Abstract
Autophagy is an evolutionarily conserved cellular process that primarily participates in lysosome-mediated protein degradation. Although autophagy is a cytoplasmic event, how epigenetic pathways are involved in the regulation of autophagy remains incompletely understood. Here, we found that H2B monoubiquitination (H2Bub1) is down-regulated in cells under starvation conditions and that the decrease in H2Bub1 results in the activation of autophagy. We also identified that the deubiquitinase USP44 is responsible for the starvation-induced decrease in H2Bub1. Furthermore, the changes in H2Bub1 affect the transcription of genes involved in the regulation of autophagy. Therefore, this study reveals a novel epigenetic pathway for the regulation of autophagy through H2Bub1.
INTRODUCTION
Autophagy is an evolutionarily conserved catabolic process that is involved in the regulation of lysosome-mediated degradation of abnormal proteins or other cellular components (1,2). Autophagy is believed to be essential for cell survival, especially when cells are exposed to different stressors, such as nutrient deprivation. The activation of autophagy under starvation allows cells to survive by providing essential crude components for cell construction via the degradation of intracellular substrates (3–5). In addition, autophagy has been shown to be critical for the maintenance of cellular homeostasis because of its role in the clearance of abnormal proteins or factors that are no longer needed (1). Furthermore, increasing evidence suggests that the dysregulation of autophagy is tightly related to many types of diseases, such as tumorigenesis, neurodegenerative disorders and pathogenic infections (6–11).
The activation of autophagy involves several membrane-related components and their rearrangements, such as autophagosome formation and elongation, autophagosome-lysosome fusion and mature autolysosome formation (5,12). Following the stepwise activation processes, autophagy eventually results in the degradation of its substrates into useful biomolecules, allowing cells to construct essential cellular organelles or coordinate responses to different cellular stressors (5).
Autophagy is primarily recognized as a cytoplasmic event, and most of its regulators are cytoplasmically localized (1,3–5). The cytoplasmic machinery responsible for the regulation of autophagy has been widely studied. However, two recent studies indicated that both the hMof-H4K16ac and G9a-H3K9me2 axes are involved in autophagy-related cell fate determination and autophagy activation (13,14), providing direct evidence that epigenetic regulators may also play a critical role in the regulation of autophagy. The levels of H4K16ac are decreased during autophagy activation, which results from autophagy-mediated Mof degradation (the acetyltransferase for H4K16ac). H4K16ac regulates the outcome of autophagy predominantly by controlling the expression of a series of autophagy-related genes (14). Additionally, G9a, a histone H3K9 methyltransferase, regulates the expression of several autophagosome formation-related genes by remodeling the chromatin landscape. Loss of G9a activity results in elevated expression and lipidation of LC3B, suggesting that enhanced autophagosome formation occurred (13). Together, these studies directly indicate that epigenetic-regulated gene expression events likely play significant roles in the control of autophagy activity.
Histone H2B monoubiquitination (H2Bub1) is an important histone modification in gene transcriptional regulation and higher-order chromatin organization (15). H2Bub1 is mainly catalyzed by the RAD6–RNF20 ubiquitination machinery at lysine 120 of H2B in mammals (16–19), although other E3 ligases, such as RNF8, BAF250B, MDM2 and BRCA1–BARD1, have also been implicated (20–22). However, aside from the RAD6–RNF20 complex, information regarding other ubiquitin ligases is limited or has been challenged (20–22). For instance, the role of RNF8 in controlling H2Bub1 has been challenged by a recent report (20,23), and MDM2-mediatedH2B monoubiquitination only occurs in free H2B rather than in native nucleosome conditions (20,24). Moreover, the BRCA1–BARD1 complex has been shown to monoubiquitinate all nucleosome core histones, including H2A/H2Ax, H2B, H3 and H4 (22,25,26). However, a recent study has revealed that H2B is only modestly ubiquitinated by the BRCA1–BARD1 complex compared with H2A (22,26). Therefore, the RAD6–RNF20 ubiquitination complex is likely the only well-recognized set of ubiquitination enzymes for H2Bub1.
H2Bub1 is typically associated with both the promoter and coding regions of highly expressed genes (17,27); several studies demonstrated that H2Bub1 is a modulator of subsequent histone H3 methylations, such as H3K4 methylation and H3K79 methylation (15,17,28–30). H3K4me3 is essential for transcriptional gene activation (31), while the roles of H3K79me3 are still controversial (32). In addition, recent studies have further indicated that the loss of H2Bub1 prevents embryonic stem cell differentiation (33–35).
In this work, we show that histone H2Bub1 functions as a critical switch between autophagy and epigenetic pathways. Our results indicated that the loss of histone H2Bub1 results in autophagy and that the levels of H2Bub1 are decreased significantly during starvation. Furthermore, the starvation-induced H2Bub1 decrease and autophagy activation are shown to be regulated by the deubiquitinase USP44, which is transcriptionally targeted by the de novo DNA methyltransferases DNMT3a and DNMT3b. The depletion of H2Bub1 via the knockdown of RNF20 and mutations in the H2Bub1 site alters the transcription of genes involved in autophagy. In summary, our work reveals that appropriate H2Bub1 levels are essential for controlling autophagy in mammals.
MATERIALS AND METHODS
Cell culture and transfection
The HEK293T human embryonic kidney cell line and HeLa human cervical carcinoma cell line were cultured at 37°C in DMEM (Gibco, #11960-044) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Gibco, #15070-063) in a 5% CO2 incubator. The transfection of constructs into the cells was performed using Lipofectamine 2000 (Invitrogen, #11668-019) according to the manufacturer's standard protocol.
Mouse ESCs were cultured under standard conditions according to previous reports (36,37).
RNAi knockdown in cultured human cell lines
The siRNAs against ATG5, ATGA7, ATG12, ATG3, ATG13 and USP44 were all designed and synthesized by the GenePharm Company (Shanghai, China). The transfection of the siRNAs into the cultured HEK293T or HeLa cells was achieved using Lipofectamine 2000 (Invitrogen, #11668-019) according to the manufacturer's protocol.
Co-immunoprecipitation (Co-IP)
Cells were transfected with Flag-tagged ATG5 or Flag-tagged eIF4A using Lipofectamine 2000 (Invitrogen, #11668-019). After 48 h, the cells were harvested, washed with ice-cold PBS, resuspended in ATM lysis buffer (containing 100 mM Tris–Cl, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween-20 and 0.2 mM PMSF) and sonicated on ice 10 times (3 s each), with 20% efficiency. The cell lysates were incubated with normal mouse IgG (Santa Cruz Biotechnology, #sc-2025, as a negative control) or anti-Flag (Abmart) antibody at 4°C overnight. Protein A/G agarose beads (Santa Cruz Biotechnology, #sc-2003) were subsequently added, and the solution was incubated for an additional 3 h, followed by centrifugation to harvest the agarose beads after they had been washed five times with lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved via SDS-PAGE. Western blot analyses were performed using the relevant antibodies.
Antibodies and western blot analysis
The antibody against RNF20 was purchased from Novus (#NB100-2242). The anti-H2Bub1 antibody was purchased from Medimabs (#MM-0029). The anti-ubiquitin antibody was purchased from R&D (#MAB701). The antibodies against RAD6 (#4944), H2B (#2934), LC3B (#2775), ATG5 (#2630), ATG7 (#2631), p62 (#5114), H3K4m3 (#9727), H3K79me3 (#4260), H3K9me1 (#7538), H3K9me2 (#9753) and H4K16ac (#13534) were purchased from Cell Signaling Technology. The translation initiation complex antibody kit was purchased from Cell Signaling Technology (#4763). The antibodies against H3K9me3 (#ab8898), hMof (#ab83502), DNMT1 (#ab13537), DNMT3a(#ab2850) and DNMT3b (#ab2851) were purchased from Abcam. All of the HRP-conjugated secondary antibodies were purchased from Zhongshan Golden Bridge. The antibodies against USP7 (#GTX125894), USP12 (#GTX115609), USP22 (#GTX120048), USP46 (#GTX117994), and USP49 (#GTX119646) were purchased from GeneTex. The anti-USP44 antibody was purchased from Santa Cruz (#sc-377203).
The cells were lysed in ATM lysis buffer (containing 100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween-20 and 0.2 mM PMSF). The protein concentration in the supernatant was measured using a BCA Assay Kit (Novagen, #71285-3). Then, the samples were loaded into a 10% or 15% gel to resolve the proteins. Different amounts of total protein were loaded in each experiment to facilitate the detection of different target proteins. After electrophoresis, the proteins were transferred to PVDF membranes (Amersham, #10600021) and hybridized with primary antibodies at a dilution of 1:2000. The HRP-labeled secondary antibodies (Zhongshan Golden Bridge) were applied at a dilution of 1:4000. An ECL detection system (Calbiochem, #345818) was used to detect the signals on the membranes.
All the western blot analyses in this work were repeated more than three times, and the results were consistent.
RT–PCR assay
The cells were lysed to isolate the total RNA using TRIzol reagent (Invitrogen, #15596-026) according to the manufacturer's instructions. Reverse transcription was performed using a reverse transcription kit (Takara, #2641A). Briefly, total RNA (5 μg) was reverse transcribed to synthesize cDNA in a volume of 20 μl using M-MLV reverse transcriptase. In each 25-μl PCR mixture, 1 μl of cDNA was used for real-time PCR or semi-quantitative PCR analyses. For semi-quantitative PCR, the PCR products were loaded onto a 2% agarose gel, stained with ethidium bromide and imaged. Real-time PCR was performed with SYBR Advantage qPCR Premix (Clontech) on an iQ5 Real-Time PCR System (Bio-Rad). Fold differences in gene expression levels were calculated and normalized against the internal control GAPDH. All the sequences of primers used in this work are listed in Supplementary Table S1.
DNA methylation profiling analysis
Bisulfite modification was carried out using a Direct Bisulfite Modification Kit (Millipore, #17-10451) followed by PCR amplification with specific primers for a USP44 CpG island. The PCR products were then cloned into a T vector and transformed into Escherichia coli. Six independent clones were selected for sequencing.
In vivo ubiquitination assay
HEK293T cells were treated with or without starvation for 10 h along with incubation with MG132. The in vivo ubiquitination assay was performed under denaturing conditions. The cells were lysed with 100 μl of an SDS lysis buffer containing 1% SDS, and the lysate was then boiled for 15 min. The resulting lysate was centrifuged for 15 min at 12 000 rpm at 4°C. The supernatant was diluted to 0.1% SDS with 900 μl of ATM lysis buffer. The lysate was subsequently incubated with normal mouse IgG antibody (Santa Cruz Biotechnology, #sc-2025) or antibodies against DNMT3a and DNMT3b at 4°C overnight. Protein A/G agarose beads were then added to precipitate the bound proteins. The ubiquitination levels were detected by western blot analysis using anti-ubiquitin antibody (R&D, #MAB701).
Autophagy analysis
To induce autophagy, the cells were washed three times with PBS and incubated for different periods in Hanks’ balanced salt solution (HBSS, Invitrogen, #14025-076). To stimulate rapamycin-induced autophagy, the cells were treated with complete medium containing 2 μM rapamycin at 37°C for 10 h. The autophagy was assessed by detecting the LC3 cleavage. To inhibit starvation-mediated autophagy, the cells were treated with HBSS containing 10 mM 3-MA (Sigma, #M9281).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to published protocols from Upstate and our previous work (38,39). The ChIP products were then used for real-time PCR assays with specific primers. For ChIP-real-time PCR assays, the enrichment of the levels of H2Bub1 was calculated relative to the input DNA, presented as the value of 2−ΔCt × p, in which ΔCt is the difference of Ct values of histone modification IP and input DNA for a given primer pair, and p is the ratio of the volume of the input over IP used in the ChIP assays. The primers were designed to amplify 150–200 bp products according to the sequences of the coding region of each gene immediately after the TSS. The intergenic region was used as the negative control for the H2Bub1 ChIP assay according to a previous study (40).
RESULTS
The loss of H2Bub1 induces autophagy
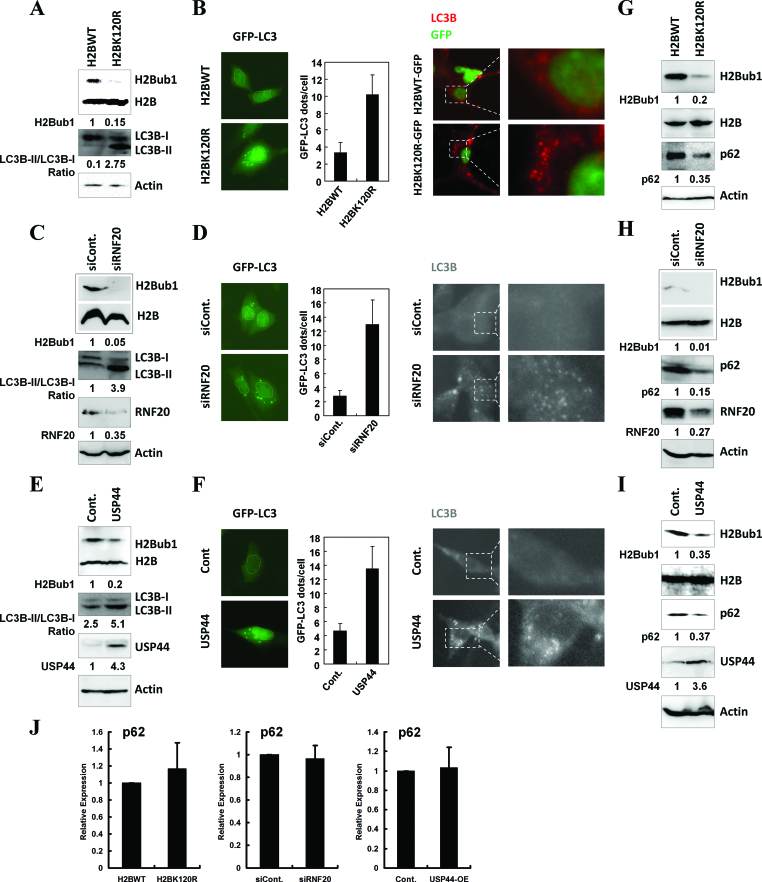
To explore the roles of epigenetic regulation in the control of autophagy, we performed a screen with various histone modification site-specific mutation constructs (data not shown). Briefly, HEK293T cells were transfected with the mutants or with the corresponding wild-type histones (used as a control), and then the cells were harvested and subjected to western blot and immunofluorescence analyses with anti-LC3B antibody to determine the effects of the histone mutants on the levels of autophagosomes. We found that H2Bub1 is likely a critical epigenetic regulator of autophagy. To determine the functions of H2Bub1 in the regulation of autophagy, we disrupted the endogenous levels of H2Bub1 by using cells with overexpression of the H2BK120R mutant, with knockdown of the E3 ligase RNF20, or with overexpression of the H2Bub1 deubiquitinase USP44 (34). With down-regulation of H2Bub1 (Figure 1A, C and E), the levels of LC3B-II in all three samples were consistently increased compared to the controls, suggesting that autophagosome formation is regulated by H2Bub1 (Figure 1A, C and E). Microscopy analyses detecting the subcellular distribution of LC3B at both the exogenous (GFP-LC3B transfection) and endogenous (immunostaining of LC3B with LC3B-specific antibodies) levels showed that the cytoplasmic foci of LC3B were increased significantly in the cells with reduced H2Bub1 (Figure 1B, D and F), further supporting the role of H2Bub1 in the control of autophagosome formation under basal conditions.
Figure 1.
The inhibition of H2Bub1 induces autophagy activation. (A) 293T cells were transfected with a control or H2BK120R mutant plasmid for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (B) Control plasmid- or H2BK120R mutant plasmid-transfected 293T cells were cotransfected with GFP-LC3 or stained with anti-LC3B antibody. The cells were then subjected to microscopy analysis. The experiments were repeated more than three times (n > 3), and 500 cells were analyzed each time. (C) 293T cells were transfected with a control siRNA or an RNF20-specific siRNA for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (D) Control siRNA- or RNF20-specific siRNA-transfected 293T cells were cotransfected with GFP-LC3 or stained with anti-LC3B antibody. The cells were then subjected to microscopy analysis. The experiments were repeated more than three times (n > 3), and 500 cells were analyzed each time. (E) 293T cells were transfected with a control or USP44 plasmid for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (F) Control plasmid- or USP44 plasmid-transfected 293T cells were cotransfected with GFP-LC3 or stained with anti-LC3B antibody. The cells were then subjected to microscopy analysis. The experiments were repeated more than three times (n > 3), and 500 cells were analyzed each time. (G) 293T cells were transfected with a control or H2BK120R mutant plasmid for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (H) 293T cells were transfected with a control siRNA or an RNF20-specific siRNA for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (I) 293T cells were transfected with a control or USP44 plasmid for 48 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (J) Inhibition of H2Bub1 does not affect the mRNA levels of p62. H2BK120R mutant plasmid-, RNF20-specific siRNA- and USP44 plasmid-transfected 293T cells and their related control cells were lysed, and total RNA was prepared and subjected to RT-PCR assays using the indicated primers (n = 3).
To determine whether autophagy activity was affected after the reduction in H2Bub1, we then used western blot analysis to examine the protein levels of p62, whose degradation is a marker for the eventual activation of autophagy (41–44). Based on the results in Figure 1G, H and I, we show that the down-regulation of H2Bub1 leads to a decrease in the protein levels of p62 (Figure 1G, H and I and Supplementary Figure S1). The reduction in p62 did not appear to be due to changes in the transcriptional regulation of p62, as our RT-PCR assays showed no changes in the p62 mRNA levels in all three groups of cells (Figure 1J), confirming that the loss of H2Bub1 activates autophagy in human cells under basal conditions.
Starvation-induced autophagy activation down-regulates H2Bub1 and is mediated by the deubiquitinase USP44
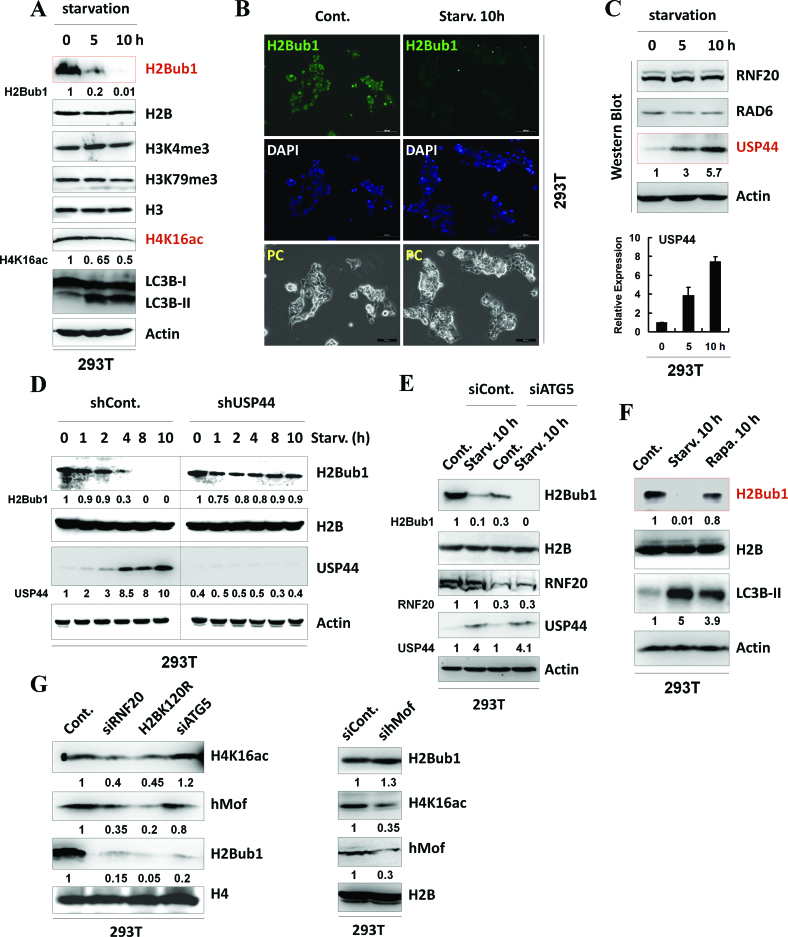
We next examined the dynamic changes in H2Bub1 levels in response to autophagy activation. Upon starvation, we found that the levels of H2Bub1 were significantly decreased, while the levels of H3K4me3 and H3K79me3 showed no obvious changes (Figure 2A and Supplementary Figure S2A), supporting the notion that the change in H2Bub1 is not always coupled with changes in H3K4me3 and H3K79me3 (33–35,45,46). H4K16ac was also shown to be decreased and involved in starvation-induced autophagy activation (14). However, the decrease in H2Bub1 in the cells under starvation seemed to be much more obvious than the changes in H4K16ac (Figure 2A and Supplementary Figure S2A). The decrease in H2Bub1 was also confirmed by immunofluorescence assay upon starvation (Figure 2B).
Figure 2.
H2Bub1 levels are decreased during starvation-induced autophagy activation mediated by its deubiquitinase USP44. (A) H2Bub1 is decreased in response to starvation as analyzed by western blot assay. 293T cells were starved by HBSS treatment for the indicated periods to activate autophagy. Cell extracts were then prepared and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (B) H2Bub1 is decreased in response to starvation as analyzed by immunofluorescence assay. Immunofluorescence assays were performed using control or starved 293T cells with anti-H2Bub1 antibody. DAPI staining was used to indicate the cell nucleus. (C) USP44 is increased in response to starvation. Control or HBSS-treated (starvation) 293T cells were lysed and subjected to western blot and RT-PCR analyses using specific antibodies or primers, as indicated. The experiments were repeated more than three times (n > 3). (D) USP44 regulates the starvation-induced down-regulation of H2Bub1. 293T cells were transfected with control or USP44-specific shRNAs for 48 h. The cells were then treated with or without HBSS (starvation) for the indicated periods. Cell extracts were prepared and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (E) Loss of H2Bub1 does not depend on autophagy activity. Control and ATG5 RNAi 293T cells were treated with or without starvation for 10 h. The cells were then lysed and subjected to western blot assays using the indicated antibodies. The experiments were repeated more than three times (n > 3). (F) H2Bub1 does not respond to rapamycin treatment. 293T cells were treated with starvation or 2 μM rapamycin for 10 h. The cells were then lysed and subjected to western blot assays using the indicated antibodies. The experiments were repeated more than three times (n > 3). (G) H2Bub1 likely functions upstream of H4K16ac in the regulation of autophagy. Cell extracts from 293T cells transfected with RNF20 siRNA, H2BK120R mutant plasmid, ATG5 siRNA, or hMof siRNA were prepared. Western blot analysis was then performed using the indicated antibodies. The experiments were repeated more than three times (n > 3).
To understand how H2Bub1 is regulated in response to starvation, we next examined the protein and mRNA levels of the major regulators of H2Bub1, including the ubiquitination-related enzymes RNF20 (16) and RAD6 (17) and the deubiquitinases USP7 (47), USP12 (48), USP22 (49,50), USP44 (21,34), USP46 (48) and USP49 (51). Interestingly, only the level of the deubiquitinase USP44 was significantly increased at both the protein and mRNA levels upon starvation, suggesting that USP44 is likely the cause of the starvation-induced decrease in H2Bub1 (Figure 2C, and Supplementary Figure S2B and C). This hypothesis is also supported by the effects of knockdown of USP44 expression, which inhibits the starvation-induced decrease in H2Bub1 levels (Figure 2D). Neither the formation of the RAD6–RNF20 ubiquitination complex nor the subcellular localization of these two proteins was affected during starvation, further indicating that USP44, but not RAD6–RNF20, is responsible for the starvation-induced down-regulation of H2Bub1 (Supplementary Figure S2D and E).
We next tested whether the reduction in H2Bub1 induced by starvation was simply due to cytological autophagy activation. We therefore depleted the expression of ATG5 and ATG7 using RNAi (Supplementary Figure S2F), which disrupts the activation of autophagy (14). As shown in Figure 2E and Supplementary Figure S2G, our results showed that the reduction in H2Bub1 could also be observed upon starvation in both ATG5 and ATG7 RNAi cells, indicating that the reduction in H2Bub1 is independent of the activation of autophagy (Figure 2E and Supplementary Figure S2G) and further suggesting that loss of H2Bub1 is likely an upstream regulator of autophagy.
In addition, intriguingly, we found that the loss of ATG5, but not ATG7, decreases the levels of H2Bub1 in both basal and starved conditions, suggesting that ATG5 is likely a novel regulator of H2Bub1 (Figure 2E and Supplementary Figure S2G). We also examined whether rapamycin, another inducer of autophagy, can affect the H2Bub1 levels. However, treatment with rapamycin, which did promote the activation of autophagy, failed to show an obvious effect on the levels of H2Bub1 as observed in starvation conditions in the western blotting analysis, supporting the idea that multiple mechanisms are probably involved in starvation- and rapamycin-induced autophagy activation (Figure 2F, Supplementary Figure S2H and Supplementary Figure S3).
Together, the above results suggest that H2Bub1 is down-regulated during starvation and that this decrease is regulated by the deubiquitinase USP44.
The crosstalk between H2Bub1 and H4K16ac in the regulation of autophagy
A recent study suggested that the presence of H4K16ac is involved in the regulation of autophagy outcomes (14). We therefore further tested the relationship between H2Bub1 and H4K16ac in the regulation of autophagy. Cells with a reduction in H2Bub1 via knockdown of RNF20 expression, overexpression of the H2BK120R mutant or knockdown of ATG5 expression were used (Figure 2G, left). Our results showed that the loss of H2Bub1 by RNF20 RNAi or H2BK120R overexpression resulted in the down-regulation of H4K16ac and the H4K16ac acetyltransferase hMof. The results suggest that the alteration in H2Bub1 modulates the subsequent changes in H4K16ac. However, the reduction in H2Bub1 via the depletion of ATG5 seemed to show no obvious effect on H4K16ac or hMof (Figure 2G, left). This result is expected, considering that the reported down-regulation of H4K16ac during the activation of autophagy is achieved by the autophagy-mediated protein degradation of hMof (14); therefore, the absence of ATG5 likely disrupts the activity of autophagy, and fails to result in hMof degradation or H4K16ac alterations. In contrast, our observations further indicate that the reduction in H4K16ac via knockdown of the expression of hMof has no obvious effect on the levels of H2Bub1 (Figure 2G, right). Together, these results at least partially suggest that H2Bub1 likely functions upstream of H4K16ac in the regulation of autophagy.
Starvation-induced USP44 up-regulation is mediated by DNMT3a and DNMT3b
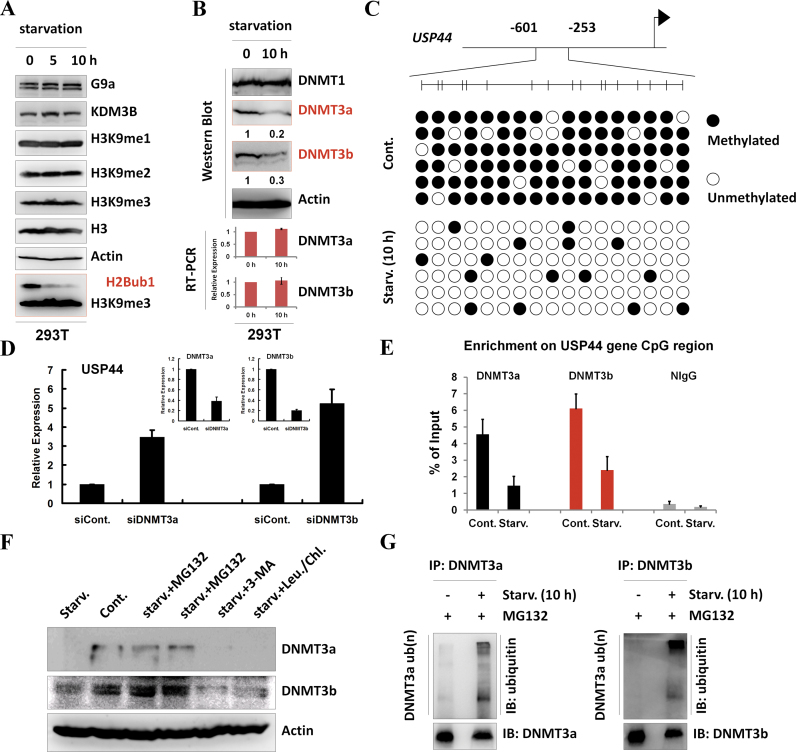
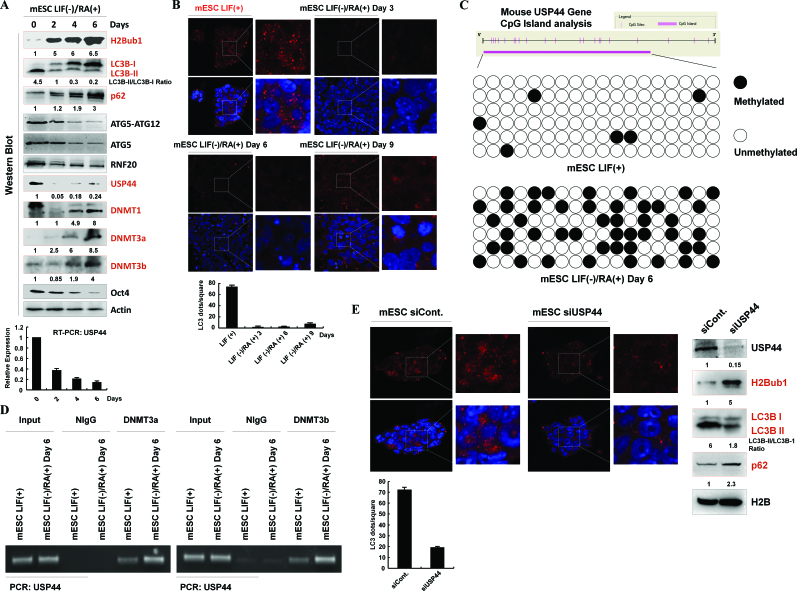
As the up-regulation of USP44 is critical for the starvation-induced reduction of H2Bub1 (Figure 2D), we next examined how USP44 is up-regulated during starvation. Our RT-PCR assay suggested that USP44 is regulated at the transcriptional level following starvation (Figure 2C and Supplementary Figure S2B). The transcriptional silencing regulators in the G9a-H3K9me2 axis participate in the regulation of autophagy (13) but do not seem to be involved in the transcriptional regulation of USP44, as there was no obvious change in H3K9 methylation, including H3K9me1/2/3 and their related methyltransferase G9a and demethylase KDM3B, upon starvation (Figure 3A). Notably, the protein levels of two de novo DNA methyltransferases, DNMT3a and DNMT3b, but not the maintenance methyltransferase DNMT1, were shown to be down-regulated after starvation (Figure 3B), indicating that the decrease in DNMT3a/3b was possibly the cause of USP44 up-regulation. Consistent with the dynamic relationship between USP44 and DNMT3a/DNMT3b, bisulfite sequencing analysis indeed showed a significant reduction in the DNA methylation levels of the USP44 CpG island after starvation (Figure 3C).
Figure 3.
Starvation-induced USP44 up-regulation is mediated by DNA methylation. (A) 293T cells were starved by treating with HBSS for the indicated periods. Cell extracts were then prepared and subjected to western blot analysis using the indicated antibodies. The experiments were repeated more than three times (n > 3). (B) 293T cells were treated with or without starvation for 10 h by incubating with HBSS. Protein extracts and total RNA were then prepared and subjected to western blot and RT-PCR analyses usingspecific antibodies and primers,as indicated. The experiments were repeated more than three times (n > 3). (C) Methylation of the USP44 gene is decreased in response to starvation. DNA methylation profiling analysis was employed in control and HBSS-starved 293T cells. The detailed procedure was performed according to the manufacturer's instructions (Millipore, Lot#: 17-10451). (D) 293T cells were transfected with a control or a DNMT3a- or DNMT3b-specific siRNA for 48 h. The total RNA was then isolated and subjected to RT-PCR analysis using the indicated primers. At least three biological replicates were analyzed (n > 3). (E) 293T cells were treated with or without starvation for 10 h. The cells were then lysed and subjected to chromatin immunoprecipitation (ChIP) assay using antibodies against DNMT3a and DNMT3b or normal control IgG (as a negative control), followed by real-time PCR with specific primers for the USP44 gene CpG region, as indicated. At least three biological replicates were analyzed (n > 3). (F) 293T cells were treated with starvation or starvation together with MG132 (25 μM), 3-MA (10 mM) or chloroquine (100 μM) and leupeptin (50 μM) for 10 h. The cells were then lysed and subjected to western blot analysis using the indicated antibodies. The experiments were repeated three times (n = 3). (G) In vivo ubiquitination assays were performed in control and HBSS-starved 293T cells together with MG132 incubation. The immunoprecipitation was performed under denaturing conditions using antibodies against DNMT3a and DNMT3b, followed by western blot analysis using the indicated antibodies. The experiments were repeated three times (n = 3).
To test whether DNMT3a and DNMT3b are indeed involved in the starvation-induced up-regulation of the USP44 gene, we performed DNMT3a and DNMT3b knockdown assays using RNAi. The results showed that the depletion of DNMT3a and DNMT3b expression significantly increased the transcriptional levels of USP44, confirming that DNMT3a and DNMT3b are transcriptional silencers of the USP44 gene (Figure 3D). To further identify whether DNMT3aand DNMT3b are directly involved in the transcriptional repression of USP44, we next performed chromatin immunoprecipitation (ChIP) assays. The results showed that both DNMT3a and DNMT3b bound to the promoter region of the USP44 gene and that the binding was significantly decreased after starvation (Figure 3E).
Our RT-PCR analysis suggested that the down-regulation of DNMT3a and DNMT3b was likely due to post-transcriptional regulation, as the mRNA levels of both genes were not affected upon starvation (Figure 3B, lower panels). To test whether the decrease in the DNMT3a and DNMT3b protein levels was due to protein degradation, proteasome, lysosome and autophagy inhibitors were used to treat cells under starvation (Figure 3F). We found that only the proteasome inhibitor MG132, but not the lysosome inhibitors leupeptin and chloroquine or the autophagy inhibitor 3-MA, rescued the down-regulation of the DNMT3a and DNMT3b protein levels induced by starvation (Figure 3F), supporting the notion that starvation-induced DNMT3a and DNMT3b down-regulation occurs via the ubiquitin-proteasome-mediated protein degradation pathway. Moreover, our in vivo ubiquitination analysis confirmed that starvation promotes the ubiquitination of both DNMT3a and DNMT3b (Figure 3G).
To further examine the correlation between USP44 and DNMT3a/b, we performed clinically based bioinformatic analysis using the Oncomine online database (www.oncomine.org). We found that the expression of DNMT3a/DNMT3b and USP44 showed an inverse correlation between cancer and normal tissues: DNMT3a and DNMT3b appear to be up-regulated in breast and lung cancers, while USP44 is down-regulated (Supplementary Figure S4). This result supports our biochemical analysis showing that DNMT3a and DNMT3b suppress the transcription of USP44 by regulating the DNA methylation of the gene (Figure 3B–E).
H2Bub1regulates the transcription of genes involved in autophagy
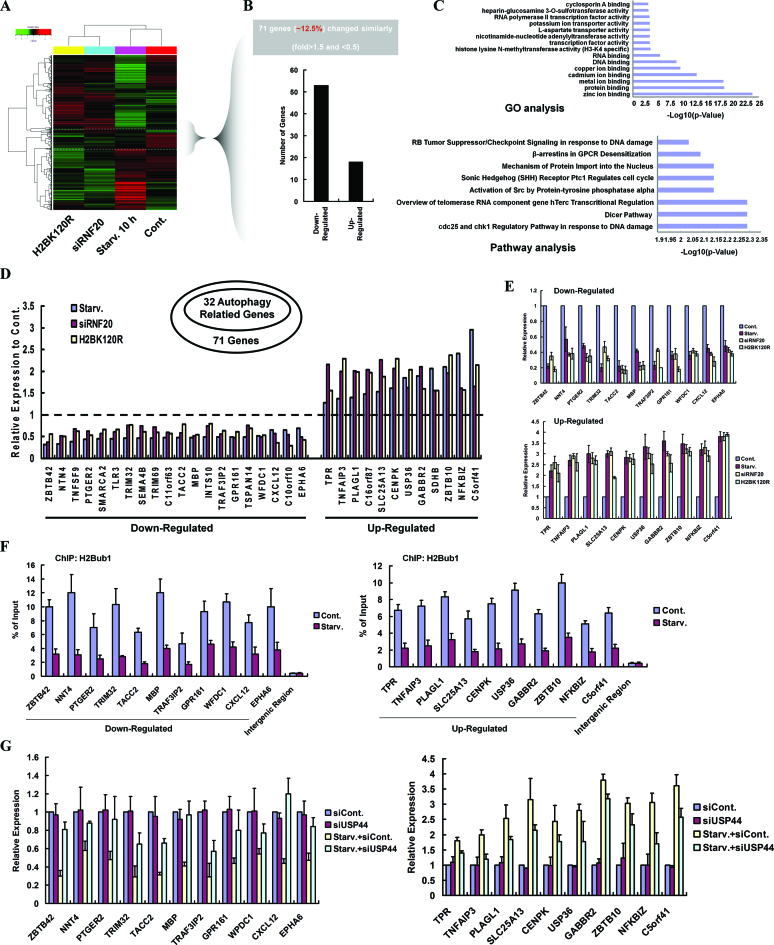
To further understand the impact of H2Bub1 in the regulation of autophagy, we next performed genome-wide expression assays to test whether changes in H2Bub1 have any impact on the transcription of autophagy-related genes. HEK293T cells transfected with RNF20-specific siRNA or H2BK120R mutant plasmid or treated with starvation for 10h (these three different treatments all resulted in a decrease in H2Bub1 levels and autophagy activation) were used, and total RNA was harvested from each group for further microarray analyses. Among the 71 genes (∼12.5% of the total affected genes) with a similar pattern in all three sample groups, we found that over two-thirds were down-regulated (Figure 4A and B). GO and Pathway annotations of these differentially expressed genes were also performed (Figure 4C). Interestingly, 32 of the 71 affected genes are associated with the regulation of autophagy (Figure 4D), which was determined by comparison with a reported resource that listed autophagy-related genes (52). RT-PCR validation of the randomly selected autophagy-related genes was also performed (Figure 4E), which confirmed the changes in the transcription of these genes. In addition, ChIP assay results demonstrated that H2Bub1 was enriched in the promoter regions of the selected genes in the control cells and that this enrichment was abolished upon starvation (Figure 4F), although a number of genes were up-regulated after starvation, possibly because the global levels of H2Bub1 are decreased significantly after starvation (Figure 2A and B, and Supplementary Figure S2A). Therefore, our ChIP assay results suggest that H2Bub1 likely plays a direct role in the control of the expression of autophagy-related genes. Additionally, as USP44 regulates the H2Bub1 levels especially under starvation conditions (Figure 2D), we examined the effects of USP44 on the expression of the selected autophagy-related genes. Consistent with our expectation, we found that knockdown of USP44 expression (Supplementary Figure S5) significantly abolished the starvation-induced expression changes of the selected genes (Figure 4G).
Figure 4.
H2Bub1 modulates the transcription of autophagy-related genes. (A) Total RNA from 293T cells transfected with a control siRNA, an RNF20-specific siRNA or an H2BK120R mutant plasmid, or cells treated with starvation for 10 h was prepared and then subjected to Affymetrix microarray analysis. (B) Differentially expressed genes with similar change patterns were selected and counted. (C) Genes with similar change patterns were selected and subjected to GO and Pathway annotation analyses. (D) Thirty two autophagy-related genes were selected from the genes with similar change patterns. (E) Total RNA was isolated from control, RNF20 RNAi-transfected and H2BK120R mutant plasmid-transfected 293T cells, and HBSS-starved 293T cells. RT-PCR analyses were performed using specific primers, as indicated. At least three biological replicates were analyzed (n > 3). (F) Chromatin immunoprecipitation (ChIP) assays were performed in control or HBSS-starved HEK293T cells using anti-H2Bub1 antibody, followed by a PCR analysis using the indicated primers. At least three biological replicates were analyzed (n > 3). (G) Total RNAs from control, USP44 RNAi, and HBSS-starved control 293T cells, and HBSS-starved USP44 RNAi 293T cells were isolated. RT-PCR analyses were then performed using specific primers, as indicated. At least three biological replicates were analyzed (n > 3).
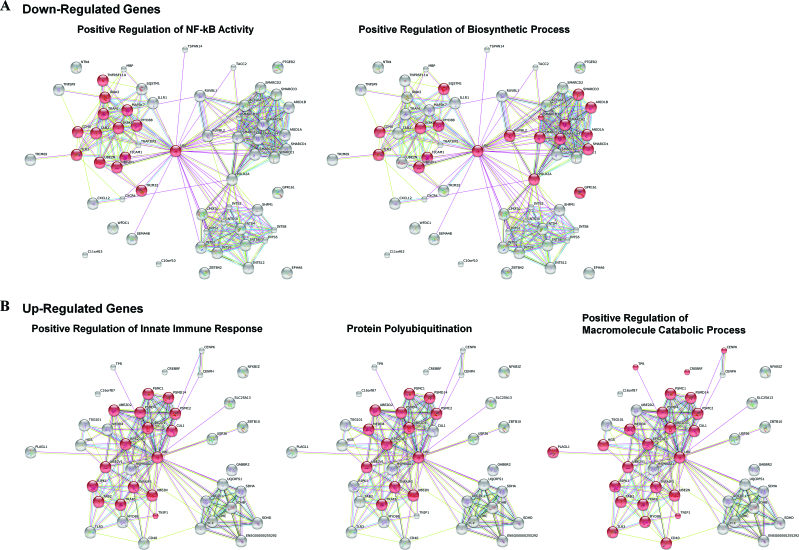
To more fully understand how the 32 differentially expressed genes affect autophagy, we further performed a gene network analysis by using the STRING online database (http://string-db.org/). Wefound that the 20 down-regulated genes predominantly function in the positive regulation of NF-κB activity and biosynthetic processes (Figure 5A, red balls), indicating that loss of H2Bub1 likely results in the inhibition of NF-κB activity and biosynthetic processes. NF-κB has been recognized as a negative regulator of autophagy in multiple conditions (53,54), and lower biosynthetic activity is a known characteristic of autophagy (55). Therefore, this result indicated a positive effect of the 20 down-regulated genes on autophagy. In addition, the 12 up-regulated genes are tightly involved in three areas of biological events, positive regulation of the innate immune response, protein polyubiquitination and the macromolecular catabolic process (Figure 5B, red balls), suggesting that loss of H2Bub1 likely positively regulates innate immunity, protein polyubiquitination, and macromolecular metabolism. All three biological events are also significantly related to autophagy. For instance, many innate immune signaling pathways activate autophagy in response to pathogen infection (10,11). Protein ubiquitination is essential for the recognition and degradation of the substrates by autophagy-mediated protein degradation processes (56).
Figure 5.
Network analysis of the 32 differentially expressed genes involved in autophagy. (A) Twenty down-regulated and (B) 12 up-regulated genes from Figure 4D were subjected to STRING network analysis (http://string-db.org/).
Taken together, our microarray study suggests that H2Bub1 regulates the activation of autophagy likely through the transcriptional controlof genes involved in autophagy.
USP44 is a key regulator of autophagy during mouse embryonic stem cell (mESC) differentiation
Because H2Bub1 was shown to be present at very low levels in mESCs and to increase significantly after the induction of differentiation (33–35), we next tested whether there is a correlation between the presence of H2Bub1 and autophagy in mESCs, as well as during mESC differentiation. We found that the autophagy activity of the mESCs is much higher than that of differentiated cells, as our western blot analyses indicated that the LC3B-II levels were dramatically decreased during ESC differentiation, combined with the accumulation of p62 protein levels (Figure 6A). Furthermore, our immunofluorescence assays using anti-LC3B antibody also confirmed that the autophagosome levels are significantly higher in stem cells than in differentiated cells (Figure 6B).
Figure 6.
H2Bub1 correlates with the autophagy activity in mESCs and the differentiation of mESCs. (A) H2Bub1 levels correlates with autophagy in mESCs and their differentiation. mESCs were cultured with RA (RA(+)) and without LIF (LIF(−)) for different periods and were lysed for western blot analysis using different antibodies, as indicated. The experiments were repeated more than three times (n > 3). (B) Autophagosomes in mESCs and differentiated mESCs. mESCs and mESCs cultured with RA (RA(+)) and without LIF (LIF(−)) for different periods were subjected to immunostaining analysis using anti-LC3B antibody. DAPI was used to detect the cell nucleus. (C) Methylation of the mouse USP44 gene is increased after mESC differentiation. DNA methylation profiling analysis was employed in control and differentiated mESCs. The detailed procedure was performed according to the manufacturer's instructions (Millipore, Lot#: 17-10451). (D) Control and differentiated mESCs were lysed and subjected to a chromatin immunoprecipitation (ChIP) assay using antibodies against DNMT3a and DNMT3b or normal control IgG (as a negative control), followed by PCR with specific primers for USP44, as indicated. The experiments were repeated more than three times (n > 3). (E) USP44 regulates autophagy in mESCs. Control siRNA- and USP44-specific siRNA-transfected mESCs were subjected to immunostaining analysis using an anti-LC3B antibody, or subjected to western blot assays with antibodies as indicated. DAPI was used to detect the cell nucleus. The experiments were repeated more than three times (n > 3).
USP44 was reported as the key regulator of H2Bub1 in mESCs and their differentiation (34); therefore, we next analyzed the dynamics of the H2Bub1-related regulators USP44, RNF20 and ATG5 during mESC differentiation. Consistent with previous studies, USP44 decreased significantly at both the protein and mRNA levels following mESC differentiation and showed an inverse correlation with the changes in H2Bub1 (Figure 6A). However, the changes in the RNF20 and ATG5 protein levels were not as obvious as those of USP44 during differentiation. We next examined whether the expression of USP44 is also regulated by DNA methylation during mESC differentiation, as previously indicated in human cell lines (Figure 3C–E). In parallel with the decrease in USP44 RNA and protein levels during mESC differentiation, the CpG island of the USP44 gene was hypermethylated upon differentiation (Figure 6C). Furthermore, consistent with the results from human cell lines (Figure 3E), the methylation of the USP44 gene promoter in mESCs is also likely regulated by DNMT3a and DNMT3b, as our chromatin immunoprecipitation assay indicated that differentiation promotes the binding of both DNMT3a and DNMT3b to the USP44 gene promoter (Figure 6D). Since both a previous report (34) and our current data suggested that USP44 likely regulates the H2Bub1 dynamics in response to differentiation and since H2Bub1 levels are correlated to autophagy activity in mESCs, we wondered whether USP44 also controls autophagy activity in mESCs. Indeed, depletion of USP44 expression by RNAi increased the levels of H2Bub1 and decreased the activity of autophagy significantly, as the levels of LC3B II were decreased and the p62 levels were increased after USP44 RNAi (Figure 6E, right). Immunofluorescence assay also supported our conclusion, as the LC3B puncta (autophagosomes) were decreased significantly after USP44 depletion (Figure 6E, left). Together, the above results suggest a conserved relationship between the down-regulation of H2Bub1 and the presence of autophagy in mESCs, indicating that H2Bub1 is a critical epigenetic regulator of autophagy at basal and physiological conditions.
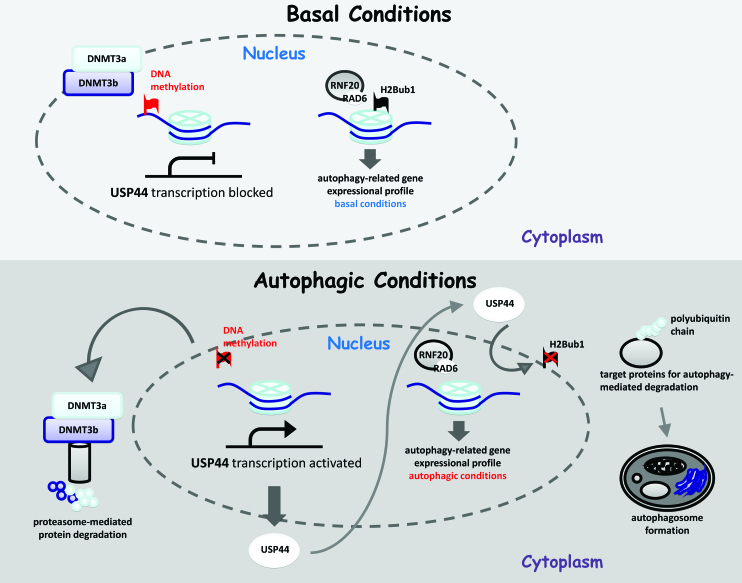
In summary, the results of this study indicate that H2Bub1 is a key epigenetic switch for the control of autophagy. We further revealed the upstream regulators of H2Bub1 during autophagy activation as well as their roles in the regulation of autophagy. Finally, a crosstalk model between H2Bub1 and the regulation of autophagy was proposed (Figure 7).
Figure 7.
A working model for H2Bub1 regulation and autophagy under different cellular conditions. Under basal conditions, DNMT3a and DNMT3b maintain USP44 at a very low level by regulating DNA methylation, and further control H2Bub1 at relatively high levels, resulting in a basal gene expressional profile with lower autophagy activity. However, when the cells are exposed to starvation, both DNMT3a and DNMT3b are degraded via the ubiquitin-proteasome pathway, resulting in the activation of USP44 expression. The elevated USP44 protein further deubiquitinates H2Bub1, leading to a decrease in the H2Bub1 levels. The alteration of the H2Bub1 levels results in global changes in the expression of genes, especially those genes involved in autophagy. These changes in gene expression eventually contribute to the activation of autophagy after starvation.
DISCUSSION
It has long been thought that the regulation of autophagy is a post-translational process; however, more recent studies have indicated that transcriptional networks (57,58) and histone modifications are also involved (59). The exact picture of how epigenetic pathways and transcriptional networks are coordinated to control autophagy under different cellular conditions is far from clear.
In this study, we demonstrate that H2Bub1 plays a critical role in the control of autophagy. H2Bub1 is decreased during starvation-induced autophagy activation (Figure 2A and B, and Supplementary Figure S2A), and the down-regulation of H2Bub1 determines autophagy activity (Figure 1). Moreover, ATG5 seems to be a novel regulator of H2Bub1, as we observed that depletion of ATG5 resulted in the decrease of H2Bub1 in both basal and starved conditions (Figure 2E and Supplementary Figure S2G). However, more investigations will be required to illustrate the exact mechanism of ATG5 in the regulation of H2Bub1. Consistent with our observation that nutrient starvation results in the loss of H2Bub1, a previous study suggested that H2Bub1 is a semi-quantitative marker of environmental glucose levels. This study found that H2Bub1 levels were significantly increased when the cells were cultured in higher glucose conditions (60), supporting our observations in another aspect. Together with our data, these results indicate that H2Bub1 is likely an immediate response factor to environmental nutrition changes achieved through controlling the activities of autophagy.
When cells were exposed to starvation, there was no change in either the RNF20 and RAD6 levels (Figure 2C and Supplementary Figure S2B), or the interaction between RNF20 and RAD6 (Supplementary Figure S2D), implying that the decrease in the H2Bub1 must therefore be due to the presence of other mechanisms. Indeed, we found that two de novo DNA methyltransferases, DNMT3a and DNMT3b, are degraded via the ubiquitin-proteasome pathway, leading to the activation of the USP44 gene (Figure 3B–G). The increased USP44 levels then result in the depletion of H2Bub1, causing a global decrease in H2Bub1, as was observed (Figure 2C, Supplementary Figure S2B, and Figure 2D). These results indicated that the DNMT3-USP44 axis is involved in the regulation of H2Bub1 and the subsequent activation of autophagy.
The ectopic expression of the H2Bub1 mutant (H2BK120R) caused an alteration of the transcription of genes involved in autophagy and a remarkable increase in autophagy, which is perhaps the most direct evidence that this specific H2B modification is critical for the activation of autophagy. The depletion of RNF20 and the overexpression of USP44 both resulted in a reduction in H2Bub1 that had the same effect on autophagy, further supporting this conclusion (Figure 1A–I). However, H2Bub1 failed to respond to the rapamycin-induced autophagy activation process (Figure 2F, Supplementary Figure S2H and Supplementary Figure S3), indicating that alternative modifications/mechanisms must be involved in the rapamycin-induced activation of autophagy. Several recent studies have indicated that starvation-induced and rapamycin-induced autophagy also seem to use different regulation machinery (14,61–63). Overall, we discovered a novel epigenetic pathway that regulates both H2Bub1 and autophagy: the DNMT3-USP44 axis.
Our study also indicates that different epigenetic modifications can interact with each other to regulate autophagy. We found that the reduction in H2Bub1 via either RNF20 RNAi or overexpression of the H2B mutant H2BK120R down-regulates the levels of H4K16ac and the acetyltransferase hMof, supporting crosstalk between epigenetic pathways. However, ATG5 RNAi caused a reduction in H2Bub1 but had no obvious effect on H4K16ac or hMof, as demonstrated by the absence of autophagy-mediated hMof degradation in ATG5-depleted cells (Figure 2G, left). Moreover, the reduction in H4K16ac by knocking down the expression of hMof had no obvious effect on the H2Bub1 levels (Figure 2G, right). These findings all indicate that H2Bub1 is possibly upstream of the hMOF-H4K16ac axis. However, a global understanding of the epigenetic regulation network of autophagy is far from clear, and further studies are recommended.
The relationship between H2Bub1 and autophagy was also examined in mESCs because we and other groups observed extremely low levels of H2Bub1 in undifferentiated mESCs (33–35). Indeed, an inverse correlation between H2Bub1 and autophagy was observed (Figure 6A and B). It has been reported that autophagy is required for the generation of induced pluripotent stem cells (iPSCs) (64). It is likely that the increased presence of autophagy in mESCs may be utilized to maintain the self-renewal/proliferation of ESCs requiring rapid energy recycling. However, more detailed studies of stem cells will be required to elucidate the functional impact of H2Bub1-induced autophagy in stem cell self-renewal or differentiation.
In conclusion, our work revealed that H2Bub1 is a critical regulator that links autophagy to epigenetics. The findings from this work and others also argue that multiple pathways must be involved in the regulation of autophagy, which ensures that subcellular protein degradation can be executed to support the survival of the cells, as well as to maintain the cellular identity or programming/reprogramming of the cells. It will be of interest to further elucidate the difference between these pathways as well as their functional implications.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Li Yu from Tsinghua University for kindly providing the GFP-LC3 plasmid.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
973 program of the Ministry of Science and Technology of China [2015CB856204, 2015CB964802, 2016YFA0100400, 2014CB964603, 2014CB965001]; National Natural Science Foundation of China [91419304, 31330043, 31271534]. Funding for open access charge: Ministry of Science and Technology of China [2015CB856204, 2015CB964802, 2016YFA0100400, 2014CB964603, 2014CB965001]; National Natural Science Foundation of China [91419304, 31330043, 31271534].
Conflict of interest statement. None declared.
REFERENCES
- 1.Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007; 8:931–937. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z., Klionsky D.J.. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010; 12:814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fimia G.M., Kroemer G., Piacentini M.. Molecular mechanisms of selective autophagy. Cell Death Differ. 2013; 20:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansen T., Lamark T.. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011; 7:279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehrpour M., Esclatine A., Beau I., Codogno P.. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010; 20:748–762. [DOI] [PubMed] [Google Scholar]
- 6.Choi A.M., Ryter S.W., Levine B.. Autophagy in human health and disease. N. Engl. J. Med. 2013; 368:651–662. [DOI] [PubMed] [Google Scholar]
- 7.Levine B., Kroemer G.. Autophagy in the pathogenesis of disease. Cell. 2008; 132:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B., Yuan J.. Autophagy in cell death: an innocent convict?. J. Clin. Invest. 2005; 115:2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J.. Autophagy fights disease through cellular self-digestion. Nature. 2008; 451:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva L.M., Jung J.U.. Modulation of the autophagy pathway by human tumor viruses. Semin. Cancer Biol. 2013; 23:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H.J., Lee S., Jung J.U.. When autophagy meets viruses: a double-edged sword with functions in defense and offense. Semin. Immunopathol. 2010; 32:323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Klionsky D.J.. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010b; 22:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artal-Martinez de Narvajas A, Gomez T.S., Zhang J.S., Mann A.O., Taoda Y., Gorman J.A., Herreros-Villanueva M., Gress T.M., Ellenrieder V., Bujanda L. et al. . Epigenetic regulation of autophagy by the methyltransferase G9a. Mol. Cell Biol. 2013; 33:3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Füllgrabe J., Lynch-Day M.A., Heldring N., Li W., Struijk R.B., Ma Q., Hermanson O., Rosenfeld M.G., Klionsky D.J., Joseph B.. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013; 500:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrasekharan M.B., Huang F., Sun Z.W.. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010; 5:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J., Hake S.B., Roeder R.G.. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005; 20:759–770. [DOI] [PubMed] [Google Scholar]
- 17.Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., Roeder R.G.. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009; 137:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robzyk K., Recht J., Osley M.A.. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000; 287:501–504. [DOI] [PubMed] [Google Scholar]
- 19.Wood A., Krogan N.J., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F. et al. . Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003; 11:267–274. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen S.A. The enigmatic role of H2Bub1 in cancer. FEBS Lett. 2012; 586:1592–1601. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs G., Oren M.. Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta. 2014; 1839:694–701. [DOI] [PubMed] [Google Scholar]
- 22.Cole A.J., Clifton-Bligh R., Marsh D.J.. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr. Relat. Cancer. 2015; 22:T19–T33. [DOI] [PubMed] [Google Scholar]
- 23.Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.Y., Eppink B., Chung Y.M., Shalev G., Shema E. et al. . Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 2011; 41:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D.. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005; 20:601–611. [DOI] [PubMed] [Google Scholar]
- 25.Mallery D.L., Vandenberg C.J., Hiom K.. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002; 21:6755–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakar A., Parvin J., Zlatanova J.. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J. Biomol. Struct. Dyn. 2010; 27:399–406. [DOI] [PubMed] [Google Scholar]
- 27.Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M.. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008; 10:483–488. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.S., Shukla A., Schneider J., Swanson S.K., Washburn M.P., Florens L., Bhaumik S.R., Shilatifard A.. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007; 131:1084–1096. [DOI] [PubMed] [Google Scholar]
- 29.Nakanishi S., Lee J.S., Gardner K.E., Gardner J.M., Takahashi Y.H., Chandrasekharan M.B., Sun Z.W., Osley M.A., Strahl B.D., Jaspersen S.L. et al. . Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 2009; 186:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z.W., Allis C.D.. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002; 418:104–108. [DOI] [PubMed] [Google Scholar]
- 31.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008; 20:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen A.T., Zhang Y.. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011; 25:1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S., Li J., Wang D.L., Sun F.L.. Histone H2B lysine 120 monoubiquitination is required for embryonic stem cell differentiation. Cell Res. 2012; 22:1402–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs G., Shema E., Vesterman R., Kotler E., Wolchinsky Z., Wilder S., Golomb L., Pribluda A., Zhang F., Haj-Yahya M. et al. . RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol. Cell. 2012; 46:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpiuk O., Najafova Z., Kramer F., Hennion M., Galonska C., König A., Snaidero N., Vogel T., Shchebet A., Begus-Nahrmann Y. et al. . The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol. Cell. 2012; 46:705–713. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures bydefined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 37.Zhong X., Jin Y.. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J. Biol. Chem. 2009; 284:9168–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L.P., Ni J.Q., Shi Y.D., Oakeley E.J., Sun F.L.. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat. Genet. 2005; 37:1361–1366. [DOI] [PubMed] [Google Scholar]
- 39.Ni J.Q., Liu L.P., Hess D., Rietdorf J., Sun F.L.. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006; 20:1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H., Shukla A., Wang X., Chen W.Y., Bernstein B.E., Roeder R.G.. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011; 144:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X. et al. . Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 2010; 12:781–790. [DOI] [PubMed] [Google Scholar]
- 42.He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q. et al. . Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012; 481:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moresi V., Carrer M., Grueter C.E., Rifki O.F., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N.. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C., Wei Y., Sun K., Li B., Dong X., Zou Z., Liu Y., Kinch L.N., Khan S., Sinha S. et al. . Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013; 154:1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vethantham V., Yang Y., Bowman C., Asp P., Lee J.H., Skalnik D.G., Dynlacht B.D.. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol. Cell Biol. 2012; 32:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn M.A., Dickson K.A., Jackson S., Clarkson A., Gill A.J., Marsh D.J.. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum. Mol. Genet. 2012; 21:559–568. [DOI] [PubMed] [Google Scholar]
- 47.van der Knaap J.A., Kumar B.R., Moshkin Y.M., Langenberg K., Krijgsveld J., Heck A.J., Karch F., Verrijzer C.P.. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell. 2005; 17:695–707. [DOI] [PubMed] [Google Scholar]
- 48.Joo H.Y., Jones A., Yang C., Zhai L., Smith AD 4th, Zhang Z., Chandrasekharan M.B., Sun Z.W., Renfrow M.B., Wang Y. et al. . Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 2011; 286:7190–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A. et al. . A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008; 29:92–101. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., McMahon S.B.. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008; 29:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z., Jones A., Joo H.Y., Zhou D., Cao Y., Chen S., Erdjument-Bromage H., Renfrow M., He H., Tempst P. et al. . USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013; 27:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipinski M.M., Hoffman G., Ng A., Zhou W., Py B.F., Hsu E., Liu X., Eisenberg J., Liu J., Blenis J. et al. . A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010; 18:1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J., Niu Z., Shi Y., Gao C., Wang X., Han J., Li J., Gao Z., Zhu X., Song X. et al. . Bcl-3, induced by Tax and HTLV-1, inhibits NF-κB activation and promotes autophagy. Cell Signal. 2013; 25:2797–2804. [DOI] [PubMed] [Google Scholar]
- 54.Zhu B.S., Xing C.G., Lin F., Fan X.Q., Zhao K., Qin Z.H.. Blocking NF-κB nuclear translocation leads to p53-related autophagy activation and cell apoptosis. World J. Gastroenterol. 2011; 17:478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singletary K., Milner J.. Diet, autophagy, and cancer: a review. Cancer Epidemiol. Biomarkers Prev. 2008; 17:1596–1610. [DOI] [PubMed] [Google Scholar]
- 56.Shaid S., Brandts C.H., Serve H., Dikic I.. Ubiquitination and selective autophagy. Cell Death Differ. 2013; 20:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W. et al. . Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014; 516:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J.M., Wagner M., Xiao R., Kim K.H., Feng D., Lazar M.A., Moore D.D.. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014; 516:112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Füllgrabe J., Klionsky D.J., Joseph B.. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014; 15:65–74. [DOI] [PubMed] [Google Scholar]
- 60.Urasaki Y., Heath L., Xu C.W.. Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PLoS One. 2012; 7:e36775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T.. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morselli E., Maiuri M.C., Markaki M., Megalou E., Pasparaki A., Palikaras K., Criollo A., Galluzzi L., Malik S.A., Vitale I. et al. . Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010; 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morselli E., Mariño G., Bennetzen M.V., Eisenberg T., Megalou E., Schroeder S., Cabrera S., Bénit P., Rustin P., Criollo A. et al. . Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011; 192:615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S., Xia P., Ye B., Huang G., Liu J., Fan Z.. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013; 13:617–625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.