The aim of this study was to assess the rate of change of MGMT methylation status during the clinical course of glioblastoma and the potential implications of these changes.
Keywords: Glioblastoma, Surgery, MGMT methylation, Heterogeneity, Recurrent glioblastoma
Abstract
Background.
MGMT methylation status represents a powerful prognostic factor in newly diagnosed glioblastoma (GBM). Recently, its role in recurrent tumors has also been suggested; however, few data investigating the stability of this biomarker during the clinical course of the disease are available. In this study, we evaluated the rate of change of MGMT methylation status between diagnosis and first recurrence in patients who received tumor resection for recurrent GBM.
Methods.
We included patients who received temozolomide concurrent with and adjuvant to radiotherapy after diagnosis of GBM and had a second surgery performed at least 3 months after radiotherapy completion. Other eligibility criteria were age ≥18 years and Eastern Cooperative Oncology Group performance status 0–2. We evaluated the MGMT methylation status by methylation‐specific polymerase chain reaction.
Results.
From our institutional data warehouse, 295 patients with recurrent GBM who underwent second surgery were evaluated. MGMT methylation status at both first and second surgery was available for 108 patients. MGMT was methylated in both surgeries in 38 patients (35.2%), while it was unmethylated in 43 patients (39.8%). We found a significant concordance between the first and the second MGMT methylation assessments (K = 0.500, p < .001), MGMT methylation being stable in 75% of the cases.
Conclusion.
MGMT methylation presents relative stability during the clinical course of GBM.
Implications for Practice.
MGMT methylation is a prognostic factor in newly diagnosed glioblastoma. In this study, we evaluated the rate of change of MGMT methylation during the clinical course of the disease, and we found a significant concordance between the first and the second MGMT methylation assessments, with MGMT methylation being stable in 75% of the cases. Thus, re‐testing this biomarker at recurrence does not provide further information for clinicians. MGMT methylation at first surgery, extent of resection at second surgery, and time between first and second surgery are significantly correlated with overall survival. Age and extent of resection are correlated with post‐progression survival.
Introduction
Current standard treatment for glioblastoma (GBM) includes surgery followed by radiation therapy (RT) and chemotherapy with temozolomide (TMZ) [1]. Despite the improvement in overall survival (OS) achieved with combined TMZ with and adjuvant to radiotherapy (RT/TMZ), most of the patients experienced disease progression and median survival did not exceed 12–14 months, with a 5‐year survival rate of 10% [1], [2].
Methylation of the O‐6‐methylguanine‐DNA methyltransferase (MGMT) gene promoter has emerged as a strong prognostic factor for newly diagnosed GBM [3], [4]. MGMT encodes for a DNA repair enzyme that provides resistance to alkylating chemotherapies such as TMZ. Because MGMT transcription can be silenced by promoter methylation in tumor cells [4], it is widely assumed that MGMT promoter methylation in patient tumors causes decreased MGMT protein expression, thereby abrogating the DNA repair activity necessary for TMZ resistance.
Few studies have evaluated if MGMT methylation status of GBM might change during the course of care, and some have found contrasting results and variable rates of change (5%–40%) [5], [6], [7], [8], even if different techniques for assessment are used (methylation‐specific polymerase chain reaction [MSP], immunohistochemistry). Moreover, it remains unclear if MGMT methylation retains its role after disease progression [9]. We performed an analysis on our institutional data warehouse evaluating all consecutive GBM patients who underwent second surgery for recurrence after standard treatment with RT/TMZ [1] in order to investigate the rate of change of MGMT methylation status at the time of disease progression and the impact of MGMT methylation status on the clinical outcome in the recurrent setting.
Subjects, Materials, and Methods
Patients
We analyzed all consecutive GBM patients from our institutional data warehouse, which was built in 2006 and captured information about clinical characteristics, histology, and molecular biology and survival. Inclusion criteria were age ≥18 years Eastern Cooperative Oncology Group performance status 0–2, and recurrence after at least 3 months from combined RT/TMZ; patients must also have undergone a second surgical procedure for recurrent disease.
All patients underwent a postoperative computed tomography (CT) scan within 48 hours of surgery to determine the extent of tumor removal. Extent of resection for each patient was classified as complete (>95% resection by volume) or partial (≤95% resection by volume). Patients who underwent biopsy (at first or second surgery) were also included. A review of patient charts was conducted to obtain demographic information, including age, sex, and description of surgical procedure. Histological evaluations were made on formalin‐fixed, paraffin embedded tissues. Tumor tissue was classified and graded as GBM according to WHO 2007 guidelines. The MGMT methylation status was evaluated with the MSP [10].
We aimed to reduce the risk that treatment‐related changes may hamper the results of the analysis of MGMT methylation on second surgical samples. After microscopic evaluation, the most representative block was selected between those used for diagnosis, and six 10‐µm‐thick sections were cut, followed by one Hematoxylin and Eosin control slide. The tumor area was marked on the control slide by a pathologist, and material was manually dissected under microscopic guidance from the corresponding 10‐µm sections using a sterile blade. The total amount of neoplastic cells and the proportion of neoplastic cells versus “contaminant” non‐neoplastic cells (i.e., endothelial, stromal, and inflammatory cells) was estimated by the pathologist in the area marked on the control slide. Only samples with at least 100 neoplastic cells and a proportion of neoplastic cells versus “contaminant” non‐neoplastic cells greater than 5%, in the marked area, were then processed for the molecular analysis. The study was approved by the institutional review board of the Azienda USL of Bologna, Italy.
Objectives.
The aim of this study was to assess the rate of change of MGMT methylation status during the clinical course of GBM and the potential implications of these changes.
Statistical Analysis.
Data are reported as means, ranges, and frequencies. Survival data (median survival times with 95% confidence interval [CI]) were computed by the Kaplan‐Meier procedure and were analyzed by the means of the log‐rank test and the Cox proportional hazards model. The significance level required to keep a variable in the multivariate forward stepwise model was 10%. Analyses were not corrected for multiple testing. The hazard ratios were computed together with their 95% CIs. Differences between MGMT methylation status obtained at first and second surgery were evaluated by the means of the McNemar test, and the concordance was evaluated with Cohen's Kappa Coefficient. Fisher's exact test was applied in order to evaluate if the rate of MGMT methylation status changes was similar among the two groups of methylated and unmethylated patients assessed at the first surgery. MGMT methylation status, extent of second surgery, chemotherapy after re‐surgery, type of chemotherapy after re‐surgery, time between first and second surgery, age, and performance status were considered in univariate and multivariate analysis. The time between first and second surgery was evaluated as a dichotomous variable with a 6‐month cut‐off and a 12‐month cut‐off; we reported and considered only the 12‐month cut‐off because it was the variable selected by the forward stepwise procedure and it was the most correlated with the outcomes. The SPSS version 13.0 for Windows (SPSS Inc., Chicago, IL, USA) was used as a statistical package. Two‐tailed p values less than .05 were considered significant.
Results
Patients' Characteristics
Two hundred ninety‐five consecutive patients at first recurrence for GBM who underwent second surgery were evaluated (Fig. 1). Histology at the time of second surgery confirmed GBM in all the cases, with enough viable tumor tissue for accurate testing.
Figure 1.

CONSORT flow chart.
Abbreviations: GBM, glioblastoma; MGMT, 0‐6‐Methylguanine DNA‐methyltransferase; MSP, methylation‐specific polymerase chain reaction.
MGMT Methylation Changes
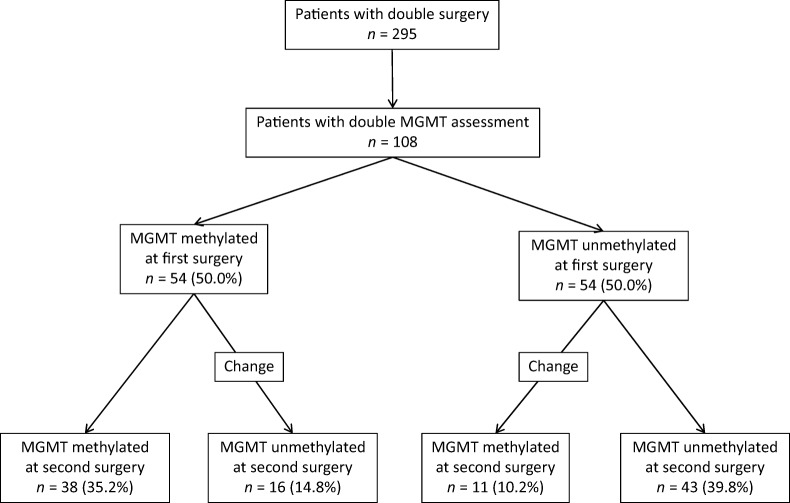
The MGMT methylation status was determined for 178 patients (60.4%) at the time of first surgery, with 79 methylated (44.4%) and 99 unmethylated (55.6%) tumors (Fig. 1). At the time of second surgery, MGMT methylation was evaluated for 137 patients (46.4%); it was methylated in 64 patients (46.7%) and unmethylated in 73 patients (53.3%). MGMT methylation status obtained both at first and second surgery was available for 108 patients (37%, Fig. 1). The patients' characteristics are summarized in Table 1. At the evaluation of second surgery samples, MGMT methylation status was changed in 16 of the 54 methylated patients (29.6%) and in 11 of the 54 unmethylated patients (20.4%); the changes were equally balanced in methylated and unmethylated patients at first surgery (p = .374, Fig. 2).
Table 1. Patient characteristics.

Figure 2.
Changes in MGMT methylation status between first and second surgery.
We found no differences between the first and the second assessment (p = .441), confirmed by the significant and positive concordance between the first and the second MGMT methylation assessments (K = 0.500, p < .001). MGMT methylation was stable in 75.0% of cases. To reduce the possibility that discrepancies are due to suboptimal tissue, we also analyzed concordance excluding 16 patients whose biopsy at first or second surgery may have yielded suboptimal samples. However, the rate of MGMT methylation stability was superimposable, 72.8%, and concordance between the first and the second MGMT methylation assessments was confirmed (K = 0.459, p < .001). Due to the small sample number of patients with discordance in MGMT methylation status, we were not able to find any peculiar characteristic of this population (age, original or relapse side).
OS.
Among the 108 patients who have MGMT methylation status obtained both at first and second surgery, median survival from first surgery was 24.4 months (95% CI: 21.3–27.5), being 35.2 months (95% CI: 18.9–51.5) in patients with MGMT methylated at both surgeries, 27.3 months (95% CI: 21.4–33.2) in patients with MGMT methylated at first surgery and unmethylated at the second surgery, 23.3 months (95% CI 19.9–26.7) in patients with MGMT unmethylated at first surgery and methylated at the second surgery, and 20.1 months (95% CI 16.6–23.3) in patients with MGMT unmethylated at both surgeries.
For all 295 patients, in univariate analysis, MGMT methylation at first (p < .001) and second surgery (p = .008), extent of resection at second surgery (p < .001), and time between first and second surgery (p < .001) were correlated with OS. Median OS was 32.1 months (95% CI: 27.1–37.1) and 21.1 months (95% CI: 19.1–23.1) in patients with MGMT methylated and MGMT unmethylated, respectively. Performance status was not significant (p = .761). In multivariate analysis, MGMT methylation at first surgery (p < .001), extent of resection at second surgery (p < .001), and time between first and second surgery (p < .001) were significantly correlated with survival (Table 2).
Table 2. Multivariate analysis (only significant variables were included in this table).

Abbreviations: —, not assessable; CI, confidence interval; HR, hazard ratio.
Survival from Time of Second Surgery
For all 295 patients, median survival after second surgery was 10.3 months (95% CI: 9.1–11.4). In univariate analysis, median survival after second surgery was not correlated with MGMT methylation obtained at second surgery (10.3 and 9.5 months in patients with MGMT methylated and MGMT unmethylated, respectively, p = .205), although it was correlated with MGMT methylation obtained at first surgery (11.9 and 9.3 months in patients with MGMT methylated and MGMT unmethylated, respectively, p = .016). In multivariate analysis, extent of resection at second surgery (p < .001) and age (p = .001) were significantly correlated with survival (Table 2).
Discussion
Change in biomarker expression in tumors is a phenomenon widely described in oncology. In breast cancer, a lack of concordance in receptor status between primary and recurrent tumors has been described in up to 40% of the cases [11]. However, similar solid data are not available for brain tumors, but some evidences come from a trial with anti‐EGFRvIII immunotherapy showing the role of treatment in changing the pattern of biomarker expression [12].
MGMT methylation has a prognostic role in GBM patients [3]. What remains unclear is the stability of this epigenetic alteration during the clinical course of the disease and thus if re‐testing MGMT methylation could provide useful information.
In our study, we included a large cohort of consecutive GBM patients who underwent a second surgical procedure for recurrent disease in order to analyze the role of MGMT methylation assessment of at the time of recurrence. Because postsurgical treatment may have an impact on genetic and epigenetic alterations, we included only GBM patients who received the same treatment, TMZ concurrent with and adjuvant to RT, and we avoided pseudoprogressions because second surgery was performed at least 3 months after RT completion. Moreover, we evaluated the stability of MGMT methylation status according to the extent of second surgery (complete or partial versus biopsy). Our study presents some limitations, particularly the retrospective nature of the data, as well as the use of postoperative CT with and without contrast enhancement instead of postoperative magnetic resonance imaging to assess the extent of surgery.
We showed that MGMT methylation status remains stable during the clinical course of GBM in the majority of patients, like in other findings from smaller cohorts [6], [7], and thus re‐testing this biomarker at recurrence does not provide further information. Moreover, in our study, OS from diagnosis is correlated with MGMT methylation status obtained at first surgery, but not at recurrence, confirming the role of MGMT methylation at diagnosis. Univariate analysis suggested that MGMT methylation at diagnosis could also predict survival after disease progression, as suggested by other groups [9]. However, this finding was not confirmed by multivariate analysis. Potential explanations could be that MGMT methylation affects OS by increasing progression‐free survival but not post‐progression survival, or that our sample size was not sufficient to show a role in multivariate analysis. However, 25% of patients showed discordant MGMT methylation status at diagnosis and at recurrence.
Different mechanisms can explain change in biomarker expression during the clinical course of neoplasms, such as pre‐analytical and analytical errors, intratumoral heterogeneity, and selective pressure of cytotoxic treatments. We excluded that the availability of tissue could be the reason of these discrepancies, because when only patients with greater tissue available were considered (partial or complete resections at both surgeries), the rate of discordant cases was similar (27.2%).
Other potential explanations of this phenomenon could be tumor heterogeneity or technical issues in MSP (i.e., the time from resection to fixation and the process of fixation itself). Tumor heterogeneity for MGMT methylation has been investigated by different groups [13], [14], [15], [16], [17], with contrasting results. However, larger studies suggested that MGMT methylation was homogeneous in tumors [15], at least in frozen samples [13]. Therefore, a technical issue could contribute to discordant results in some cases.
Moreover, it should be considered that MSP could suffer from two technical issues: (a) the number of investigated CpG islands is limited: even if the primers and the beacon probes used for MSP are designed for evaluating the methylation status of the main clinically relevant CpG islands [3], [18], not all the CpG islands in the MGMT promoter region are investigated; for this reason, we are not aware of the methylation status of the entire MGMT promoter, and (b) the amount/quality of input DNA: the MSP is a real‐time technique, and the results are evaluated during a “log‐linear phase”; if on one hand this approach allows a semiquantitative evaluation of the methylation level, then on the other hand, the results could be influenced by low quantity or low quality of input DNA (e.g., due to over‐fixation). Because promoter methylation‐mediated gene silencing depends strongly on the location of the methylated CpGs, further improvement in MGMT methylation techniques (i.e., HumanMethylation450 ‐ HM‐450K BeadChip) [19] could improve results and concordance between samples.
Conclusion
Our study suggests that MGMT methylation is stable over time in the majority of the patients. Moreover, the assessment of this biomarker at recurrence barely seems informative for GBM patients.
Acknowledgments
We thank the patients and their families. This project was supported by Fondazione Giovanni Celeghin Onlus (www.fondazioneceleghin.it) through its research funding. We are indebted to Dr. Carlo Descovich and the Azienda USL of Bologna for their continuous support for clinical research.
Footnotes
For Further Reading: Alba A. Brandes, Marco Bartolotti, Alicia Tosoni et al. Practical Management of Bevacizumab‐Related Toxicities in Glioblastoma. The Oncologist 2015;20:166‐175.
Implications for Practice: Given the widespread use of bevacizumab in clinical practice, it is important to raise clinicians' awareness of the potential risks of this treatment. Our aim was to provide an overview of the most common side effects of bevacizumab and to suggest a practical approach for their management.
Author Contributions
Conception/Design: Alba A. Brandes
Provision of study material or patients: Alba A. Brandes, Enrico Franceschi, Giovanni Tallini, Dario De Biase, Claudio Ghimenton, Daniela Danieli, Elena Zunarelli, Giovanni Lanza, Enrico Maria Silini, Carmelo Sturiale, Lorenzo Volpin, Franco Servadei, Andrea Talacchi, Antonio Fioravanti, Maria Pia Foschini, Annalisa Pession
Collection and/or assembly of data: Alba A. Brandes, Enrico Franceschi, Alexandro Paccapelo, Stefania Bartolini
Data analysis and interpretation: Alba A. Brandes, Alexandro Paccapelo, Mario Ermani
Manuscript writing: Alba A. Brandes
Final approval of manuscript: Alba A. Brandes, Enrico Franceschi, Alexandro Paccapelo, Giovanni Tallini, Dario De Biase, Claudio Ghimenton, Daniela Danieli, Elena Zunarelli, Giovanni Lanza, Enrico Maria Silini, Carmelo Sturiale, Lorenzo Volpin, Franco Servadei, Andrea Talacchi, Antonio Fioravanti, Maria Pia Foschini, Stefania Bartolini, Annalisa Pession, Mario Ermani
Disclosures
Maria Pia Foschini: Roche, Devicor Mammotome (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Stupp R, Mason WP, van den Bent MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–996. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Mason WP et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5‐year analysis of the EORTC‐NCIC trial. Lancet Oncol 2009;10:459–466. [DOI] [PubMed] [Google Scholar]
- 3. Hegi ME, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997–1003. [DOI] [PubMed] [Google Scholar]
- 4. Esteller M, Garcia‐Foncillas J, Andion E et al. Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350–1354. [DOI] [PubMed] [Google Scholar]
- 5. Brandes AA, Franceschi E, Tosoni A et al. O(6)‐methylguanine DNA‐methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: Clinical implications. Neuro Oncol 2010;12:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Felsberg J, Thon N, Eigenbrod S et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer 2011;129:659–670. [DOI] [PubMed] [Google Scholar]
- 7. Metellus P, Coulibaly B, Nanni I et al. Prognostic impact of O6‐methylguanine‐DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: A prospective patient cohort. Cancer 2009;115:4783–4794. [DOI] [PubMed] [Google Scholar]
- 8. Park CK, Kim JE, Kim JY et al. The changes in MGMT promoter methylation status in initial and recurrent glioblastomas. Transl Oncol 2012;5:393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weller M, Tabatabai G, Kästner B et al. MGMT promoter methylation is a strong prognostic biomarker for benefit from dose‐intensified temozolomide rechallenge in progressive glioblastoma: The DIRECTOR trial. Clin Cancer Res 2015;21:2057–2064. [DOI] [PubMed] [Google Scholar]
- 10. Herman JG, Graff JR, Myöhänen S et al. Methylation‐specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996;93:9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dieci MV, Barbieri E, Piacentini F et al. Discordance in receptor status between primary and recurrent breast cancer has a prognostic impact: A single‐institution analysis. Ann Oncol 2013;24:101–108. [DOI] [PubMed] [Google Scholar]
- 12. Sampson JH, Heimberger AB, Archer GE et al. Immunologic escape after prolonged progression‐free survival with epidermal growth factor receptor variant iii peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol 2010;28:4722–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamilton MG, Roldán G, Magliocco A et al. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol 2011;102:255–260. [DOI] [PubMed] [Google Scholar]
- 14. Grasbon‐Frodl EM, Kreth FW, Ruiter M et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer 2007;121:2458–2464. [DOI] [PubMed] [Google Scholar]
- 15. Cao VT, Jung TY, Jung S et al. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery 2009;65:866–875; discussion 875 [DOI] [PubMed] [Google Scholar]
- 16. Parkinson JF, Wheeler HR, Clarkson A et al. Variation of O(6)‐methylguanine‐DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J Neurooncol 2008;87:71–78. [DOI] [PubMed] [Google Scholar]
- 17. Dunn J, Baborie A, Alam F et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer 2009;101:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morandi L, Franceschi E, de Biase D et al. Promoter methylation analysis of O6‐methylguanine‐DNA methyltransferase in glioblastoma: Detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer 2010;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bady P, Sciuscio D, Diserens AC et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP‐status. Acta Neuropathol 2012;124:547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]