Abstract
Controllable rotational manipulation enables multi-dimensional imaging and rapid screening of single cells and model organisms. Current approaches to rotationally maneuver small objects depend on optical, magnetic, or electrical properties of the sample under investigation. This dependence renders the existing methods sample-specific which limits their applicability. Here we present a new rotational manipulation method based on oscillating sidewall sharp-edge microstructures and thin glass slides in a microchannel. This method is independent of the intrinsic properties of sample under investigation and can be effectively applied to particles, cells, and multicellular organisms.
Keywords: acoustofluidics, rotational manipulations, C. elegans, microfluidics
Graphical Abstract
Controllable rotational manipulation of microscale objects benefits various fields including single cell analysis,[1–3] model organism studies,[4,5] and nanotechnology.[6–8] For example, in single cell studies, heterogeneity in cell populations necessitates additional capabilities in single cell manipulation in order to acquire comprehensive phenotypic, genotypic, and functional intercellular characteristics.[9,10] Furthermore, in model animal studies including small organism microsurgery and phenotyping, multi-dimensional optical imaging is crucial to obtain precise cellular and subcellular positional identities. [11–13]
Different approaches have been practiced to generate rotational manipulation at the microscale.[14–19] Optical tweezers were used to rotate birefringent particles and single cells using complex optical setups.[15,20,21] Nonetheless, laser-generated heating renders the optical methods unreliable for biological samples.[22] Magneto rotation of particles and biological cells require magnetic property which is provided through an additional step of insertion of nanoparticles into the cells.[1,7] Electric field based methods have also been applied to various nanotubes and single cells but they require special sample and/or medium properties that are often unavailable.[8,23] For rotational orientation of model organisms, only few methods have been proposed including stepper motors[4,5] and lateral orientation through predefined microchannel geometries.[13] Stepper motor based approaches have advantages in rapid operation and dynamic control but this technique lacks simplicity, dexterity, and on-chip adaptability. Passively orienting worms using curve channels is highly limited in control, and lacks the precision in angle of rotation. Recently, we have developed an acoustofluidic based rotational manipulation method utilizing acoustic bubble generated microstreaming flows.[24] Even though this method has various advantages over the existing rotational manipulation technologies, the variation of the acoustic parameters due to the changing microbubble size and geometry is undesired for longer period of operation. High-frequency travelling waves have also been reported to enable fluidic actuation and manipulation of suspended objects; however, this method has not been used in manipulating model organisms yet.[25–27] Therefore, there is an unmet need for a robust and simple method for on-chip rotational manipulation of specimens from single cell level entities to complex model organisms.
Here, we present an acoustofluidic[28–34] device that achieves highly tunable on-chip rotational manipulation of single HeLa cells and model organism Caenorhabditis elegans (C. elegans) using acoustic waves. In particular, we employ steady streaming microvortices generated by oscillating solid structures in an acoustic field where the cells or the organisms are being rotated by the torque generated via the streaming flows. Compared with the existing technologies, the presented acoustofluidic rotation method is robust, biocompatible, independent of optical, magnetic or electrical properties of specimen and adoptable to existing microfluidic platforms.
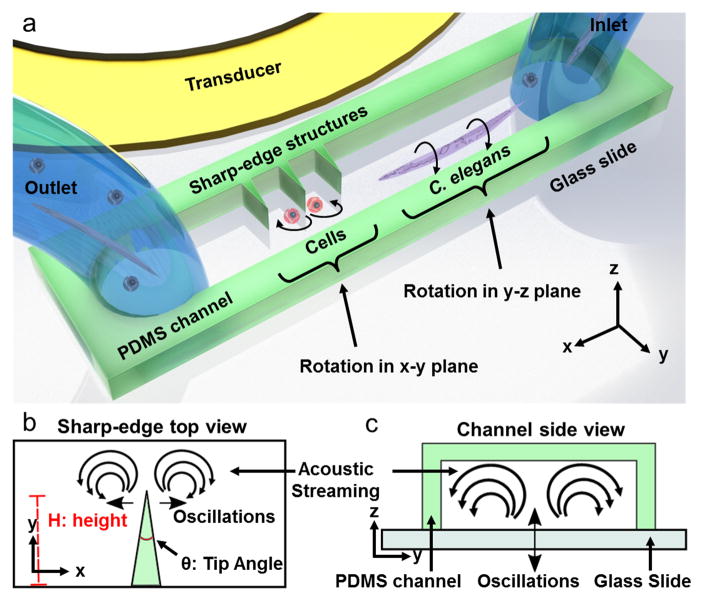
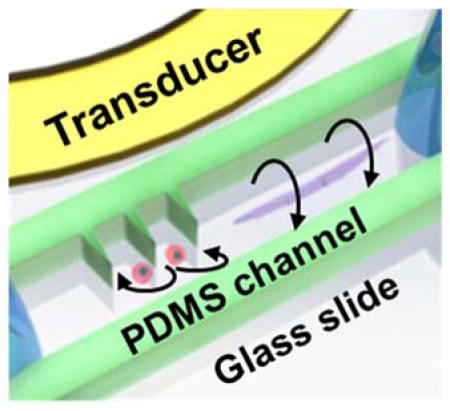
Our acoustofluidic device (Figure 1a) includes a polydimethylsiloxane (PDMS) microfluidic device adjacent to a piezoelectric transducer which provides the acoustic field. The PDMS channel features one inlet and one outlet for loading and unloading the samples into the device. Both the microchannel and the piezoelectric transducer are bonded on a glass slide of thickness ~170 μm. The rotational manipulation device consists of side wall sharp-edge structures[35–37] and bare channel parts (see Figure S1). In a single device, using the same method of acoustic actuation, we aim to generate in-plane (x-y plane) and out-of-plane (y-z plane) rotational manipulation using oscillations of the sharp-edges (Figure 1b) and the glass slide (Figure 1c), respectively.
Figure 1.
Schematic and the working principle of the acoustofluidic rotational manipulation device. (a) A simple PDMS channel contains both sharp-edge and bare regions for the cell and worm rotation, respectively. Oscillations of (b) PDMS sharp-edge structures and (c) glass slide generate circulating streaming flows that are used for rotational manipulation.
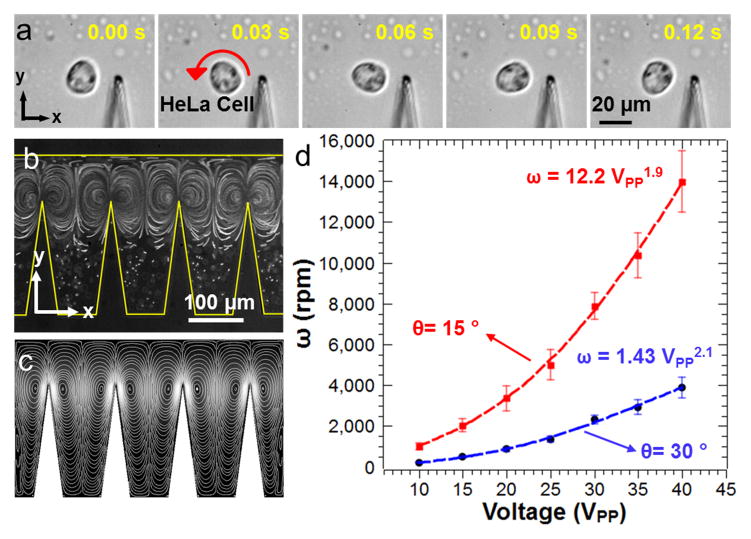
In-plane rotational manipulation of one HeLa cell around the tip of a sharp-edge structure is shown in Figure 2a. The oscillating sharp-edge structures generate two counter rotating microvortices in the surrounding fluid due to viscous dissipation in the microchannel (Figure 2b,c). To demonstrate acoustic streaming generated by the oscillating sharp-edges, we performed numerical simulations, by following an approach similar to the one reported previously.[35] Specifically, we employ a perturbation approach to solve the governing equations (i.e. balance of mass and balance of linear momentum) for the motion of a linear viscous compressible fluid, such that the flow variables are split into two parts: (1) the first-order components, which are assumed to be time-harmonic and are indicative of the acoustic response of the fluid, and (2) the second-order components, which are assumed to be time-independent and are indicative of the acoustic streaming response of the fluid. Further details about this approach are provided in the Supporting Information. Once the circulating streaming flows are established, nearby HeLa cells are trapped at the center of the microvortices. Depending on the initial concentration of the cell solution, it is possible to trap and rotate two HeLa cells by one sharp-edge structure (Movie 1, Supporting Information). Rotation rate of the trapped HeLa cells mainly depends on the peak-to-peak voltage (VPP) applied to the piezoelectric transducer. Oscillation amplitude of the sharp-edge structures also increases with increasing length (see the Supporting Information) and decreasing tip angle. Rotational rates of HeLa cells exceeding 14,000 RPM are demonstrated using 15° tip angle and 200 μm height for the sharp-edge structures (Figure 2d). As the tip angle was increased to 30°, the speed of the circulating flows decreased which in turn reduced the rotational speed of the HeLa cells. Height of the sharp-edge structures also affects the rotational speeds of the cells; the 100 μm sharp-edge structure results in lower rotational speed for the HeLa cells compared to the 200 μm structure with the same tip angle of 30° (see Figure S2). The flexibility in rotational speed renders this method an excellent tool for single cell studies where shear force on the rotating cells can be tuned on-demand. Rooney and Nyborg et al estimated the shear stress near oscillating wires and air bubbles that caused hemolysis above certain oscillation threshold.[38,39] A similar estimation of the shear stress generated near the sharp-edge structures oscillating at ~5 kHz frequency driven at 10 VPP (~0.5 μm oscillation amplitude) and 40 VPP (~5 μm oscillation amplitude) resulted roughly in 10 and 987 dynes/cm2, respectively. Magnitudes of the shear stresses on this order as continuous or pulsatile were reported to be sufficient to alter endothelial cell function and result in DNA fragmentation in human SH-SY5Y neuroblastoma cell line.[40,41] In addition, fluid shear stress applied to suspended cells was found to be an important parameter for cell growth rate, volume, and metabolite production.[42,43] As a potential application, our acoustofluidic rotational manipulation method could be used in related single cell studies in order to investigate the effect of rotation on cell fate and drug response in variable hydrodynamic shear conditions. Another benefit of using our acoustofluidic rotational manipulation in single cell mechanobiology studies is the easy tracking and observation of individual cells thanks to the fixed position rotation at each sharp-edge structure.
Figure 2.
In-plane rotation of HeLa cells. (a) The oscillating sharp-edge structure traps and rotates a HeLa cell through acoustic streaming vortices that are visualized (b) experimentally using 1 μm fluorescent polystyrene beads and (c) numerically by a perturbation approach. (d) The rotation rate of the HeLa cells can be tuned by the applied peak-to-peak voltage and the geometry of the sharp-edge structures. Smaller sharp-edge angles and longer sharp edges yield higher rotation rates.
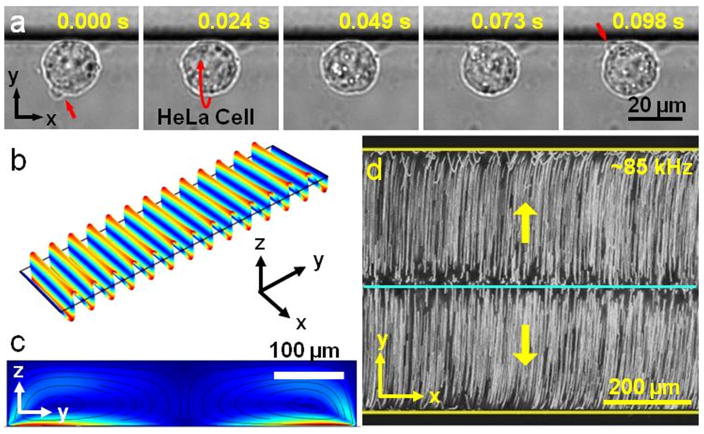
Figure 3a shows the out-of-plane rotational manipulation of one HeLa cell in the bare section of the microfluidic device at a relative higher driving frequency (~85 kHz) compared to the one used in sharp edge induced in-plane rotational manipulation (Movie 2, Supporting Information). Here, the HeLa cell is positioned near the microchannel wall due to the wall effect,[31] and rotated by the torque generated through the out-of-plane streaming flows. To generate the out-of-plane streaming flows, different vibrational modes of the glass slide are utilized. We performed numerical simulations in order to identify the vibration modes of the glass slide and associated Eigen frequencies. As shown in Figure 3b, at 86,937 Hz, we observe a mode where the displacement field is almost purely axially varying, with no significant variations along the width and the thickness of the glass slide. Since we bind the PDMS microchannel parallel to the shorter edge of the glass slide, these displacement lines become parallel to the microchannel as well. This observation implies that there is no variation of the displacement field along the length of the channel, which is confirmed by our experimental observations where the streaming vortices are seen to be entirely planar (residing in the plane parallel to the width of the channel). The minor variation in the numerically predicted and experimentally employed frequency may be due to the frequency shift arising out of the viscous damping associated with the loading of microchannel onto the glass slide. We also performed the numerical simulations to observe the streaming field inside the microchannel at this frequency. The glass slide is modeled as a linear elastic solid, while the acoustic streaming patterns inside the microchannel are obtained by using the same approach as employed to perform the numerical simulations for investigating sharp-edge induced streaming, as described earlier.[35] To model the walls of the microchannel, we utilized impedance boundary conditions at the channel walls, while the continuity of velocity and traction is enforced at the fluid-substrate interface. The numerical results obtained (Figure 3c) are seen to be in good qualitative agreement with the experimental observations (Figure 3d, and Movie 3, Supporting Information), both showing the presence of two acoustic streaming vortices along the width of the channel.
Figure 3.
Out-of-plane streaming vortices are generated via the oscillating glass slide. (a) Out-of-plane rotation of a HeLa cell is demonstrated at ~85 kHz and 30 VPP, respectively. (b) Numerical simulation of the vibration mode of the glass slide at 86,937 Hz yields parallel displacement lines. (c) Simulated streaming profile reveals two vortices inside the PDMS microchannel that is positioned along the parallel displacement lines on the glass slide. (d) 1 μm fluorescent beads are used to visualize the experimental streaming profile inside the microchannel at ~85 kHz and 30 VPP.
We note that since the vibration profile of the glass slide depends on the actuation frequency, a variety of displacement profiles can be generated at the microchannel-substrate interface (see Figure S3), depending on the frequency used as well as the orientation and position of the microchannel. Thus, in principle, one can obtain a myriad of acoustic streaming patterns inside the microchannel. We expect that certain modes of the glass slide oscillations in this particular geometry could yield out-of-plane streaming vortices inside the device as shown in Figure 3d. It is also important to note that as the thickness of the glass slide increases, the eigenfrequency which corresponds to the purely axial glass displacement mode observed in Figure 3d shifts to higher frequencies as demonstrated by a numerical simulation (see Figure S4). In this study, we use ~170 μm thick glass slides for all the experiments. By sweeping the excitation frequency of the transducer, different streaming profiles are investigated. In addition to the parallel out-of-plane streaming vortices, some of more irregular streaming profiles are also observed featuring combination of in-plane and out-of-plane streaming flows (see Figure S5). These vortex formations are considered to be the result of more complex vibrational Eigen modes of the glass slide shown in Figure S3.
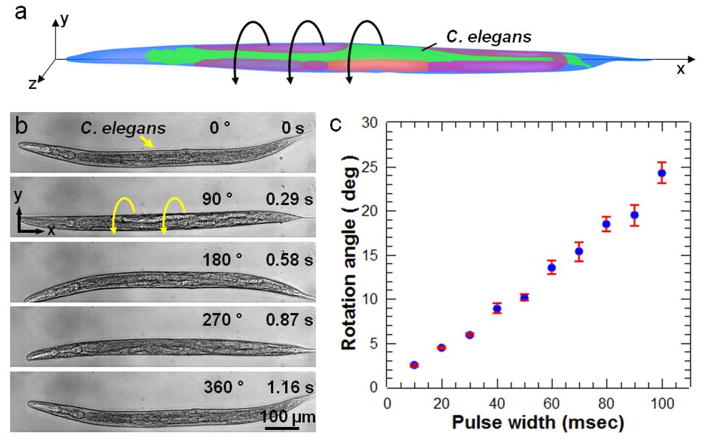
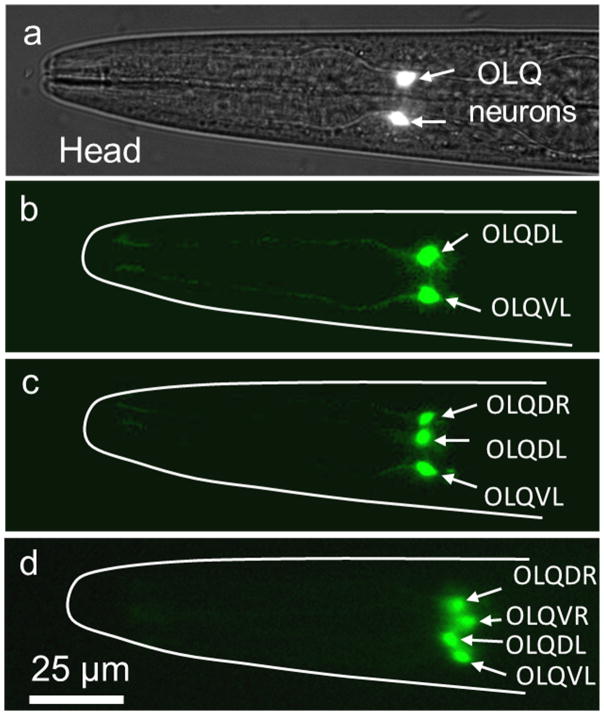
Figure 4 shows out-of-plane rotational manipulation of C. elegans. The C. elegans at the developmental stage of late L4 is rotated around its long axis as illustrated in Figure 4a. C. elegans are first treated with Levamisole (a widely used anesthetic for worms) to reduce physical activity. Figure 4b demonstrates a 360° rotation of the worm by applying a continuous RF sine signal to the transducer (Movie 4, Supporting Information). To precisely tune the angle of rotation, we utilized very short pulses of sine waves from 10 milliseconds to 100 milliseconds (Figure 4c). Low Reynolds number (Re < 10) inside the microfluidic channel stops the worm as soon as the transducer is turned off (Movie 5, Supporting Information). Using this effect, the angle of rotation can be controlled down to 2.5° which corresponds to less than 2 μm lateral displacement on the surface of a worm of 40 μm diameter. Worms rotated in the device resume normal activity after they recover from the effect of the anesthetic. As a demonstration of the capability of our acoustofluidic rotational manipulation method, we used the method to simultaneously image left/right (L/R) pairs of neurons in the C. elegans head. The OLQ cells in C. elegans are a set of four mechanosensory neurons consisting of dorsal and ventral L/R pairs. Animals typically position themselves on a microscopy slide so that imaging is done from a lateral view and neurons or other cells with L/R pairs are difficult to visualize simultaneously (Figure 5a,b). By gradually adjusting the rotation angle, we simultaneously imaged all four OLQ cells in the head of a fourth larval stage (L4) C. elegans under the standard fluorescence microscopy (Figure 5b–d). Such imaging capability is advantageous for easily scoring differentiation or cell death decisions, for example, within a cell type.
Figure 4.
Rotational manipulation of C. elegans. (a) A cartoon schematic of a C. elegans demonstrates rotational manipulation. (b) An L4 stage C. elegans is being rotated 360° in the bare section of the microchannel. (c) Characterization of the stepwise rotation showing the rotation angle at different pulse width of acoustic power.
Figure 5.
Observation of OLQ head neuron cells of an L4 stage C. elegans. (a) Bright-field and fluorescence images of ocr-4::GFP transgenic animal are overlapped in order to show the position of (b) the OLQ dorsal L (OLQDL) and ventral L (OLQVL) neurons in the head of the worm. By rotational adjustment, the paired R OLQ neurons are gradually visualized simultaneously: (c) OLQDL, OLQVL and OLQDR, and (d) OLQDL, OLQVL, OLQDL and OLQDR.
In conclusion, we have demonstrated precise in-plane and out-of-plane rotational manipulation of single HeLa cells and C. elegans using acoustic microstreaming flows generated by oscillating sharp-edge structures and glass slide under the microchannel. Dynamic control of rotational manipulation is achieved using short pulses of sine waves to adjust the angular position. The OLQ neuron cells located in the head of an L4 stage C. elegans are observed through the rotational manipulation. Using sharp-edge structures, single cell rotation rates exceeding 14,000 rpm are realized. As a potential application, effective tailoring of the rotational speed could be utilized to apply adjustable shear forces to single cells. Furthermore, out-of-plane rotation of single cells and model organisms like C. elegans is an indispensable tool for comprehensive bio-imaging capabilities. Our acoustofluidic rotational manipulation device is simple-to-fabricate and easy-to-operate. It can be conveniently integrated into the existing microfluidic devices[5,44–46] designed for handling cells and small model organisms.
Experimental Section
Device design and fabrication
The acoustofluidic rotational manipulation devices were designed with sharp-edge and bare channel regions to generate both in-plane and out-of-plane rotational manipulation. For sharp-edge and bare channel regions, tip angle, sharp-edge length, sharp-edge period, and bare channel width were experimentally optimized as 15°, 200 μm, 150 μm, and 500 μm, respectively (Figure S1).
The acoustofluidic device was fabricated using PDMS soft lithography and replica molding technique. For master mold preparation, a silicon wafer was patterned and anisotropically etched using deep reactive ion etching (DRIE) process. The surface of the silicon master mold was treated with silane vapor using 1H,1H,2H,2H-perfluorooctyl-trichlorosilane (Sigma Aldrich, USA) for approximately 1 hour. SylgardTM 184 Silicone Elastomer Base was mixed with SylgardTM 184 Silicone Elastomer Curing Agent (Dow Corning, USA) at a ratio of 10:1 and cured at 65 °C for 2 hours to form the PDMS channels. Once completely cured, inlet and outlet holes were punched into the designated ports of the PDMS micro channel using a hand-operated puncher (Harris Uni-Core, Ted Pella, USA). Then, the PDMS channel surface and a 25 × 50 × 0.170 (width × length × thickness) mm glass slide (SuperSlips, VWR, USA) were treated with oxygen plasma for 1 minute, and bonded in 65 °C overnight. Finally, a piezoelectric transducer (81-7BB-27-4L0, Murata Electronics, Japan) was bonded on top of the glass slide next to the PDMS channel using an epoxy (84101, Permatex, USA).
Device operation
HeLa cells (5 × 105 ml−1) and C. elegans were stably injected into the microchannel through the inlet tubing using a 1 ml syringe (309659, Becton Dickinson, USA) ) via syringe pumps (Nemesys, Cetoni, Germany). Preparation of HeLa cells and C. elegans were previously discussed.[47–49] For In-plane rotation, once the HeLa cells were positioned in the sharp-edge region, the piezoelectric transducer was driven around 5 kHz with varying peak-to-peak voltages from 10 to 40 VPP using a function generator (AFG 3011, Tektronix, USA) and an RF power amplifier (25A250A, Amplifier Research, USA). The working frequency for in-plane rotation was found to be close to the resonant frequency of the transducer. For out-of-plane rotation, C. elegans were positioned in the bare section of the microchannel using the user-controlled syringe pump which provided better positioning precision compared to manual injection. Working frequency for this mode of rotation was found to be between 70 and 100 kHz. HeLa cell and C. elegans imaging were performed using an inverted microscope (Eclipse TE 2000-U, Nikon, Japan). Images were captured with Photron FASTCAM Viewer (PFV, Photron, USA) connected to a fast camera (Fastcam SA4, Photron, USA). For fluorescence imaging, a Nikon filter cube (excitation: 470 nm, emission: 515 nm), a CCD digital camera (CoolSNAP HQ2, Photometrics, USA), and a fibre optic illumination system (Intensilight, Nikon, Japan) were used.
Supplementary Material
Acknowledgments
We thank Dr. Awani Upadhyay and Dr. Nagagireesh Bojonala for their help with the worm preparation. We gratefully acknowledge financial support from National Institutes of Health (R01 GM112048 and R33 EB019785), National Science Foundation (IIP-1534645, CBET-1438126, and IDBR-1455658), and the Penn State Center for Nanoscale Science (MRSEC) under grant DMR-1420620. This work was completed in the Penn State Nanofab Facility. Adem Ozcelik and Murat Kaynak acknowledge the support from Turkey’s Ministry of National Education.
Footnotes
Supporting Information is available from the Wiley Online Library.
Contributor Information
Dr. Adem Ozcelik, Department of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
Nitesh Nama, Department of Engineering Science and Mechanics, The Pennsylvania State University, University Park, PA 16802, USA.
Dr. Po-Hsun Huang, Department of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
Murat Kaynak, Department of Engineering Science and Mechanics, The Pennsylvania State University, University Park, PA 16802, USA.
Melanie R. Mcreynolds, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA
Prof. Wendy Hanna-Rose, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA
Prof. Tony Jun Huang, Department of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
References
- 1.Elbez R, McNaughton BH, Patel L, Pienta KJ, Kopelman R. PLoS One. 2011;6:e28475. doi: 10.1371/journal.pone.0028475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohanty SK, Uppal A, Gupta PK. Biotechnol Lett. 2004;26:971. doi: 10.1023/b:bile.0000030041.94322.71. [DOI] [PubMed] [Google Scholar]
- 3.Tanner K, Mori H, Mroue R, Bruni-Cardoso A, Bissell MJ. Proc Natl Acad Sci. 2012;109:1973. doi: 10.1073/pnas.1119578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardo-Martin C, Allalou A, Medina J, Eimon PM, Wählby C, Fatih Yanik M. Nat Commun. 2013;4:1467. doi: 10.1038/ncomms2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Proc Natl Acad Sci U S A. 2007;104:13891. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan D, Zhu F, Cammarata R, Chien C. Phys Rev Lett. 2005;94:247208. [Google Scholar]
- 7.Bentley AK, Trethewey JS, Ellis AB, Crone WC. Nano Lett. 2004;4:487. [Google Scholar]
- 8.Fan DL, Zhu FQ, Xu X, Cammarata RC, Chien CL. Proc Natl Acad Sci U S A. 2012;109:9309. doi: 10.1073/pnas.1200342109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junker JP, van Oudenaarden A. Cell. 2014;157:8. doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Cell. 2009;138:822. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Chung K, Crane MM, Lu H. Nat Methods. 2008;5:637. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 12.Crane MM, Stirman JN, Ou CY, Kurshan PT, Rehg JM, Shen K, Lu H. Nat Methods. 2012;9:977. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de I, Cáceres C, Valmas N, Hilliard MA, Lu H. PLoS One. 2012;7:e35037. doi: 10.1371/journal.pone.0035037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanty SK, Gupta PK. Rev Sci Instrum. 2004;75:2320. [Google Scholar]
- 15.Friese MEJ, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Nature. 2003;395:4. [Google Scholar]
- 16.Xi W, Solovev AA, Ananth AN, Gracias DH, Sanchez S, Schmidt OG. Nanoscale. 2013;5:1294. doi: 10.1039/c2nr32798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Zentgraf T, Liu Y, Bartal G, Zhang X. Nat Nanotechnol. 2010;5:570. doi: 10.1038/nnano.2010.128. [DOI] [PubMed] [Google Scholar]
- 18.Black BJ, Mohanty SK. Opt Lett. 2012;37:5030. doi: 10.1364/OL.37.005030. [DOI] [PubMed] [Google Scholar]
- 19.Wu T, Nieminen Ta, Mohanty S, Miotke J, Meyer RL, Rubinsztein-Dunlop H, Berns MW. Nat Photonics. 2011;6:62. [Google Scholar]
- 20.Shelby JP, Mutch Sa, Chiu DT. Anal Chem. 2004;76:2492. doi: 10.1021/ac035531v. [DOI] [PubMed] [Google Scholar]
- 21.Paterson L, MacDonald MP, Arlt J, Sibbett W, Bryant PE, Dholakia K. Science. 2001;292:912. doi: 10.1126/science.1058591. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen MB, Oddershede LB, Siegumfeldt H. Appl Environ Microbiol. 2008;74:2441. doi: 10.1128/AEM.02265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chau LH, Liang W, Cheung FWK, Liu WK, Li WJ, Chen SC, Lee GB. PLoS One. 2013;8:e51577. doi: 10.1371/journal.pone.0051577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed D, Ozcelik A, Bojanala N, Nama N, Upadhyay A, Chen Y, Hanna-Rose W, Huang TJ. Nat Commun. 2016;7:11085. doi: 10.1038/ncomms11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Destgeer G, Sung HJ. Lab Chip. 2015;15:2722. doi: 10.1039/c5lc00265f. [DOI] [PubMed] [Google Scholar]
- 26.Destgeer G, Ha B, Park J, Sung HJ. Anal Chem. 2016;88:3976. doi: 10.1021/acs.analchem.6b00213. [DOI] [PubMed] [Google Scholar]
- 27.Destgeer G, Cho H, Ha BH, Jung JH, Park J, Sung HJ. Lab Chip. 2016;16:660. doi: 10.1039/c5lc01104c. [DOI] [PubMed] [Google Scholar]
- 28.Ozcelik A, Ahmed D, Xie Y, Nama N, Qu Z, Nawaz AA, Huang TJ. Anal Chem. 2014;86:5083. doi: 10.1021/ac5007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding X, Li P, Lin SCS, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ. Lab Chip. 2013;13:3626. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed D, Peng X, Ozcelik A, Zheng Y, Huang TJ. AIP Adv. 2015;5:097161. doi: 10.1063/1.4931641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Z, Xie Y, Guo F, Ren L, Huang PH, Chen Y, Rufo J, Costanzo F, Huang TJ. Lab Chip. 2016;16:515. doi: 10.1039/c5lc00707k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo F, Li P, French JB, Mao Z, Zhao H, Li S, Nama N, Fick JR, Benkovic SJ, Huang TJ. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1422068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang P, Truica CI. Proc Natl Acad Sci U S A. 2015;112:4970. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, Mao Z, Chen Y, Xie Z, Lata JP, Li P, Ren L, Liu J, Yang J, Dao M, Suresh S, Huang TJ. Proc Natl Acad Sci. 2016;113:1522. doi: 10.1073/pnas.1524813113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nama N, Huang PH, Huang TJ, Costanzo F. Lab Chip. 2014;14:2824. doi: 10.1039/c4lc00191e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang PH, Xie Y, Ahmed D, Rufo J, Nama N, Chen Y, Chan CY, Huang TJ. Lab Chip. 2013;13:3847. doi: 10.1039/c3lc50568e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang PH, Nama N, Mao Z, Li P, Rufo J, Chen Y, Xie Y, Wei CH, Wang L, Huang TJ. Lab Chip. 2014;14:4319. doi: 10.1039/c4lc00806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rooney JA. Science. 1970;169:869. doi: 10.1126/science.169.3948.869. [DOI] [PubMed] [Google Scholar]
- 39.Williams AR, Hughes DE, Nyborg WL. Science. 1970;169:871. doi: 10.1126/science.169.3948.871. [DOI] [PubMed] [Google Scholar]
- 40.Triyoso DH, Good TA. J Physiol. 1999;515:355. doi: 10.1111/j.1469-7793.1999.355ac.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dardik A, Chen L, Frattini J, Asada H, Aziz F, Kudo FA, Sumpio BE. J Vasc Surg. 2005;41:869. doi: 10.1016/j.jvs.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Merchuk JC. Adv Biochem Eng Biotechnol. 1991;44:65. doi: 10.1007/BFb0000748. [DOI] [PubMed] [Google Scholar]
- 43.Zhong JJ, Fujiyama K, Seki T, Yoshida T. Biotechnol Bioeng. 1994;44:649. doi: 10.1002/bit.260440512. [DOI] [PubMed] [Google Scholar]
- 44.Shi W, Wen H, Lin B, Qin J. Top Curr Chem. 2011;304:323. doi: 10.1007/128_2011_145. [DOI] [PubMed] [Google Scholar]
- 45.San-Miguel A, Lu H. WormBook. 2013:1. doi: 10.1895/wormbook.1.162.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornaglia M, Mouchiroud L, Marette A, Narasimhan S, Lehnert T, Jovaisaite V, Auwerx J, Gijs MAM. Sci Rep. 2015;5:10192. doi: 10.1038/srep10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed D, Muddana HS, Lu M, French JB, Ozcelik A, Fang Y, Butler PJ, Benkovic SJ, Manz A, Huang TJ. Anal Chem. 2014;86:11803. doi: 10.1021/ac5033676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L, Hanna-Rose W. Dev Biol. 2006;300:599. doi: 10.1016/j.ydbio.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Crook M, Mcreynolds MR, Wang W, Hanna-Rose W. Dev Dyn. 2014;243:965. doi: 10.1002/dvdy.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.