-1 and bRho in vitro. Nevertheless, as it is difficult to estimate the endogenous concentration of H2O2, whether and how H2O2 affects LITE-1 function in vivo remains to be determined.
Genetic screens identify residues critical for LITE-1 function
To obtain a better understanding of LITE-1 photoabsorption, we attempted to identify residues critical for LITE-1 function. In a genetic screen for mutant animals defective in UV light-induced avoidance behavior, we isolated several lite-1 mutant alleles (Liu et al., 2010). We hypothesized that mutations in transmembrane domains are more likely to affect the photoabsorption of LITE-1 rather than its coupling to downstream signaling molecules. Two mutants, lite-1(xu8) and lite-1(xu10), thus came to our attention, as the residues mutated (A332V and S226F, respectively) reside in putative transmembrane domains (Figure 7I). The objective was to purify these mutant forms of LITE-1 protein and then characterize their photoabsorption in vitro. We first tested their role in vivo, and found that as expected, A332V and S226F mutations disrupted LITE-1 function in vivo. Specifically, worms ectopically expressing LITE-1 harboring either mutation were no longer sensitive to UVA light in behavioral assays (Figure 4A and S5A). In addition, these two point mutations nearly abolished UVA light-evoked calcium transients in muscle cells ectopically expressing LITE-1 (Figure 4B–E). We successfully purified LITE-1A332V and LITE-1S226F proteins to homogeneity (Figure 4F–G). LITE-1A332V and LITE-1S226F displayed an absorbance spectrum distinct from wild-type LITE-1: they both lost the 320 nm peak but retained normal absorption at 280 nm (Figure 5A–B). Thus, the two mutations disrupted LITE-1’s absorption of UVA but not UVB light. This is consistent with the fact that our genetic screen was targeted for isolating mutants defective in responding to UVA but not UVB light, since the optical system of the microscope used to evoke and assay phototaxis behavior did not transmit UVB light (Liu et al., 2010).
Figure 7. Genetic engineering of a photoreceptor by introducing a tryptophan residue into another GR family member GUR-3.
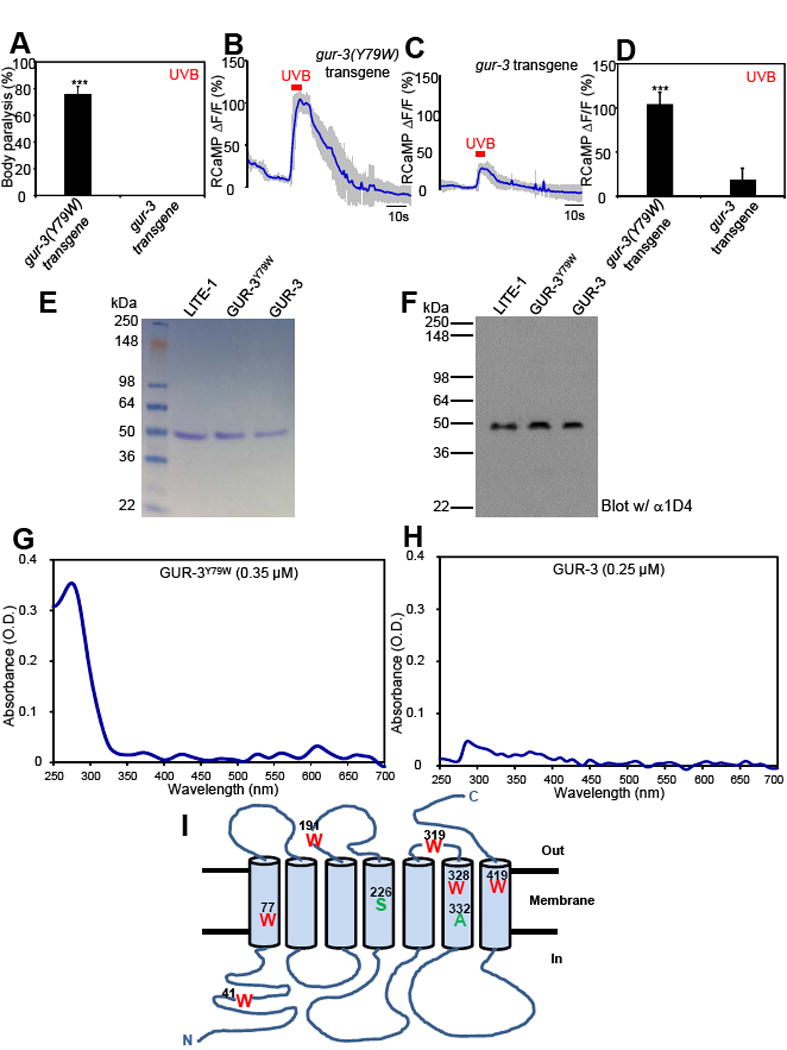
(A) Mutating Y79 to W in GUR-3 promotes photosensitivity in vivo shown by behavioral assays. GUR-3Y79W and GUR-3 were expressed as a transgene in muscle cells. Worms were exposed to a 20 sec pulse of UVB light (280±10 nm, 0.03 mW/mm2), and those showing muscle contraction-induced paralysis during light illumination were scored positive. n=50. Error bars: SEM. ***p<0.00001 (t test).
(B–D) Mutating Y79 to W in GUR-3 promotes photosensitivity in vivo shown by calcium imaging. The experiments were done as described in Figure 2B. A 5 sec pulse of UVB light (280±10 nm, 0.02 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (B–C) represent SEM. (D) Bar graph. n=20. ***p<0.00001 (t test).
(E–F) Purification of GUR-3Y79W and GUR-3. Shown in (E) is an SDS-PAGE gel stained with coomassie blue. Shown in (F) is a Western blot probed with anti-1D4 that recognizes the 1D4 tag attached to the C-terminus of GUR-3Y79W and GUR-3, as well as LITE-1. LITE-1 was purified side-by-side as a reference. As predicted, GUR-3 showed a slightly larger molecular weight than LITE-1. The amount of each sample loaded in (F) was 1/10 of that in (E). Samples for SDS-PAGE and Western were prepared at room temperature under non-reducing conditions (free of β-ME and DTT) to avoid aggregation of LITE-1.
(G–H) Mutating Y79 to W in GUR-3 greatly potentiates the absorption of UVB light (280 nm) in vitro.
(I) A schematic model denoting LITE-1 membrane topology and the position of residues investigated in this study.
Also see Figure S7.
Figure 4. Residues S226 and A332 in LITE-1 are critical for its sensitivity to UVA light in vivo.
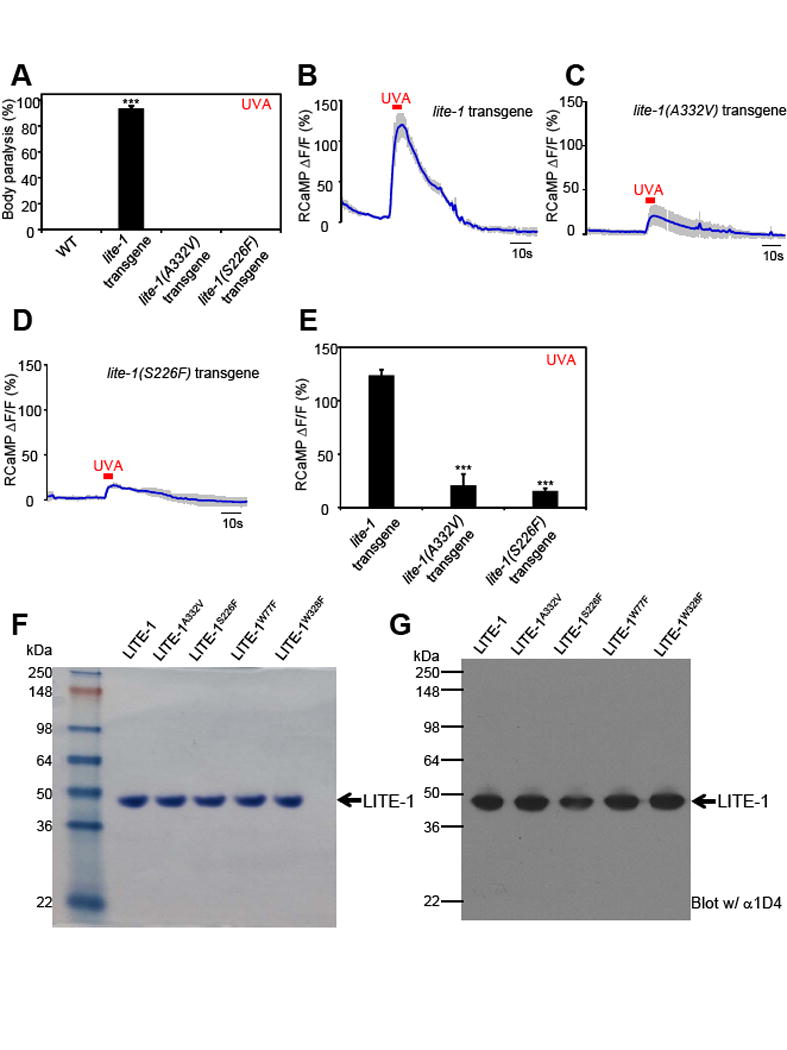
(A) S226F and A332V mutations disrupt the function of LITE-1 in vivo shown by behavioral assays. LITE-1 harboring S226F or A332V was expressed as a transgene in muscles under the myo-3 promoter. WT (wild-type) and transgenic worms were exposed to a 20 sec pulse of UVA light (350±20 nm, 0.8 mW/mm2). Animals showing muscle contraction-induced paralysis during light illumination were scored positive. Some genotypes had all data points as zero, and thus no statistical analysis was performed on them. n=20. Error bars: SEM. ***p<0.00001 (ANOVA with Bonferroni test).
(B–E) S226F and A332V mutations disrupt the function of LITE-1 in vivo shown by calcium imaging. The experiments were done as described in Figure 2B. A 5 sec pulse of UVA light (340±20 nm, 0.7 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (B–D) represent SEM. (E) Bar graph. n≥7. ***p<0.00001 (ANOVA with Bonferroni test).
(F–G) Purification of mutant forms LITE-1. Shown in (F) is an SDS-PAGE gel stained with coomassie blue. Shown in (G) is a Western blot probed with anti-1D4 that recognizes the 1D4 tag attached to the C-terminus of LITE-1 variants. The amount of each sample loaded in (G) was 1/10 of that in (F). Samples for SDS-PAGE and Western were prepared at room temperature under non-reducing conditions (free of β-ME and DTT) to avoid aggregation of LITE-1.
Also see Figure S5.
Figure 5. Residues S226 and A332 in LITE-1 are required for its absorption of UVA but not UVB light in vitro.
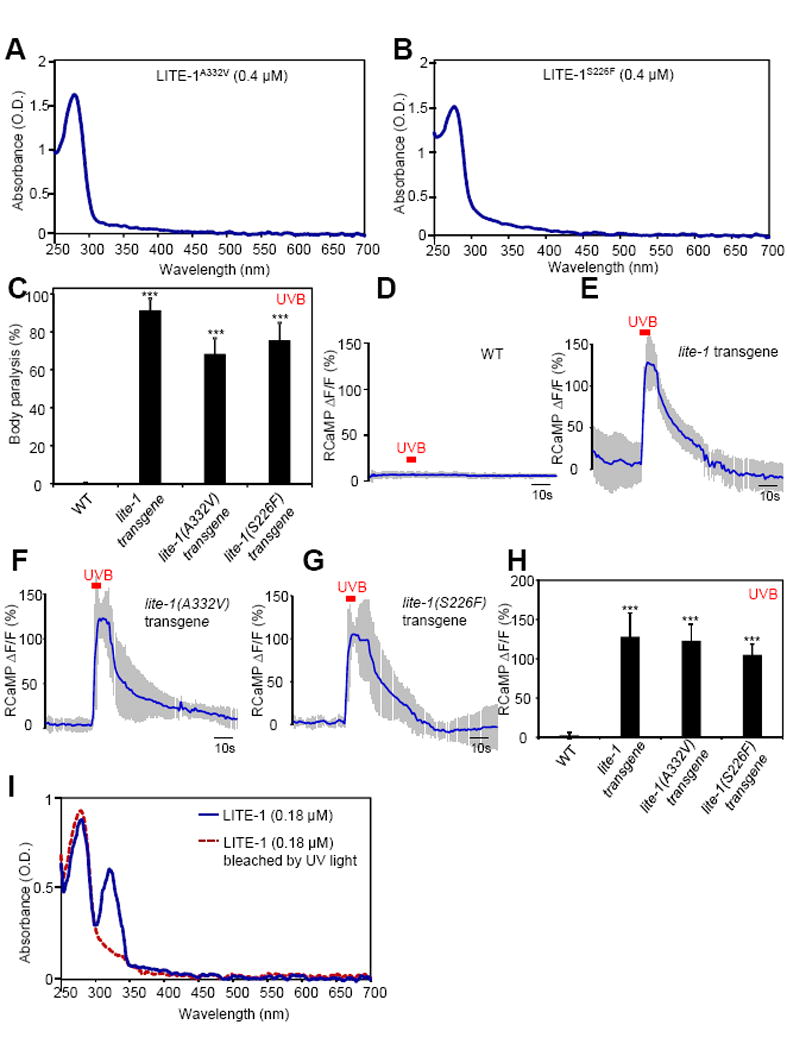
(A–B) S226F and A332V mutations disrupt LITE-1’s absorption of UVA but not UVB light in vitro. The extinction coefficient at 280 nm for LITE-1A332V and LITE-1226F is: 4.0×106 M−1cm−1 and 3.75×106 M−1cm−1, respectively, which are similar to wild-type LITE-1 (Figure 2G).
(C) S226F and A332V mutations do not disrupt the sensitivity of LITE-1 to UVB light in vivo shown by behavioral assays. LITE-1 harboring S226F or A332V was expressed as a transgene in muscle cells under the myo-3 promoter. WT (wild-type) and transgenic worms were exposed to a 20 sec pulse of UVB light (280±10 nm, 0.03 mW/mm2). Animals showing muscle contraction-induced paralysis during light illumination were scored positive. n=20. Error bars: SEM. ***p<0.00001 (ANOVA with Bonferroni test).
(D–H) S226F and A332V mutations do not disrupt the sensitivity of LITE-1 to UVB light in vivo shown by calcium imaging. The experiments were done as described in Figure 2B. A 5 sec pulse of UVB light (280±10 nm, 0.02 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (D–G) represent SEM. (H) Bar graph. n≥10. ***p<0.00001 (ANOVA with Bonferroni test).
(I) LITE-1 absorption of UVB but not UVA light shows resistance to photobleaching. LITE-1 was pre-exposed to UV light for 5 min (17 μW/mm2, 302 nm) at room temperature prior to spectrophotometric analysis. Pre-exposure to UV light for 30 min still did not notably affect the UVB photoabsorption. The photoabsorption at 280 nm was eventually lost after 1 hour of pre-exposure, probably because LITE-1 was denatured. As a direct comparison, bRho, when tested under the same condition, showed photobleaching of its 568 nm peak in less than 5 min of pre-exposure to ambient light, and such photobleaching became complete at 10 min.
Also see Figure S5.
Given that LITE-1A332V and LITE-1S226F proteins retained normal absorption of UVB light in vitro, one would predict that these two mutant forms of LITE-1 shall preserve the sensitivity to UVB light in vivo. To test this idea, we set up an optical path through which UV light was directed to the worm directly. Indeed, though transgenic worms expressing these two mutant forms of LITE-1 were insensitive to UVA light (Figure 4A and S5A), they were nevertheless sensitive to UVB light (Figure 5C and S5B). In addition, as was the case with wild-type LITE-1, UVB light also induced robust calcium transients in muscle cells ectopically expressing these two mutant forms of LITE-1 (Figure 5D–H). These results are in line with the data from spectral analysis (Figure 5A–B). Thus, it appears that the absorption of UVA and UVB light by LITE-1 can be separated, and also provides further evidence demonstrating the specificity of LITE-1 photoabsorption.
LITE-1 absorption of UVB but not UVA light shows resistance to photobleaching
Prolonged light illumination bleaches photoreceptors (Wang and Montell, 2007; Yau and Hardie, 2009). We tested this property of LITE-1, and found that pre-exposure to UV light can readily bleach LITE-1’s ability to absorb UVA light by eliminating its 320 nm peak (Figure 5I). Surprisingly, such treatment spared the 280 nm peak (Figure 5I), indicating that the ability for LITE-1 to capture UVB light was more stable and relatively resistant to photobleaching. This experiment reveals an additional feature that distinguishes LITE-1 absorption of UVA and UVB light.
Two tryptophan residues are required for LITE-1 function
Our success in identifying residues critical for LITE-1’s absorption of UVA light encouraged us to explore what may underlie its absorption of UVB light. Tryptophan residues show intrinsic absorption of UVB light, peaking at 280 nm. It is also known that light absorption by tryptophan is quite resistant to photobleaching (Wu et al., 2008). These two features together led us to question whether tryptophan residues in LITE-1 play a role in mediating its absorption of UVB light. Six tryptophan residues are found in LITE-1 (Figure 7I). However, should any of these tryptophan residues be important for LITE-1 function, they would not be expected to be picked up by our genetic screen, as the mutagen (EMS) used in the screen would typically mutate a tryptophan residue to a stop codon rather than generate a missense mutation.
Therefore, to test the above hypothesis, we mutated each of the six tryptophan residues to alanine through site-directed mutagenesis and expressed the corresponding mutant forms of LITE-1 as a transgene in muscle cells. We first examined their function in vivo. Two tryptophan residues, W77 and W328, when mutated to alanine, abolished the sensitivity of LITE-1 to UVA light in vivo in behavioral assays (Figure 6A and S6A), whereas mutating the other four tryptophan residues did not elicit a notable effect (Figure 6A). We obtained a similar result when mutating W77 and W328 to F (phenylalanine) (Figure 6A). Furthermore, the two tryptophan mutations W77F and W328F nearly eliminated UVA light-induced calcium transients in muscle cells ectopically expressing LITE-1 (Figure 6C–F). These data identify a critical role for W77 and W328 in LITE-1 function in vivo.
Figure 6. The two tryptophan residues W77 and W328 in LITE-1 are required for LITE-1 function both in vivo and in vitro.
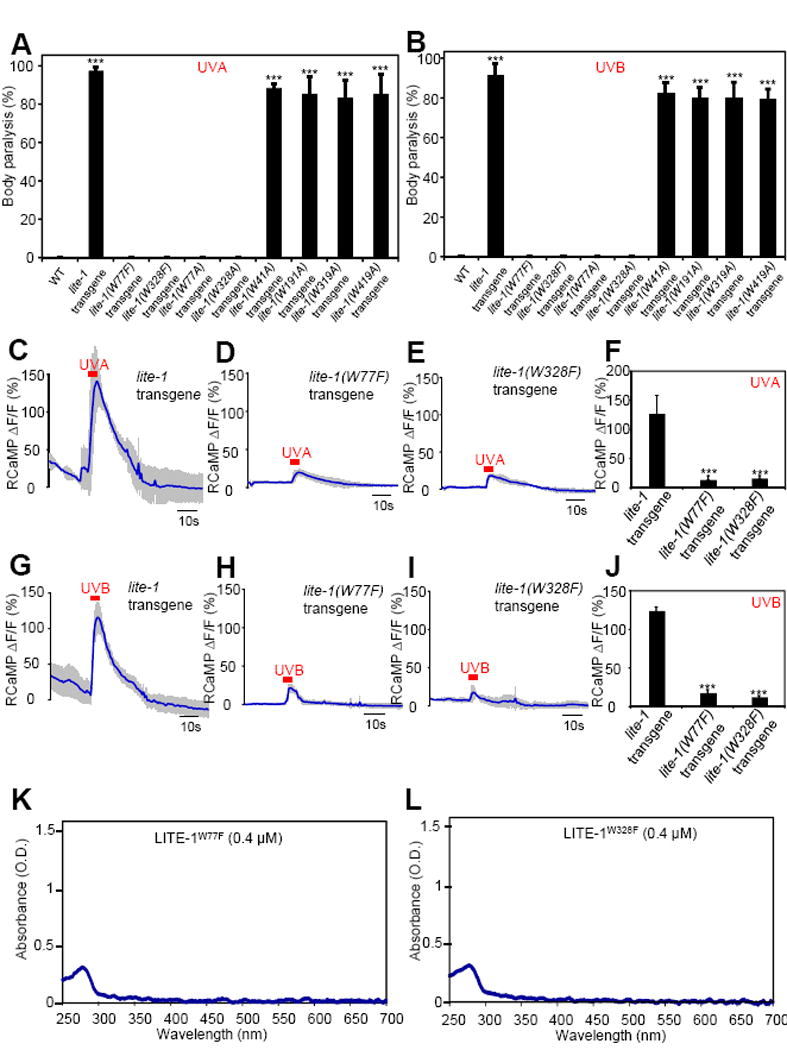
(A–B) Mutating W77 and W328 but not the other four W residues disrupts the sensitivity of LITE-1 to both UVA and UVB light in vivo shown by behavioral assays. LITE-1 variants harboring mutations in each W residue were expressed as a transgene in muscle cells. Wild-type (WT) and transgenic worms were exposed to a 20 sec pulse of UVA light (A), or UVB light (B). Animals showing muscle contraction-induced paralysis during light illumination were scored positive. n=20. Error bars: SEM. ***p<0.00001 (ANOVA with Bonferroni test).
(C–F) W77F and W328F mutations disrupt the sensitivity of LITE-1 to UVA light in vivo shown by calcium imaging. The experiments were done as described in Figure 2B. A 5 sec pulse of UVA light (340±20 nm, 0.7 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (C–E) represent SEM. (F) Bar graph. n≥6. ***p<0.00001 (ANOVA with Bonferroni test).
(G–J) W77F and W328F mutations disrupt the sensitivity of LITE-1 to UVB light in vivo shown by calcium imaging. A 5 sec pulse of UVB light (280±10 nm, 0.02 mW/mm2) was applied to muscles to elicit calcium transients. Shades along the traces in (G–I) represent SEM. (J) Bar graph. n≥10. ***p<0.00001 (ANOVA with Bonferroni test).
(K–L) W77F and W328F mutations disrupt LITE-1’s absorption of both UVA and UVB light in vitro.
Also see Figure S6.
Lastly, we purified the two mutant forms of LITE-1, LITE-1W77F and LITE-1W328F, to homogeneity (Figure 4F–G), and examined their photoabsorption in vitro. Strikingly, W77F and W328F mutations not only abolished LITE-1’s absorption of UVA light at 320 nm, but also nearly eliminated its absorption of UVB light at 280 nm (Figure 6K–L). Consistently with this spectral data, we found that these two tryptophan mutations abolished the sensitivity of LITE-1 to UVB light in vivo in behavioral assays (Figure 6B and S6B). In addition, UVB light elicited little, if any, calcium transients in muscle cells ectopically expressing these two mutant forms of LITE-1 (Figure 6G–J). The residual calcium response arose from the other tryptophan residue, as mutating both tryptophan residues eliminated the response (J.G. and X.Z.S.X. unpub.). Thus, the two tryptophan residues W77 and W328 are critical for LITE-1 function both in vivo and in vitro. These experiments identify key molecular determinants required for LITE-1 function in vivo and in vitro.
Genetic engineering of photoreceptors
To provide further evidence supporting a critical role for the two tryptophan residues in mediating photoabsorption, we wondered if introducing such tryptophan residues into another protein would promote photoabsorption. On the other hand, tryptophan residues alone are unlikely to underpin the high photoabsorption capacity of LITE-1, and other parts of LITE-1 must be involved, which may serve as a “backbone” to support the function of the two tryptophan residues in capturing photons. We thus reasoned that those proteins related to LITE-1, such as other GR genes, may possess such a backbone structure and thereby would have a higher likelihood to be engineered as a photoreceptor. The C. elegans GR family contains five members. With the exception of LITE-1, no other GR genes have both tryptophan residues at the corresponding positions (Figure S7A). We noticed that although GUR-3 is not that similar to LITE-1 at the sequence level (40% sequence identity with LITE-1), it has one tryptophan residue in place, which corresponds to W328 in LITE-1 (Figure S7A). GUR-3 was suggested to function as a chemoreceptor (Bhatla and Horvitz, 2015). As expected, ectopic expression of GUR-3 in muscle cells did not promote their sensitivity to UV light in behavioral assays (Figure 7A, S7B, and S7F–G), suggesting that GUR-3 is not photosensitive. Indeed, calcium imaging revealed that UV light evoked little, if any, calcium response in muscle cells ectopically expressing GUR-3 (Figure 7B–D and S7C–E). We then mutated residue Y79 in GUR-3 to W (i.e. GUR-3Y79W), which corresponds to W77 in LITE-1 (Figure S7A). Strikingly, worms ectopically expressing the tryptophan-bearing GUR-3Y79W then became sensitive to UVB light (Figure 7A and S7G). UVA light was not effective on these worms (Figure S7B–F). This result was expected, as UVA absorption by LITE-1 apparently requires additional key elements such as residues A226 and S332 and perhaps others (Figure 4A–E). We also examined UVB light-evoked calcium transients in muscle cells, and found that ectopic expression of the tryptophan-bearing GUR-3Y79W greatly potentiated UVB light-induced calcium response in these cells (Figure 7B–D). Thus, introducing a tryptophan residue into GUR-3 promotes photosensitivity.
Having characterized the photosensitivity of GUR-3Y79W and GUR-3 in vivo, we then purified both proteins to homogeneity (Figure 7E–F), and examined their photoabsorption in vitro (Figure 7G–H). As expected, GUR-3 showed little, if any, absorption of UVB light (Figure 7H). By contrast, strong absorption of UVB light at 280 nm was observed in GUR-3Y79W (Figure 7G). The extinction coefficient of this tryptophan-bearing GUR-3Y79W protein reached the level of 106 M−1cm−1 (1.1×106 M−1cm−1), which is about one-third of that found for LITE-1. This data provides a biochemical basis for the observed photosensitivity of GUR-3Y79W. This set of experiments also raises the intriguing prospect that it might be possible to genetically engineer new photoreceptors.
Discussion
In summary, our results demonstrate that the C. elegans taste receptor homolog LITE-1 is a bona fide photoreceptor. As some photoreceptors such as rhodopsin also respond to other types of stimuli such as heat (Shen et al., 2011), we do not exclude the possibility that LITE-1 may be polymodal and may sense additional cues, including chemical cues such as H2O2 (Bhatla and Horvitz, 2015). Several features distinguish LITE-1 from known photoreceptors, including an exceptionally high efficiency in photoabsorption, an ability to sense both UVA and UVB light, a strict dependence on protein conformation for photoabsorption, a strong resistance to photobleaching, and a reversed membrane topology compared to opsins. LITE-1 also bears no sequence homology with the two known metazoan photoreceptors (i.e. opsins and cryptochromes) or any other photoreceptors in microbes and plants. Apparently, LITE-1 represents a distinct type of photoreceptor in nature.
While it is easy to appreciate the requirement of tryptophan residues for the absorption of UVB light at 280 nm, it is a bit surprising that the absorption of UVA light at 320 nm also depend on the same tryptophan residues. On the other hand, some mutations (e.g. S226F and A332V) only affect the absorption of UVA but not UVB light (Figure 5A–B). As such, we suggest that the two tryptophan residues W77 and W328 regulate the absorption of both UVB and UVA light, while the absorption of UVA light requires additional residues such as S226 and A332. The W77F and W328F data, together with the unusually strict dependence of LITE-1 photoabsorption on its protein conformation, raises the intriguing possibility that LITE-1 may not have a prosthetic chromophore. Interestingly, the plant-specific protein UVR8, a soluble protein which is completely unrelated to LITE-1, also requires tryptophan residues for UVB light detection and lacks a prosthetic chromophore (Christie et al., 2012; Rizzini et al., 2011; Wu et al., 2012). This prompted us to speculate that the two tryptophan residues W77 and W328 may contribute to the formation of the chromophore of LITE-1, which may underlie its high photon-capturing efficiency. Though the definitive answer shall await the determination of the atomic structure of LITE-1, our finding that introducing such a tryptophan residue into another GR family protein can promote photosensitivity lends support to this model. This experiment also raises the intriguing prospect that it might be possible to genetically engineer new photoreceptors.
LITE-1 is a member of the invertebrate GR gene family, which contains 5 homologs in worms and over 60 members in insects (Clyne et al., 2000; Liu et al., 2010; Scott et al., 2001). Some of them in fact do not act as chemoreceptors (Thorne and Amrein, 2008). For example, Drosophila Gr28b(d) encodes a thermosensor (Ni et al., 2013), while another Gr28b isoform has been implicated in UV light-induced avoidance behavior (Xiang et al., 2010). The inverted membrane topology makes it unlikely for LITE-1 to function as a GPCR. Interestingly, LITE-1 can functionally interact with G protein signaling (Liu et al., 2010); but given the atypical topology of LITE-1, its interaction with G protein signaling is likely to be indirect (Liu et al., 2010). It is also unclear whether LITE-1 possesses ion channel activity like some OR and GR members. At the sequence level, no clear mammalian LITE-1 homologs could be identified. This, however, does not necessarily imply a lack of LITE-1 orthologs in mammals, as 7-TM receptors tend to share limited homologies even among those within the same subfamilies. In fact, 7-TM receptors with a reversed membrane topology are present in the mammalian genome (Iwabu et al., 2010). For example, the 7-TM adiponectin receptors AdipR1 and AdipR2, which play a pivotal role in diabetes, obesity and insulin resistance in mammals, also bear a membrane topology opposite to classical GPCRs (Iwabu et al., 2010). Some mammalian tissues/cells (e.g. skin keratinocytes and melanocytes) are sensitive to UV light, but the underlying photoreceptors have not been definitively identified (Bellono et al., 2013; Moore et al., 2013). It is conceivable that some receptor proteins functionally related to LITE-1 may sense UV light in these skin cells.
Ectopic expression of LITE-1 can confer photosensitivity to photo-insensitive cells by triggering neuronal excitation and muscle contraction (Edwards et al., 2008; Liu et al., 2010). LITE-1’s exceptionally high efficiency in photon absorption, sensitivity to both UVA and UVB light, and strong resistance to photobleaching make it a promising candidate as an optogenetic tool. These features also demonstrate the potential for developing LITE-1 as an organic additive to sunscreens for skin protection against harmful UV in the sunlight. The current study provides an entry point to characterize this interesting photoreceptor.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for reagents and resources may be directed to X.Z. Shawn Xu (shawnxu@umich.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained at 20 °C on nematode growth medium (NGM) plates seeded with OP50 bacteria. Liquid culture was used to produce large quantities of worms for protein purification (see Methods Details). Transgenic lines were generated by injecting plasmid DNA directly into hermaphrodite gonad. Integrated transgenic strains were outcrossed at least six times before used for protein purification.
METHODS DETAILS
Immunostaining to determine the membrane topology of LITE-1
Immunostaining was performed on primary cultured cells using standard protocols (Christensen et al., 2002). Muscle cells co-express LITE-1 and GFP or express GFP alone as a transgene driven by the muscle-specific promoter myo-3. Gravid hermaphrodites were lysed to release eggs, and embryos were dissociated by chitinase treatment and trituration, filtered through a 5 μm membrane, plated on cover glasses coated with peanut lectin, and cultured in L15 with 10% serum (340–345 mOsm) at 20 °C. To perform non-permeablized surface staining, live cells were first blocked with 3% BSA and 5% normal goat serum (NGS) in PBS for 30 min, and then incubated with primary antibodies (1 μg/ml) for one hour in PBS (1.5% BSA) at room temperature. Following three washes with PBS, cells were fixed for 10 min with 1.5% paraformaldehyde (PFA) in PBS followed by three washes with PBS and one hour incubation in second antibodies (1:2000, Cy3 conjugated). After five washes with PBS, cover glasses were mounted for imaging analysis. To perform permeabilized staining, cells were first fixed with 1.5% PFA in PBS for 10 min at room temperature, rinsed three times with PBS, and permeabilized with 0.5% Trition X-100 in PBS for 5 min. After three washes with PBS, cells were blocked with BSA and NGS, incubated with primary antibodies, and washed five times. Following one hour incubation with secondary antibodies, cover glasses were rinsed five times before mounting. The N- and C-terminal end peptides (15 residues) were used to immunize rabbits to generate LITE-1 antibodies which were affinity-purified before use for staining (YenZym Antibodies).
Purification and spectrophotometric analysis of LITE-1 and control proteins
Worms were cultured in the dark. They were first cultured on NGM plates and then transferred to 10 litter of S medium for liquid culture using a fermenter (New Brunswick, 20 °C, 50% dissolved oxyg en, 300 rpm agitation, pH7.2) with the support from concentrated HB101 bacteria. After 2 generations (about 7–8 days) in the fermenter, worms were harvested and suspended in 80 ml of 25 mM bis-trsi-propane BTP buffer (pH7.2) supplemented with proteinase inhibitor cocktail (Complete Mini, EDTA-free). All purification steps were carried out in the dark. A microfluidizer (Microfluidics Inc.) was used to break the worms (120 psi, 5 cycles). After removing the debris by low speed centrifugation at 1,000g for 10 min at 4 °C, the su pernatant was collected and centrifuged again at high speed (100,000g) for 1 h at 4 °C to pellet cell membranes, which were solubilized with 20 mM n-dodecyl-β-D-maltopyranoside (DDM; Affymentrix) in BTP buffer (pH7.2) containing 500mM NaCl. After removing unsolubilized materials by centrifugation at 40,000g for 30 min, we loaded the extract to an α1D4 affinity column. Note: we attached a 1D4 tag to the C-terminus of LITE-1 and GUR-3 expressed as a transgene in the worm muscle, as described for A2A receptor (Salom et al., 2012). Bovine rhodopsin (Rho) has this tag sequence at its C-terminus. After washing with the washing buffer (10 mM DDM in 25 mM BTP buffer [pH7.2] and 500mM NaCl), we eluded LITE-1 with 1.5 mg/ml of 1D4 peptide diluted in this buffer. Purified LITE-1 was loaded onto a molecular size separation column (GE healthcare Bio-Sciences) to remove 1D4 peptide before spectrophotometric analysis. When purifying bovine rhodopsin (Rho), 2 mM 9-cis-retinal was used to resuspend pelleted cell membranes and incubate for 30 min prior to solubilization with DDM. This treatment was not performed when purifying LITE-1, GUR-3, or A2A receptor. Purified protein samples used for SDS-PAGE were prepared under non-reducing conditions at room temperature (no heating) to avoid aggregation.
The concentration of purified proteins was first determined by the Bradford assay (Bio-Rad Inc.), and then verified by SDS-PAGE followed by coomassie staining using rhodopsin as a standard. The concentration data were also independently verified by silver staining following SDS-PAGE using rhodopsin as a standard.
Spectrophotometric analysis was performed on a UV-Vis spectrophotometer (Varian Cary 50) in a quartz cuvette. Samples and reference blanks were all diluted in the same washing buffer. Note: 1D4 peptide was removed from samples prior to spectrophotometric analysis (see above). For those experiments involving treatment with denaturing agents or H2O2, LITE-1 was incubated with these agents for 5 min at room temperature prior to spectrophotometric analysis. All the assays were carried out in the dark.
Behavior assays to quantify LITE-1 function
Body paralysis assay was performed on day 1 gravid adult hermaphrodites, which were raised on NGM plates, under a Zeiss fluorescence dissection scope (Zeiss Discovery) coupled with an M2Bio lens system from Kramer Scientifics. The assay was done on NGM plates without OP50 using a protocol similar to that for assaying phototaxis behavior (Liu et al., 2010; Ward et al., 2008). UVA light pulses (350±20 nm, 0.8 mW/mm2, up to 20 sec) were delivered from an Arc lamp (X-Cite 120) to the worm through a 10× lens in combination with 2.5× zoom. To deliver UVB light, we attached a 280±10 nm excitation filter (from Semrock, 0.03 mW/mm2) to the end of the liquid light guide of the lamp, which was then directly pointed to the worm using a micromanipulator. We manually moved the dish to keep the worm in the view field. In another assay, we quantified body paralysis by monitoring locomotion speed decrease over time using the Wormlab system (MBF Bioscience). UVA and UVB light was directed to the worm using a liquid light guide as described above. To minimize the effect of endogenous lite-1 gene on locomotion speed under UV light (Liu et al., 2010), this assay was performed in lite-1(xu7) mutant background for all genotypes. A total of 20–50 animals were assayed for each genotype in each experiment unless otherwise indicated. The sample size of each assay was found to be adequate after running power analysis (P>0.8). Each worm was assayed five times, and once the worm was paralyzed, we stopped the assay to let it recover for next round of test.
Calcium imaging to quantify LITE-1 function
Calcium imaging of muscle cells was performed on an inverted microscope (Olympus IX73) under a 60× lens as previously described (Li et al., 2014; Xiao et al., 2013). RCaMP was expressed as a transgene in muscle cells using the myo-3 promoter. Transgenic worms expressing LITE-1 or control worms were glued on an agarose pad and bathed in solution (10 mM HEPES [pH 7.4], 5 mM KCl, 145 mM NaCl,1.2 mM MgCl2, 2.5 mM CaC12, and 10 mM glucose). UV light (UVA: 340±20 nm, 0.7 mW/mm2; UVB: 280±10 nm, 0.02 mW/mm2; 5 sec) was directly projected to the worm through a liquid light guide mounted on a micromanipulator. Images were acquired with a Roper CoolSnap CCD camera and processed with MetaFluor software (Molecular Devices). To minimize the contribution from endogenous photosensation system, all genotypes, including WT, carried lite-1(xu7) mutation in the background (Liu et al., 2010). The peak percentage change in the intensity of RCaMP (ΔF/F) fluorescence was quantified.
Molecular biology
All the plasmids are listed in the KEY RESOURCES TABLE. All the LITE-1 and GUR-3 constructs carry a 1D4 tag at the C-terminus, with the exception in Figure 1 where no such a tag was included to LITE-1. Myc tag was only included in the construct used in Figure 1B. As listed in the KEY RESOURCES TABLE, some plasmids contain an SL2∷YFP fragment, which directs expression of YFP as a separate transcript under the control of the same promoter of its upstream gene in an operon-like fashion. This enables expression of YFP as a co-expression marker in muscle cells under the control of the same muscle-specific myo-3 promoter that drives expression of LITE-1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical parameters were indicated in the legends of each figure, including error bars (SEM), n numbers, and p values. For those involving multiple group comparisons, we applied ANOVA followed by a post hoc test. We considered p values of <0.05 significant.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-1D4 | Polgenix, Inc. | N/A |
| Rabbit monoclonal anti-C-LITE-1 | This paper | N/A |
| Rabbit monoclonal anti-N-LITE-1 | This paper | N/A |
| Mouse monoclonal anti-N-Myc | ThermoFisher | CAT# MA1-16638 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| bis-trsi-propane | Sigma | CAT# 79-97-0 |
| proteinase inhibitor cocktail | Roche | CAT# 11836170001 |
| n-dodecyl-β-D-maltopyranoside | Affymentrix | CAT# 69227-93-6 |
| 9-cis-retinal | Toronto Research Chemicals | CAT# 514-85-2 |
| Hydrogen peroxide solution | Sigma | CAT# 7722-84-1 |
| Urea | Sigma | CAT# 57-31-6 |
| 1D4 peptide | Genscript | Lot# 89521380001 |
| Critical Commercial Assays | ||
| Bradford protein assay | Bio-Rad Inc. | CAT# 5000001 |
| ECL Western blotting kit | ThermoFisher | CAT# 35050 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: lite-1(xu7) | Ward et al., 2008; Caenorhabditis Genetics Center | TQ800 |
| C. elegans: N2 | Caenorhabditis Genetics Center | WormBase: N2 |
| C. elegans: xuIs98 [Pmyo-3∷lite-1∷1D4∷SL2∷YFP] | This paper | TQ2518 |
| C. elegans: xuIs397 [Pmyo-3∷lite-1(W328F)∷1D4∷SL2∷YFP] | This paper | TQ6448 |
| C. elegans: xuIs399 [Pmyo-3∷lite-1(W77F)∷1D4∷SL2∷YFP] | This paper | TQ6450 |
| C. elegans: xuIs403 [Pmyo-3∷lite-1(A332V)∷1D4∷SL2∷YFP] | This paper | TQ6454 |
| C. elegans: xuIs404 [Pmyo-3∷lite-1(S226F)∷1D4∷SL2∷YFP] | This paper | TQ6455 |
| C. elegans: xuEx1623 [Pmyo-3∷lite-1(W77A)∷1D4∷SL2∷YFP] | This paper | TQ5143 |
| C. elegans: xuEx1627 [Pmyo-3∷lite-1(W191A)∷1D4∷SL2∷YFP] | This paper | TQ5148 |
| C. elegans: xuEx1628 [Pmyo-3∷lite-1(W319A)∷1D4∷SL2∷YFP] | This paper | TQ5149 |
| C. elegans: xuEx1629 [Pmyo-3∷lite-1(W328A)∷1D4∷SL2∷YFP] | This paper | TQ5150 |
| C. elegans: xuEx1632 [Pmyo-3∷lite-1(W41A)∷1D4∷SL2∷YFP] | This paper | TQ5152 |
| C. elegans: xuEx1633 [Pmyo-3∷lite-1(W419A)∷1D4∷SL2∷YFP] | This paper | TQ5153 |
| C. elegans: xuEx2430[Pmyo-3∷bJun∷N-YFP∷lite-1∷SL2∷DsRed] | This paper | TQ6688 |
| C. elegans: xuEx2431[Pmyo-3∷bFos∷C-YFP] | This paper | TQ6689 |
| C. elegans: xuEx2432[Pmyo-3∷ ΔbFos∷C-YFP] | This paper | TQ6690 |
| C. elegans: xuEx386[Pmyo-3∷myc∷lite-1∷SL2∷YFP] | This paper | TQ1353 |
| C. elegans: xuIs32[Pmyo-3∷lite-1∷SL2∷YFP] | This paper | TQ1230 |
| C. elegans: xuIs442[Pmyo-3∷GUR-3(Y79W)∷1D4∷SL2∷YFP] | This paper | TQ7405 |
| C. elegans: xuIs441[Pmyo-3∷GUR-3∷1D4∷SL2∷YFP] | This paper | TQ7404 |
| C. elegans: xuIs444[Pmyo-3∷RCaMP]; lite-1(xu7) | This paper | TQ7428 |
| Recombinant DNA | ||
| Pmyo-3∷lite-1∷1D4∷SL2∷YFP | This paper | pSX1580 |
| Pmyo-3∷lite-1(W328F)∷1D4∷SL2∷YFP | This paper | pSX1712 |
| Pmyo-3∷lite-1(W77F)∷1D4∷SL2∷YFP | This paper | pSX1713 |
| Pmyo-3∷lite-1(A332V)∷1D4∷SL2∷YFP | This paper | pSX1710 |
| Pmyo-3∷lite-1(S226F)∷1D4∷SL2∷YFP | This paper | pSX1711 |
| Pmyo-3∷lite-1(W77A)∷1D4∷SL2∷YFP | This paper | pSX1714 |
| Pmyo-3∷lite-1(W191A)∷1D4∷SL2∷YFP | This paper | pSX1715 |
| Pmyo-3∷lite-1(W319A)∷1D4∷SL2∷YFP | This paper | pSX1716 |
| Pmyo-3∷lite-1(W328A)∷1D4∷SL2∷YFP | This paper | pSX1717 |
| Pmyo-3∷lite-1(W41A)∷1D4∷SL2∷YFP | This paper | pSX1718 |
| Pmyo-3∷lite-1(W419A)∷1D4∷SL2∷YFP | This paper | pSX1719 |
| Pmyo-3∷bJun∷N-YFP∷lite-1∷SL2∷DsRed | This paper | pSX1740 |
| Pmyo-3∷bFos∷C-YFP | This paper | pSX1741 |
| Pmyo-3∷ ΔbFos∷C-YFP | This paper | pSX1742 |
| Pmyo-3∷myc∷lite-1∷SL2∷YFP | This paper | pSX153 |
| Pmyo-3∷GUR-3(Y79W)∷1D4∷SL2∷YFP | This paper | pSX1902 |
| Pmyo-3∷GUR-3∷1D4∷SL2∷YFP | This paper | pSX1903 |
| Pmyo-3∷RCaMP | This paper | pSX1904 |
| Software and Algorithms | ||
| Wormlab system | MBF Bioscience | N/A |
| MetaFluor | Molecular Devices | N/A |
Supplementary Material
Highlights.
LITE-1, a taste receptor homolog, is a bona fide photoreceptor that senses UV light
LITE-1 has a high efficiency of photon capturing
Photoabsorption by LITE-1 relies on its conformation and requires two Trp residues
Introducing such a Trp residue into a related protein promotes photosensitivity
Acknowledgments
We thank Tom Kerppola for BiFC plasmids, and David Salom and Kris Palczewski for technical assistance and providing strains, Zhaohui Xu for helpful discussions, and Wenyuang Zhang, Jiejun Zhou, John Tesmer, Frederick Stull, and James Bardwell for technical assistance. Some strains were obtained from the CGC. This work utilized the Core Center for Vision Research funded by P30 EY007003 from the NEI. A.W. was supported by a predoctoral training grant from the NEI (T32EY013934). This work was supported by the NSFC (31130028, 31225011, and 31420103909 to J.L.), the Program of Introducing Talents of Discipline to Universities from the Ministry of Education of China (B08029 to J.L.), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT: IRT13016), and grants from the NEI and NIGMS (X.Z.S.X).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J.G. performed the experiments and analyzed the data. Y.Y. B.Z. Z.W., J.P., and Z.F. assisted J.G. in performing the experiments. A.W. initiated the project and generated reagents. L.K. performed immunostaining on primary cultured muscle cells and analyzed the data. J.G., J.L., and X.Z.S.X. wrote the paper.
References
- Bellono NW, Kammel LG, Zimmerman AL, Oancea E. UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2383–2388. doi: 10.1073/pnas.1215555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatla N, Horvitz HR. Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron. 2015;85:804–818. doi: 10.1016/j.neuron.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Estevez A, Yin X, Fox R, Morrison R, McDonnell M, Gleason C, Miller DM, 3rd, Strange K. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron. 2002;33:503–514. doi: 10.1016/s0896-6273(02)00591-3. [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- Dutta A, Kim TY, Moeller M, Wu J, Alexiev U, Klein-Seetharaman J. Characterization of membrane protein non-native states. 2. The SDS-unfolded states of rhodopsin. Biochemistry. 2010;49:6329–6340. doi: 10.1021/bi100339x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Charlie NK, Milfort MC, Brown BS, Gravlin CN, Knecht JE, Miller KG. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 2008;6:e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A, Bowler C. The evolution and function of blue and red light photoreceptors. Curr Top Dev Biol. 2005;68:317–350. doi: 10.1016/S0070-2153(05)68011-8. [DOI] [PubMed] [Google Scholar]
- Foster RG, Soni BG. Extraretinal photoreceptors and their regulation of temporal physiology. Rev Reprod. 1998;3:145–150. doi: 10.1530/ror.0.0030145. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Oxygen: how do we stand it? Med Princ Pract. 2013;22:131–137. doi: 10.1159/000339212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagins FM. Purification and partial characterization of the protein component of squid rhodopsin. J Biol Chem. 1973;248:3298–3304. [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Hubbard R. Absorption spectrum of rhodopsin: 500 nm absorption band. Nature. 1969;221:432–435. doi: 10.1038/221432a0. [DOI] [PubMed] [Google Scholar]
- Insinna C, Daniele LL, Davis JA, Larsen DD, Kuemmel C, Wang J, Nikonov SS, Knox BE, Pugh EN., Jr An S-opsin knock-in mouse (F81Y) reveals a role for the native ligand 11–cis-retinal in cone opsin biosynthesis. J Neurosci. 2012;32:8094–8104. doi: 10.1523/JNEUROSCI.0131-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- Kolesnikov AV, Kisselev OG, Kefalov VJ. Signaling by Rod and Cone Photoreceptors: Opsin Properties, G-protein Assembly, and Mechanisms of Activation. In: Martemyanov KA, Sampath AP, editors. G Protein Signaling Mechanisms in the Retina. Springer; 2014. [Google Scholar]
- Li Z, Liu J, Zheng M, Xu XZ. Encoding of both analog- and digital-like behavioral outputs by one C. elegans interneuron. Cell. 2014;159:751–765. doi: 10.1016/j.cell.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ward A, Gao J, Dong Y, Nishio N, Inada H, Kang L, Yu Y, Ma D, Xu T, et al. C. elegans phototransduction requires a G protein–dependent cGMP pathway and a taste receptor homolog. Nat Neurosci. 2010;13:715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang B, Lei H, Feng Z, Liu J, Hsu AL, Xu XZ. Functional Aging in the Nervous System Contributes to Age-Dependent Motor Activity Decline in C. elegans. Cell metabolism. 2013;18:392–402. doi: 10.1016/j.cmet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglova L, Atanasov B, Keszthelyi L. Unfolding of Monomeric Bacteriorhodopsin in Water-Urea Solution. Biochimica Et Biophysica Acta. 1989;975:271–276. [Google Scholar]
- Marti T, Rosselet SJ, Otto H, Heyn MP, Khorana HG. The retinylidene Schiff base counterion in bacteriorhodopsin. J Biol Chem. 1991;266:18674–18683. [PubMed] [Google Scholar]
- Matsuyama T, Yamashita T, Imamoto Y, Shichida Y. Photochemical properties of mammalian melanopsin. Biochemistry. 2012;51:5454–5462. doi: 10.1021/bi3004999. [DOI] [PubMed] [Google Scholar]
- Moore C, Cevikbas F, Pasolli HA, Chen Y, Kong W, Kempkes C, Parekh P, Lee SH, Kontchou NA, Yeh I, et al. UVB radiation generates sunburn pain and affects skin by activating epidermal TRPV4 ion channels and triggering endothelin-1 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3225–3234. doi: 10.1073/pnas.1312933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano VC, Theobald DL, Griffith LC, Garrity PA. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature. 2013;500:580–584. doi: 10.1038/nature12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Hess B. Reversible photolysis of the purple complex in the purple membrane of Halobacterium halobium. Eur J Biochem. 1973;37:316–326. doi: 10.1111/j.1432-1033.1973.tb02990.x. [DOI] [PubMed] [Google Scholar]
- Okano T, Fukada Y, Shichida Y, Yoshizawa T. Photosensitivities of iodopsin and rhodopsins. Photochem Photobiol. 1992;56:995–1001. doi: 10.1111/j.1751-1097.1992.tb09722.x. [DOI] [PubMed] [Google Scholar]
- Radding CM, Wald G. Acid-base properties of rhodopsin and opsin. J Gen Physiol. 1956;39:909–922. doi: 10.1085/jgp.39.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- Salom D, Cao P, Sun W, Kramp K, Jastrzebska B, Jin H, Feng Z, Palczewski K. Heterologous expression of functional G-protein-coupled receptors in Caenorhabditis elegans. FASEB J. 2012;26:492–502. doi: 10.1096/fj.11-197780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Shen WL, Kwon Y, Adegbola AA, Luo J, Chess A, Montell C. Function of rhodopsin in temperature discrimination in Drosophila. Science. 2011;331:1333–1336. doi: 10.1126/science.1198904. [DOI] [PubMed] [Google Scholar]
- Sperling W, Rafferty CN. Relationship between absorption spectrum and molecular conformations of 11–cis-retinal. Nature. 1969;224:590–594. doi: 10.1038/224591a0. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Sancar A. Photolyase/cryptochrome blue-light photoreceptors use photon energy to repair DNA and reset the circadian clock. Oncogene. 2002;21:9043–9056. doi: 10.1038/sj.onc.1205958. [DOI] [PubMed] [Google Scholar]
- Thorne N, Amrein H. Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J Comp Neurol. 2008;506:548–568. doi: 10.1002/cne.21547. [DOI] [PubMed] [Google Scholar]
- Vought BW, Dukkipatti A, Max M, Knox BE, Birge RR. Photochemistry of the primary event in short-wavelength visual opsins at low temperature. Biochemistry. 1999;38:11287–11297. doi: 10.1021/bi990968b. [DOI] [PubMed] [Google Scholar]
- Wang R, Mellem JE, Jensen M, Brockie PJ, Walker CS, Hoerndli FJ, Hauth L, Madsen DM, Maricq AV. The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron. 2012;75:838–850. doi: 10.1016/j.neuron.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nature Neurosci. 2008;11:916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- Wu LZ, Sheng YB, Xie JB, Wang W. Photoexcitation of tryptophan groups induced reduction of disulfide bonds in hen egg white lysozyme. Journal of Molecular Structure. 2008;882:101–106. [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Zhang B, Dong Y, Gong J, Xu T, Liu J, Xu XZS. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. 2013;152:806–817. doi: 10.1016/j.cell.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139:246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HJ, Anderson AR, Trowell SC, Luo AR, Xiang ZH, Xia QY. Topological and functional characterization of an insect gustatory receptor. PloS one. 2011;6:e24111. doi: 10.1371/journal.pone.0024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


