Abstract
The rules of engagement between zinc finger transcription factors and DNA have been partly defined by in vitro DNA-binding and structural studies, but less is known about how these rules apply in vivo. Here, we demonstrate how a missense mutation in the second zinc finger of Krüppel-like factor-1 (KLF1) leads to degenerate DNA-binding specificity in vivo, resulting in ectopic transcription and anemia in the Nan mouse model. We employed ChIP-seq and 4sU-RNA-seq to identify aberrant DNA-binding events genome wide and ectopic transcriptional consequences of this binding. We confirmed novel sequence specificity of the mutant recombinant zinc finger domain by performing biophysical measurements of in vitro DNA-binding affinity. Together, these results shed new light on the mechanisms by which missense mutations in DNA-binding domains of transcription factors can lead to autosomal dominant diseases.
INTRODUCTION
Zinc finger (ZF)-containing proteins (ZFPs) are the largest class of proteins encoded by the human genome (1). The C2H2-ZF subfamily is the most common. While some members function as RNA-binding or protein-binding modules, the majority are thought to function as DNA-binding modules. These have been the focus of extensive in vitro studies, both experimental and computational, in an effort to better understand the sequence specificity encoded by each ZF; i.e. the DNA-interaction code (2–4). This code underpins the ability to design artificial sequence specific ZFPs for manipulation of gene expression in vivo (5,6).
Arguably, most is known about the transcription factors (TFs) with three tandem C2H2 type ZFs; i.e. the related Egr (also known as Krox and Zif), Sp and Klf families (5). The current model for DNA binding is based primarily on structural studies of EGR1, SP1 and KLF4 (7–10); Four key residues located relative to the start of the C-terminal α-helix (−1, +2, +3 and +6) of each ZF mediate nucleotide contact. Residues at −1, +3 and +6 bind to consecutive nucleotides and the residue at +2 makes contact with the opposite strand or plays an indirect role in binding (8–13). Thus, a single ZF essentially confers ∼3 nucleotide sequence specificity based on a code defined by the amino acids at positions −1, +3, +6 and to a lesser extent, +2 (14). Based on this code each ZF in KLF1 recognizes a triplet nucleotide sequence that together form KLF1's cognate 9 bp binding motif, CCM-CRC-CCN (15,16). In fact, KLFs and SP family proteins actually make direct contact with the opposite G-rich strand of DNA (8–14); i.e. NGG-GYG-KGG. We have employed this G-rich strand nomenclature in the figures and text from hereon. Based on genome wide ChIP-seq data either a T or C at position 5 (A or G on C-rich strand) is permissive for KLF1, KLF2, KLF3, KLF4, KLF5 and KLF11 DNA binding (17–22). Indeed, all SP/KLFs studied to date bind an identical motif in vivo.
Many KLFs are expressed broadly, but KLF1 is restricted to the erythroid lineage (15,23). Klf1 null mice die by E14.5 due to disruption of the second (or definitive) wave of erythropoiesis (24,25). Detailed transcriptome and ChIP-seq studies showed KLF1 directly regulates nearly all aspects of erythroid differentiation, including iron procurement, heme biosynthesis, globin production, cytoskeletal structure and the cell cycle (26,27). It functions primarily as a transcriptional activator in vivo (28), but can function as a repressor in some contexts (29).
The neonatal anemia (Nan) mouse carries a point-mutation in Klf1 leading to a conservative amino acid change (Glu339Asp; position +3 relative to the α-helix) in ZF2 (30). This mutation leads to semi-dominant hemolytic anemia that persists throughout life and is transferable to wild type recipients via bone marrow transplantation; i.e. it is cell autonomous (30–32). Heterozygous Nan mice display hallmarks of hereditary spherocystosis and thalassemia, with splenomegaly, iron deposition in the liver and spleen, abnormal erythrocyte morphology and elevated zinc protoporphyrin (30,32). We previously showed Glu339Asp results in reduced affinity at endogenous KLF1 target sites for which position 5 is an adenine, such as at the Hbb gene promoter and E2f2 enhancer, but not where position 5 is a guanine, such as at the Cdkn1a (p21) enhancer (30). Binding was tightly associated with expression of these genes. This potentially creates two categories of binding sites/target genes across the genome with respect to wild type and Nan versions of KLF1: (i) sites that are co-occupied by both KLF1 and Nan-KLF1 (Category I sites) and (ii) sites which are bound only by KLF1 (Category II sites). However, this hypomorphic model of Nan-KLF1 biochemical function cannot fully explain the dominant phenotype of heterozygous Nan mice, as it would predict less disease severity than that generated by a complete loss of function allele; in fact, the opposite is true. Surprisingly, homozygous Nan mice die much earlier in gestation (at E10-11) than Klf1 null mice (24–25,30,32–33). Klf1+/− mice (and humans) display no overt phenotype. Thus, the Nan mutation represents a rare class of mutation whereby the resulting phenotype is more severe than a loss-of-function mutation. This is particularly remarkable given the conservative glutamate to aspartate change.
In this study, we test the hypothesis that the Nan mutation confers aberrant DNA-binding specificity that is detrimental to cell viability. Using ChIP-seq we show Nan-KLF1 binds promiscuously to degenerate motifs at genomic loci not normally occupied by wild-type KLF1. Using a sensitive RNA labeling approach (4sU) combined with RNA-seq, we show Nan drives expression of a corrupted transcriptome that is detrimental to erythroid cell survival. This study provides insight into how an unexpected neomorphic function can occur in ZFPs when loss of DNA-binding specificity leads to DNA-binding promiscuity. To our knowledge this is the first description of this disease mechanism. Our work suggests a one to one interaction between amino acid residues at −1, +3 and +6 with single nucleotides in the recognition motif is an oversimplification of the binding mechanism. Rather, we suggest the amino acid at the +3 position can indirectly influence DNA-binding affinity and specificity between the −1, +2 residues and DNA.
METHODS
Detailed information of protocols and reagents are further described in Supplementary Material.
ChIP-seq
K1-ER and K1-NanER cells were generated from an immortalized Klf1−/− fetal liver cell line (K1) by retroviral transfection with plasmids containing ER™ fusions of KLF1 and Nan-KLF1 as described previously (34). Three clonally independent K1-ER and K1-NanER cell lines (1 × 107 cells of each) were induced with 2 mM tamoxifen (or ethanol control) for 3 h before crosslinking with 1% formaldehyde. ChIP was performed with a rabbit polyclonal antibody raised against the N-terminal domain of KLF1 (24). Enriched DNA samples were validated by qPCR and pooled prior to generating Ion Torrent fragment libraries.
RNA-seq
Total RNA was collected from three independent biological samples of Klf1Nan/+ and wild-type mouse fetal liver (E14.5). Illumina TruSeq Stranded mRNA libraries were generated from globin mRNA-cleared samples as previously described. Replicate parental cell lines (K1) and two clonally independent K1-ER and K1-NanER cell lines were incubated with 2 mM tamoxifen and 500 μM 4sU (to label newly synthesized RNA) for 1 h prior to isolation of 4sU-labeled RNA as described previously (35). Purified 4sU-labeled RNA was used to generate Ion Xpress™ Plus fragment libraries.
Protein–DNA binding assays
Purified recombinant ZF's of KLF1 and Nan-KLF1 (600 nM) were hybridized with 32P-labeled oligos (5 nM) listed in Supplementary Table S7 for electromagnetic shift assay (EMSA) as previously described (36). Fluorescence polarization assays were performed using 2 nM Fluorescein labeled probes (Supplementary Table S7) as previously described (37,38).
Structural modeling
Protein structure visualization was performed using the UCSF Chimera software (39) based on the structure of KLF4 bound to DNA (PDB ID:2WBU) (8). The Nan mutation was modeled from a backbone-dependent rotamer library (40).
RESULTS
Fetal liver erythroid cells from Klf1Nan/+ mice display an aberrant transcriptome
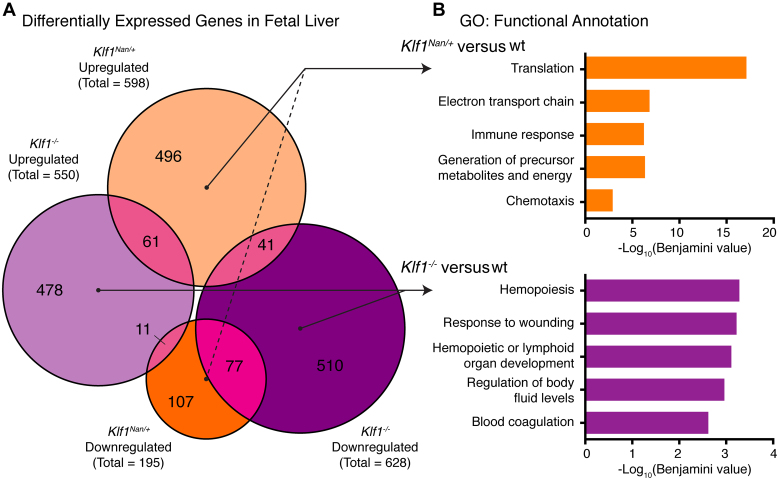
To identify how the Nan mutation disrupts erythropoiesis, we performed mRNA sequencing (RNA-seq) on Klf1Nan/+ versus wild-type fetal livers (n = 3 per group). We obtained >35 million reads for each sample and performed differential expression (DE) analysis using the Tuxedo Suite pipeline (41). After filtering (see Materials and Methods), we found 598 upregulated and 195 downregulated genes in Klf1Nan/+ compared to wild type fetal liver (Figure 1A). Upon comparison with differentially expressed genes (DEGs) in Klf1−/− fetal liver (550 upregulated and 628 downregulated (28), it is striking that the DEGs upregulated in the Klf1Nan/+ fetal liver have little overlap with either of the Klf1−/− categories, suggesting a high level of ectopic expression. There was a small set of just 190 shared DEGs between data sets, and a quarter of these responded in opposing directions. This suggests Nan-KLF1 is both hypomorphic with respect to some KLF1 target genes and neomorphic, since many non-KLF1 target genes are activated or repressed in Klf1Nan/+ fetal liver.
Figure 1.
Transcriptional dysregulation in Klf1Nan/+ fetal liver. (A) Proportional Venn diagram illustrating overlap between DEGs from Klf1Nan/+ and Klf1−/− fetal liver compared to wild type controls. (B) Top five functional annotation gene ontology (GO) terms for each unique set of DEGs from Klf1Nan/+ and Klf1−/− fetal liver. GO terms are ranked and displayed as -log10(Benjamini corrected P-value).
To explore the possible functions of the Klf1Nan/+ and Klf1−/− DEGs, we performed gene ontology (GO) analysis using DAVID (42). We found the majority of DEGs from Klf1Nan/+ fetal liver are not normally associated with hematopoietic functions (Figure 1B), which is consistent with a neomorphic function for the Nan mutation. The full list of genes dysregulated in Klf1Nan/+ FL is provided in Supplementary Table S1.
An in vitro model of the Nan mutation mimics the in vivo phenotype
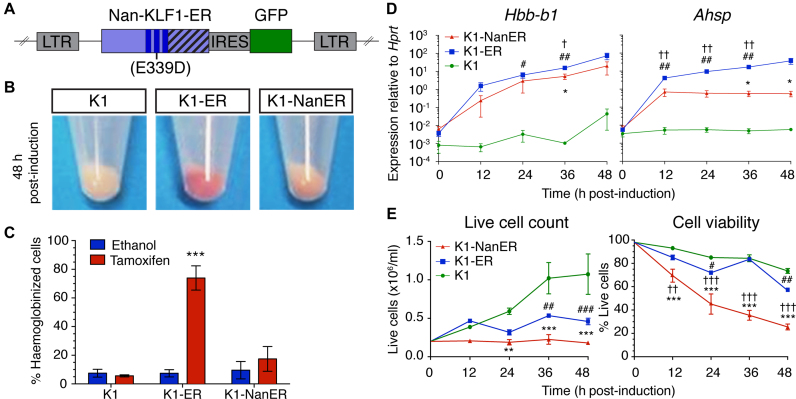
We could not obtain definitive erythroid cells in which only the Nan mutation is present, since Klf1Nan/Nan mice die at ∼E10.5 (33). Also, ChIP-seq in primary fetal liver cells from Klf1Nan/+ mice would be difficult to interpret, since we would not be able to tell whether the signal was from the wild-type or mutant KLF1 binding to DNA, or both. We needed a ‘cleaner’ system if we were to compare the sites occupied by KLF1 versus Nan-KLF1, one in which we could examine the transcriptional consequences of binding events without competition for binding. To this end, we developed an in vitro model of the Nan mutation using a Klf1 null erythroid cell line (K1) generated from Klf1−/− E14.5 FL (34). Retroviruses consisting of either a Nan or wild type Klf1 transgene fused to a modified Estrogen Receptor (ERTM) were introduced into K1 cells to generate stable cell lines, which we will refer to as K1-ER and K1-NanER cells (Figure 2A). Tamoxifen-inducible nuclear localization and equivalent expression amongst independent clones of each construct was confirmed by Western blotting (Supplementary Figure S1).
Figure 2.
K1-NanER cell lines provide an in vitro model of the Nan phenotype. (A) Schematic overview of the retroviral vector MSCV-Nan-KLF1-ER™-IRES-GFP used to transduce K1 cells. The introduced Nan mutation (Glu339Asp) is indicated within the second ZF (dark blue bars) of KLF1. Nan-KLF1 is expressed as an ER™ (striped bars) fusion protein. GFP; Green fluorescent protein, LTR; long-terminal repeat. (B) K1-NanER cells undergo less hemoglobinization (pink pellet) than K1-ER cells (red pellet) following induction by 4-OHT for 48 h. (C) K1-ER cells show significant hemoglobinization compared to ethanol treated controls, determined by the percentage of cells staining DAF positive within 48 h. Data are represented as mean ± SEM from 3 clonally independent cell lines. ***P < 0.001 (Student's t-test). (D) Expression of Hbb-b1 and Ahsp following induction K1-ER and K1-NanER cells by 4-OHT as determined by qRT-PCR. Each point shown is the mean ± SEM for three clonally independent replicates normalized to the expression of Hprt compared to parental K1 cells. *P < 0.05 for K1-NanER; #P < 0.05, ##P < 0.01 for K1-ER; †P < 0.05 for comparison between K1-NanER and K1-ER (Student's t-test). (E) Activation of Nan-KLF1 and KLF1 halts cell proliferation while Nan-KLF1 also reduces cell viability (n = 3 clonally independent cell lines). Live cell count was performed using trypan blue exclusion. A two-way ANOVA with Bonferroni post-test correction was used to determine significant differences between groups. Graphs show mean ± SEM. *P < 0.05, **P < 0.01 ***P < 0.001 for K1-NanER; #P < 0.05, ##P < 0.01 ###P < 0.001 for K1-ER; †P < 0.05, ††P < 0.01 †††P < 0.001 for comparison between K1-NanER and K1-ER.
We have previously shown that K1-ER cells replicate many of the processes involved in terminal erythroid differentiation such as hemoglobinization (34), chromatin condensation and activation of KLF1 target genes (e.g. Hbb-b1, Ahsp and Dmnt) (43,44). In contrast, K1-NanER cell lines make limited amounts of hemoglobin upon tamoxifen induction (Figure 2B and C) and activate Hbb-b1 and Ahsp ∼10-fold and ∼30-fold less than for K1-ER cells compared to parental K1 cells (Figure 2D). These results are consistent with a hypomorphic function for the Nan-KLF1 protein. Proliferation is halted but viability is maintained upon KLF1 induction in K1-ER cells, presumably via direct up-regulation of cell cycle inhibitors such as Cdkn2c (p18) (45) and Cdkn1a (46) (Figure 2E). However, K1-NanER cells exhibit rapid loss of viability within the first 24 h following induction. This suggests that Nan-KLF1 is somehow toxic to erythroid cells.
The Nan mutation confers altered DNA-binding specificity
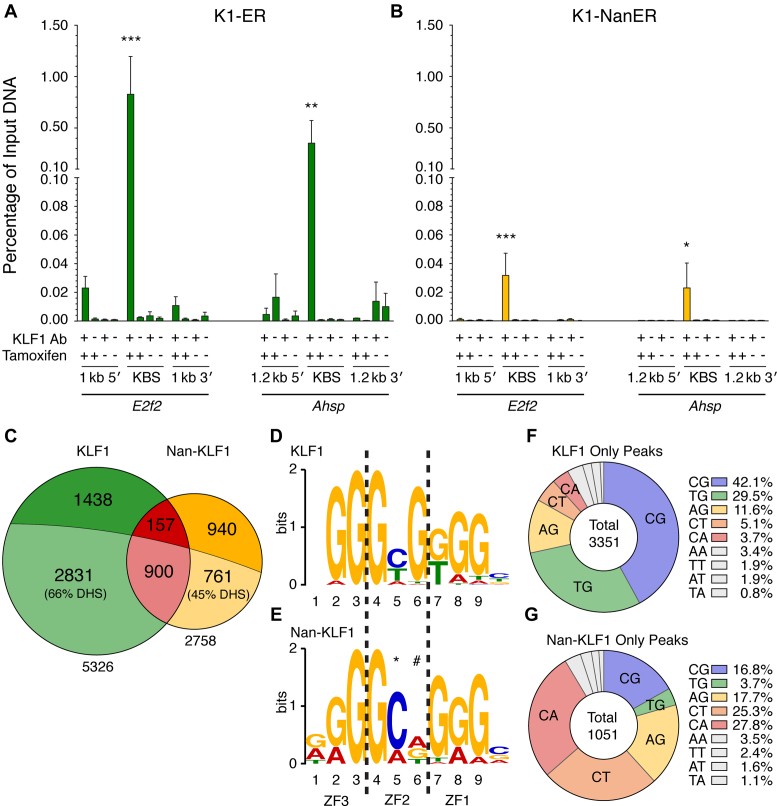
We hypothesized that the Nan mutation would lead to loss of DNA-binding affinity through destabilization of the ZF2-DNA interaction. Indeed, we previously reported reduced Nan-KLF1 binding to some established KLF1 target sites in vitro by EMSA (30). We performed ChIP with 3 clonally independent K1-NanER and K1-ER cell lines following induction with tamoxifen for 3 h and confirmed in vivo occupancy at validated KLF1 binding sites in the intronic enhancer of E2f2 (47) and the promoter of Ahsp (45) for both KLF1 and Nan-KLF1 (Figure 3A and B). It is difficult to quantify independent ChIP experiments but comparison to input DNA provides some idea of efficiency.
Figure 3.
The Nan mutation (Glu339Asp) in ZF2 of KLF1 alters genome binding specificity. The E2f2 intronic enhancer and Ahsp promoter are by bound (A) KLF1 and (B) Nan-KLF1. ChIP was performed on 3 clonally independent K1-ER and K1-NanER cell lines with the following treatments serving as controls: KLF1 Ab (+) or pre-immune serum (−), and treatment with 4-OHT (+) or ethanol (−) for 3 h. Enrichment of bound DNA was determined by qPCR using primers targeted to KLF1 occupied sites (KBS). Regions ∼1 kb upstream and downstream serve as controls to ensure specificity for Ab binding, and local chromatin state. (C) Proportional Venn diagram comparing the total number and overlap (red) of declared ChIP-seq peaks from KLF1 (green) versus Nan-KLF1 (yellow). Lightly-shaded portions represent the intersection with DNase I hypersensitivity sites (DHS) contributed from erythroid tissues/cells (combined total DHS = 295 322). Consensus binding motifs (or position weight matrix (PWM)) of (D) KLF1 and (E) Nan-KLF1 derived from declared peaks using de novo motif discovery (MEME). The Nan-KLF1 binding motif displays increased specificity at position 5 (*) and a loss of specificity at nucleotide position 6 (#). Pie graphs displaying the proportions of each di-nucleotide found at position 5 and 6 in the central NGG-GNN-KGG motif (found nearest to the summit) of unique (F) KLF1 and unique (G) Nan-KLF1 peaks. Only peaks containing an identified motif are shown. The legend lists the nine most highly represented di-nucleotide combinations in each case. The color coding is maintained in subsequent figures.
We sequenced fragment libraries generated from ChIP and input DNA samples to determine genome-wide Nan-KLF1 versus KLF1 occupancy (see Materials and Methods). In brief, uniquely mapped reads were used for peak calling by MACS2 (48) and declared peaks were filtered and ranked based on fold enrichment (>30-fold compared with input) and a low false-discovery rate (FDR < 0.1%). This yielded a list of high-confidence ChIP-seq peaks: 2758 in K1-NanER cells and 5326 in K1-ER cells (Figure 3C). Supplementary Table S2 lists the top 30 peaks for each ranked by fold enrichment and the entire lists can be found in Supplementary Table S3 and S4. Although some Nan-KLF1 occupied sites were also bound by KLF1 (1057), the majority were not. Also, fewer unique Nan-KLF1 peaks than KLF1 peaks overlap DNase I hypersensitivity sites (DHS) from erythroid cells (∼45% cf. ∼66%) (Figure 3C) (49,50). This observation suggests Nan-KLF1 binds to some genomic sites that are not normal erythroid enhancers or promoters. Yet, Nan-KLF1 peaks were distributed similarly to KLF1 peaks with respect to the nearest transcriptional start site (TSS), being located primarily within introns, promoters or distally (Supplementary Figure S2A).
De novo motif discovery using MEME (51) identified highly enriched motifs for both KLF1 and Nan-KLF1 (E-values = 3.0e-2939 and 6.5e-1405, respectively; Figure 3D). The position weight matrix (PWM) for KLF1 binding is in perfect agreement with the motif identified in fetal liver (CCM-CRC-CCN) (18) and other KLF ChIP-seq data sets (17,20–22). We have reported the PWM in Figure 3D based on the complementary strand (NGG-GYG-KGG) to be consistent with structural studies of binding (8–14). Thus, our system provides an accurate model of chromatin-dependent KLF genome occupancy. Furthermore, CentriMo analysis (52) shows a central enrichment of the identified PWM surrounding the peak summit for both Nan-KLF1 and KLF1-occupied sites, which indicates the ChIP has enriched for real binding events (Supplementary Figure S2B and C).
Close inspection of the Nan-KLF1-occupied consensus sequence (NGG-GMN-KGG) reveals aberrant specificity at nucleotide positions 5 and 6 on the G-rich strand (underlined above), consistent with the position of the mutation in ZF2. There is altered specificity at position 5 from Y (cytosine, thymine) to M (cytosine, adenosine) with a particular preference for C and a loss of specificity at position 6 (or position 4 on the complementary C-rich strand) from predominantly G (guanine) to any nucleotide (Figure 3E). Overall these results suggest a 2-fold increased specificity at position 5, but 4-fold redundancy at position 6 for Nan-KLF1 versus KLF1.
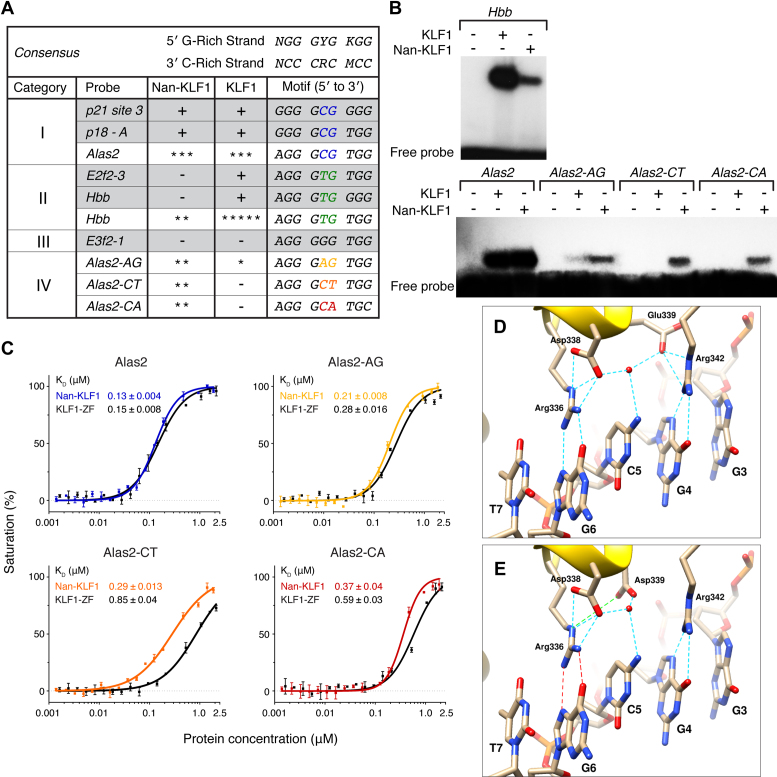
To further analyze the altered sequence specificity at positions 5 and 6, the central motif of every occupied site from both Nan-KLF1 and KLF1 peak lists were placed into one of 16 ‘bins’ based on the di-nucleotide sequence (Figure 3F and G; Supplementary Table S2–S4). The CG di-nucleotide was present in 42% and TG in 30% of the 3351 KLF1-only occupied sites in vivo (Figure 3F), consistent with the PWM derived from MEME (Figure 3D) and our previous EMSA data (26,30). In contrast, position 5–6 was CG in 17% and TG in just 4% of the Nan-KLF1-only bound peaks (Figure 3G). This result supports a normal or mildly hypomorphic function of Nan-KLF1 at some sites in the genome (Category I sites such as the Alas2 first intron enhancer), and severely hypomorphic or amorphic function at other sites (Category II sites such as the Hbb gene promoter) (Figure 4A). Surprisingly, Nan-KLF1 peaks also exhibited a dramatic specificity shift to include any of AG, CA and CT di-nucleotides at positions 5–6 (Figure 3G). This was unexpected and is consistent with a neomorphic biochemical function for the Nan-KLF1 protein.
Figure 4.
Differential binding by recombinant Nan-KLF1 and KLF1 zinc fingers in vitro. (A) Four categories of binding sites are differentially recognized by Nan-KLF1 and KLF1. An expanded Table from Siatecka et al. (PNAS, 2010) reports binding (+) or lack of binding (−) to Category I–III sequences as determined by electromagnetic shift assay (EMSA) (shaded rows). Binding of new Category IV sequences are reported using a five-star scale (* = very weak to ***** = very strong). (B) EMSA comparing complex formation between Nan-KLF1 or KLF1 ZF and radiolabeled dsDNA for known KLF1 binding sites at Alas2 enhancer (Category I), Hbb promoter (Category II) and di-nucleotide variants of position 4–5 in Alas2 (Category IV). Equivalent amounts of recombinant protein were loaded for each probe. (C) Binding affinities of Nan-KLF1 and KLF1 with oligos containing variant nucleotides at positions 5–6, as measured by fluorescence polarization assays. KLF1 affinities and curves are indicated in black, while Nan-KLF1 are color-coded according to the probe used in A. (D) Model of the interaction of KLF1 ZF2 with DNA based on the structure of KLF4-DNA interactions (PDB ID: 2WBU). The basic amino acids at positions −1 (Arg336) and +6 (Arg342) of the α-helix form hydrogen bonds (blue dotted lines) with G6 and G4, respectively. Glu339 does not make direct contact with C5. Instead, it forms a salt bridge with Arg342 and a hydrogen bond with an ordered water molecule found in the center of the binding site (red sphere). Asp338 forms a hydrogen bond with the same water and a salt bridge with the sidechain of Arg336. (E) Model proposing the likely structural consequence of the Glu339Asp Nan KLF1 mutation. The preferred sidechain orientation selected from a backbone dependent rotamer library search suggests a significant change in orientation, such that residue 339 no longer interacts with Arg342 but instead is brought into close proximity to Arg336 (green line), disrupting its interaction with G6 (red lines).
Nan-KLF1 binds an altered sequence motif
To validate the aberrant binding specificity suggested by ChIP-seq, we purified recombinant Nan-KLF1 and KLF1 ZF domains (see Methods). Correct folding of protein was verified using circular dichroism (CD; Supplementary Figure S3). Data below 195 nm were too noisy for reliable analysis due to buffer interference. Nevertheless, the CD spectra indicated that both proteins are folded. Estimates of secondary structure, determined using the Dichroweb Server (53) running the K2D method (54) from multiple data sets, yielded estimates ranging between 20–30% helix and 20–30% β-sheet, consistent with the secondary structure content of similar proteins with the same folds.
We then tested the ability of the two ZF domains to bind consensus sequences in vitro using EMSA. Binding to the Hbb (Category II) probe by Nan-KLF1 was diminished in comparison to KLF1, whereas binding to the Alas2 (Category 1) probe was about equivalent (Figure 4A and B). This result is consistent with our ChIP-seq data and previous analyses (26,30). We also compared variants of the Alas2 probe where AG, CT and CA di-nucleotides were introduced at position 5–6. Moderate binding to all Alas2 variants was observed for Nan-KLF1, while KLF1 bound only AG weakly. Fluorescence polarization was used to measure the dissociation constant (KD) for each variant (Figure 4C). Nan-KLF1 (KD = 0.13 μM) and KLF1 (KD = 0.15 μM) showed equivalent affinity for the Alas2 probe. However, Nan-KLF1 affinity for AG, CT and CA was higher by factors of ∼1.3, ∼2.9 and ∼1.6, respectively. This result is consistent with the ChIP-seq data and demonstrates Nan-KLF1 is able to bind these new sequences in vitro with an enhanced affinity. Collectively, we refer to these new sequences as category IV sites (Figure 4A). Although binding to aberrant sequences is weak in vitro, the binding affinity is sufficient to detect as a ChIP-seq signal in many cases, but probably not all, as evidenced by the smaller number of MACS2 peaks passing our filters.
Glu339Asp is a conservative amino acid substitution and so the profound impacts seen on KLF1 function associated with this mutation were surprising. Two structures of KLF4 bound to a Category I DNA motif are available (i.e. C at position 5 on the G-rich strand) (8,55). There is no reported structure of KLF4-Category II DNA complex. KLF4 is identical to KLF1 at all of the DNA-binding residues within each of ZFs, and so serves as a good model for KLF1-DNA interactions. Arg336 and Arg342 in ZF2 of murine KLF1 (= Arg443 and Arg449 in KLF4) make hydrogen bonds with the G-rich strand of the DNA consensus sequence at G6 and G4, respectively (Figure 4D). Glu339 does not make direct contact with the C5 nucleotide but rather participates in an extensive electrostatic network with Arg342, Asp338 (+2), a bound water molecule and the C5 nucleotide. To assess the possible structural consequences of the Glu339Asp mutation on DNA binding, we introduced this amino acid substitution in silico into the KLF1-DNA structure (PDB ID:2WBU). The orientation of the side chain was selected from a rotamer library (40) to minimize steric clashes with surrounding residues. As shown in Figure 4E, the shorter aspartate sidechain is unlikely to be able to simultaneously make all of the same interactions as Glu339. While it is not possible to predict definitively the structural consequences of this change, it is very likely that some reorganization of the Arg336, Asp338 and Arg342 will take place. In turn, such a reorganization could easily alter the DNA-binding specificity of ZF2, consistent with the observed ChIP-seq data.
Ectopic transcription is activated following ectopic Nan-KLF1 binding to DNA
We next asked whether altered DNA-binding specificity leads to an aberrant transcriptional response. To test this, we employed 4-thiouridine (4sU)-RNA-seq, a robust and reproducible method that captures newly synthesized RNA through biotinylation of the incorporated uridine analog, 4sU (35). We utilized 4sU-RNA-seq to examine the immediate transcriptional response to a tamoxifen-induced KLF1-DNA binding event (see Materials and Methods). We performed qRT-PCR for primary pre-spliced RNA species and fully processed RNA species derived from the Hbb-b1 and Hprt genes that convincingly demonstrates that the 4sU-labeling and enrichment captures newly synthesized RNA (Supplementary Figure S4A). Libraries were generated from 4sU-enriched RNA for sequencing on the Ion Torrent Proton system. We collected greater than 25 million sequence reads from each sample including: (i) two clonally independent replicates for K1-ER and K1-NanER cell lines and (ii) two replicates of the parental line, K1 (as a negative control). Mapping of reads to mm9 identified levels of intron coverage consistent with prior studies using a similar 4sU-labeling period (35,56) and mRNA-seq (Supplementary Figure S4B). The alignment of sequenced tags was largely to exons, suggesting co-transcriptional splicing is efficient within the 1 h labeling window, consistent with prior studies on variability of rates of co-transcriptional splicing (56–58).
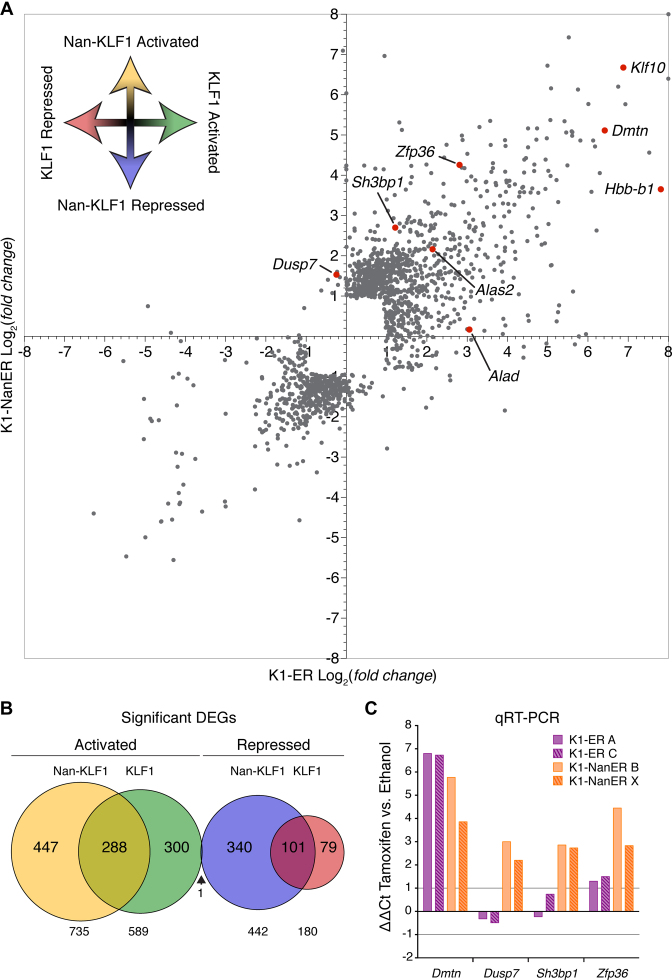
To simplify the analysis, we counted tags over exons and employed Cuffdiff (41) to find DEGs between parental (K1) and K1-ER or K1-NanER cell lines. The two replicates for each cell line were highly correlated (Supplementary Figure S4C), with a Spearman's correlation coefficient great than 0.95 across sample groups. Comparing the magnitude of fold change between KLF1 and Nan-KLF1 DEGs shows most DEGs are differentially responsive between sample groups (Figure 5A). For example, some known KLF1 target genes, such as Alas2 (26) and Klf10 (59), are activated equally (4.4-fold and ∼100-fold up-regulated respectively) by KLF1 and Nan-KLF1, while other KLF1 target genes such as Alad (60) and Hbb-b1 (15) are more responsive to KLF1. Finally a third set of genes, not regulated by KLF1, are responsive to Nan-KLF1; this set includes Dusp7, Sh3bp1 and Zfp36. Expression of these genes is consistent with a neomorphic function for Nan-KLF1.
Figure 5.
4sU-RNA-seq reveals a corrupted transcriptome in K1-NanER cells. (A) Scatter plot representation of DEGs in K1-ER and K1-NanER cells compared to the parental K1 cells. Each data point represents a single RefSeq gene plotted according to the log2(fold change) between K1-ER/K1 (x-axis) and the log2(fold change) between K1-NanER/K1 (y-axis). Specific data point referring to genes of interest have been highlighted in red and annotated accordingly. (B) Proportional Venn diagram displaying the overlap between DEGs in two clonally independent K1-ER and K1-NanER cell lines compared to replicate parental K1 cells following 1 h of tamoxifen induction. (C) Validation of primary transcript expression by qRT-PCR from two independent K1-ER (clone A and C) and two K1-NanER (clone B and X) cell lines illustrates both hypomorphic and neomorphic regulation of target genes by Nan-KLF1. Expression was normalized to Hprt primary transcript (that does not change in absolute FPKM values between groups) using the ΔΔCt method comparing tamoxifen and ethanol treated controls.
In total, we found significant activation of 589 genes and repression of 180 genes by KLF1, whereas Nan-KLF1 induced 735 genes and repressed 442 genes (Supplementary Table S5 and S6). The overlap between these DEGs have been represented by a Venn diagram in Figure 5B. A further 8227 non-differentially expressed genes were identified (using a cutoff for expression of >1 FPKM), suggesting KLF1 regulates about 9% of the erythroid gene repertoire in proerythroblasts.
We validated the 4sU-RNA-seq data by qRT-PCR on 4sU-RNA using primers designed to detect primary pre-spliced transcripts (Figure 5C). The trends observed for the qRT-PCR data mirror those observed in the 4sU-RNA-seq data, and highlight both the hypomorphic and neomorphic functions of Nan-KLF1 at different genome locations or contexts.
Differential Nan-KLF1 occupancy is co-incident with ectopic gene expression
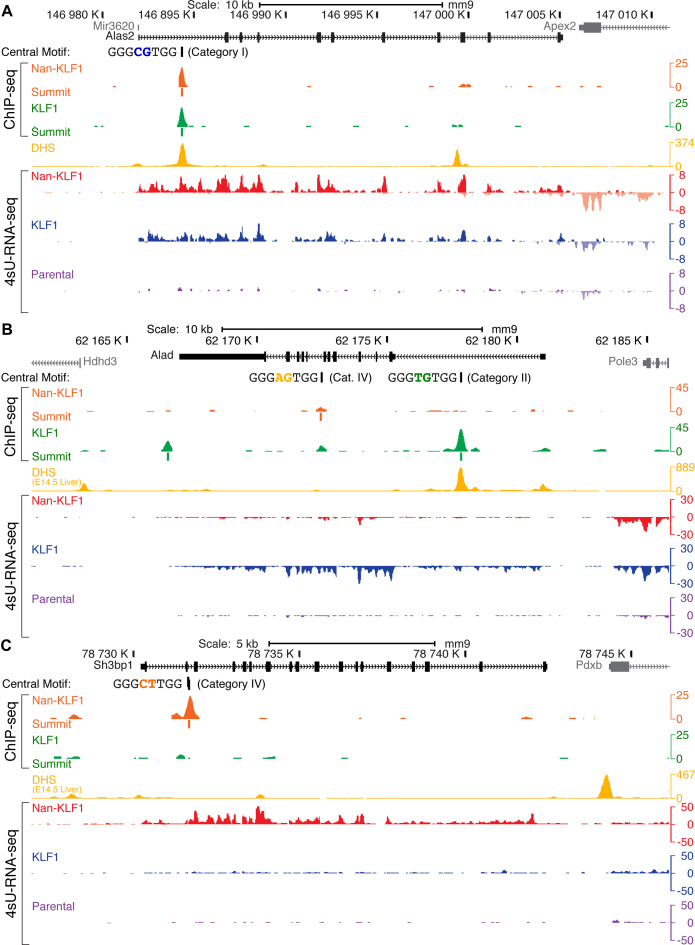
To gain further insight at specific gene loci, we inspected genes from each of the activated groups using the UCSC Genome Browser (Figure 6). Alas2 is the first enzyme in the heme biosynthetic pathway and a known KLF1 target gene (26). Nan-KLF1 and KLF1 both bound strongly in the first intron of Alas2, co-incident with strong transcriptional activation (∼4.5-fold) by both (Figure 6A). The central motif identified in both ChIP-seq data sets contains the CG di-nucleotide (Category I motif), consistent with the ability of both Nan-KLF1 and KLF1 to bind this sequence by ChIP-seq.
Figure 6.
Nan-KLF1 binding is co-incident with ectopic gene regulation at specific loci. Images from the murine UCSC Genome Browser with read density graphs (wiggle plots) displaying KLF1 ChIP-seq, and 4sU-RNA-seq from representative K1, K1-ER and K1-NanER cell lines following 4-OHT treatment and DHS tracks from E14.5 FL (yellow). Additional tracks depict the di-nucleotide sequence and peak summit location for significantly enriched peaks. 4sU-RNA-seq reads mapping to the Crick strand are displayed as negative values in wiggle plots. (A) Alas2 locus depicting transcriptional activation by both Nan-KLF1 and KLF1 co-incident with binding of both Nan-KLF1 (orange track) and KLF1 (green track) within the first intron. ChIP peaks at this location share a common central motif containing the di-nucleotide CG (Category I site), and they overlap a DHS in fetal liver (yellow track). There is robust induction of transcription for both K1-ER and K1-NanER cells (blue and red tracts) compared to parental K1 cells (purple track). (B) The Alad locus is activated and primarily bound by KLF1. The largest peak in the first intron overlaps with DHS and contains the TG central di-nucleotide (Category II site). It is not bound by Nan-KLF1 and there is minimal activation of Alad transcription by Nan-KLF1. (C) Ectopic binding and transcriptional activation of Sh3bp1 by Nan-KLF1. The peak in the second intron (Category IV) does not display DHS in fetal liver nor is it bound by KLF1. Nevertheless, Nan-KLF1 binds this site robustly in vivo and results in Nan-KLF1-specific transcriptional activation.
Alad encodes the second enzyme of the heme biosynthetic pathway and is also directly activated by KLF1 (60). KLF1 binds strongly to the middle of intron 1, and this overlaps a DHS in fetal liver (Figure 6B). We found no evidence for Nan-KLF1 binding. Furthermore, KLF1 activates transcription robustly (blue track) whereas Nan-KLF1 does so poorly (red track), similar to parental K1 cells (purple track). The motif bound by KLF1 in the Alad intron is an example of a Category II motif, containing the di-nucleotide TG. Again, this result is consistent with reduced binding and a hypomorphic function of Nan-KLF1 at Category II sites.
Lastly, we find the altered motif specificity of Nan-KLF1 leads to aberrant binding within the second intron of Sh3bp1. This is a novel binding site, with no evidence of DHS in erythroid cells, nor KLF1 binding (Figure 6C). The central motif of this peak contains an CT di-nucleotide; i.e. it is an example of a Category IV motif (Figure 4A). Sh3bp1 is ectopically activated by Nan-KLF1; it is a RhoGAP protein normally expressed by neural tissues (Figure 5C) (61). Expression in erythroid cells has not previously been reported and aberrant expression could lead to dysregulated signaling. Similarly, upregulation of the dual-specificity phosphatase, Dusp7 (Figure 5C), could lead to downregulation of p38-MAP kinase signaling (an EpoR-activated pathway). Also, upregulation of Zfp36 (Figure 5C) could induce a signal to proliferate as it might mimic a steroid hormone receptor signaling pathway that works via a related protein, ZFP36L2 (62). We propose that a suite of such Nan-KLF1-activated genes are together responsible for erythroid cell toxicity and the semi-dominant anemia. Although these genes are sometimes upregulated in stress responses, they are still likely to be direct targets of Nan-KLF1 with differential occupancy found in the promoter or upstream of both genes (Supplementary Figure S5).
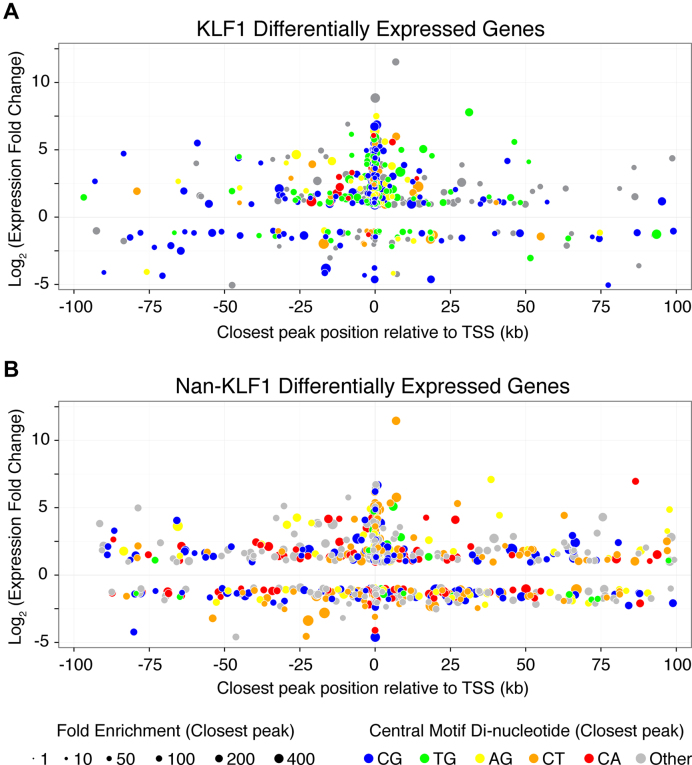
Exploring the relationship between aberrant Nan-KLF1 binding and ectopic gene regulation at the genome-wide level we find 205 KLF1 promoter peaks within KLF1 DEGs (28%) and 102 Nan-KLF1 promoter peaks within the Nan-KLF1 DEGs (9%). This strongly implies direct regulation of these genes. However, KLF1 is also known to bind enhancers that may be located distally (>1 kb) or within other genes (18). To investigate possible distal regulation of DEGs, we plotted the fold change of each DEG, together with the distance to the nearest to ChIP-seq peak as a ‘four-dimension bubble plot’ for both Nan-KLF1 and KLF1. The concept is similar to that employed by Soler et al. (63). The fold enrichment and di-nucleotides at positions 5–6 of each peak's central motif were included by adjusting the size and color of each data point (Figure 7).
Figure 7.
Global changes in aberrant gene regulation are associated with genome-wide corruption of DNA-binding specificity. Bubble plot representation of (A) KLF1 and (B) Nan-KLF1 binding sites in the neighborhood of DEGs between (A) K1-ER or (B) K1-NanER compared to K1 (parental) cell lines. Each data point is displayed to represent four parameters of a DEG: distance to the nearest ChIP-seq peak (x-axis), log2(fold change) (y-axis), fold enrichment of tag count over the ChIP-seq peak compared with input DNA (bubble diameter) and central motif di-nucleotide (bubble color).
This multi-dimensional comparison provides useful insight into the underlying mechanism by which the Nan mutation exerts both repressive and neomorphic effects on the erythroid genome. The most obvious perturbation is a shift in the central motif category (color). For example, peaks closest to Nan-KLF1 DEGs show a marked increase in the proportion of CT, or CA di-nucleotides (Category IV sites) and a loss of the TG di-nucleotide (Category II sites) compared with peaks closest to KLF1 DEGs. This is illustrated by the shift in bubble color from blue and green for KLF1-responsive genes to blue, orange and red for Nan-KLF1-responsive genes. Also, while KLF1 is almost exclusively a transcriptional activator, a large proportion of genes are repressed by Nan-KLF1 (found below x-axis). The majority of these genes are not KLF1 targets. Lastly, Nan-KLF1 binding sites are sometimes found more distant to the nearest regulated TSS (both activated and repressed). This suggests they are not directly regulated or are distally regulated by Nan-KLF1. This could occur if Nan-KLF1 blocked binding of other erythroid transcription factors, or disrupted enhancer-promoter interactions (e.g. via blocking of DNA looping). We propose a model whereby ectopic binding of Nan-KLF1 not only directly drives aberrant gene transcription, but sometimes indirectly disrupts the activity of KLF1-independent erythroid genes or housekeeping genes.
DISCUSSION
We have demonstrated the transcriptome of Klf1Nan/+ fetal liver is aberrant, since the majority of DEGs are not normal KLF1 target genes. This lead to the hypothesis that Nan-KLF1 must have a detrimental gain of function. To dissect this without confounding effects from wild-type KLF1, we developed an in vitro model system where we could induce expression of either version of KLF1 on a Klf1−/− genetic background but in an erythroid context. Using ChIP-seq, we showed the Glu339Asp mutation alters the DNA-binding specificity of KLF1 in a complex and context-dependent manner. We confirmed Nan-KLF1 has less affinity for a T at position 5 in the consensus motif in vivo as it does in vitro (30). We also demonstrated a dramatic and surprising loss of specificity at position 6. Whereas KLF1 only binds G, Nan-KLF1 binds A, G or T at this position in vitro and in vivo. The use of 4sU-labeling of newly synthesized RNA facilitated the detection of immediate transcriptional events following specific activation of either version of KLF1. In combination with the tamoxifen-inducible expression system and ChIP-seq, 4sU-RNA-seq is a powerful and sensitive technique for interrogating the direct transcriptional consequences of altering the TF-DNA-interaction code. It will be generally applicable to the analysis of any rapid transcriptional responses to external signals, transcription factor or splicing factor modifications.
The motif degeneracy observed at position 6 in the Nan-KLF1 PWM should theoretically increase the number of possible binding sites genome-wide (by 4-fold, since each nucleotide could be observed at this position), but in practice it leads to a reduction in total detectable peaks called from ChIP-seq data. The most likely explanation is that there is a reduction in overall binding affinity, as suggested by our EMSA data, such that many binding events fall below the detection threshold of ChIP. Nevertheless, the transcriptional changes are consistent with meaningful low affinity occupancy at aberrant sites that outnumber normal sites due to degenerate binding specificity. This has important implications for our understanding of disease-causing mutations in ZF transcription factors.
A dominant mutation in human KLF1 at the equivalent residue, Glu325Lys (DNA-contacting position +3 in ZF2) results in congenital dyserythropoietic anemia (64). Although not yet tested, it is likely this mutation also causes a gain of function altering DNA-binding specificity. There are other reported missense mutations in KLF1 at key DNA-contact residues in ZF1 at +6 (Ala298Pro) and ZF2 at +6 (Arg328His and Arg328Leu), which could also result in neomorphic as well as hypomorphic functions (65–67). Since ZFPs are the most common class of proteins encoded in the mammalian genome, it is likely that this type of neomorphic mutation occurs in other transcription factors. Indeed, there is a reported dominant mutation in ZF1 of murine KLF3 (H275R at +3), which leads to dominant congenital heart disease (68). Barrera et al. recently identified thousands of human alleles from publicly available genome sequencing projects that are likely to alter DNA binding of various TFs (69). Many of these alleles are Mendelian disease mutations (69). We believe our analysis of Nan-KLF1 in vivo binding site preferences and the consequences for cell function is the first for such a disease-associated allele. It will provide a template for further similar studies of the effects of DNA-contact mutations in other ZFPs and in different cellular contexts.
Our initial prediction of the disease mechanism underpinning the Nan mutation, based upon extensive in vitro binding data, turned out to be incorrect. We found the +3 amino acid in ZF2 of KLF1 plays an important indirect role in determining the interaction between the −1 amino acid and DNA. However, our data are, in fact, consistent with previous studies that were designed to select the most favorable amino acid combinations at positions −1, +2, +3 and +6 in C2H2 fingers for binding to DNA triplets. These studies suggest amino acids at these four positions work together to confer DNA-binding specificity; i.e. they work as a module to define DNA-binding specificity rather than work independently (4). We suggest there may be different ZF-DNA conformations which reflect the precise preferred DNA-sequence; i.e. whether an T or C resides at position 6 (for ZF2) and whether an T or G resides at position 8 (for ZF1). Although a great deal can be predicted (3), further ChIP-seq studies are required to investigate natural or engineered mutations at other critical DNA-contacting amino acids to fully understand the DNA interaction code of ZFPs. These studies will complement the extensive in vitro data we currently use for predicting binding specificity of mutant TFs and the design of engineered ZF (3,70).
ACCESSION NUMBERS
All sequencing data generated by this study have been deposited in the Gene Expression Omnibus (GEO) under the accession GSE71396.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Planutis and L. Xue for technical assistance.
Author contributions: Conceptualization, A.C.P.; Methodology, K.R.G. and M.R.T.; Formal Analysis, K.R.G., M.L., T.B., M.W.P. and J.H.G.; Investigation, K.R.G., M.D.I., D.N., R.S., G.W.M., L.A.M. and A.C.P.; Writing – Original Draft, K.R.G. and A.C.P.; Writing – Review and Editing, K.R.G., M.L., T.B., M.J.L., J.M., L.L.P., J.J.B. and A.C.P.; Visualization, K.R.G., L.A.M., M.J.L. and J.M.; Supervision, M.R.T. and A.C.P.; Funding Acquisition, L.L.P., J.J.B. and A.C.P. None of the authors have any competing interests with respect to this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [APP1082429 to A.C.P.]; United States Public Health Service [R01 GM103544 to T.B., R01 DK100692 to L.L.P. and R01 DK46865 to J.J.B.]; Victorian Government Operational Infrastructure Support Scheme [St Vincent's Institute of Medical Research to M.W.P]. Funding for open access charge: Mater Research - UQ [APP1082429].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ravasi T., Huber T., Zavolan M., Forrest A., Gaasterland T., Grimmond S., Hume D.A., RIKEN GER GroupGSL Members . Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 2003; 13:1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enuameh M.S., Asriyan Y., Richards A., Christensen R.G., Hall V.L., Kazemian M., Zhu C., Pham H., Cheng Q., Blatti C. et al. . Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013; 23:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M. et al. . C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015; 33:1–12. [DOI] [PubMed] [Google Scholar]
- 4.Persikov A.V., Wetzel J.L., Rowland E.F., Oakes B.L., Xu D.J., Singh M., Noyes M.B.. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015; 43:1965–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010; 79:213–231. [DOI] [PubMed] [Google Scholar]
- 6.Deng W., Rupon J.W., Krivega I., Breda L., Motta I., Jahn K.S., Reik A., Gregory P.D., Rivella S., Dean A. et al. . Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014; 158:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka S., Shiraishi Y., Yoshida T., Ohkubo T., Sugiura Y., Kobayashi Y.. NMR structure of transcription factor Sp1 DNA binding domain †,‡. Biochemistry. 2004; 43:16027–16035. [DOI] [PubMed] [Google Scholar]
- 8.Schuetz A., Nana D., Rose C., Zocher G., Milanovic M., Koenigsmann J., Blasig R., Heinemann U., Carstanjen D.. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 2011; 68:3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavletich N.P., Pabo C.O.. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991; 252:809–817. [DOI] [PubMed] [Google Scholar]
- 10.Narayan V.A., Kriwacki R.W., Caradonna J.P.. Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J. Biol. Chem. 1997; 272:7801–7809. [DOI] [PubMed] [Google Scholar]
- 11.Kriwacki R.W., Schultz S.C., Steitz T.A., Caradonna J.P.. Sequence-specific recognition of DNA by zinc-finger peptides derived from the transcription factor Sp1. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:9759–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavletich N.P., Pabo C.O.. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993; 261:1701–1707. [DOI] [PubMed] [Google Scholar]
- 13.Elrod-Erickson M., Benson T.E., Pabo C.O.. High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998; 6:451–464. [DOI] [PubMed] [Google Scholar]
- 14.Elrod-Erickson M., Rould M.A., Nekludova L., Pabo C.O.. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996; 4:1171–1180. [DOI] [PubMed] [Google Scholar]
- 15.Miller I.J., Bieker J.J.. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 1993; 13:2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory R.C., Taxman D.J., Seshasayee D., Kensinger M.H., Bieker J.J., Wojchowski D.M.. Functional interaction of GATA1 with erythroid Krüppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996; 87:1793–1801. [PubMed] [Google Scholar]
- 17.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. et al. . Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 18.Tallack M.R., Whitington T., Yuen W.S., Wainwright E.N., Keys J.R., Gardiner B.B., Nourbakhsh E., Cloonan N., Grimmond S.M., Bailey T.L. et al. . A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010; 20:1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiwari N., Meyer-Schaller N., Arnold P., Antoniadis H., Pachkov M., van Nimwegen E., Christofori G.. Klf4 Is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8). PLoS One. 2013; 8:e57329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aksoy I., Giudice V., Delahaye E., Wianny F., Aubry M., Mure M., Chen J., Jauch R., Bogu G.K., Nolden T. et al. . Klf4 and Klf5 differentially inhibit mesoderm and endoderm differentiation in embryonic stem cells. Nat. Commun. 2014; 5:3719. [DOI] [PubMed] [Google Scholar]
- 21.Burdach J., Funnell A.P.W., Mak K.S., Artuz C.M., Wienert B., Lim W.F., Tan L.Y., Pearson R.C.M., Crossley M.. Regions outside the DNA-binding domain are critical for proper in vivo specificity of an archetypal zinc finger transcription factor. Nucleic Acids Res. 2014; 42:276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loft A., Forss I., Siersbæk M.S., Schmidt S.F., Larsen A.-S.B., Madsen J.G.S., Pisani D.F., Nielsen R., Aagaard M.M., Mathison A. et al. . Browning of human adipocytes requires KLF11 and reprogramming of PPARγ superenhancers. Genes Dev. 2015; 29:7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins A. Erythroid Kruppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell Biol. 1999; 31:1175–1192. [DOI] [PubMed] [Google Scholar]
- 24.Perkins A.C., Sharpe A.H., Orkin S.H.. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995; 375:318–322. [DOI] [PubMed] [Google Scholar]
- 25.Nuez B., Grosveld F.. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995; 375:316–318. [DOI] [PubMed] [Google Scholar]
- 26.Tallack M.R., Perkins A.C.. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life. 2010; 62:886–890. [DOI] [PubMed] [Google Scholar]
- 27.Siatecka M., Bieker J.J.. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011; 118:2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tallack M.R., Magor G.W., Dartigues B., Sun L., Huang S., Fittock J.M., Fry S.V., Glazov E.A., Bailey T.L., Perkins A.C.. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 2012; 22:2385–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frontelo P., Manwani D., Galdass M., Karsunky H., Lohmann F., Gallagher P.G., Bieker J.J.. Novel role for EKLF in megakaryocyte lineage commitment. Blood. 2007; 110:3871–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siatecka M., Sahr K.E., Andersen S.G., Mezei M., Bieker J.J., Peters L.L.. Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15151–15156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon M.F., Glenister P.H., Loutit J.F., Peters J.. Dominant haemolytic anemia. Mouse News Lett. 1983; 68:68. [Google Scholar]
- 32.White R.A., Sokolovsky I.V., Britt M.I., Nsumu N.N., Logsdon D.P., McNulty S.G., Wilmes L.A., Brewer B.P., Wirtz E., Joyce H.R. et al. . Hematologic characterization and chromosomal localization of the novel dominantly inherited mouse hemolytic anemia, neonatal anemia (Nan). Blood Cells Mol. Dis. 2009; 43:141–148. [DOI] [PubMed] [Google Scholar]
- 33.Heruth D.P., Hawkins T., Logsdon D.P., Gibson M.I., Sokolovsky I.V., Nsumu N.N., Major S.L., Fegley B., Woods G.M., Lewing K.B. et al. . Mutation in erythroid specific transcription factor KLF1 causes hereditary spherocytosis in the nan hemolytic anemia mouse model. Genomics. 2010; 96:303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coghill E., Eccleston S., Fox V., Cerruti L., Brown C., Cunningham J., Jane S., Perkins A.. Erythroid kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood. 2001; 97:1861–1868. [DOI] [PubMed] [Google Scholar]
- 35.Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N. et al. . Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011; 29:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunez N., Clifton M.M.K., Funnell A.P.W., Artuz C., Hallal S., Quinlan K.G.R., Font J., Vandevenne M., Setiyaputra S., Pearson R.C.M. et al. . The multi-zinc finger protein ZNF217 contacts DNA through a two-finger domain. J. Biol. Chem. 2011; 286:38190–38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi A.M., Taylor C.W.. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011; 6:365–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandevenne M., Jacques D.A., Artuz C., Nguyen C.D., Kwan A.H.Y., Segal D.J., Matthews J.M., Crossley M., Guss J.M., Mackay J.P.. New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217. J. Biol. Chem. 2013; 288:10616–10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 40.Dunbrack R.L., Karplus M.. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 1993; 230:543–574. [DOI] [PubMed] [Google Scholar]
- 41.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A.. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003; 4:P3. [PubMed] [Google Scholar]
- 43.Hodge D., Coghill E., Keys J., Maguire T., Hartmann B., McDowall A., Weiss M., Grimmond S., Perkins A.. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006; 107:3359–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keys J.R., Tallack M.R., Hodge D.J., Cridland S.O., David R., Perkins A.C.. Genomic organisation and regulation of murine alpha haemoglobin stabilising protein by erythroid kruppel-like factor. Br. J. Haematol. 2007; 136:150–157. [DOI] [PubMed] [Google Scholar]
- 45.Tallack M.R., Keys J.R., Perkins A.C.. Erythroid Kruppel-like factor regulates the G1 cyclin dependent kinase inhibitor p18INK4c. J. Mol. Biol. 2007; 369:313–321. [DOI] [PubMed] [Google Scholar]
- 46.Siatecka M., Lohmann F., Bao S., Bieker J.J.. EKLF directly activates the p21WAF1/CIP1 gene by proximal promoter and novel intronic regulatory regions during erythroid differentiation. Mol. Cell. Biol. 2010; 30:2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tallack M.R., Keys J.R., Humbert P.O., Perkins A.C.. EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J. Biol. Chem. 2009; 284:20966–20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kowalczyk M.S., Hughes J.R., Garrick D., Lynch M.D., Sharpe J.A., Sloane-Stanley J.A., McGowan S.J., De Gobbi M., Hosseini M., Vernimmen D. et al. . Intragenic enhancers act as alternative promoters. Mol. Cell. 2012; 45:447–458. [DOI] [PubMed] [Google Scholar]
- 50.Mouse ENCODE Consortium. Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R. et al. . An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012; 13:418–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 52.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitmore L., Wallace B.A.. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004; 32:W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrade M.A., Chacón P., Merelo J.J., Morán F.. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993; 6:383–390. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y., Olanrewaju Y.O., Zheng Y., Hashimoto H., Blumenthal R.M., Zhang X., Cheng X.. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014; 42:4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Windhager L., Bonfert T., Burger K., Ruzsics Z., Krebs S., Kaufmann S., Malterer G., L'Hernault A., Schilhabel M., Schreiber S. et al. . Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 2012; 22:2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wuarin J., Schibler U.. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994; 14:7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ameur A., Zaghlool A., Halvardson J., Wetterbom A., Gyllensten U., Cavelier L., Feuk L.. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011; 18:1435–1440. [DOI] [PubMed] [Google Scholar]
- 59.Funnell A.P.W., Maloney C.A., Thompson L.J., Keys J., Tallack M.R., Perkins A.C., Crossley M.. Erythroid kruppel-like factor directly activates the basic kruppel-like factor gene in erythroid cells. Mol. Cell. Biol. 2007; 27:2777–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desgardin A.D., Abramova T., Rosanwo T.O., Kartha S., Shim E.-H., Jane S.M., Cunningham J.M.. Regulation of Delta-Aminolevulinic Acid Dehydratase by Krüppel-Like Factor 1. PLoS One. 2012; 7:e46482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tata A., Stoppel D.C., Hong S., Ben-Zvi A., Xie T., Gu C.. An image-based RNAi screen identifies SH3BP1 as a key effector of Semaphorin 3E-PlexinD1 signaling. J. Cell Biol. 2014; 205:573–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L., Prak L., Rayon-Estrada V., Thiru P., Flygare J., Lim B., Lodish H.F.. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013; 499:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soler E., Andrieu-Soler C., de Boer E., Bryne J.C., Thongjuea S., Stadhouders R., Palstra R.J., Stevens M., Kockx C., van IJcken W. et al. . The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010; 24:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arnaud L., Saison C., Helias V., Lucien N., Steschenko D., Giarratana M.-C., Prehu C., Foliguet B., Montout L., de Brevern A.G. et al. . A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am. J. Hum. Genet. 2010; 87:721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singleton B.K., Lau W., Fairweather V.S.S., Burton N.M., Wilson M.C., Parsons S.F., Richardson B.M., Trakarnsanga K., Brady R.L., Anstee D.J. et al. . Mutations in the second zinc finger of human EKLF reduce promoter affinity but give rise to benign and disease phenotypes. Blood. 2011; 118:3137–3145. [DOI] [PubMed] [Google Scholar]
- 66.Liu D., Zhang X., Yu L., Cai R., Ma X., Zheng C., Zhou Y., Liu Q., Wei X., Lin L. et al. . Erythroid kruppel-like factor mutations are relatively more common in a thalassemia endemic region and ameliorate the clinical and hematological severity of -thalassemia. Blood. 2014; 124:803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viprakasit V., Ekwattanakit S., Riolueang S., Chalaow N., Fisher C., Lower K., Kanno H., Tachavanich K., Bejrachandra S., Saipin J. et al. . Mutations in kruppel-like factor 1 cause transfusion-dependent hemolytic anemia and persistence of embryonic globin gene expression. Blood. 2014; 123:1586–1595. [DOI] [PubMed] [Google Scholar]
- 68.Kelsey L., Flenniken A.M., Qu D., Funnell A.P.W., Pearson R., Zhou Y.-Q., Voronina I., Berberovic Z., Wood G., Newbigging S. et al. . ENU-induced mutation in the DNA-binding domain of KLF3 reveals important roles for KLF3 in cardiovascular development and function in mice. PLoS Genet. 2013; 9:e1003612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrera L.A., Vedenko A., Kurland J.V., Rogers J.M., Gisselbrecht S.S., Rossin E.J., Woodard J., Mariani L., Kock K.H., Inukai S. et al. . Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016; 351:1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V. et al. . TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003; 31:374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.