Abstract
Gene expression profiles of Escherichia coli K-12 W3110 were compared as a function of steady-state external pH. Cultures were grown to an optical density at 600 nm of 0.3 in potassium-modified Luria-Bertani medium buffered at pH 5.0, 7.0, and 8.7. For each of the three pH conditions, cDNA from RNA of five independent cultures was hybridized to Affymetrix E. coli arrays. Analysis of variance with an α level of 0.001 resulted in 98% power to detect genes showing a twofold difference in expression. Normalized expression indices were calculated for each gene and intergenic region (IG). Differential expression among the three pH classes was observed for 763 genes and 353 IGs. Hierarchical clustering yielded six well-defined clusters of pH profiles, designated Acid High (highest expression at pH 5.0), Acid Low (lowest expression at pH 5.0), Base High (highest at pH 8.7), Base Low (lowest at pH 8.7), Neutral High (highest at pH 7.0, lower in acid or base), and Neutral Low (lowest at pH 7.0, higher at both pH extremes). Flagellar and chemotaxis genes were repressed at pH 8.7 (Base Low cluster), where the cell's transmembrane proton potential is diminished by the maintenance of an inverted pH gradient. High pH also repressed the proton pumps cytochrome o (cyo) and NADH dehydrogenases I and II. By contrast, the proton-importing ATP synthase F1Fo and the microaerophilic cytochrome d (cyd), which minimizes proton export, were induced at pH 8.7. These observations are consistent with a model in which high pH represses synthesis of flagella, which expend proton motive force, while stepping up electron transport and ATPase components that keep protons inside the cell. Acid-induced genes, on the other hand, were coinduced by conditions associated with increased metabolic rate, such as oxidative stress. All six pH-dependent clusters included envelope and periplasmic proteins, which directly experience external pH. Overall, this study showed that (i) low pH accelerates acid consumption and proton export, while coinducing oxidative stress and heat shock regulons; (ii) high pH accelerates proton import, while repressing the energy-expensive flagellar and chemotaxis regulons; and (iii) pH differentially regulates a large number of periplasmic and envelope proteins.
Escherichia coli and related enteric bacteria respond to a wide range of pH stresses by regulating gene expression (for reviews see references 21 and 68) and protein profiles (73, 82). Enteric bacteria encounter a wide range of external pHs in their natural habitat, the human digestive tract (17). Colonization of the intestine requires transient survival through the stomach at pH 1 to 2 (fasting) or 2 to 7 (transiently, during feeding) (18), as well as exposure to pancreatic secretions at pH 10 (25) followed by growth and persistence at a range of external pHs of 5 to 8 (20). Growth at a pH substantially higher or lower than the cytoplasmic pH 7.6 induces protective responses with two fundamental aims: to maintain internal pH homeostasis and to prepare the cell to survive future exposure to more extreme pH conditions (below pH 5 or above pH 9) that no longer permit growth (11, 41, 70).
The effects of pH on enteric bacteria contribute to disease. Low pH enhances expression of numerous virulence factors, such as the ToxR-ToxT virulence regulon in Vibrio cholerae (7), the phoP-phoQ regulon of Salmonella enterica (6), and the pH 6 antigen of Yersinia pestis (50). Acid stress contributes to food preservation; many food preservatives are membrane-permeant acids whose uptake is enhanced by acid (60), and acid interacts in complex ways with both temperature and organic food preservatives (65).
While growth in acid challenges pH homeostasis, the pH difference across the inner cell membrane (ΔpH) nevertheless contributes cell energy in the form of proton potential or proton motive force (Δp). The proton potential powers motility, ATP synthesis, and catabolite transport (for a review see reference 29). But low pH also amplifies the uptake of membrane-permeant acids that dissipate the proton potential (59). Thus, we expect low pH to induce a combination of positive and negative responses.
Much of bacterial catabolism affects pH, and in E. coli a growing number of catabolic enzymes and catabolite transporters are known to be regulated by pH (21, 73). Sugar fermentation initially generates short-chain acids that are excreted but accumulate and reenter the cytoplasm, causing acidification. Thus, it is not surprising that sugar transporters such as OmpF and the maltose regulon are down-regulated at low pH (13). Consumption of acids by the tricarboxylic acid (TCA) cycle causes alkalinization, a common result of growth to stationary phase in tryptone-based media (66, 73). Catabolism of amino acids by decarboxylases generates alkaline amines, which help the cell counteract external acidification, for example, the lysine and arginine decarboxylases (4, 27, 45, 47, 71). High pH, however, induces deaminases that generate acids, such as tryptophan deaminase (tnaAB) and serine deaminase (sda) (9, 73, 82).
A complicated case is that of the glutamic acid decarboxylase genes gadA and gadBC (12, 44). The gad system enables cells to survive extreme acid (77), but its expression is induced mainly at high pH, or in Luria-Bertani medium grown to stationary phase, where pH naturally increases (73, 82). An alternative role of gad, particularly under anaerobiosis, may be to channel its product γ-aminobutyric acid into fermentation acids.
Even mild acid (pH 6 to 7) greatly amplifies the uptake of membrane-permeant weak acids such as acetate. Permeant acids pass through the bacterial membrane and dissociate in the cytoplasm, causing accumulation of anions and depression of internal pH (34, 56). Acetate concentrations rise as cell density increases, and acetate induces a large number of genes and proteins (3, 35). Growth inhibition occurs as a result of both lower internal pH and the differential ability of anions to inhibit metabolism (60). The effect of permeant acids is critical in the human colon, where the concentration of short-chain fatty acids totals approximately 100 mM (15).
While numerous responses to pH stress are known, the mechanisms by which E. coli maintains its internal pH at 7.6 remain poorly understood. The electron transport chain pumps protons outside the cell, and the H+-ATPase either exports or imports protons, but mutants in these components maintain pH homeostasis. There is evidence that potassium exchange contributes to pH homeostasis in external acid (5, 10, 52, 80), but the precise mechanisms remain unclear. At high pH, the electrical potential (Δψ) is diminished in order to compensate for the inverted ΔpH. The sodium-proton antiporterNhaA contributes to internal pH maintenance under sodium stress (24, 75). High pH also induces major stress systems such as heat shock response (1, 28, 74), the SOS regulon (63), and the CpxP envelope stress response (16).
At more extreme pH values, well below the growth range (as low as pH 1.5 for clinical isolates) E. coli can retain viability for many hours, a phenomenon termed acid survival or acid resistance. Acid resistance is enhanced by many genes induced during growth at the acid end of the pH range (pH 5) or growth to stationary phase. Acid-induced acid resistance factors include periplasmic chaperones such as the hdeA product (23), envelope proteins such as OsmY, and redox modulators such as Tpx (73, 78). A complex acid resistance regulon including the gad system is regulated by transcription factors GadX-GadW and EvgA-YdeO, as well as by RpoS, H-NS, and cyclic AMP (11, 12, 44, 79). E. coli also exhibits base resistance, the ability to survive at or above pH 10 (58, 70). Base resistance requires rpoS and components of the gad system (30).
Finally, pH may affect flagellar motility, although the present picture is unclear. According to one report, growth in acid represses flagellar genes and eliminates motility (72), whereas another group finds motility enhanced by acetate and propionate, which cause acid stress (53).
To investigate acid and base response, we used microarrays to compare E. coli gene expression at low, neutral, or high external pH. Past microarray studies of pH response have been limited by their absence of pH conditions above pH 7 (44, 78); their use of glucose minimal medium (78), in which many catabolic genes are repressed; and their focus on only a single acid resistance regulon (44). Our experimental design included both acid and base conditions, as well as pH 7.0. For each growth condition, five independent cultures were hybridized separately, a number of replicates that ensured detection of virtually all expression ratios of at least twofold. The coregulation of numerous genes within operons confirmed the biological relevance of our expression ratios. Our study revealed unexpected patterns of pH response and clarified the overlap of pH stress with other stress responses.
MATERIALS AND METHODS
Growth conditions.
E. coli K-12 strain W3110 (R. VanBogelen and F. Neidhardt) was grown overnight in unbuffered potassium-modified Luria broth (LBK) (10 g of tryptone/liter, 5 g of yeast extract/liter, 7.45 g of KCl/liter). For pH-controlled growth, media were buffered with 100 mM homopiperazine-N,N′-bis-2-(ethanesulfonic acid) (HOMOPIPES) (pKa, 4.55 and 8.12). The pH of the media were adjusted to 5.0, 7.0, or 8.7 with KOH solution to avoid extra sodium ions, which stress cells at high pH (24). To maximize aeration and maintain logarithmic growth, the overnight culture was diluted 1,000-fold into 12 ml of buffered medium in a 125-ml baffled flask and rotated at 240 rpm. Cultures were grown at 37°C to an optical density at 600 nm of 0.3. For all cultures, the pH was tested after growth to ensure that the values were maintained at ±0.2 pH unit of the pH of the original uninoculated medium.
To observe motility, we used E. coli K-12 strain RP437 and S. enterica serovar Typhimurium SJW1103 from a laboratory in which strains are maintained for motility (M. Macnab). Culture was spotted on tryptone-KCl soft-agar plates (0.35% Bacto Agar) and incubated at 37°C until cells swam out. Culture was picked from the leading edge of the swimming cells and inoculated into LBK for overnight growth. For quantitative assay of motility, 5 μl of culture was spotted in triplicate on plates containing tryptone-KCl with 100 mM sulfonate buffer of appropriate pKa (73). After growth for 8 h, the diameter of motile cell growth was measured.
RNA isolation.
Bacterial RNA was isolated using the Qiagen RNeasy kit with on-column DNA digestion (Qiagen), with additional DNA removal with Ambion DNase. To perform this additional DNase digestion, RNA was precipitated and redissolved in 85 μl of nuclease-free water. We then added 10 μl of 10× DNase I buffer and 5 μl of (1-U/μl) DNase I (Ambion). The DNase reaction mixture was incubated at 37°C for 30 min and then chilled on ice. A second RNeasy column purification was performed.
cDNA preparation and array hybridization.
For microarrays, standard methods were used for cDNA synthesis, fragmentation, and end-terminus biotin labeling, based on Affymetrix protocols. Labeled cDNA was hybridized to E. coli Affymetrix Antisense Genome Arrays. Hybridized arrays were stained with streptavidin-phycoerythrin with the use of the Affymetrix Fluidic Station. After staining, arrays were scanned with a GC2500 scanner.
Statistical analysis of gene expression.
The experiment was designed so as to minimize both false-positive and false-negative results for expressed genes. Five full replicates (with respect to E. coli growth, RNA isolation, sample preparation, and array hybridization) were performed for each pH condition.
The median within-group variance in expression for all genes in the data set was 0.031 (or standard deviation, 0.175). To test for significant differences in expression between the pH classes, one-way analysis of variance (ANOVA) was performed at a significance level of 0.001; thus, of every thousand genes tested, only one false positive would be expected. For a gene with average within-group variability, our sample size provided statistical power of 98% to detect a twofold difference in gene expression among pH groups. That is, only 2% of genes that show a twofold difference in expression between any two pH groups would be missed (false negatives).
Model-based expression analysis with dChip software (40) was performed on the probe-level data from Affymetrix's DAT files. The model relates target RNA levels to the probe signals by a linear function that weights the significance of all oligonucleotide probes for each gene. The analysis includes normalization, which rescales data from different arrays so that comparisons can be made among arrays. Each array was normalized to a baseline array from a pH 7 culture, by using local regression on an invariant set of probes (62). Model-based expression indices were calculated for each gene on each array by using only the perfect match probes (61), and outlier detection was performed (39). Only probe sets that received an Affymetrix call of “present” on greater than 50% of the arrays were used in subsequent analyses. “Present” or “absent” calls use information from paired perfect-match and single-base-mismatch probes. Four thousand six hundred fifty probe sets passed this criterion.
For genes whose probe sets passed the 50% screen, one-way ANOVA was performed on the log2-transformed model-based expression indices, on a gene-by-gene basis. For each gene that displayed significant differences in expression among the classes, pairwise comparisons of pH classes were determined using Tukey's multiple comparisons procedure to control the familywise error rate for the t test.
Additional analyses were performed to explore categories of differential gene expression. Global relationships among arrays were visualized by performing a principal component analysis (81) on the expression data and plotting arrays in two-dimensional space corresponding to the first two principal components. The gene expression profiles of the arrays were visualized in two-dimensional Euclidian space, by using BRB ArrayTools software. In addition, categories of differential expression profiles across the pH classes were generated by a hierarchical cluster analysis of differentially expressed genes, based on the average linkage method (19) with BRB ArrayTools.
RESULTS
Growth range of pH.
To study the full range of pH response, we selected the widest pH range (pH 5.0 to 8.7) in which cultures maintained reasonable doubling times and approximately constant pH throughout growth. Culture media were adjusted to pH 5.0, 7.0, and 8.7. The doubling time for E. coli cultured at pH 5.0 and 8.7 was approximately 25 min and at pH 7.0 was 18 min. All cultures were grown to an optical density of 0.3 in order to facilitate at least five complete replications. The final pH of growth cultures was found to be within ±0.2 of the initial pH. The internal pH of the cytoplasm is approximately 7.6 (69); thus, growth at external pH 7.0 might induce some acid response.
Probe hybridization.
To determine differential gene expression, the log2 transforms of normalized model-based expression values of genes were compared. Of the 7,231 genes and intergenic regions (IGs) on the array, 4,650 loci were detected on more than half (eight or more) of the 15 arrays. These loci, constituting about 70% of the total array, were taken for further analysis.
Principal component analysis.
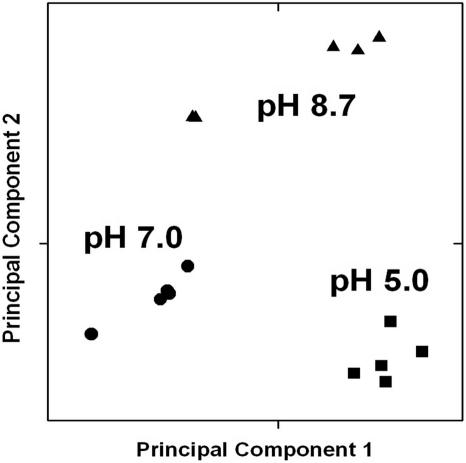
Global relationships among arrays were visualized by performing a principal component analysis (81) on the expression data (Fig. 1). Before dimensional reduction, each array existed in 4,650-dimensional space (one dimension for each of the 4,650 intensity values). The array comparisons were plotted in two-dimensional space, corresponding to the first and second principal components of variation. The first principal component for each array is the weighted linear combination of intensity values that shows maximum variation, whereas the second principal component is a weighted linear combination orthogonal to the first component that has maximum variance.
FIG. 1.
Principal component analysis. The gene expression profiles of the arrays were visualized in two-dimensional Euclidian space, by using BRB ArrayTools software as described under Materials and Methods. The first and second principal components are shown. pH 5.0, squares; pH 7.0, circles; pH 8.7, triangles.
The principal component analysis indicated that the microarrays from each of the three pH conditions appeared in distinct groups (Fig. 1). Within-class variability was small relative to variability among pH levels. The pH 8.7 arrays showed the greatest degree of separation, clustering into two groups based on the date on which the arrays were hybridized, but this difference was small compared to the differences between pH classes.
ANOVA for significance of expression profiles.
We compared gene expression among the three pH groups on a gene-by-gene basis using one-way ANOVA at a significance level of 0.001. The significance level indicates the probability of a false positive, and we therefore expect 0.001 × 4,650 = 4.65 false-positive genes (i.e., genes that are not truly differentially expressed but that appear in our differentially expressed list) in our full analysis. Of the 4,650 loci with eight or more “present” calls on arrays, 761 genes and 353 IGs showed a significant F value for differential expression among the three pH classes. Thus, about 17% of E. coli genes showed significant modulation of expression as a function of pH.
Cluster analysis.
As a first attempt at categorizing differentially expressed genes, we performed a hierarchical cluster analysis (19) of differentially expressed genes (Fig. 2). We used average linkage and one minus the centered Pearson correlation as the distance metric. At a correlation value of approximately 0.6, the dendrogram generated six clusters of gene expression profiles.
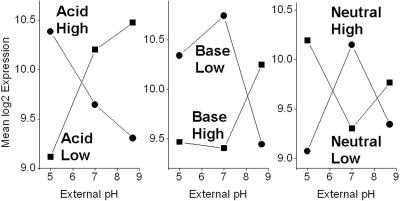
FIG. 2.
Cluster analysis of differentially expressed genes. The dendrogram was generated based on the average linkage method (19) with BRB ArrayTools. At a correlation of 0.6, six clusters of related gene expression were designated Acid High (AH), Acid Low (AL), Base High (BH), Base Low (BL), Neutral High (NH), and Neutral Low (NL).
Within each of the six clusters, the average profiles were determined for all the gene expression indices (log2 intensity values) across the three pH conditions (Fig. 3). The clusters were defined by their mean expression profiles across the three pH conditions. The Acid High cluster showed highest expression at pH 5.0, declining at pH 7.0 and 8.7. It included 160 genes and 49 IGs. Acid Low (113 genes, 57 IGs) showed approximately the reverse profile, with its lowest expression at pH 5.0, rising at pH 7.0 and 8.7. Base High (93 genes, 70 IGs) showed low expression at pH 5.0 and 7.0 and higher expression at pH 8.7, whereas Base Low (123 genes, 40 IGs) showed the reverse, higher expression at pH 5.0 and 7.0 than at pH 8.7. The Neutral High cluster (93 genes, 14 IGs) showed highest expression at pH 7.0 and lower expression at both pH extremes. The Neutral Low cluster (181 genes 123 IGs) showed the lowest expression at pH 7.0 and higher expression at both pH extremes, although the mean expression was substantially greater at pH 5.0 than at pH 8.7; a number of acid-induced genes fell in this category.
FIG. 3.
Cluster mean expression profiles. The mean expression profiles over pH are plotted for the six clusters defined in Fig. 2.
Table 1 lists the genes that fell into each cluster; details of description and Blattner open reading frame (ORF) numbers are available online in Table S1 in the supplemental material. In many cases, all or most of the ORFs in a given operon were induced in the same cluster; see, for example, the atp operon (Base High cluster) and the flg and fli operons (Base Low cluster).
TABLE 1.
Clusters of pH-dependent genes
Known acid-induced genes and acid resistance genes such as sucBC and hdeA (73) generally fell under Acid High, Base Low, or Neutral Low, a cluster whose mean expression indices were actually twofold higher in acid than in base (Fig. 3). These results are generally consistent with the cluster pH profiles and with the structure of the cluster dendrogram, in which the Acid High profile correlates most closely with the Neutral Low profile. Most known base-induced genes, such as alx (ygjT) (8, 73) and tnaA (9), fell under Base High or Acid Low.
For IGs, the cluster assignment and expression ratios are presented online in Table S2 in the supplemental material. Expression of an IG may result from a small regulatory RNA that lies between protein-encoding genes (2, 43), or it may indicate the tail end of mRNA containing pH-regulated genes. For example, the IGs upstream of tnaC (tnaA leader peptide) and downstream of tnaB both were repressed in acid, as are tnaA and tnaB.
Individual gene expression ratios.
For genes whose overall expression profile yielded a significant F value (one-way ANOVA), we used the Tukey procedure to determine ratios of average model-based expression indices from cultures at pH 5.0 versus pH 7.0, at pH 8.7 versus pH 7.0, and at pH 8.7 versus pH 5.0. The full list of individual log2 expression ratios for all analyzed genes is presented in Table S1 in the supplemental material and for IGs is presented in Table S2 in the supplemental material; for genes of particular interest grouped in functional categories, the data are presented in Tables 3 through 7. Expression ratios that are significant at α = 0.001 are shown in boldface.
TABLE 3.
Flagellar and chemotaxis genes
| Gene | Function | Log2 pH ratioa
|
Classb | ||
|---|---|---|---|---|---|
| 5/7 | 8.7/7 | 8.7/5 | |||
| cheA | Chemotaxis sensor kinase | −1.125 | −3.199 | −2.0474 | BL |
| cheB | Protein methylesterase | −1.050 | −1.578 | −0.528 | BL |
| cheR | Chemotaxis MCPc methyltransferase | −0.564 | −1.013 | −0.448 | BL |
| cheW | Chemotaxis signal transducer | −1.336 | −2.785 | −1.449 | BL |
| cheY | Response regulator for chemotactic signal | −1.089 | −1.310 | −0.221 | BL |
| cheZ | CheY-P phosphatase | −1.505 | −3.199 | −1.694 | BL |
| flgA | Flagellar synthesis | −0.261 | −1.056 | −0.795 | BL |
| flgB | Basal body rod subunit | −0.192 | −1.120 | −0.928 | BL |
| flgC | Basal body rod subunit | −0.257 | −1.241 | −0.984 | BL |
| flgD | Basal body rod modification | 0.133 | −1.241 | −1.107 | BL |
| flgE | Hook subunit | 0.272 | −0.856 | −1.128 | BL |
| flgF | Basal body rod subunit | 0.109 | −1.239 | −1.348 | BL |
| flgG | Basal body rod major subunit | 0.220 | −1.116 | −1.335 | BL |
| flgI | Basal body P-ring | −0.011 | −1.218 | −1.207 | BL |
| flgJ | Flagellum-specific muramidase | 0.030 | −0.857 | −0.886 | BL |
| flgK | Flagellar synthesis | −0.211 | −1.875 | −1.664 | BL |
| flgL | Flagellar synthesis | 0.111 | −1.165 | −1.276 | BL |
| flgM | Anti-sigma 28 (FliA); regulates FlhD | −0.292 | −1.424 | −1.132 | BL |
| flgN | Flagellar synthesis | −0.295 | −1.567 | −1.272 | BL |
| flhA | Flagellar export pore protein | 0.333 | −0.528 | −0.861 | AH |
| flhE | Function unknown | 0.492 | −0.454 | −0.946 | AH |
| fliA | Sigma 28; regulates class III flagellar genes | −0.150 | −1.127 | −0.976 | BL |
| fliC | Flagellin subunit, H-antigen | −1.165 | −4.561 | −3.396 | BL |
| fliD | Hook-associated protein | −0.311 | −1.953 | −1.641 | BL |
| fliE | Flagellar synthesis; basal body component | −0.478 | −1.861 | −1.203 | BL |
| fliF | Flagellar basal body M-ring | −0.159 | −1.216 | −1.057 | BL |
| fliG | Motor switching and energizing | 0.182 | −1.313 | −1.495 | BL |
| fliH | Negative regulator of FliI; flagellar assembly and export | 0.389 | −1.245 | −1.634 | BL |
| fliI | Membrane ATPase, flagellar, axial subunit export | 0.270 | −1.238 | 1.508 | BL |
| fliJ | Flagellar biosynthesis | −0.068 | −1.404 | −1.336 | BL |
| fliK | Hook filament junction | 0.278 | −1.186 | −1.464 | BL |
| fliL | Rotational direction of flagella | −0.102 | −1.083 | −0.981 | BL |
| fliM | Flagellar synthesis, motor switching and energizing | 0.029 | −1.053 | −1.082 | BL |
| fliN | Flagellar switch | 0.273 | −0.875 | −1.148 | BL |
| fliO | Flagellar synthesis | 0.191 | −0.921 | −1.112 | BL |
| fliP | Flagellar synthesis | 0.419 | −0.669 | −1.087 | AH |
| fliQ | Flagellar synthesis | 0.463 | −0.980 | −1.444 | AH |
| fliR | Flagellar synthesis | 0.924 | −0.899 | −1.822 | AH |
| fliS | Cytosolic chaperone inhibits premature FliC assembly | −0.174 | −1.947 | −1.772 | BL |
| fliT | Flagellar synthesis | −0.323 | −1.021 | −0.698 | BL |
| fliY | Cystine-binding protein, periplasmic; may regulate FliA (sigma 28) | 0.233 | −0.252 | −0.484 | AH |
| fliZ | Not required for motility; may regulate FliA (sigma 28) | 0.144 | −1.165 | −1.309 | BL |
| motA | Flagellar rotation | −0.682 | −1.847 | −1.166 | BL |
| motB | Flagellar rotation | −1.241 | −3.238 | −1.997 | BL |
| tap | Dipeptide chemoreceptor | −0.622 | −1.089 | −0.466 | BL |
| tar | Aspartate, maltose chemoreceptor | −0.519 | −2.213 | −1.694 | BL |
| tsr | Serine chemoreceptor | −1.061 | −1.826 | −0.765 | BL |
| ycgR | Suppresses hns motility defect | −0.531 | −0.783 | −0.252 | BL |
| yhjH | Suppresses hns motility defect | −0.972 | −2.522 | −1.550 | BL |
Boldface for ratios indicates significance (α = 0.001).
BL, Base Low; AH, Acid High.
MCP, methyl-accepting chemotaxis protein.
The genes most strongly regulated by pH are summarized in Table 2. These genes each showed an expression ratio of at least fourfold (log2 = 2) between two of the pH classes. Note that the two genes most strongly induced in acid are ORFs with no known function, yhcN and yagU. Other acid-induced genes include those for catabolic enzymes in pathways that consume acids, such as sdhCD (succinate dehydrogenase). Genes repressed at high pH include several members of the flagellar regulon, including the main flagellar subunit fliC (for a review see reference 42).
TABLE 2.
Strongest pH-dependent expression ratios (fourfold or higher)
| pH dependence and pH ratio | Gene | Log2 ratio | pH dependence and pH ratio | Gene | Log2 ratio | |
|---|---|---|---|---|---|---|
| Acid induced | ||||||
| 5.0/7.0 | yagU | 3.220 | ||||
| yhcN | 3.064 | |||||
| sdhC | 2.728 | |||||
| lysP | 2.662 | |||||
| sdhD | 2.349 | |||||
| cfa | 2.075 | |||||
| nemA | 2.060 | |||||
| 5.0/8.7 | yhcN | 4.199 | ||||
| yagU | 3.962 | |||||
| fliC | 3.396 | |||||
| fimA | 2.579 | |||||
| cfa | 2.555 | |||||
| gltB | 2.271 | |||||
| ydiY | 2.193 | |||||
| ycdN | 2.147 | |||||
| yncD | 2.117 | |||||
| mqo | 2.075 | |||||
| dhaH | 2.074 | |||||
| cheA | 2.074 | |||||
| motB | 1.997 | |||||
| 7.0/8.7 | fliC | 4.561 | ||||
| malM | 3.780 | |||||
| malK | 3.748 | |||||
| lamB | 3.735 | |||||
| malP | 3.373 | |||||
| motB | 3.238 | |||||
| cheA | 3.199 | |||||
| cheZ | 3.199 | |||||
| flxA | 3.007 | |||||
| malE | 2.933 | |||||
| malQ | 2.883 | |||||
| cheW | 2.785 | |||||
| ibpB | 2.618 | |||||
| yhjH | 2.522 | |||||
| htpG | 2.439 | |||||
| deoC | 2.267 | |||||
| pstS | 2.262 | |||||
| dnaK | 2.249 | |||||
| tar | 2.213 | |||||
| yjdA | 2.208 | |||||
| dnaJ | 2.155 | |||||
| yrfG | 2.055 | |||||
| yheL | 2.031 | |||||
| deoA | 2.003 | |||||
| Base induced | ||||||
| 8.7/7.0 | yifO | 2.769 | ||||
| ymgD | 2.221 | |||||
| 8.7/5.0 | tnaC | 5.517 | ||||
| cpxP | 4.234 | |||||
| tnaA | 4.028 | |||||
| nmpC | 3.961 | |||||
| treB | 3.895 | |||||
| yjiY | 3.665 | |||||
| treC | 3.233 | |||||
| b3913 | 3.176 | |||||
| yifO | 3.095 | |||||
| borD | 3.088 | |||||
| tnaB | 2.993 | |||||
| ycfS | 2.820 | |||||
| yghJ | 2.762 | |||||
| ymgD | 2.433 | |||||
| yccA | 2.378 | |||||
| yfiA | 2.364 | |||||
| yebE | 2.343 | |||||
| yjiX | 2.292 | |||||
| nrdD | 2.182 | |||||
| dniR | 2.081 | |||||
| alx | 2.009 | |||||
| mutL | 2.000 | |||||
| 7.0/5.0 | tnaC | 5.026 | ||||
| lamB | 4.881 | |||||
| malK | 4.790 | |||||
| malM | 4.643 | |||||
| yjiY | 4.359 | |||||
| nmpC | 4.012 | |||||
| malP | 4.000 | |||||
| tnaA | 3.805 | |||||
| malE | 3.425 | |||||
| borD | 3.378 | |||||
| malQ | 3.322 | |||||
| cpxP | 3.232 | |||||
| treB | 3.156 | |||||
| yghJ | 3.081 | |||||
| yjiX | 2.959 | |||||
| fecB | 2.842 | |||||
| ompF | 2.834 | |||||
| treC | 2.613 | |||||
| fecA | 2.608 | |||||
| pstS | 2.479 | |||||
| b3913 | 2.177 | |||||
| fecE | 2.098 |
The genes most strongly induced at high pH included tnaC, encoding the tryptophanase leader peptide (26), as well as tnaA (tryptophanase) and the Trp transporter gene tnaB, with its leader peptide gene tnaC. Previously in proteomic gels, we found tryptophanase to be the most highly expressed protein observed at high pH (9). The alkali-inducible protease gene cpxP (16) was also strongly induced. Members of the maltose transport regulon (malEKM) were strongly repressed by acid, consistent with previous reports (31, 73). But proteins strongly induced by base also included those from genes of unknown function, such as yifO and ymgD.
Flagellar and chemotaxis regulons.
Motility in E. coli is governed by the flagellar chemotaxis regulon including 50 components in 19 operons, governed by the major regulators FlhC and FlhD (42, 76). The expression of the regulatory operon flhCD is controlled by numerous environmental response systems, such as adenylate cyclase (37), RcsCDB (22), and ClpXP (76).
Nearly all the genes of the flagellar regulons (47 genes) were repressed at high pH (Table 2). Forty-one genes fell in the Base Low cluster, which means that the bulk of significant expression difference occurred between pH 7.0 and 8.7. (The other six genes were Acid High.) These genes were among the most strongly base-repressed genes in the arrays (Table 2); for instance, fliC, encoding the flagellin monomer, had the lowest pH 8.7/pH 7.0 ratio observed, down-regulated about 20-fold (Table 3). Some of the che and mot genes showed a relatively small degree of repression in acid compared to that at pH 7.0 but overall were repressed at high pH.
The major regulator operon flhCD, however, showed no effect of pH. Thus, either the flhCD probes failed to show up in our arrays or pH may affect expression posttranscriptionally.
Motility assays.
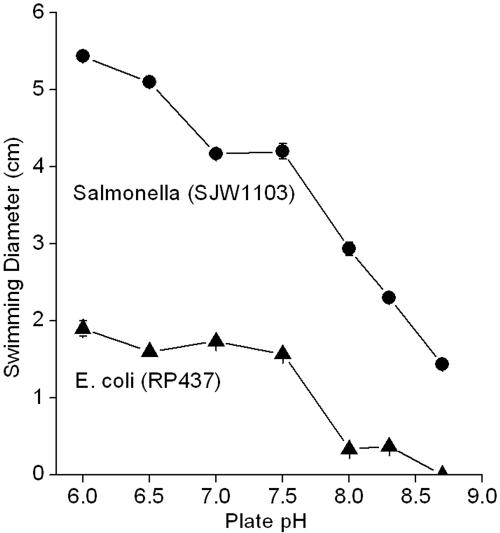
The effect of pH on motility was tested by spotting motile cultures of E. coli K-12 RP437 and S. enterica serovar Typhimurium SJW1103 on motility agar buffered at a range of pH values (Fig. 4). Both species showed a steady decline of motility as pH increased. The decline was particularly steep between pH 7.5 and 8.7.
FIG. 4.
Swimming distance as a function of pH. E. coli K-12 RP437 and S. enterica serovar Typhimurium SJW1103 were spotted on soft-agar plates as described under Materials and Methods. Error bars represent standard errors of the means (n = 3); in most cases their size was smaller than the symbol.
Catabolism and proton transport.
Several enzymes for catabolism of sugars and amino acids show a pH dependence that minimizes acid production at low external pH or maximizes acids at high pH (68, 73). Our microarrays revealed many new components, showing the broad scope of pH regulation of catabolism (Table 4).
TABLE 4.
Catabolism and respiration
| Group | Gene | Function | Log2 pH ratioa
|
Classb | ||
|---|---|---|---|---|---|---|
| 5/7 | 8.7/7 | 8.7/5 | ||||
| Sugar catabolism and TCA cycle | aceE | Pyruvate dehydrogenase | 1.928 | 0.527 | −1.401 | NL |
| aceF | Pyruvate dehydrogenase dihydrolipoamide acetyltransferase | 1.802 | 0.402 | 1.401 | NL | |
| aceK | Isocitrate dehydrogenase kinase/phosphatase | 0.667 | 0.038 | −0.629 | AH | |
| acnA | Aconitase A, stationary phase induced | 0.769 | −0.143 | −0.912 | AH | |
| acnB | Aconitase B; 2-methylaconitate hydratase | 1.036 | −0.041 | −1.077 | NL | |
| acrR | Regulator for acrA and acrB | −0.873 | 0.164 | 1.037 | AL | |
| dcuR (yjdG) | Fumurate respiration regulator (anaerobic) | −0.220 | 0.055 | 0.275 | AL | |
| dcuS (yjdH) | Fumurate respiration regulator (anaerobic) | −0.324 | 0.065 | 0.390 | AL | |
| dhaK | Dihydroxyacetone kinase, subunit I | 1.041 | −0.379 | −1.420 | AH | |
| dhaL | Dihydroxyacetone kinase, subunit II | 0.742 | −0.385 | −1.127 | AH | |
| dld | d-Lactate dehydrogenase | 0.919 | 0.566 | −0.353 | NL | |
| eno | Enolase; RNA degradosome | 0.299 | 0.052 | −0.247 | AH | |
| fucI | l-Fucose isomerase | −0.693 | −0.173 | 0.520 | AL | |
| fucK | l-Fuculose kinase | −1.259 | −0.519 | 0.740 | NH | |
| fucR | Positive regulator, fuc operon | −0.046 | 0.495 | 0.541 | BH | |
| galF | Putative regulator of galU | NL | ||||
| galK | Galactokinase | −0.616 | −0.562 | 0.053 | BL | |
| galM | Galactose mutarotase; aldose-1-epimerase | −0.619 | −0.647 | −0.028 | ||
| gapA | Glyceraldehyde 3-P dehydrogenase A | 0.093 | −0.752 | −0.844 | BL | |
| gatA | Galactitol-specific enzyme IIA of PTSd | 0.929 | −0.626 | −1.555 | AH | |
| gatB | Galactitol-specific enzyme IIB of PTS | 0.632 | −0.895 | −1.528 | AH | |
| gatC | Galactitol-specific enzyme IIC of PTS | 0.973 | −0.949 | −1.922 | AH | |
| gatD | Galactitol-1-phosphate dehydrogenase | 1.037 | −0.893 | −1.930 | AH | |
| gatY | d-Tagatose-1,6-bisphosphate aldolase, class II | 0.846 | −0.483 | −1.329 | AH | |
| gatZ | Enhances GatY activity | 0.774 | −0.605 | −1.380 | AH | |
| glpA | Glycerol-3-phosphate dehydrogenase large subunit (anaerobic) | −0.200 | 0.558 | 0.758 | AL | |
| glpB | Glycerol-3-phosphate membrane anchor (anaerobic) | −0.124 | 0.437 | 0.562 | BH | |
| glpC | Glycerol-3-phosphate dehydrogenase (anaerobic) small subunit | −0.169 | 0.469 | 0.638 | BH | |
| glpX | Fructose 1,6-bisphosphatase | −0.162 | 0.375 | 0.537 | BH | |
| gltA | Citrate synthase | 0.288 | −0.559 | −0.846 | AH | |
| gnd | Gluconate-6-phosphate dehydrogenase | 0.895 | 0.351 | −0.544 | NL | |
| gntT | High-affinity gluconate transport | −0.613 | −0.362 | 0.251 | NH | |
| gpsA | Glycerol-3-phosphate dehydrogenase | −0.492 | −0.534 | −0.041 | NH | |
| gpmA | Phosphoglycerate mutase I | 0.303 | −0.118 | −0.421 | AH | |
| icdA | Isocitrate dehydrogenase | 1.211 | 0.061 | −1.272 | AH | |
| lldD | l-Lactate dehydrogenase | 0.613 | −0.554 | −1.167 | AH | |
| lldP | l-Lactate permease; glycolate uptake | 1.527 | 0.219 | −1.307 | NL | |
| lpdA | Lipoamide dehydrogenase; E3 component of pyruvate and 2-oxoglutarate dehydrogenase complexes | 1.507 | 0.281 | −1.226 | NL | |
| malE | Maltose-binding protein, periplasmic | −3.425 | −2.933 | 0.491 | NH | |
| malF | Maltose transport, inner membrane | −1.016 | −0.935 | 0.082 | NH | |
| malG | Maltose transport, inner membrane subunit | −1.577 | −1.502 | 0.075 | NH | |
| malK | Maltose transport, ATP-binding subunit | −4.790 | −3.748 | 1.041 | NH | |
| malM | Periplasmic protein, mal regulon | −4.643 | −3.780 | 0.863 | NH | |
| malP | Maltodextrin phosphorylase | −4.000 | −3.373 | 0.627 | NH | |
| malQ | Amylomaltase | −3.322 | −2.883 | 0.439 | NH | |
| malT | mal positive regulator | −0.103 | 0.617 | 0.720 | BH | |
| pdhR | Pyruvate dehydrogenase operon repressor | 0.755 | −0.109 | −0.863 | AH | |
| pfkB | 6-Phosphofructokinase-2 | 0.363 | 0.095 | −0.268 | NL | |
| pflA | Pyruvate formate lyase I activase | 0.964 | 0.667 | −0.297 | NL | |
| pflB | Pyruvate formate lyase I (anaerobic) | 0.812 | 0.578 | −0.234 | NL | |
| pgi | Glucose phosphate isomerase | 0.654 | 0.177 | −0.477 | NL | |
| pta | Phosphotransacetylase | 1.167 | 0.639 | −0.528 | NL | |
| ptsG | Glucose PTS enzyme IIBC | 0.841 | 0.544 | −0.287 | NL | |
| ptsH | PTS system histidine phosphocarrier protein Hpr | 0.369 | 0.564 | 0.195 | NL | |
| ptsI | PTS system enzyme I | 0.241 | 0.339 | 0.098 | NL | |
| ptsO | NPr, N-regulated HPr-like protein | 0.639 | 0.168 | −0.470 | NL | |
| rpiA | Ribose-5-phosphate isomerase A | 0.180 | 0.364 | 0.184 | BH | |
| srlA | Sorbitol-specific enzyme II of PTS | −1.156 | −1.720 | −0.564 | BL | |
| srlB | Sorbitol-specific enzyme III of PTS | −0.908 | −1.140 | −0.232 | BL | |
| srlD | Sorbitol-6-phosphate dehydrogenase | −0.660 | −1.334 | −0.674 | BL | |
| srlE | srl operon protein | −1.013 | −1.613 | −0.599 | BL | |
| srlR | srl regulator | −0.151 | 0.804 | −0.654 | BL | |
| sucA | 2-Oxoglutarate dehydrogenase, E1 component | 0.850 | −0.851 | −1.701 | AH | |
| sucB | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex (E2) | 0.566 | −1.073 | −1.638 | AH | |
| sucC | Succinyl-CoAc synthase beta subunit | 0.494 | −1.019 | −1.513 | AH | |
| sucD | Succinyl-CoA synthase alpha subunit | 0.526 | −1.225 | −1.751 | AH | |
| tpiA | Triosephosphate isomerase | 0.772 | 0.245 | −0.526 | NL | |
| treB | Trehalose-specific PTS enzyme II | −3.156 | 0.739 | 3.895 | AL | |
| treC | Trehalose-6-phosphate hydrolase | −2.613 | 0.620 | 3.233 | AL | |
| Proton transport and electron transport chain | atpA | ATP synthase subunit alpha, F1 | 0.111 | 0.460 | 0.349 | BH |
| atpB | ATP synthase subunit a, F0 | 0.125 | 0.484 | 0.609 | BH | |
| atpC | ATP synthase subunit epsilon, F1 | 0.266 | 1.005 | 0.739 | BH | |
| atpD | ATP synthase subunit beta, F1 | 0.542 | 1.172 | 0.630 | BH | |
| atpE | ATP synthase subunit c, F0 | 0.182 | 0.503 | 0.321 | BH | |
| atpF | ATP synthase subunit b, F0 | −0.055 | 0.321 | 0.377 | BH | |
| atpG | ATP synthase subunit gamma, F1 | 0.372 | 0.632 | 0.260 | BH | |
| atpH | ATP synthase subunit delta, F1 | −0.172 | 0.186 | 0.358 | AL | |
| atpI | ATP synthase subunit, F1F0-type proton-ATPase | 0.017 | 0.499 | 0.482 | BH | |
| cydA | Cytochrome d (bd-I) terminal oxidase subunit I (microaerobic) | −0.268 | 0.872 | 1.140 | BH | |
| cydB | Cytochrome d (bd-I) terminal oxidase subunit II (microaerobic) | −0.097 | 0.777 | 0.874 | BH | |
| cydC | Cysteine exporter to periplasm required for Cyd assembly | 0.135 | 0.490 | 0.355 | BH | |
| cydD | Cysteine exporter to periplasm required for cytochrome assembly | 0.232 | 0.574 | 0.342 | BH | |
| cyoA | Cytochrome o oxidase subunit II | 1.160 | 0.217 | −0.943 | NL | |
| cyoB | Cytochrome o oxidase subunit I | 0.812 | −0.204 | −1.016 | NL | |
| cyoC | Cytochrome o oxidase subunit III | 1.026 | 0.019 | −1.007 | NL | |
| cyoD | Cytochrome o oxidase subunit IV | 0.829 | −0.192 | −1.021 | NL | |
| cyoE | Cytochrome o oxidase subunit protoheme IX farnesyltransferase | 1.094 | 0.225 | −0.869 | NL | |
| fdoG | Formate dehydrogenase-O, major selenopeptide subunit | 0.353 | −0.138 | −0.491 | AH | |
| fdoH | Formate dehydrogenase-O Fe-S subunit | 0.003 | −0.414 | −0.416 | BL | |
| frdA | Fumarate reductase flavoprotein subunit | −0.305 | 0.046 | 0.351 | AL | |
| frdC | Fumarate reductase membrane anchor polypeptide | −0.378 | −0.008 | 0.370 | AL | |
| fumA | Fumarase A | 0.595 | −0.647 | −1.242 | AH | |
| nfsA (mdaA) | Nitroreductase A | 0.561 | −0.419 | −0.980 | AH | |
| mdaB | Probable nitroreductase or quinone reductase | 0.444 | −0.223 | −0.667 | AH | |
| napC | Cytochrome electron source for NapAB, membrane bound | −0.797 | −0.382 | 0.416 | NH | |
| ndh | Respiratory NADH dehydrogenase II; NADH:ubiquinone oxidoreductase II | 0.789 | 0.161 | −0.628 | NL | |
| nuoC | NADH:ubiquinone oxidoreductase subunit C | 0.440 | −0.084 | −0.524 | AH | |
| nuoG | NADH:ubiquinone oxidoreductase subunit G; NADH dehydrogenase I | 0.525 | 0.103 | −0.442 | NL | |
| nuoH | NADH:ubiquinone oxidoreductase subunit H; NADH dehydrogenase I | 0.564 | 0.185 | −0.379 | NL | |
| nuoI | NADH:ubiquinone oxidoreductase subunit I; NADH dehydrogenase I | 0.792 | 0.310 | −0.481 | NL | |
| nuoJ | NADH:ubiquinone oxidoreductase subunit J; NADH dehydrogenase I | 0.468 | 0.234 | −0.234 | NL | |
| nuoK | NADH:ubiquinone oxidoreductase subunit K; NADH dehydrogenase I | 0.766 | 0.460 | −0.306 | NL | |
| nuoL | NADH:ubiquinone oxidoreductase subunit L; NADH dehydrogenase I | 0.311 | 0.078 | −0.233 | NL | |
| nuoN | NADH:ubiquinone oxidoreductase subunit N; NADH dehydrogenase I | 0.126 | −0.210 | −0.336 | AH | |
| sdhA | Succinate dehydrogenase flavoprotein subunit | 1.438 | −0.018 | −1.458 | AH | |
| sdhB | Succinate dehydrogenase iron-sulfur protein | 1.833 | 0.329 | −1.505 | AH | |
| sdhC | Succinate dehydrogenase membrane anchor subunit, cytochrome b556 | 2.728 | 0.890 | −1.838 | NL | |
| sdhD | Succinate dehydrogenase hydrophobic subunit | 2.349 | 0.595 | −1.754 | NL | |
| Amino acid catabolism and transport | artI | Arginine periplasmic binding protein | 0.286 | 0.655 | 0.369 | BH |
| artM | Arginine periplasmic binding protein | −0.115 | 0.293 | 0.407 | BH | |
| cadA | Lysine decarboxylase, degradative | 1.024 | 0.137 | −0.887 | AH | |
| cysK | O-Acetylserine sulfhydrylase A (cysteine synthase) | 1.204 | 1.351 | 0.147 | NL | |
| dadA | d-Amino acid dehydrogenase | 1.273 | −0.229 | −1.501 | AH | |
| dadX | d-Amino acid dehydrogenase | 0.683 | −0.393 | −1.076 | AH | |
| dppC | Dipeptide permease system | −0.346 | −0.032 | 0.315 | AL | |
| gdhA | Glutamate dehydrogenase | −0.080 | −0.496 | −0.416 | BL | |
| hisC | Histidinol-phosphate aminotransferase | 0.253 | 0.984 | 0.731 | BH | |
| hisF | Imidazole glycerol phosphate synthase (cyclase) | 0.150 | 0.703 | 0.553 | BH | |
| hisH | Amidotransferase of imidazole glycerol phosphate synthase | 0.034 | 0.393 | 0.359 | BH | |
| hisI | PR-ATP pyrophosphatase and PR-AMP cyclohydrolase | 0.234 | 0.641 | 0.407 | BH | |
| hisJ | Histidine-binding protein | −0.149 | 0.400 | 0.549 | BH | |
| lysC | Aspartokinase III | 1.464 | −0.226 | −1.690 | AH | |
| lysP | Lysine permease | 2.662 | 0.963 | −1.698 | NL | |
| lysU | Lysine-tRNA ligase | 0.544 | −0.077 | −0.621 | AH | |
| potD | Putrescine-ornithine transporter | 0.053 | 0.509 | 0.456 | BH | |
| sdaA | l-Serine deaminase, degradative | −0.409 | 0.638 | 1.048 | BH | |
| sdaB | l-Serine deaminase | −1.111 | −0.434 | 0.678 | AL | |
| sdaC | H+/serine symporter; regulator of serine deaminase | −1.205 | −0.410 | 0.794 | AL | |
| tnaA | Tryptophan deaminase, degradative; also deaminases serine and cysteine | −3.805 | 0.223 | 4.028 | AL | |
| tnaB | Tryptophan transporter | −1.840 | 1.153 | 2.993 | AL | |
| tnaC | tnaA leader peptide | −5.026 | 0.490 | 5.517 | AL | |
| tdcB | Threonine dehydratase, degradative | −0.849 | −0.296 | 0.553 | NH | |
| ydfG | l-allo-Threonine, l-serine, d-serine dehydrogenase | 0.407 | 0.287 | −0.120 | NL | |
Values in boldface are significant (α = 0.001).
NL, Neutral Low; AH, Acid High; AL, Acid Low; BH, Base High; BL, Base Low; NH, Neutral High.
CoA, coenzyme A.
PTS, phosphotransferase.
Many operons encoding processes of glycolysis and the TCA cycle, such as aceEF (pyruvate dehydrogenase), dhaKL (dihydroxyacetone kinase), pta (phosphotransacetylase), and pts (glucose phosphotransferase), showed elevated expression in acid. Others, however, were elevated at high pH. Operons elevated at high pH tended to be those induced by anaerobiosis, such as glpABC (anaerobic glycerol-3-phosphate dehydrogenase), pflBA (anaerobic pyruvate formate lyase), and dcu (anaerobic fumarate respiration). The mal system, however, is strongly repressed by acid (13, 31) and showed up as such in our arrays.
Membrane-bound systems for proton and electron transport were regulated by acid or base along lines largely consistent with their relative degree of export or import of H+. An example is the atp operon encoding F1Fo ATP synthase (32), which imports H+ during oxidative respiration. Most of the atp genes were strongly upregulated at high pH, whereas ndh and nuo (the NADH dehydrogenases I and II), which export H+, were down-regulated. The sdh gene (succinate dehydrogenase), which contributes electrons for proton export, is also down-regulated at high pH. On the other hand, cytochrome d oxidase (cyd) is expressed in preference to cytochrome o oxidase (cyo) at high pH, presumably because it exports half as many H+ per electron (14).
Enzymes for degradation of amino acids showed pH regulation as expected, with high pH favoring deaminase operons such as tna (tryptophan deaminase), sda (serine deaminase), and tdcB (threonine dehydratase). Acid induced only one of the decarboxylase operons, cad (lysine decarboxylase). Several decarboxylases are known to be induced by acid, but their induction is repressed by oxygen (4, 30), which may explain their absence in our highly aerobic cultures.
Oxidative stress and salicylate stress.
Several acid stress genes are known to overlap with oxidative stress, for example, the alkyl hydroperoxide reductase ahpC (9, 84), and certain permeant acids such as salicylate are considered oxidative stress agents (54). We surveyed our pH-regulated genes for overlap with response to H2O2, paraquat, and salicylate, as reported in references 54 and 84 (Table 5).
TABLE 5.
pH-regulated oxidative stress responsea
| Gene | Function | Log2 pH ratiob
|
PQ, Sal, or H2O2 | Classc | ||
|---|---|---|---|---|---|---|
| 5/7 | 8.7/7 | 8.7/5 | ||||
| acnA | Aconitase A | 0.769 | −0.143 | −0.912 | Sal | AH |
| adhE | Acetaldehyde-coenzyme A dehydrogenase | 0.043 | −0.380 | −0.422 | Sal | BL |
| ahpC | Alkyl hydroperoxide reductase small subunit | 1.003 | 0.436 | −0.568 | PQ, H2O2 | NL |
| ahpF | Alkyl hydroperoxide reductase large subunit | 0.777 | −0.222 | −0.999 | H2O2 | AH |
| aldA | Aldehyde dehydrogenase, NAD linked | 0.773 | 0.764 | −0.009 | PQ | NL |
| alx (ygjT) | Membrane protein, alkali induced | −1.317 | 0.692 | 2.009 | PQ− | AL |
| artI | Periplasmic arginine binding protein | 0.286 | 0.655 | 0.369 | PQ | BH |
| aspA | Aspartate ammonia-lyase (aspartase) | −0.592 | 0.770 | 1.362 | PQ− | BH |
| carA | Carbamoylphosphate synthase small subunit | 0.029 | 1.059 | 1.030 | PQ− | BH |
| cfa | Cyclopropane fatty acid synthase | 2.075 | −0.480 | −2.555 | Sal | AH |
| cyaA | Adenylate cyclase | 0.691 | −0.156 | −0.846 | Sal | AH |
| cyoD | Cytochrome o oxidase subunit IV | 0.829 | −0.192 | −1.021 | PQ | NL |
| cysK | Cysteine synthase | 1.204 | 1.351 | 0.147 | PQ, Sal, H2O2 | NL |
| dadX | Alanine racemase | 0.683 | −0.393 | −1.076 | PQ | AH |
| deoA | Thymidine phosphorylase | −1.031 | −2.003 | −0.972 | Sal | BL |
| deoB | Deoxyribouratase, phosphopentomutase | −0.731 | −1.502 | −0.771 | PQ, Sal | BL |
| dhaH | Dihydroxyacetone phosphoryl donor | 1.547 | −0.527 | −2.074 | Sal | AH |
| dhaK | Dihydroxyacetone kinase | 1.041 | −0.379 | −1.420 | Sal | AH |
| dnaK | HSP-70-type molecular chaperone | −0.894 | −2.249 | −1.356 | Sal | BL |
| dps | Stress response DNA-binding protein | 1.130 | 0.105 | −1.025 | PQ, Sal, H2O2 | NL |
| fliS | Flagellar synthesis; flagellar regulon member | −0.174 | −1.947 | −1.772 | PQ− | BL |
| fpr | Ferredoxin NADP+ reductase; anaerobic | 0.565 | 0.275 | −0.289 | PQ, H2O2 | NL |
| gapA | GAPDHd A | 0.093 | −0.752 | −0.844 | Sal | BL |
| gatA | Galactitol-specific enzyme IIA of PTSe | 0.929 | −0.626 | −1.555 | PQ, Sal | AH |
| gatB | Galactitol-specific enzyme IIB of PTS | 0.632 | −0.895 | −1.528 | PQ, Sal | AH |
| gatC | Galactitol-specific enzyme IIC of PTS | 0.973 | −0.949 | −1.922 | Sal | AH |
| gatD | Galactitol-1-phosphate dehydrogenase | 1.037 | −0.893 | −1.930 | PQ, Sal | AH |
| gatZ | Tagatose 6-phosphate aldolase 2 | 0.774 | −0.605 | −1.380 | Sal | AH |
| gltA | Citrate synthase | 0.288 | −0.559 | −0.846 | PQ, Sal | AH |
| gltB | Glutamate synthase, large subunit | 0.846 | −1.425 | −2.271 | Sal | AH |
| grxA | Glutaredoxin 1 | 1.666 | −0.106 | −1.772 | H2O2 | AH |
| gshB | Glutathione synthetase | 0.768 | −0.081 | −0.848 | Sal | AH |
| hdeA | Periplasmic acid chaperone | 0.841 | −0.326 | −1.167 | Sal | AH |
| hdeB | Periplasmic acid chaperone | 0.782 | −0.622 | −1.404 | Sal | AH |
| hisF | Cyclase component of IGP synthase | 0.150 | 0.703 | 0.553 | PQ− | BH |
| ibpB | Chaperone, HSP20 family | −1.691 | −2.618 | −0.928 | H2O2 | BL |
| katG | Catalase hydrogen peroxidase 1 | −0.578 | −0.313 | 0.265 | H2O2 | AL |
| lamB | Maltose high-affinity uptake | −4.881 | −3.735 | 1.146 | PQ | NH |
| lldP | l-Lactate permease | 1.527 | 0.219 | −1.307 | Sal | NL |
| lysU | Lysyl tRNA synthetase, inducible | 0.544 | −0.077 | −0.621 | Sal | AH |
| malE | Maltose-binding protein, periplasmic | −3.425 | −2.933 | 0.491 | PQ | NH |
| malK | Maltose transport complex, ATP-binding subunit | −4.790 | −3.748 | 1.041 | PQ | NH |
| manX | PTS family, mannose-specific enzyme IIA component | 0.000 | 0.517 | 0.517 | Sal | AL |
| map | Methionine aminopeptidase | 0.510 | 0.056 | −0.454 | PQ | AH |
| marA | Multiple antibiotic resistance | 1.321 | −0.516 | −1.836 | PQ, Sal | AH |
| marB | Regulator for mar | 0.894 | −0.172 | −1.066 | Sal | AH |
| marR | Repressor of mar | 0.352 | −0.437 | −0.789 | Sal | AH |
| mdaB | Drug activity modulator | 0.444 | −0.223 | −0.667 | Sal | AH |
| murF | d-Alanyl:d-alanine adding to cell wall | 0.216 | −0.170 | −0.386 | PQ | AH |
| nfnB | Nitrofurantoin resistance; nitroreductase | 0.818 | −0.204 | −1.022 | PQ, Sal | AH |
| nuoI | NADH dehydrogenase I subunit | 0.792 | 0.310 | −0.481 | PQ | NL |
| nuoK | NADH dehydrogenase I subunit | 0.766 | 0.460 | −0.306 | PQ | NL |
| ompF | Outer membrane porin | −2.834 | −0.892 | 1.942 | PQ− | AL |
| ompT | Outer membrane protease VII | −0.815 | −1.204 | −0.389 | Sal− | BL |
| pdhR | Repressor of pdh | 0.755 | −0.109 | −0.863 | PQ | AH |
| pepN | Aminopeptidase N | 0.671 | 0.118 | −0.553 | Sal | NL |
| pflB | Pyruvate formate lyase I (anaerobic) | 0.812 | 0.578 | −0.234 | Sal | NL |
| pgi | Glucose phosphate isomerase | 0.654 | 0.177 | −0.477 | PQ | NL |
| ptsG | PTS family IIC, glucose specific | 0.841 | 0.554 | −0.287 | PQ | NL |
| putA | Proline dehydrogenase | −1.062 | −0.353 | 0.708 | Sal | AL |
| pyrB | Aspartate transcarbamylase | 0.133 | 0.608 | 0.475 | PQ−, Sal− | BH |
| sdhB | Succinate dehydrogenase | 1.833 | 0.329 | −1.505 | PQ | AH |
| tnaA | Tryptophanase | −3.805 | 0.223 | 4.028 | H2O2 | AL |
| treB | Tre-specific PTS enzyme II | −3.156 | 0.739 | 3.895 | Sal− | AL |
| yahA | Putative repressor | −0.667 | 0.422 | 1.089 | PQ− | AL |
| yaiA | Function unknown | 1.154 | 0.416 | −0.738 | H2O2 | NL |
| ybjC | Function unknown | 0.463 | −0.447 | −0.910 | PQ, Sal | AH |
| ycfR | Function unknown | 0.537 | −0.819 | −1.356 | H2O2 | AH |
| yfiA | Stabilizes ribosome against dissociation | −1.782 | 0.581 | 2.364 | PQ−, Sal, H2O2 | AL |
| yggJ | Function unknown | 0.576 | −0.178 | −0.755 | Sal | AH |
| yqjD | Function unknown | −0.347 | 0.378 | 0.725 | Sal | AL |
| yncE | Function unknown | −0.476 | 0.331 | 0.808 | PQ, Sal | AL |
Oxidative response is based on data in references 54 and 84. Induction was by H2O2, paraquat (PQ), or sodium salicylate (Sal). Repression is indicated by minus sign (Sal−, PQ−).
Values in boldface indicate significance (α = 0.001).
AH, Acid High; AL, Acid Low; BH, Base High; BL, Base Low; NH, Neutral High; NL, Neutral Low.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
PTS, phosphotransferase.
Of the 73 pH-dependent genes known to be induced by H2O2, paraquat, or salicylate, virtually all were induced by acid or repressed by base. This finding confirms our hypothesis of a strong connection between acid stress and oxidative stress. It may be that low pH amplifies the toxicity of oxygen radicals. Genes repressed by paraquat or salicylate were repressed in acid or induced at high pH, such as the base-inducible membrane protein gene alx, the histidine cyclase gene hisF, and outer membrane protein gene ompF. An exception to these generalizations was the maltose regulon (lamB, malE, and malK), which was repressed by acid but induced by paraquat.
Envelope and periplasmic stress.
A large part of E. coli function takes place in the outer membrane and envelope (48) and the periplasm (49), compartments essentially exposed to “extracellular” pH. Thus, it is not surprising that several envelope and periplasmic components show pH-dependent expression (16, 23, 73, 82). Our microarrays revealed an even greater number of such responses (Table 6). Both acid and base induction were observed. Acid-induced periplasmic proteins included the well-known acid chaperone from hdeAB (23), as well as the newly observed TolA-binding protein (ybgF) and the lipoprotein from pal. High pH induced the ferric transporters from fecAB and fhuD, possibly due to low iron solubility at high pH. At high pH, various transport proteins and redox modulators such as that from dsbA are known to be induced. In addition, several additional base-induced periplasmic and envelope proteins appeared, including the vitamin B12 transporter from btuB, the outer membrane protein from nmpC, and the peptidylprolyl-cis-trans-isomerase from ppiA.
TABLE 6.
Envelope and periplasmic genes
| Gene | Function | Log2 pH ratioa
|
Classb | ||
|---|---|---|---|---|---|
| 5/7 | 8.7/7 | 8.7/5 | |||
| artI | Periplasmic binding protein of Arg transport system | 0.286 | 0.655 | 0.369 | BH |
| artM | Arginine periplasmic binding protein | −0.115 | 0.293 | 0.407 | BH |
| btuB | B-12 transporter, outer membrane receptor | −0.667 | 0.037 | 0.704 | AL |
| cirA | Colicin I receptor production | −0.885 | 0.260 | 1.145 | AL |
| clpX | ATPase subunit of ClpXP protease | −0.361 | −0.625 | −0.264 | BL |
| cpxA | Periplasmic stress sensor (CpxAR) | −0.461 | 0.230 | 0.691 | AL |
| cpxP | CpxAR-regulated periplasmic stress protein | −3.232 | 1.002 | 4.234 | AL |
| cpxR | Periplasmic stress response regulator (CpxAR) | −0.662 | 0.590 | 1.251 | AL |
| dsbA | Thiol:disulfide interchange, periplasmic | 0.086 | 1.117 | 1.031 | BH |
| dsbC | Disulfide bond isomerase, periplasmic chaperone | −0.288 | −0.534 | −0.246 | BL |
| fadL | Fatty acid transport, outer membrane | −0.861 | 1.069 | 1.931 | BH |
| fecA | Outer membrane ferric citrate receptor | −2.608 | −1.128 | 1.480 | NH |
| fecB | Periplasmic ferric citrate-binding protein | −2.842 | −1.538 | 1.304 | NH |
| fepA | Ferrienterobactin outer membrane receptor | −0.966 | −0.289 | 0.676 | NH |
| fhuD | Ferric hydroxamate binding protein; hydroxamate-dependent iron uptake | −0.767 | −0.180 | 0.587 | AL |
| fliY | Cystine-binding protein, periplasmic | 0.233 | −0.252 | −0.484 | AH |
| hdeA | Acid periplasmic chaperone | 0.841 | −0.326 | −1.167 | AH |
| hdeB | Acid periplasmic protein | 0.782 | −0.622 | −1.404 | AH |
| hisJ | High-affinity histidine-binding protein | −0.149 | 0.400 | 0.549 | BH |
| hlpA | Periplasmic chaperone for OMPsc | 0.099 | −0.661 | −0.759 | BL |
| lamB | Maltoporin, maltose high-affinity uptake; phage lambda receptor | −4.881 | −3.735 | 1.146 | NH |
| lon | DNA-binding, ATP-dependent protease | −0.600 | −1.666 | −1.065 | BL |
| malE | Maltose-binding protein, periplasmic | −3.425 | −2.933 | 0.491 | NH |
| malM | Maltose operon periplasmic protein | −4.643 | −3.780 | 0.863 | NH |
| mltB | Membrane-bound murein hydrolase | −0.739 | −0.419 | 0.320 | NH |
| nmpC | Outer membrane | −4.012 | −0.051 | 3.961 | AL |
| ompF | Outer membrane porin protein 1a | −2.834 | −0.892 | 1.942 | AL |
| ompT | Outer membrane protease VII | −0.815 | −1.204 | −0.389 | BL |
| ompX | OMP, induces RNAP-sigma E | 1.523 | 0.439 | −1.083 | NL |
| oppA | Periplasmic oligopeptide binding protein | 1.358 | 0.406 | −0.953 | NL |
| pal | Lipoprotein associated with peptidoglycan | 0.532 | 0.379 | −0.153 | NL |
| potD | Spermidine-binding membrane protein; regulates pot | 0.053 | 0.509 | 0.456 | BH |
| ppiA | Rotamase; peptidylprolyl-cis-trans-isomerase A | −0.526 | 0.231 | 0.757 | AL |
| pstS | High-affinity, periplasmic phosphate binding protein | −2.479 | −2.262 | 0.217 | NH |
| rbsB | d-Ribose binding protein, periplasmic | −0.749 | −1.150 | −0.401 | BL |
| rseB | Periplasmic, binds RseA; enhances RpoE-RseA cytoplasmic complex formation | 0.631 | −0.105 | −0.736 | AH |
| secD | SecDF-YajC inner membrane secretion complex | −0.165 | 0.287 | 0.452 | AL |
| surA | Periplasmic outer membrane porin chaperone, stationary phase | 0.310 | 0.209 | −0.101 | NL |
| tatA | Twin arginine translocation | 0.043 | 0.679 | −0.636 | BH |
| tatB | Twin arginine translocation | 0.075 | 0.446 | 0.372 | BH |
| tolB | Group A colicin uptake and tolerance | 0.498 | 0.421 | −0.077 | NL |
| tpX | Thiol peroxidase, antioxidant | 0.637 | 0.178 | −0.459 | NL |
| tsx | Phage T6, colicin K resistance; nucleoside channel | −0.707 | −0.904 | −0.196 | BL |
| ybgF | TolA-binding periplasmic protein | 0.312 | −0.007 | −0.318 | AH |
| yceI | Function unknown; periplasmic protein | −0.036 | 1.038 | 1.074 | BH |
| yhcN | Periplasmic protein | 3.064 | −1.136 | −4.199 | AH |
Values in boldface indicate significance (α = 0.001).
AH, Acid High; AL, Acid Low; BH, Base High; BL, Base Low; NH, Neutral High; NL, Neutral Low.
OMPs, outer membrane proteins.
RNAP, RNA polymerase.
Universal stress and heat shock.
Various heat shock and universal stress proteins are inducible by the permeant acid benzoate, such as the products of clpB, htpG, dnaK, groS, and uspA (38). Some of these showed pH response in our microarrays (Table 7). The DNA damage response gene uspD was acid induced, as was dps, encoding the DNA-binding protein involved in stationary phase and acid resistance. Acid induced rseAB, the antisigma regulators of the rpoE envelope heat stress system (1). High pH induced the rpoH heat shock sigma 32 gene (28) as well as heat shock proteasome genes hslUV and regulators hslOR.
TABLE 7.
Universal stress and heat shock response genes
| Gene | Function | Log2 pH ratioa
|
Classb | ||
|---|---|---|---|---|---|
| 5/7 | 8.7/7 | 8.7/5 | |||
| ahpC | Alkyl hydroperoxide reductase | 1.003 | 0.436 | −0.568 | NL |
| ahpF | NAD(P)H:peroxiredoxin oxidoreductase | 0.777 | −0.222 | −0.999 | AH |
| cfa | Cyclopropane fatty acid synthase; acid resistance in stationary phase | 2.075 | −0.480 | −2.555 | AH |
| clpB | ClpB protease, ATP-dependent chaperone | −0.219 | −1.963 | −1.744 | BL |
| cysK | Cysteine synthase, o-acetylserine sulfhydrylase A | 1.204 | 1.351 | 0.147 | NL |
| cysZ | Unknown function | 0.440 | 0.212 | −0.228 | NL |
| dinI | Inhibits RecA coprotease | 0.132 | 0.568 | 0.436 | BH |
| dinJ | Induced by DNA damage | 0.683 | 1.192 | 0.508 | BH |
| dnaJ | DnaK cochaperone | −0.908 | −2.155 | −1.247 | BL |
| dnaK | HSP-70-type molecular chaperone | −0.894 | −2.249 | −1.356 | BL |
| dps | Stress response DNA-binding protein | 1.130 | 0.105 | −1.025 | NL |
| grpE | Nucleotide exchange factor for DnaKJ | −0.491 | −1.141 | −0.650 | BL |
| grxA | Glutaredoxin 1 | 1.666 | −0.106 | −1.772 | AH |
| hdeA | Acid periplasmic chaperone | 0.841 | −0.326 | −1.167 | AH |
| hdeB | Acid periplasmic chaperone | 0.782 | −0.622 | −1.404 | AH |
| hslJ | Heat-inducible novobiocin resistance | 0.368 | −0.676 | −1.044 | AH |
| hslU | Heat-inducible ATP-dependent protease | −0.745 | −1.688 | −0.946 | BL |
| hslV | Heat-inducible ATP-dependent protease | −1.125 | −1.913 | −0.788 | BL |
| ibpB | Heat-inducible chaperone, HSP20 family | −1.691 | −2.618 | −0.928 | BL |
| hslO | Hsp33, cytoplasmic heat shock chaperone activated by disulfide bond formation | −1.453 | −1.737 | −0.284 | BL |
| hslR (yrfH) | Hsp15, heat shock, binds RNA and DNA | −1.773 | −1.940 | −0.168 | NH |
| katG | Catalase-hydrogen peroxidase I | −0.578 | −0.313 | 0.265 | AL |
| rpoE | RNAPc sigma E, envelope heat stress | 0.310 | −0.427 | −0.738 | AH |
| rpoH | RNAP sigma 32, heat shock regulons | −0.378 | 0.084 | 0.462 | AL |
| rseA | Anti-RpoE sigma factor, spans inner membrane | 0.419 | −0.159 | −0.578 | AH |
| rseB | Binds periplasmic domain of anti-RpoE sigma RseA | 0.631 | −0.105 | −0.736 | AH |
| sodB | Superoxide dismutase, Fe; acid inducible | 0.752 | 0.280 | −0.472 | NL |
| ycdB | Function unknown, peroxidase homolog | 0.654 | −0.329 | −0.983 | AH |
| ycdO | Acid inducible, function unknown | 0.842 | −0.625 | −1.467 | AH |
| uspD (yiiT) | UV resistance | 1.333 | −0.497 | −1.830 | AH |
Values in boldface indicate significance (α = 0.001).
AH, Acid High; AL, Acid Low; BH, Base High; BL, Base Low; NH, Neutral High; NL, Neutral Low.
RNAP, RNA polymerase.
DISCUSSION
Overall, our work revealed a large number of genes not previously known to be regulated by pH. Furthermore, many of these genes had no previously known function or response, such as yhcN and yagU (induced by acid) and yifO and ymcG (induced by base).
An important question is to assess the biological relevance of the expression ratios reported (36, 51). Most of the ratios we reported as significant (boldface in Tables 3 through 7) are greater than twofold (log2 = 1). In many cases, all or most members of an operon fell in the same cluster and show similar expression profiles; the flagellar regulon was particularly consistent (Table 3). The gene probes are synthesized on the array independently of their operon map; thus, parallel expression profiles within operons do not reflect array position. Note that even genes with significant expression ratios of less than 2 (log2 = 1) tend to group with their operons. In previous studies, comparison with quantitative reverse transcriptase real-time PCR shows that microarray ratios, while quantitatively consistent, generally underestimate the actual differences in mRNA levels between the biological systems compared (83).
Flagellar biosynthesis and motility.
The effects of pH in flagellar biosynthesis and motility remain poorly understood. It has long been known that low external pH (thus, large ΔpH) contributes to the proton motive force that drives flagellar rotation (33). The cytoplasmic pH, however, must remain high; permeant acids such as acetate and benzoate, which depress internal pH and decrease proton motive force, are chemotactic repellents (67) and impair rotation of the flagellar motor (46). Low pH elicits negative chemotaxis (55, 67), whereas a pH increase up to 8.3 elicits a positive response (55).
In recent reports acid stress is associated with low motility (72), yet acetate has been reported to induce the flagellar regulon and enhance motility (53). We believe that the previous reports are limited in several ways. Reference 72 does not compare pH conditions directly but notes repression of flagellar genes in an hns mutant in which acid resistance is increased. The motility assay is not clearly described, and the acid dependence of flhDC-cat expression was observed on plasmids, not in the genome. Reference 53 reports induction of chromosomal flhDC-lacZ fusions by acetate. Those authors' assays of motility, however, show relatively small differences between pH conditions.
Our microarrays showed strong evidence for suppression of motility and chemotaxis at high pH. This evidence was supported by the decrease in motility at high pH, observed for both E. coli and S. enterica serovar Typhimurium, which swims twice as fast as E. coli. We also found weaker evidence for repression of che and mot genes at pH 5, but the flagellar synthesis genes were strongly induced at low pH. Overall, our data point to alkaline suppression of flagellar motility. Work in progress shows that, at high pH, the number of flagella per cell is decreased to one to three per cell (about 20% of normal) (S. Aizawa and J. Slonczewski, unpublished data).
No pH dependence was observed for the flagellar regulators flhD and flhC. On the other hand, in a microarray study of anaerobic cultures, flhD and flhC are induced by acid (E. Hayes and J. L. Slonczewski, unpublished data). Acid induction of these regulators would be consistent with the report of their induction by acetate (53). We did see acid induction of two known activators of flhDC: adenylate cyclase cyaA (37) (Acid High) and dnaK-dnaJ-grpE (64) (Base Low). We saw no acid induction of other flagellar activators such as crp (37), nor did we see alkaline induction of the negative flagellar regulator rcsCDB (22).
An alternative model is that pH regulation of the flagellar regulon is mediated by proteolysis, as in the case of ClpXP proteolysis of FlhD and FlhC (76). We find that ClpX is down-regulated at high pH (Base Low cluster), but a different protease could be involved.
Catabolism.
The picture of catabolism is more complicated, but in general our expression ratios confirm our present hypotheses of pH regulation while extending our knowledge to many more components. Systems that consume acids are enhanced at low pH. On the other hand, initial import and breakdown of some sugars, such as maltose, are favored at high pH, where they may quickly generate a large burst of fermentation acids.
With respect to proton export, E. coli appears to prefer components such as ATP synthase that import protons at high pH (counteracting the alkaline stress on cytoplasmic pH) and prefers to minimize proton export associated with the terminal oxidase cyd in preference to cyo. This observation is consistent with the previous report that cyd expression is higher at pH 7.5 than at pH 5.0 in an fnr mutant (14), although in those experiments cyo expression also increased with pH. It is likely that our broader range of pH classes (up to pH 8.7) provided a clearer picture of pH regulation of cyo and cyd.
Under amino acid catabolism, relatively few new components of pH response were observed. This makes sense, because most amino acid decarboxylases are repressed by oxygen (4, 68), as are deaminases such as sdaA (82). In preliminary experiments, we have repeated our microarray study on cultures grown anaerobically. Under anaerobiosis, several amino acid decarboxylases and deaminases show pH-dependent expression (Hayes and Slonczewski, unpublished).
Stress responses.
Several stress responses are known to interact with pH stress and pH resistance, including oxidative stress, heat shock, and envelope stress (for reviews see references 21 and 68). The overlap with salicylate stress could be explained in part by salicylate's effect as a permeant acid, stressing internal pH (60). The mar drug resistance operon is known to be coinduced by aromatic permeant acids and low pH (69) under regulation by MarR as well as by the superoxide regulator SoxRA (57).
Beyond salicylate, however, a large number of oxidative stress genes inducible by H2O2 or by paraquat showed significant pH-dependent expression, nearly all induced by acid or repressed by base. This finding confirms our hypothesis of a strong connection between acid stress and oxidative stress. Since so much of aerobic respiration is stepped up at pH 5, including cytochrome o oxidase, it is likely that acid conditions accelerate the production of oxygen radicals, thus inducing a partial oxidative stress response.
Various envelope and periplasmic stress responses are induced by acid, contributing to acid resistance; the best characterized in terms of mechanism is the acid-induced periplasmic chaperone HdeA (23). Extracellular acid induces a dimer-to-monomer transition in HdeA, which then suppresses aggregation by acid-denatured proteins. Our study reveals additional potential contributors to acid resistance and base resistance, including genes of unknown function such as yhcN, induced by acid, and yceI, induced by base.
Our study presents the most comprehensive picture to date of acid and base response by E. coli grown aerobically in complex medium. Overall, low pH accelerates acid consumption and proton export, while coinducing oxidative stress, possibly through increased production of oxygen radicals. High pH accelerates proton import while repressing the energy-expensive systems of flagellar biosynthesis and chemotaxis. Finally, pH differentially regulates a large number of periplasmic and envelope stress systems, as well as transporters, chaperones, and redox regulators.
Supplementary Material
Acknowledgments
The class comparison and cluster analysis were performed using BRB ArrayTools v3.1 developed by Richard Simon and Amy Peng Lam. We thank Bryan Lin and Ariel Kahrl for excellent technical assistance.
This work was supported by grant MCB-0234732 from the National Science Foundation and by undergraduate research funds from the Kenyon College grant from the Howard Hughes Medical Institute Biological Sciences Education Program.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E., I. L. Grigorova, and C. A. Gross. 2003. Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. Gerhart, H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auger, E. A., K. E. Redding, T. Plumb, L. C. Childs, S. Y. Meng, and G. N. Bennett. 1989. Construction of lac fusions to the inducible arginine and lysine decarboxylase genes of Escherichia coli K-12. Mol. Microbiol. 3:609-620. [DOI] [PubMed] [Google Scholar]
- 5.Bakker, E. P., and W. E. Mangerich. 1981. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 147:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behari, J., L. Stagon, and S. B. Calderwood. 2001. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J. Bacteriol. 183:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingham, R. J., K. S. Hall, and J. L. Slonczewski. 1990. Alkaline induction of a novel gene locus, alx, in Escherichia coli. J. Bacteriol. 172:2184-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buurman, E. T., D. McLaggan, J. Naprstek, and W. Epstein. 2004. Multiple paths for nonphysiological transport of K+ in Escherichia coli. J. Bacteriol. 186:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanie-Cornet, M., T. A. Penfound, D. Smith, J. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castanie-Cornet, M., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 13.Chagneau, C., M. Heyde, S. Alonso, R. Portalier, and P. Laloi. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 183:5675-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1997. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drasar, B. S., M. Shiner, and G. M. McLeod. 1969. Studies on the intestinal flora. I. The bacterial flora of the gastrointestinal tract in healthy and achlorhydric persons. Gastroenterology 56:71-79. [PubMed] [Google Scholar]
- 18.Dressman, J. B., R. R. Berardi, L. C. Dermentzoglou, T. L. Russel, S. P. Schmaltz, J. L. Barnett, and K. M. Jarvenpaa. 1990. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm. Res. 7:756-761. [DOI] [PubMed] [Google Scholar]
- 19.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, D. F., G. Pye, R. Bramley, A. G. Clark, T. J. Dyson, and J. D. Hardcastle. 1988. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29:1035-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 22.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 23.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 24.Gerchman, Y., Y. Olami, A. Rimon, D. Taglicht, and S. Schuldiner. 1993. Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1212-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannella, R. A., S. A. Broitman, and N. Zamcheck. 1973. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Ann. Intern. Med. 78:271-276. [DOI] [PubMed] [Google Scholar]
- 26.Gong, F., and C. Yanofsky. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864-1867. [DOI] [PubMed] [Google Scholar]
- 27.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 29.Harold, F. M., and P. C. Maloney. 1996. Energy transduction by ion currents, p. 283-306. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heyde, M., P. Laloi, and R. Portalier. 2000. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J. Bacteriol. 182:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasimoglu, E., S. Park, J. Malek, C. P. Tseng, and R. P. Gunsalus. Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth. J. Bacteriol. 178:5563-5567. [DOI] [PMC free article] [PubMed]
- 33.Khan, S., and R. Macnab. 1980. Proton chemical potential, proton electrical potential and bacterial motility. J. Mol. Biol. 138:599-614. [DOI] [PubMed] [Google Scholar]
- 34.Kihara, M., and R. Macnab. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kothapalli, R., S. J. Yoder, S. Mane, and T. P. Loughran, Jr. 2002. Microarray results: how accurate are they? BMC Bioinformatics 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutsukake, K. 1997. Autogenous and global control of the flagellar master operon flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440-448. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, C., and W. H. Wong. 2003. DNA-chip analyzer (dChip), p. 120-141. In G. Parmigiani, E. S. Garrett, R. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software. Springer-Verlag, New York, N.Y.
- 41.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 43.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 44.Masuda, N., and G. Church. 2003. Regulatory network of acid resistance in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 45.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 46.Minamino, T., Y. Imae, F. Oosawa, Y. Kobayashi, and K. Oosawa. 2003. Effect of intracellular pH on rotational speed of bacteria flagellar motors. J. Bacteriol. 185:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neely, M. N., C. L. Dell, and E. R. Olson. 1994. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J. Bacteriol. 176:3278-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 49.Oliver, D. B. 1996. Periplasm, p. 88-103. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 50.Payne, D., D. Tatham, E. D. Williamson, and R. W. Titball. 1998. The pH 6 antigen of Yersinia pestis binds to β1-linked galactosyl residues in glycosphingolipids. Infect. Immun. 66:4545-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piper, M. D. W., P. Daran-Lipujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsens, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 52.Plack, R. H., Jr., and B. P. Rosen. 1980. Cation/proton antiport systems in Escherichia coli. Absence of potassium/proton antiporter activity in a pH-sensitive mutant. J. Biol. Chem. 255:3824-3825. [PubMed] [Google Scholar]
- 53.Polen, T., D. Rittmann, V. F. Wendisch, and H. Sahm. 2003. DNA microarray analysis of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 69:1759-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pomposiello, P., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Repaske, D., and J. Adler. 1981. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J. Bacteriol. 145:1196-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roe, A. J., D. McLaggan, I. Davidson, C. O'Byrne, and I. R. Booth. 1998. Perturbations of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosner, J. L., and J. L. Slonczewski. 1994. Dual regulation of inaA by the multiple antibiotic resistance (mar) and superoxide (soxRS) stress response systems of Escherichia coli. J. Bacteriol. 176:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowbury, R. J., Z. Lazim, and M. Goodson. 1996. Regulatory aspects of alkali tolerance induction in Escherichia coli. Lett. Appl. Microbiol. 22:429-432. [DOI] [PubMed] [Google Scholar]
- 59.Russell, J. B., and F. Diez-Gonzalez. 1998. The effect of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 60.Salmond, C. V., R. G. Kroll, and I. Booth. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 130:2845-2850. [DOI] [PubMed] [Google Scholar]
- 61.Schadt, E. E., C. Li, B. Ellis, and W. Wong. 2001. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. 84(S37):120-125. [DOI] [PubMed] [Google Scholar]
- 62.Schadt, E. E., C. Li, C. Su, and W. H. Wong. 2001. Analyzing high-density oligonucleotide gene expression array data. J. Cell. Biochem. 80:192-202. [PubMed] [Google Scholar]
- 63.Schuldiner, S., V. Agmon, J. Brandsma, A. Cohen, E. Friedman, and E. Padman. 1986. Induction of SOS function by alkaline intracellular pH in Escherichia coli. J. Bacteriol. 168:936-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi, W., Y. Zhou, J. Wild, J. Adler, and C. A. Gross. 1992. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J. Bacteriol. 174:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skandamis, P. N., and G.-J. E. Nychas. 2000. Development and evaluation of a model predicting the survival of Escherichia coli O157:H7 NCTC 12900 in homemade eggplant salad at various temperatures, pHs, and oregano essential oil concentrations. Appl. Environ. Microbiol. 66:1646-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slonczewski, J. L., T. N. Gonzalez, F. M. Bartholomew, and N. J. Holt. 1987. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J. Bacteriol. 169:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slonczewski, J. L., R. M. Macnab, J. R. Alger, and A. M. Castle. 1982. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J. Bacteriol. 152:384-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slonczewski, J. L., and J. W. Foster. 1996. pH-related genes and survival at extreme pH, p. 1539-1552. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 69.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soksawatmaekhin, W., A. Kuraishi, K. Skata, K. Kashiwagi, and K. Igarahi. 2004. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 51:1401-1412. [DOI] [PubMed] [Google Scholar]
- 72.Soutourina, O. A., E. Krin, C. Laurent-Winter, F. Hommais, A. Danchin, and P. N. Bertin. 2002. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiology 148:1543-1551. [DOI] [PubMed] [Google Scholar]
- 73.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taglicht, D., E. Padan, A. B. Oppenheim, and S. Schuldiner. 1987. An alkaline shift induces the heat shock response in Escherichia coli. J. Bacteriol. 169:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taglicht, D., E. Padan, and S. Schuldiner. 1991. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 266:11289-11294. [PubMed] [Google Scholar]
- 76.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443-452. [DOI] [PubMed] [Google Scholar]
- 77.Tramonti, A., M. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH responses in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.White, S., F. E. Tuttle, D. Blankenhorn, D. C. Dosch, and J. L. Slonczewski. 1992. pH dependence and gene structure of inaA in Escherichia coli. J. Bacteriol. 174:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeung, K. Y., and W. L. Ruzzo. 2001. Principal component analysis for clustering gene expression data. Bioinformatics 17:763-774. [DOI] [PubMed] [Google Scholar]
- 82.Yohannes, E., D. M. Barnhart, and J. L. Slonczewski. 2004. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J. Bacteriol. 186:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.