Abstract
Compartmentalized biochemical activities are essential to all cellular processes, but there is no generalizable method to visualize dynamic protein activities in living cells at a resolution commensurate with their compartmentalization. Here we introduce a new class of fluorescent biosensors that detect biochemical activities in living cells at a resolution up to three-fold better than the diffraction limit. Utilizing specific, binding-induced changes in protein fluorescence dynamics, these biosensors translate kinase activities or protein-protein interactions into changes in fluorescence fluctuations, which are quantifiable through stochastic optical fluctuation imaging. A Protein Kinase A (PKA) biosensor allowed us to resolve minute PKA activity microdomains on the plasma membrane of living cells and uncover the role of clustered anchoring proteins in organizing these activity microdomains. Together, these findings suggest that biochemical activities of the cell are spatially organized into an activity architecture, whose structural and functional characteristics can be revealed by these new biosensors.
Protein-based nanomachinery processes biochemical information and underlies all aspects of cellular function. Coordinated protein assembly and biochemical activity at specific loci in living cells ultimately leads to functional changes in cell growth, division, migration, or programmed death. An emerging picture is that these biochemical activities are dynamically regulated in various temporal waveforms1, 2 and spatially organized into distinct micro- or nano-domains3–5. The concept suggests that in addition to their physical structure, cells also maintain an “activity architecture” that is composed of organized, activated molecules and their regulatory partners. This model has not been directly tested, and critical questions about spatial organization of biochemical activities remain. The classical example of compartmentalized signaling is that of PKA, where PKA holoenzyme is anchored3 via A Kinase Anchoring Proteins (AKAPs) into signaling microdomains. However, this compartmentalization is achieved through the regulatory (R) subunits rather than the catalytic (C) subunits. PKA stimulation and subsequent release/diffusion of the C subunit would diminish this spatial compartmentalization. Therefore, even in this classical model, it is unclear whether and how the kinase activity is spatially organized in living cells. In recent years, many superresolution imaging methods that reveal the location of nanoscale cellular features6, 7 with vastly improved spatial resolution, such as STED8, PALM/STORM9, 10, SOFI/pcSOFI11, 12, and SIM13, have emerged. While there have been considerable efforts to move superresolution imaging beyond biomolecule localization14–16, currently there is no general methodology to visualize dynamic biochemical activities such as protein-protein interactions and posttranslational modifications in live cells at superresolution. Here, we address this need by introducing a new class of generalizable, genetically encodable biosensors that enabled the first direct visualization of dynamic biochemical activities at a resolution beyond the diffraction limit. Using these new biosensors in combination with existing superresolution techniques, we report direct evidence of highly active PKA activity microdomains in the plasma membrane.
Discovery and characterization of FLINC
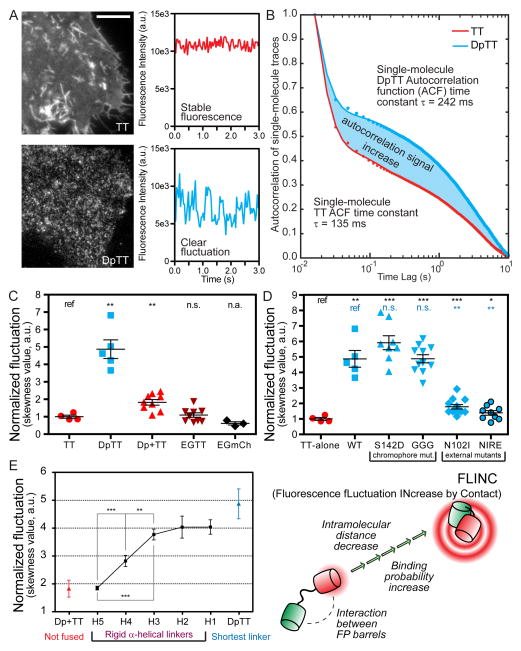
Examining the fluorescence dynamics of TagRFP-T in a series of plasma membrane-tethered constructs, we discovered that the proximity of Dronpa17 significantly increases the fluorescence fluctuations of TagRFP-T18 (Supplementary Video 1). We characterized this phenomenon using Dronpa-TagRFP-T (DpTT), where these two fluorescent proteins (FPs) are directly fused together by a short flexible linker. Several characteristics were revealed. First, fluctuations are easily detected in live cells expressing membrane targeted DpTT (Fig. 1A; Supplementary Fig. 1). The single molecule fluorescence fluctuations generated by purified DpTT is quantitatively more robust than that from TagRFP-T (Fig. 1B). Secondly, this effect occurs specifically between tethered Dronpa and TagRFP-T (Fig. 1C). Thirdly, the external residues of Dronpa, not its chromophore, are key determinants of this effect (Fig. 1D and Supplementary Fig. 1, Supplementary Note). Lastly, decreasing the distance between Dronpa and TagRFP-T by using rigid helical linkers of successively shorter lengths19 revealed a corresponding increase in TagRFP-T fluorescence fluctuations (Fig. 1E), indicating an effective range of 5–6 nm.
Figure 1. TagRFP-T (TT) red fluorescence fluctuations can be increased by proximity of Dronpa (Dp) in a distance-dependent manner.
(A) Representative images and single-pixel fluorescence intensity traces in HeLa cells expressing DpTT (Dronpa-linker-TagRFP-T) and TagRFP-T alone when excited by 561 nm laser; Scale bar: 10 μm. (B) Aggregated mean normalized autocorrelation function (ACF) of many single molecule fluorescence traces from purified fluorescent DpTT and TagRFP-T. The amplitude increase demonstrates the clear gain in autocorrelation signal from increased millisecond fluctuations of DpTT. (C) Quantified fluctuation in various constructs demonstrating the specific nature of the fluctuation increase; n (cell) numbers are: TT (n = 8), DpTT (n = 7), Dp+TT (n = 9), EGTT (EGFP-linker-TagRFP-T, n = 8), and EGFP-linker-mCherry (EGmCh, n = 9). (D) Quantified fluctuation in various mutant constructs demonstrating Dronpa’s chromophore is not involved but external Dronpa residues are important for the phenomenon. n (cell) numbers are: TT-alone (n = 8), WT (n = 7); chromophore mutants: Dp[S142D]-TT (S142D, n = 8), Dp[C62G/Y63G]-TT (GGG, n = 11); FP surface mutants: Dp[N102I]-TT (N102I, n = 12), Dp[N102I/R149E]-TT (NIRE, n = 9). (E) A variable number of rigid helical repeats permit measurement of distance dependence and sensitivity of the change in fluctuation. Hn represents the number (n) of rigid (EAAAK) repeats; DpTT (short linker) and Dp+TT (not fused) were included as comparisons. H2–H5 linkers are marked with nominal FP distances reported in literature. All constructs in (E) were targeted to the plasma membrane of HeLa cells by the lyn sequence and examined at the n (cell) numbers as follows: DpTT (n = 5), H1 (n = 7), H2 (n = 6), H3 (n = 14), H4 (n = 17), H5 (n = 10), Dp+TT (n = 9). Pair-wise t-test results in (C), (D), and (E) are marked where data were compared with the construct marked “ref”. n.s.: not-significant; *: p<0.05; **: p<0.01; ***: p<0.001 where applicable. In all dot plots and (E), center line and whiskers mark the average and s.e.m. values, respectively.
We termed this phenomenon Fluorescence fLuctuation INcrease by Contact (FLINC). Despite a low intrinsic affinity between Dronpa and TagRFP-T (Supplementary Fig. 2, Kd = 159–917 μM), the high effective concentrations in the fusions facilitate their intramolecular binding. While TagRFT-T, like many FPs, “blinks” stochastically20, 21, the specific binding of Dronpa can alter the blinking behavior of TagRFP-T, making a larger percentage (25%) of TagRFP-T undergo dark-state conversion at a 25% faster rate (Supplementary Fig. 3). This photophysical mechanism gives rise to the modulated single molecule fluorescence fluctuation that can be utilized to assess whether Dronpa is in close proximity to TagRFP-T (Supplementary Note).
FLINC-based Biosensors
FLINC therefore forms the basis of a new class of fluorescent biosensors that could provide activity information in superresolution. Engineered molecular switches can modulate the proximity between Dronpa and TagRFP-T in a manner similar to FRET-based biosensors22. The output of these FLINC-based biosensors is activity-dependent changes in the fluorescence fluctuations, which are readily quantified at superresolution using photochromic Stochastic Optical Fluctuation Imaging (pcSOFI)11. In pcSOFI, time series of fluorescence images are recorded to obtain many single molecule fluctuations across the field of view. Pair-wise cross-cumulant, calculated utilizing the appropriate pixel-pairings, yields an autocorrelation-like “pcSOFI value” at sub-pixel resolution that quantifies the strength of fluctuation23 with high signal-to-noise (Supplementary Fig. 4). The collection of pcSOFI values constitutes a quantitative image, which provides a superresolution map of the biochemical activity under study.
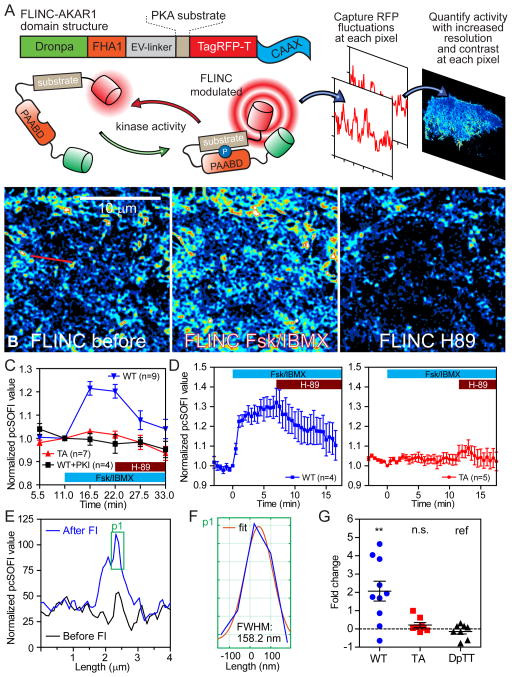
We modeled FLINC-based activity biosensors after FRET-based biosensors, which serve as surrogate substrates of the kinase of interest. Phosphorylation of these biosensors leads to a change of fluorescence properties, thereby allowing us to monitor kinase activity by imaging a reporter, without labeling or disrupting the active kinase. In the FLINC-based PKA biosensor, we combined the molecular switch in AKAR24 with an EV linker (AKARev)25 with Dronpa and TagRFP-T into FLINC-AKAR1 (targeted to the plasma membrane). Here, FLINC would be affected in a phosphorylation-dependent manner, such that high pcSOFI values indicate high PKA activities (Fig. 2A). Normalizing the pcSOFI values to correct for uneven biosensor distribution (Supplementary Note; Supplementary Fig. 5A–E), we could quantify PKA activity both at the single pixel level or averaged across the entire cell. The average normalized pcSOFI value rapidly increased in the first minute after PKA activation in HeLa cells treated with a cocktail of the adenylyl cyclase activator forskolin (Fsk) and phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX), reaching a plateau at 25–39% increase after 7–10 minutes (Fig. 2B–D) and showing a dynamic range of up to 40% increase. Addition of the PKA-specific inhibitor H-89 (20 μM) gradually decreased the average normalized pcSOFI value, demonstrating the reversibility. Additional experiments using a negative control biosensor that cannot be phosphorylated (Fig. 2C–D) and membrane-targeted PKA inhibitor PKI (Fig. 2C) demonstrated that the response was dependent on phosphorylation of the biosensor and PKA activity, respectively. Time courses with H-89 inhibition and submaximal dosage of Fsk (Supplementary Fig. 6) showed that FLINC-AKAR1 could accurately report over a range of PKA activity. We could quantify PKA activity initially every 5 minutes (Fig. 2C), and with further development every 30 s (Fig. 2D). The kinetics of PKA stimulation as monitored by FRET-based or FLINC-based AKAR showed no significant difference (FRET t1/2 = 1.2 min, n = 7; FLINC t1/2 = 0.73 min, n = 4). Importantly, our analysis of FLINC-AKAR1 fluctuations generated superresolution images of PKA activity at each time point throughout the treatment course. Monitoring the same profile line in the normalized pcSOFI images before and after Fsk/IBMX treatment (Fig. 2E) showed that we can resolve sub-diffraction-limit activity features (Fig. 2F) that have responded to stimulation. Actin-targeted FLINC-AKAR1 enabled us to distinguish the stimulated PKA activity with an average Gaussian full-width-half-maximum of 179 ± 6 nm (n = 7) and 116 ± 6 nm (n = 7), consistent with the respective use of second- and third-order analyses (Supplementary Fig. 7A/B). Furthermore, we could resolve the stimulated membrane PKA activity in converging filopodial features separated by 160 nm and 107 nm using second- and third-order cumulant analyses, respectively (Supplementary Note, Supplementary Fig. 7C–D). Under the conditions described here (Supplementary Note), biosensor diffusion does not affect the temporal and spatial resolution of pcSOFI11 and its accurate quantification of FLINC. Thus, as a reporter of kinase activity, FLINC-AKAR1 provides not only a consistent readout for PKA activity, but also allows us to monitor the dynamic changes of PKA activity with high contrast at superresolution.
Figure 2. FLINC resolves PKA activity microdomains on the plasma membrane at superresolution.
(A) Schematic of the FLINC-KAR design principle, the domain structure of FLINC-AKAR1, and the acquisition of superresolution activity images using pcSOFI. (B) FLINC-AKAR1 superresolution images clearly resolves the response to Fsk/IBMX stimulation (Fsk 50 μM, IBMX 100 μM) and inhibition (H-89 20 μM), and detailed spatial information on membrane PKA activity emerges. Color scale are identical. (C) The mean normalized pcSOFI response time course from live HeLa cells expressing wild-type (WT, n = 9 cells) FLINC-AKAR1, non-phosphorylatable mutant (TA, n = 7), and WT co-expressed with PKI (WT+PKI, n = 4) upon PKA stimulation and inhibition. (D) The normalized pcSOFI responses time course from live HeLa cells expressing wild-type (WT, n = 4) FLINC-AKAR1 and non-phosphorylatable mutant (TA, n = 5) upon PKA stimulation and inhibition using a fast acquisition imaging scheme. (E) A profile line at the same position across the pcSOFI images before and after Fsk/BIMX (“FI”) stimulation clearly demonstrate sensing of PKA activity at superresolution; the profile has been marked by a red line in B. (F) Zoom view of the active PKA feature in F showing the Gaussian fitting and FWHM size of this sub-diffraction limit PKA activity microdomain resolved. (G) A comparison of the changes in the fraction of membrane area occupied by punctate structures after stimulation across various FLINC constructs, all targeted using the same CAAX motif. In FLINC-AKAR1 experiments (WT, n = 10; TA, n = 8), cells were stimulated with Fsk/IBMX; DpTT experiments (n = 8) received no drug treatment. Unless indicated otherwise, pair-wise t-test results are marked where data were compared with the construct marked “ref”. n.s.: not-significant; *: p<0.05; **: p<0.01; ***: p<0.001 where applicable. In (C) and (D), center line and whiskers mark the average and s.e.m. values, respectively.
PKA Activity Microdomains
The PKA activity maps generated by FLINC-AKAR1 revealed many minute and highly active punctate features on the basal membrane of living cells. These activity puncta, with a mean diameter of 350 nm (Supplementary Note), were clearly resolved after Fsk/IBMX stimulation. Activating PKA induces a 2-fold increase in microdomain coverage over the basal membrane, and is not observed with the DpTT control or the non-phosphorylatable mutant (TA) (Fig. 2G). Using Stochastic Optical Reconstruction Microscopy (STORM) in total internal reflection fluorescence (TIRF) condition, we further verified the presence of these highly active PKA microdomains (Supplementary Note). Phospho-PKA-substrates (p-PKAsub) were clearly clustered (Supplementary Fig. 8A/B), forming distinct microdomains on the basal membrane with a mean diameter of approximately 250 nm (Supplementary Fig. 8C, Supplementary Note). The basal membrane of cells stimulated by Fsk/IBMX again displayed a significant 4.8 fold increase in phospho-PKA-substrate clusters per unit membrane area (Supplementary Fig. 8D). All these characteristics observed in STORM are consistent with those observed using FLINC-AKAR1 in live cells. Thus, despite the conventionally held views of rapid cAMP diffusion26–30 and mobility of PKA catalytic subunits31, 32, PKA activity is not uniform but confined within microdomains on the plasma membrane of living mammalian cells.
Clustered anchoring proteins spatially organize PKA activity microdomains
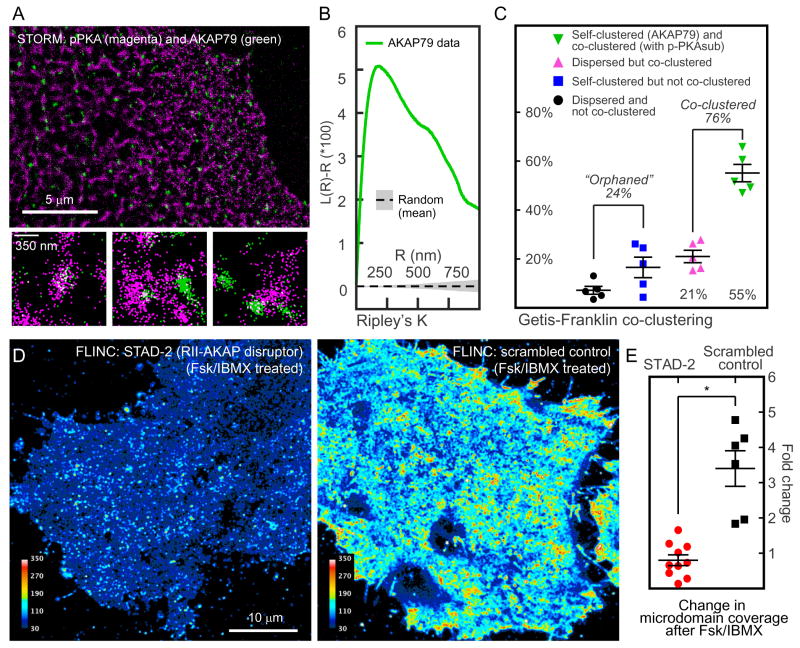
AKAPs are critical components of the PKA compartmentalization. We set out to directly determine the precise spatial relationship between highly active PKA microdomains and the loci of specific PKA anchoring. AKAP79/AKAP5 is a doubly-lipidated, predominately membrane-localized scaffold known to associate with RII PKA holoenzymes and other regulators of PKA signaling33, 34. Two-color STORM imaging in fixed cells (Supplementary Fig. 9A/B) showed that AKAP79 was itself highly clustered (Fig. 3A/B, Supplementary Fig. 9C), with a mean diameter of 127 nm. Getis-Franklin co-clustering (Supplementary Note, Supplementary Fig. 9D) demonstrated a high degree of spatial correlation between AKAP79 and p-PKAsub (Fig. 3C). 76% of total AKAP79 localizations, whether in clusters or dispersed, were associated with p-PKAsub microdomains. Surprisingly, however, a total of 24% of AKAP79 localizations could not be associated with p-PKAsub, suggesting heterogeneity of AKAP-mediated complexes and that a sub-pool of these complexes could be regulated differentially, e.g. due to a difference in phosphatase activity. Through a combination of AKAP79 localization and FLINC-AKAR1 imaging in the same cells, we observed characteristics that mirrored those reported above. Significant overlap with PKA activity microdomains was observed above random clustering (Supplementary Fig. 9C5), and we again observed so-called “orphaned” AKAP79 molecules.
Figure 3. A Kinase Anchoring Proteins (AKAPs) are co-clustered with PKA activity microdomains and are required for their formation.
(A) A representative two-color STORM image of p-PKAsub (magenta) and AKAP79 (green) with specific examples of p-PKAsub/AKAP79 relationship beneath. (B) Ripley’s K analysis of AKAP79 localizations show clear clustering above random sampling. (C) Getis-Franklin co-clustering analyses of the two-color STORM images identify populations with different p-PKAsub/AKAP79 relationships; a majority (55 + 21 = 76%) of AKAP79 are associated with p-PKAsub (n = 5 cells). (D) Representative live cell superresolution FLINC-AKAR1 images of stimulated PKA activity after the disruption of AKAP-PKA RII interactions using a synthetic AKAP disruptor, STAD-2, in comparison with its scrambled control. Scale bar: 10 μm. The color scales are identical. (E) The effect of RII-AKAP disruption on PKA activity microdomains quantified by fold change in microdomain coverage. Cells were either pretreated with STAD-2 (n = 10) or scrambled peptide control (n = 6) before Fsk/IBMX stimulation. Unless indicated otherwise, pair-wise t-test results are marked where data were compared with the construct marked “ref”; n numbers are marked inside the corresponding bars. n.s.: not-significant; *: p<0.05; **: p<0.01 where applicable. In (C) and (E), center line and whiskers mark the average and s.e.m. values, respectively.
Next we tested whether, beyond a high degree of spatial correlation, AKAP anchoring is required for the formation of highly active membrane PKA microdomains in living HeLa cells. To this end, we used the synthetic peptide (STAD-2), which specifically disrupts the interaction between AKAPs and the PKA regulatory subunit RII isoform35. Consistent with previous observations, the membrane PKA activity in living cells pretreated with STAD-2 was low after Fsk/IBMX stimulation (Supplementary Fig. 10) and devoid of highly active PKA microdomains, in contrast to cells treated with scrambled peptide (Fig. 3D and 3E). These live cell data indicated that beyond a high degree of spatial correlation, PKA activity microdomains require anchoring by AKAPs such as AKAP79.
Polarized distribution of activity microdomains
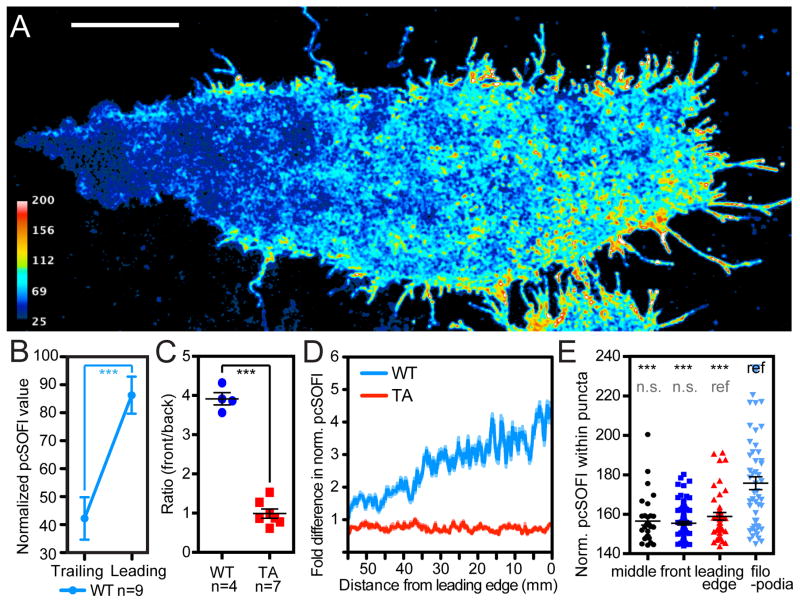
Having confirmed that cells do maintain a well-structured PKA spatial signaling architecture, we next examined the presence and organization of highly active PKA microdomains during integrin-dependent cell migration, which requires effective polarization. Chinese Hamster Ovary cells stably expressing α4 integrin (α4CHO) exhibit a gradient of PKA activity as they migrate toward wounds36. This migration-dependent PKA gradient was resolved at superresolution using live α4CHO cells expressing FLINC-AKAR1 (Fig. 4A). The difference between the normalized pcSOFI values in the leading and that in trailing regions was highly significant (WT, Fig. 4B) and absent in cells expressing the non-phosphorylatable biosensor (TA, Fig. 4C). Inhibiting PKA activity globally using H-89 markedly suppressed PKA activity in the leading front, whereas the trailing edge showed little change (Supplementary Fig. 11). The gradient observed using FLINC was approximately linear (Fig. 4D), similar to that seen using a FRET-based biosensor36. However, the superresolution images reveal that the leading front of the cell hosted many microdomains of high PKA activity, which were missing in the trailing end. With a median diameter of approximately 280 nm, these highly active microdomains in CHO cells displayed significantly different levels of PKA activity depending on their subcellular location, with most elevated activity at the filopodia. In 8/9 cells, microdomains within the filopodia displayed a significantly higher PKA activity as compared to those on the remainder of the basal membrane (Fig. 4E), consistent with the model that a pool of PKA molecules (such as integrin-anchored Type I PKA) are highly active at the very tip of migrating cells37. These data suggest that distinct PKA microdomains in the same cell can have differentially regulated amplitude and spatiotemporal characteristics.
Figure 4. PKA activity gradient of a migrating cell at superresolution.
(A) The PKA activity gradient in a migrating α4CHO cell expressing the wild-type (WT) FLINC-AKAR1. Scale bar: 10 μm. (B) Average normalized pcSOFI values within leading/trailing edge regions of migrating cells show clearly different PKA activities. (C) Migrating α4CHO cells expressing wild-type FLINC-AKAR1 show a significant gradient in terms of normalized pcSOFI values compared to cells expressing non-phosphorylatable TA mutant. Pair-wise t-test results are marked. (D) Representative PKA activity profile observed along the migration direction using FLINC-AKAR1 and its TA mutant. (E) Representative data showing that active microdomains within the filopodia harbor significantly higher PKA activity compared to elsewhere on the basal membrane of migrating CHO cells; basal membrane regions are indicated for their increasing distance from the leading edge of the migration direction (filopodia, leading-edge, front, and middle) (n = 9 cells). Unless indicated otherwise, pair-wise t-test results are marked where data were compared with the construct marked “ref”. n.s.: not-significant; *: p<0.05; **: p<0.01 where applicable. In (C) and (E), center line and whiskers mark the average and s.e.m. values, respectively.
FLINC is a general platform for biosensing
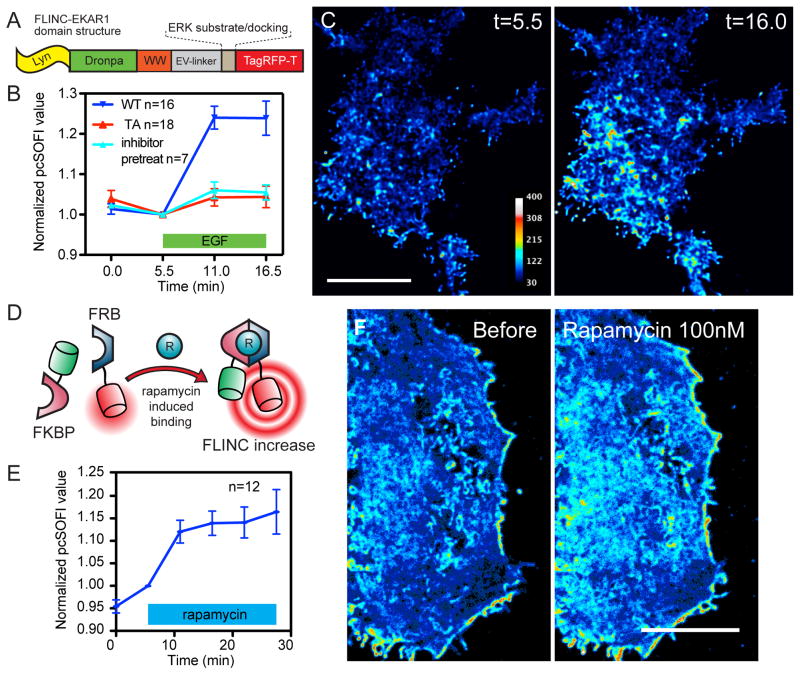
To demonstrate the generalizability of the design, we constructed a FLINC biosensor for Extracellular signal-Regulated Kinase (ERK) activity based on EKARev25 (Fig. 5A). In HEK293 cells expressing plasma membrane-targeted FLINC-EKAR1, pcSOFI values again provided consistent and robust ERK activity readouts. Stimulation of ERK using epidermal growth factor (EGF, 100 ng/mL) increased the averaged normalized pcSOFI value by 21–27% within 10 minutes (WT, Fig. 5B, n = 16). Stimulation in cells expressing a non-phosphorylatable mutant biosensor or pretreated with an upstream inhibitor of ERK (U0126, 20 μM) gave no response (TA/pretreat, Fig. 5B). The superresolution ERK activity maps (Fig. 5C) showed a similar resolution enhancement of 160 nm at second-order (Supplementary Fig. 7E).
Figure 5. FLINC-based design is generalizable.
(A) The domain structure of FLINC-EKAR1. (B) The normalized pcSOFI response time course from live HEK293 cells expressing wild-type FLINC-EKAR1 (WT, n = 16 cells), non-phosphorylatable mutant (TA, n = 18) FLINC-EKAR1, and cells expressing WT but pre-treated with the ERK upstream inhibitor U0126 (20 μM; inhibitor-pretreat, n = 7), all subjected to growth-factor stimulation (EGF 100 μM). (C) Representative superresolution images of ERK activity dynamics after growth-factor stimulation and chemical inhibition. Scale bar: 10 μm. (D) Schematic of the design and (E) Normalized pcSOFI time course of the bimolecular protein-protein interaction sensor based on FKBP-rapamycin-FRB (n = 12) in response to rapamycin (100 nM). (F) Normalized pcSOFI images of a HeLa cell before and after stimulation with rapamycin to induce the dimerization between FKBP and FRB. Scale bars: 10 μm. The color scales for the before/after images are identical within panels (C) and (F), respectively.
FLINC could also be harnessed to monitor protein-protein interactions (PPI), and we demonstrate this by using the inducible dimerization system of cytosolic FK506 Binding Protein (FKBP) and membrane targeted FKBP12-Rapamycin Binding domain (FRB, Fig. 5D)38. Addition of rapamycin (100 nM) led to the rapid heterodimerization of FKBP and FRB, which could be detected by a fast normalized pcSOFI value increase across the cell membrane that plateaued at 15% in 20 minutes (Fig. 5E/F). When optimized, FLINC can also detect weaker PPI. We split FLINC-AKAR1 into two separate constructs consisting of a diffusible FHA1-Dronpa and a membrane-targeted PKA-substrate-TagRFP-T. PKA phosphorylation of the substrate recruits cytosolic FHA1 and induce a corresponding normalized pcSOFI signal increase, even though the affinity between FHA1 and phospho-substrate is ~ 0.5 μM 39 (Supplementary Fig. 12). These examples showcase the generalizability of FLINC-based biosensors and its potential to enhance the dissection of spatially organized biochemical activities.
Discussion
The nanometer sensitivity of FLINC enabled us to emulate the powerful molecular ruler FRET40, 41 and develop new biosensors for kinase activities and PPI. Although the spatial resolution is not as high as BiFC-based superresolution methods14–16, the fact that FLINC does not require complementation and fluorophore maturation allows us to achieve reversibility and fast response in the biosensors for investigating dynamic signaling activities in live cells. FLINC-based biosensors may be created and optimized in a fashion similar to their FRET-based counterparts. We note that a FLINC biosensor utilizing the original, compact molecular switch in AKAR224 did not give a significant signal upon PKA activation, demonstrating that the linker in AKARev is necessary to maximize the dynamic range of a FLINC biosensor. In addition to intermolecular distance, the FP orientation within a biosensor will be a crucial consideration in future designs. Although FLINC is currently limited to Dronpa and TagRFP-T, future studies will test other fluorescent proteins mutants for their applicability. As in FRET-based biosensor, monomerized fluorescent proteins are preferable, although in our case we have not observed issues arising from dimerization of TagRFP-T42.
While we have shown that the relationship between pcSOFI value and FP proximity is ordinal, at this time, pcSOFI value cannot be used to directly infer molecular distance. The absolute pcSOFI values depend on biosensor expression and the optical setup (especially camera gain and excitation intensity). However, our normalization scheme largely removes the expression dependence. Therefore, normalized pcSOFI values can be compared across experiments so long as the imaging parameters remain identical. The current temporal resolution may fall short for tracking rapid changes. However, we note that pcSOFI calculations can converge to a high signal-to-noise43 very quickly (Supplementary Note). Specifically, we utilize only 4s of the 30s interval to collect data. The temporal resolution will thus improve as more sensitive and faster camera becomes available. In principle, as long as fluctuations can be observed, many types of illumination can be used to monitor FLINC, including epi-fluorescence and z-sectioning methods such as lattice light-sheet.
Under the conditions described here and previously11, diffusion does not affect the accuracy and temporal/spatial resolution of pcSOFI and its quantification of FLINC. This is because fluorophore diffusion over the total duration of pcSOFI measurement does not lead to distortions as long as the fluorophore motion within a single exposure (35 ms) is sufficiently small. Over the imaging time, we can in fact sample the local environment more thoroughly by virtue of biosensor diffusion. In our experiments, we estimated that biosensor diffusion is within a single pixel during a single exposure (Supplementary Note); simulations using published diffusion coefficients did not reveal distortions. However, depending on the local access and sensitivity to phosphatases that resets kinase biosensors, biosensor diffusion could potentially lower the spatial resolution of the kinase activity map. In this case, while the PKA activity microdomains resolved will still accurately reflect the environment experienced by any endogenous substrate, there would be a slight overestimate of the size of PKA activation zone. There is no evidence that the above issue was significant in our experiments, however, since the sizes of microdomains measured by the both superresolution methods STORM and FLINC were in good agreement.
To our knowledge, this study represents the first demonstration of superresolution imaging of dynamic enzymatic activities such as protein kinase activities in living cells. PKA substrates are known to be more rapidly phosphorylated when localized to AKAP complexes44, consistent with high PKA activity in the vicinity of AKAP-assembled signalosomes. We can now visualize the activity zone of these nanomachines, which are shown to be larger than the intrinsic flexibility of the PKA holoenzyme complex45. The significant self-clustering in AKAP79 and co-clustering of AKAP79 and phospho-PKA substrate signals suggest that the clustering of AKAP complexes, and hence an increased effective concentration of PKA regulatory subunits, may serve as an efficient means to re-capture dissociated C subunit and maintain PKA activity compartmentalization. Interestingly, 24% of total AKAP79 were orphaned and not clustered with phospho-PKA substrate. This and other data suggest that AKAPs may maintain microdomains with varying PKA activity levels, evolving dynamically based on the associated regulatory partners such as adenylyl cyclases, phosphodiesterases, kinases, and phosphatases46. As PKA represents a classical example of compartmentalized signaling, other biochemical activities may be under similarly precise spatiotemporal regulation. These spatial details and their functional roles could be illuminated with this new class of FLINC biosensors.
METHODS
Plasmid and construct generation
All mammalian constructs were cloned using the pcDNA3.0 vector with a modified multiple cloning site. All E. coli constructs were cloned using the pRSET-B vector. Plasmids were generated through typical molecular cloning methods using a combination of polymerase chain reaction (PCR) and restriction enzyme cloning. PCR was always performed using the Phusion polymerase (New England BioLabs) unless noted. Cloning and subcloning were performed using E. coli DH5α strain.
Mutagenesis (error-prone PCR and site-directed)
All site-directed mutagenesis experiments were performed following a published protocol 1 using the E. coli JM109 strain. The randomized screens at N102 of Dronpa and D159 of TagRFP-T were both performed on a wild-type tandem DpTT template using fully degenerate primers (NNN). Error-prone PCR was performed using Taq polymerase and an in-house dNTP mixture. Eight rounds of consecutive PCR were performed (on Dronpa only) using a reverse primer containing the 8-amino acid linker at the 3′ end. This ensured that the linker was not subject to mutation. The mutation rate was approximated through sequencing, and the 3rd-to-5th-round PCR products (approximately < 2% point mutation rate) were selected for further cloning. These mutant Dronpa fragments were ligated to a linearized pRSET-B plasmid containing wild-type TagRFP-T using the sites BamHI/KpnI. All of the ligate was transformed into JM109 cells and plated onto LB-Amp agar. The candidate mutants were then screened on the basis of colony fluorescence. Once a candidate was chosen, its fluorescence intensity was confirmed by averaging the fluorescence of many colonies over a larger area on the LB-Amp agar plate.
Mutant screening
Expression of TagRFP-T causes an intense salmon/pink color due to the chromophore absorption. The tandem DpTT construct showed a faint but visible color. During error-prone or site-directed mutagenesis screening, we rejected any mutant that did not display this color, thereby excluding badly folded mutants and those containing premature stop codons and frame shifts. To quantify the effects of the mutations on mutant DpTT constructs, E. coli (JM109) expressing the mutants were streaked onto LB-Amp agar plates. The mean intensity of each mutant in the GFP or RFP channel was measured using the same exposure settings on an in-house fluorescence imager, illuminated by a broad-spectrum lamp source (MAX-303, Asahi Spectra) and monitored by a Thorlabs USB digital camera mounted behind a Thorlabs emission filter wheel (maxima/bandwidth in nm; CFP: 430/40; GFP: 535/40; RFP: 630/75). For comparison, each plate was streaked with DpTT as well as TagRFP-T colonies as normalizing controls. The images were analyzed using ImageJ (v. 1.47g). The site-directed, randomized screen at N102 of Dronpa was carried out following the same criteria outlined above. Only brightly fluorescent colonies (in both GFP/RFP channels) were sequenced.
Cell culture, transfection, and pretreatment
The HeLa and HEK293 cells utilized for these experiments were between passages 20–65. Cells were maintained in DMEM growth media supplemented with 10% FBS and 1% penicillin/streptomycin. CHO cells (passage 8–22) stably expressing α4 integrin were maintained in DMEM F12 medium with 10% FBS and 1% penicillin/streptomycin. All cells were transfected at an approximate confluency of 70% using Lipofectamine 2000 and incubated for 24 hours before imaging. All cells were imaged in HBSS buffer at room temperature. To achieve AKAP-PKA RII disruption, HeLa cells were incubated with 5 μM of the synthetic disruptor peptide STAD-2 (or its scrambled control) in DMEM growth media for 6 hours at 37°C and 5% CO2. Immediately prior to imaging, the HBSS imaging buffer was supplemented with the appropriate peptide at 5 μM. For migration studies, CHO cells were plated on cover slips coated overnight with 10 μg/mL human fibronectin (in DPBS). Cells were grown to a monolayer and wounded using a 200-μL pipette tip, then washed and incubated in HBSS buffer for 1 hour prior to imaging to induce migration.
Epifluorescence imaging
All epifluorescence imaging was performed on a Zeiss Axiovert 200M Microscope equipped with a xenon lamp and a cooled CCD, under a 40X oil immersion objective. FRET microscopy of GFP/RFP biosensors such as FLINC-AKAR1 was performed using the following excitation/emission filter combinations (maxima/bandwidths in nm): GFP – EX480/30, EM535/45; RFP – EX568/55, EM653/95; FRET: EX480/30, EM653/95. All epifluorescence experiments were subsequently analyzed using the MetaFluor software.
Total Internal Reflection Fluorescence (TIRF) imaging
All pcSOFI TIRF imaging was performed on either a Nikon Eclipse Ti microscope equipped with a Photometric Evolve 512 EMCCD, a Melles Griot argon laser (GFP excitation: 488 nm), and a Coherent Sapphire solid-state laser (RFP excitation: 561 nm) or the very similar Nikon N-STORM/TIRF microscope equipped with an Andor IXON3 Ultra DU897 electron-multiplying CCD camera. Cell were always imaged under a 100X oil immersion objective (NA 1.49) without further magnification. The TIRF condition was created using a commercial TIRF mirror setup from Nikon and controlled electronically through the imaging software (Nikon NIS-Elements v3.22.00). All TIRF imaging was performed with electronic focus drift correction. All TIRF images or time-lapse series were captured in 16-bit without binning at 35-ms exposure. For the FLINC dataset, we estimate that the 561 nm (RFP) laser intensity at the sample is 24 W/cm2. The multiplier gain and laser power were kept consistent across all FLINC experiments involving wild-type Dronpa (DpTT, FLINC-AKAR1, FLINC-EKAR1, FLINC-AKAR1-TA, FLINC-EKAR1-TA, and FKBP-Dronpa/FRB-TagRFP-T-CAAX) reported in this article. However, since mutant and control constructs displayed disparate intensities, multiplier-gain and laser power were varied in order to obtain proper exposure. We performed laser and multiplier gain calibration, and found both to be linear. Therefore, the data presented for mean intensity comparison between wild-type DpTT, mutant DpTT, and controls (Supplementary Fig. 1B, D, E) were normalized to a gain of 1 and laser power of 100% by applying the appropriate correction factor. Skewness calculations were performed using home-built Matlab software, and pcSOFI calculation and normalization were performed using the Localizer software. Post-processing algorithms are described in detail in Supplementary Note.
Statistical Methods
All statistical significance was tested using Welch’s t-test (two-sided, 95% confidence interval). All error bars mark the values of the standard error of the mean (s.e.m.). For pcSOFI and STORM measurements, single-cell responses were summarized from at least two separate experiments, each with multiple cells, over at least two passages. In the case of migrating CHO cells, cells in both wild-type and TA groups were excluded if their migration direction could not be unambiguously established.
Size-exclusion chromatography (SEC)
All SEC experiments were performed on a GE ÄKTA protein purification system using a Superdex 200 column at a flow rate of 0.2 mL/min. Affinity-purified Dronpa and TagRFP-T protein stock solutions were first further purified individually using SEC. The pure fractions of each FP were then pooled, mixed at a 1:1 molar ratio, and concentrated to a total protein concentration of approximately 120 μM; the stoichiometry was verified at 1.00:1 using absorption spectroscopy followed by spectral deconvolution. A total of 150 μL of the mixed stock solution was subsequently examined with the same SEC parameters to resolve any additional oligomeric states present in the concentrated mixture (Supplementary Fig. 2A). Collected fractions were subjected to absorption spectroscopy and spectral deconvolution in order to determine the concentration of Dronpa and TagRFP-T in each. A peak at approx. 92 kDa that was not observed in the chromatographs of the individual FPs was found to contain a stoichiometric ratio of approximately 2.5 TagRFP-T molecules to 1 Dronpa. It is known that TagRFP-T forms dimers at high concentrations. Since Dronpa and TagRFP-T are each 30kDa, both size and stoichiometry verified the presence of a trimeric FP complex between a TagRFP-T dimer and a Dronpa monomer.
Analytical ultracentrifugation (AUC)
All sedimentation velocity (SV) experiments were conducted in a Beckman-Coulter Optima XL-A analytical ultracentrifuge at 50,000 rpm and 20°C. A set of initial SV experiments were performed using purified TagRFP-T, Dronpa, and their mixture at the concentrations of 12, 10, and 12/10 μM, respectively, which found that the association constant was weaker than 104 M−1. Three further SV experiments were then collected using highly concentrated proteins: (1) TagRFP-T alone at 171 μM detected at 595 nm; (2) Dronpa alone at 172 μM detected at 435 nm; and (3) a mixture of TagRFP-T (171 μM) and Dronpa (172 μM) detected at 595 nm. The individual Dronpa and TagRFP-T samples were fit to normalized g(s*) distributions using DCDT+ (version 2.4.1) with apparent weight average sedimentation coefficients equal to 2.4 S and 3.7 S, respectively. The sedimentation coefficient distribution function for the mixed sample was poorly described by a sum of non-interacting species, indicating an association between the two components. However, the sedimentation coefficient for the Dronpa+TagRFP-T complex could not be accurately determined because the population of the species was low. To obtain an estimate of the association constant, we further analyzed the data using SedAnal (version 6.01.6926), which allows direct boundary fitting using mass action equations. The above sedimentation coefficients were used in an initial fitting attempt to a simple A+B ↔ C association model, which converged poorly. We reasoned that the sedimentation coefficients may be depressed in the mixed sample due to molecular crowding and therefore set them to values 10% lower than their original estimates for subsequent fitting using a range of sedimentation coefficients for the complex to obtain estimates of the association constant. A typical fit is shown in Supplementary Fig. 2B. The calculated maximum sedimentation coefficient for a spherical shape of the Dronpa + TagRFP-T complex is 5.95 Sv and was found using the following equation:
Any asymmetry in the complex shape would lead to lower sedimentation coefficients. Because the complex sedimentation coefficient could not be experimentally determined, we fit the data by fixing it at several different values to obtain the subsequent association constant. These are shown in Table 1, in which the Ka varies between 1090 M−1 and 6280 M−1 for reasonable values of the complex sedimentation coefficient. Using these estimates, an A + B ↔ C model yields an estimated Kd in the range of 159 μM to 917 μM.
Table 1.
Summary of SedAnal best fit values of the association constant obtained using estimates of the Dronpa+TagRFP-T complex sedimentation coefficient. This range of Ka values provides limits for the association constant between Dronpa and TagRFP-T.
| s(Dronpa+TagRFP-T) (Svedbergs) | Ka (M−1) | Std. Deviation |
|---|---|---|
| 4.00 | 6.288E+3 | 1.10E-2 |
| 4.25 | 3.610E+3 | 1.14E-2 |
| 4.50 | 2.491E+3 | 1.17E-2 |
| 4.75 | 1.891E+3 | 1.19E-2 |
| 5.00 | 1.520E+3 | 1.21E-2 |
| 5.25 | 1.268E+3 | 1.23E-2 |
| 5.50 | 1.089E+3 | 1.24E-2 |
Single-molecule fluorescence characterization
For single-molecule imaging, 30-mm round cover glasses (No. 1, VWR Corporate Headquarters, PA) were sequentially cleaned by ultrasonication in 1% Alconox (Alconox Inc, NY), water, ethanol and 1M KOH. PEG and Biotin-PEG (Laysan Bio) were coated on the clean cover glass following the protocol in Roy, et al. 2008. A Hybriwell (Secure-Seal, Grace) is pasted on the cover glass and sealed by epoxy. The assembled chamber was incubated with A) 0.01mg/ml neutravidin B) 1mM Biotin-NTA (Biotium) in PBS for 10min sequentially. Each step was followed by 1X PBS wash for twice. His-tagged purified TagRFP-T or DpTT was diluted in 1X Tris buffer with different salt (300–3000mM NaCl) concentration to a final protein concentration about 100 pM. Immobilization was done by incubating the final TagRFP-T or DpTT solution in the chamber for 10 min and wash by the same buffer once. Single molecule fluorescence imaging was performed in the buffer without protein on an Olympus IX-71 inverted microscope under a 60X, 1.45 NA TIRFM objective. A home-built optical system with 561 nm solid-state excitation laser (Sapphire, Coherent, Santa Clara, CA) and an EMCCD camera (iXon DU897E, Andor Technology, Belfast, Northern Ireland) was used to examine the samples (emission: ZET488/561m, Chroma Technology, Rockingham, VT). An iris aperture was used to restrict the laser spot size to 150×150 pixels (~700 μm2). The laser power density was attenuated by a ND filter (NE10A, Thorlabs, NJ) to 12 W/cm2. Single-molecule image streams were collected with maximum EM Gain (300) in 16-ms integration time for 2000 frames. Molecules were detected by local maximum searching using ImageJ plugin ThunderSTORM 2. Aggregated molecules were filtered by removing high intensity spots (>3sigma). The spots closer than 1 pixel (267nm) were discarded to guarantee signal is from single molecules. Signal of the spots in every single frame was integrated from a 3×3 ROI centered at the ThunderSTORM detections. Background was subtracted by the whole frame averaged intensity. The real photon count is obtained per instruction from camera manufacturer (Andor). The autocorrelation curves were calculated from the intensity trace of individual spots in Matlab (xcorr function) and normalize to the autocorrelation value at time lag of zero. All autocorrelation curves were averaged to a single autocorrelation function (ACF) for each condition.
Photophysical characterizations
For dark-state conversion (DSC) measurements, HeLa cell expressing nuclear-localized TagRFP-T/DpTT were illuminated with a 561 nm solid-state diode-pumped laser (Genesis MX, Coherent) through a custom-built inverted microscope at an intensity of ~ 8–10 kW/cm2. A 629/56 nm band-pass filter (Semrock) was used to remove excitation from emission. Epi-fluorescence from the cells was collected using a PMT (Hamamatsu Photonics) and subsequently the PMT photo-current was converted to voltage using a custom-built trans-impedance operational amplifier for better signal to noise ratio. Fluorescence signal was digitized using a data acquisition card (DAQ, National Instruments) at a rate of 1 MHz. Pulsed illumination was achieved using an acousto-optic modulator (AOM, Gooch & Housego, 35210-BR). Cells expressing FPs were bleached continuously for 0.1 s and fluorescence traces were collected for further analysis. The raw fluorescence data were background-corrected and spline-fitted for smoothing. The fluorescence traces had three distinct features: a rapid decay up to 100 μs time-scale, a flat region in 100–1000 μs region, followed by a slow decay in ms time-scale. Hence, the fluorescence traces were fitted with a 3-exponetial function using equi-distant time-points in log scale. The fastest time-constants and its corresponding amplitude from the fitting results were considered as DSC time-constants and percent DSC respectively 3. To measure the ground-state recovery (GSR) time-constants, cells expressing nuclear-localized TagRFP-T/DpTT were excited with pulse-trains having 2 ms exposure time and varying inter-pulse delays (dark time) ranging from 5 μs to 10 ms. Fluorescence traces collected using the pulsed excitations were used to compute percent recoveries using the following equation without further processing: PR=(FR−FB)/(FL−FB)×100, where PR is the percent recovery. FL, FB, FR are the fluorescence intensities at initial point, after 2 ms exposure, and after the recovery between pulses, respectively. To measure the GSR time-constants, FPs were excited with pulse-trains having 2 ms exposure time and varying inter-pulse delays (dark time) ranging from 10 μs to 50 ms. A home-built software (Matlab) was used to compute percent recoveries of the fluorescence and finally percent recovery vs. dark time plots were fitted with single exponential to extract GSR time-constants 4.
Excited state lifetimes of purified TagRFP-T and fluorescent DpTT were measured on a Fluro Time 100 commercial TCSPC system (PicoQuant) using 560 nm laser excitation with a repetition rate of 2.5 MHz. Lifetime measurements were performed using purified proteins in PBS buffer (pH=7.4) at appropriate dilutions. Supplementary Fig. 3D displays the fluorescence decays of the proteins and instrument response function (IRF). The fluorescence traces of the FPs were fitted with iterative reconvolution with a bi-exponential function and using measured IRF of the system. Intensity-weighted average excited state lifetime of TagRFP-T and DpTT were 2.45 ns and 2.40 ns, respectively.
Direct Stochastic Optical Reconstruction Microscopy (dSTORM) imaging
The primary antibodies used in these experiments were mouse anti-AKAP79 (BD Biosciences, 610314) and rabbit anti-Phospho-(Ser/Thr) PKA Substrate (“pPKAsub”, Cell Signaling, 9621). The AKAP79 antibody was used 1:50 dilution; the anti-Phospho-(Ser/Thr) PKA substrate antibody was used at 1:200 after a dilution series test was conducted. The secondary antibodies used in these experiments were AlexaFluor® 568 and AlexaFluor® 647 (Life Technologies) at dilutions of 1:2000 and 1:1000, respectively. Saponin from Quillaja Bark, sodium borohydride, glutaraldehyde (GA), and normal goat serum (NGS) were from Sigma-Aldrich; paraformaldehyde (PFA) from Electron Microscopy Sciences; glycine from Fisher Scientific; bovine serum albumin from Roche Diagnostics. Cells plated in 35mm dishes were washed three times in HBSS and appropriately pre-treated (or no pre-treatment) for 15 min. at 37° C, then washed with PBS before fixation using 4% PFA, 0.2% GA PBS for 10 minutes at room temperature. All subsequent steps were performed on a shaker; saponin was always used at 0.0005%; all solutions were at pH 7.4. Cells were quickly rinsed in PBS after fixation and quenched with freshly made 0.1% NaBH4 ice-cold PBS. Following 3X PBS washes of 5 min. each, cells were permeabilized and blocked for 1 hour at room temperature in 10% NGS + saponin PBS, followed by 3X saponin PBS washes of 5 min. each. Primary antibodies were made to working concentrations in 1% BSA + saponin PBS. Cells were incubated with primary overnight at 4° C, followed by 3X saponin PBS washes of 5 min. each. Secondary antibodies were made working concentrations in 1% BSA + saponin PBS. Cells were incubated with secondary antibodies for 1 hour at room temperature in the dark. All remaining steps were performed in the dark. Cells were washed 3X saponin PBS for 5 min. each followed by washing 3X PBS for 5 min. each. All samples were post-fixed in 4% PFA PBS for 10 min. at room temperature, and quickly rinsed in PBS and quenched with 3X 100mM glycine PBS for 5 min. each. The cells were washed 3X PBS for 5 min. each and stored in PBS at 4°C in the dark until imaging within 24 hours. All STORM imaging was performed on a Nikon Ti microscope under total-internal reflection (TIR) condition, using a 100X/1.49 NA Apo TIRF objective. Images were collected within a 256 × 256 pixel ROI on an Andor IXON3 Ultra DU897 electron-multiplying CCD camera using the multicolor continuous mode setting in the Nikon Elements software. Power of the 568 nm and 647 nm excitation (as well as activating 405 nm excitation) was adjusted to enable stochastic blinking behavior within the ROI. The images were reconstructed and drift-corrected using the Nikon N-STORM software. Post-processing of the STORM data is described in Supplementary Note.
Supplementary Material
Table 2.
Summary of photophysical measurements. Average values for fluorescence lifetime, percentage of dark-state conversion possible in the sample (% DSC), time constant of DSC (τDSC), and time constant of GSR (τGSR) are reported. See also Supplementary Fig. 3.
| Constructs | Lifetime (ns) | % DSC | τ DSC (μs) | τ GSR (μs) |
|---|---|---|---|---|
| TagRFP-T | 2.45 | 19 (± 4.0) | 24 (± 5.8) | 50 |
| DpTT | 2.40 | 24 (±1.8) | 18 (± 1.4) | 51 |
Acknowledgments
The authors acknowledge Qiang Ni, L. Mario Amzel, Sohum Mehta, and Pablo A. Iglesias for critical reading of the manuscript, and Xin Zhou for aid in protein purification. This work was supported by: NIH DP1 CA174423, R35 CA197622 and R01 DK073368 (to J.Z.); R01 GM079440, T32 GM008403, and NSF MCB 141210B (to K.G.F.); R01CA74305 (to P.C.); NCI1K22CA154600 and 1R03A188439 (to E.J.K.); Research-Foundation Flanders (FWO-Vlaanderen) postdoctoral fellowship and KU Leuven Research Professorship (to P.D.); and a graduate research fellowship from the NSF DGE-1232825 (to A.M.P.). R.J. acknowledges NIST and the NSF Physics Frontier Center at JILA for support.
ACCESSION CODES
Addgene codes are available for the following constructs:
DpTT - 87704
FLINC-AKAR1 - 87705
FLINC-AKAR-TA - 87706
FLINC-EKAR1 - 87707
FLINC-EKAR-TA - 87708
FKBP-Dp - 87709
FRB-TT-CAAX - 87710
Dp-FHA1 - 87711
PKAsub-TT-CAAX - 87712
PKAsub(TA)-TT-CAAX - 87740
SOFTWARE AVAILABILITY
“Localizer” software is freely available at https://bitbucket.org/pdedecker/localizer. The Matlab software is available upon reasonable request.
AUTHOR CONTRIBUTIONS
GM and JZ made the initial discovery and designed all experiments. PD was also involved in the initial proposal of the research direction. GM, BR, FH, PM, EG, XY, CB, AMP, ZC and KGF performed experiments and analyzed data with input from JZ, JX, RJ, AP, KGF, and PC. YW and EJK provided reagents. GM, BR, EG, and PD developed and validated all post-processing algorithms; PD formalized the normalization approach. GM and JZ wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
DISCLAIMER
R.J. is a staff member in the Quantum Physics Division of the National Institute of Standards and Technology (NIST). Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
DATA AVAILABILITY
The data that support these findings are available upon reasonable request.
References
- 1.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nature Reviews Molecular Cell Biology. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 2.Ganesan A, Zhang J. How cells process information: Quantification of spatiotemporal signaling dynamics. Protein Science. 2012;21:918–928. doi: 10.1002/pro.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nature Reviews Molecular Cell Biology. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 4.Rizzuto R, Pozzan T. Microdomains of intracellular Ca2+: Molecular determinants and functional consequences. Physiol Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- 5.Cambi A, Lidke DS. Nanoscale Membrane Organization: Where Biochemistry Meets Advanced Microscopy. Acs Chemical Biology. 2012;7:139–149. doi: 10.1021/cb200326g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta P, van Engelenburg SB, Lippincott-Schwartz J. Superresolution Imaging of Biological Systems Using Photoactivated Localization Microscopy. Chemical Reviews. 2014;114:3189–3202. doi: 10.1021/cr400614m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Bates M, Zhuang X. Annual Review of Biochemistry. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dedecker P, Mo GCH, Dertinger T, Zhang J. Widely accessible method for superresolution fluorescence imaging of living systems. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10909–10914. doi: 10.1073/pnas.1204917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rego EH, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E135–E143. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, et al. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nature Communications. 2014;5 doi: 10.1038/ncomms5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickerson A, Huang T, Lin L-J, Nan X. Photoactivated Localization Microscopy with Bimolecular Fluorescence Complementation (BiFC-PALM) for Nanoscale Imaging of Protein-Protein Interactions in Cells. Plos One. 2014;9 doi: 10.1371/journal.pone.0100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertel F, Mo GCH, Duwe S, Dedecker P, Zhang J. RefSOFI for Mapping Nanoscale Organization of Protein-Protein Interactions in Living Cells. Cell Reports. 2016;14:390–400. doi: 10.1016/j.celrep.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 18.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Engineering. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 20.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 21.Bourgeois D, Adam V. Reversible photoswitching in fluorescent proteins: A mechanistic view. Iubmb Life. 2012;64:482–491. doi: 10.1002/iub.1023. [DOI] [PubMed] [Google Scholar]
- 22.Newman RH, Fosbrink MD, Zhang J. Genetically Encodable Fluorescent Biosensors for Tracking Signaling Dynamics in Living Cells. Chemical Reviews. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissbuehler S, et al. Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nature Communications. 2014;5 doi: 10.1038/ncomms6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Molecular Biology of the Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacskai BJ, et al. SPATIALLY RESOLVED DYNAMICS OF CAMP AND PROTEIN KINASE-A SUBUNITS IN APLYSIA SENSORY NEURONS. Science. 1993;260:222–226. doi: 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- 27.Chen CH, Nakamura T, Koutalos Y. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophysical Journal. 1999;76:2861–2867. doi: 10.1016/S0006-3495(99)77440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neves SR, et al. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolaev VO, Bunemann M, Hein L, Hannawacker A, Lohse MJ. Novel single chain cAMP sensors for receptor-induced signal propagation. Journal of Biological Chemistry. 2004;279:37215–37218. doi: 10.1074/jbc.C400302200. [DOI] [PubMed] [Google Scholar]
- 30.Saucerman JJ, Greenwald EC, Polanowska-Grabowska R. Mechanisms of cyclic AMP compartmentation revealed by computational models. Journal of General Physiology. 2014;143:39–48. doi: 10.1085/jgp.201311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harootunian AT, et al. Movement of the Free Catalytic Subunit of cAMP-Dependent Protein-Kinase Into and Out of the Nucleus Can Be Explained by Diffusion. Molecular Biology of the Cell. 1993;4:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen W, et al. FACTORS THAT INFLUENCE THE NUCLEAR ACCESSIBILITY OF THE CATALYTIC SUBUNIT OF THE CAMP-DEPENDENT PROTEIN-KINASE. Faseb Journal. 1994;8:A1226–A1226. [Google Scholar]
- 33.Dessauer CW. Adenylyl Cyclase-A-kinase Anchoring Protein Complexes: The Next Dimension in cAMP Signaling. Molecular Pharmacology. 2009;76:935–941. doi: 10.1124/mol.109.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodge K, Scott JD. AKAP79 and the evolution of the AKAP model. Febs Letters. 2000;476:58–61. doi: 10.1016/s0014-5793(00)01671-9. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Isoform-Selective Disruption of AKAP-Localized PKA Using Hydrocarbon Stapled Peptides. ACS Chem Biol. 2014;9:635–642. doi: 10.1021/cb400900r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim CJ, et al. Integrin-mediated Protein Kinase A Activation at the Leading Edge of Migrating Cells. Molecular Biology of the Cell. 2008;19:4930–4941. doi: 10.1091/mbc.E08-06-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim CJ, et al. alpha 4 integrins are Type I cAMP-dependent protein kinase-anchoring proteins. Nature Cell Biology. 2007;9:415–U496. doi: 10.1038/ncb1561. [DOI] [PubMed] [Google Scholar]
- 38.Inoue T, Do Heo W, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nature Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durocher D, et al. The molecular basis of FHA Domain : Phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 40.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Applications of Chimeric Genes and Hybrid Proteins Pt B. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 41.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; 2010. [Google Scholar]
- 42.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nature Methods. 2012;9:480–U498. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandenberg W, et al. Model-free uncertainty estimation in stochastical optical fluctuation imaging (SOFI) leads to a doubled temporal resolution. Biomedical optics express. 2016;7:467–480. doi: 10.1364/BOE.7.000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Ma YL, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith FD, et al. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. Elife. 2013;2:e01319. doi: 10.7554/eLife.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gold MG, et al. Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6426–6431. doi: 10.1073/pnas.1014400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 1.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic acids research. 2000;28:E78–E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ovesny M, Krizek P, Borkovec J, Svindrych ZK, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean KM, et al. Analysis of Red-Fluorescent Proteins Provides Insight into Dark-State Conversion and Photodegradation. Biophysical Journal. 2011;101:961–969. doi: 10.1016/j.bpj.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manna P, Jimenez R. Time and Frequency-Domain Measurement of Ground-State Recovery Times in Red Fluorescent Proteins. Journal of Physical Chemistry B. 2015;119:4944–4954. doi: 10.1021/acs.jpcb.5b00950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support these findings are available upon reasonable request.