Abstract
Genomic imprinting is an epigenetic regulation that leads to gene expression in a parent-of-origin specific manner. AFF3, the central component of the Super Elongation Complex-like 3 (SEC-L3), is enriched at both the intergenic-differentially methylated region (IG-DMR) and the Meg3 enhancer within the imprinted Dlk1-Dio3 locus to regulate the allele-specific gene expression in this locus. The localization of AFF3 to IG-DMR requires ZFP57. However, how AFF3 functions at the Meg3 enhancer in maintaining allele-specific gene expression remains unclear. Here, we demonstrate that AFF3 is associated with the Krüppel-like zinc finger protein ZFP281 in mouse embryonic stem (ES) cells. ZFP281 recruits AFF3 to the Meg3 enhancer within the imprinted Dlk1-Dio3 locus, thus regulating the allele-specific expression of the Meg3 polycistron. Our genome-wide analyses further identify ZFP281 as a critical factor generally associating with AFF3 at enhancers and functioning together with AFF3 in regulating the expression of a subset of genes. Our study suggests that different zinc finger proteins can recruit AFF3 to different regulatory elements and differentially regulate the function of AFF3 in a context-dependent manner.
INTRODUCTION
Genomic imprinting refers to an epigenetic regulation of gene expression in a parent-of-origin specific manner. Genes within one given imprinted locus are coordinately regulated by cis-regulatory elements, including imprinting control regions (ICRs) and enhancers, epigenetic machineries and transcriptional factors to tightly control the allele-specific expression pattern (1,2). Dysregulation of their interaction within the imprinted loci can lead to imprinted gene expression defects and numerous human congenital diseases, such as Angelman, Prader-Willi and Beckwith–Wiedemann Syndromes; and a number of cancers as well (3).
The differential methylation status of an ICR is a major factor to dictate the imprinted gene expression pattern. Within the imprinted loci, ICRs can be methylated by de novo DNA methyltransferases during gametogenesis to establish differentially methylated regions (DMRs) (4). The parent-of-origin allele specific DNA methylation acquired at ICRs is maintained after fertilization by the Krüppel-associated box (KRAB) domain-containing Cys2-His2 (C2H2) zinc finger transcription factor ZFP57, which recruits TRIM28, the H3K9 methyltransferase SETDB1 and DNA methyltransferases (DNMTs) (5,6). The removal of either H3K9 methylation or DNA methylation at ICR leads to loss of imprinting (7,8). While TRIM28 is essential for the maintenance of DNA methylation at ICRs during genome wide demethylation at early embryonic developmental stage, a recent study shows that loss of TRIM28 at later development stage after genome wide demethylation has no effect in DNA methylation at ICR (9).
Previously, we have identified the P-TEFb-containing super elongation complex (SEC) and super elongation complex-like (SEC-L2 and SEC-L3), and demonstrated their functional specificities in development and diseases (10–12). SEC plays critical roles in rapid transcriptional induction of HSP70, certain developmentally regulated genes and HIV proviral gene (10,13–15). AFF3, the scaffold protein of SEC-L3, occupies both the methylated ICRs and the enhancer regions in mouse embryonic stem (ES) cells (16). ZFP57 and/or the heterochromatin, maintained by ZFP57 recruited epigenetic regulators such as TRIM28, SETDB1 and DNMTs, are required for recruiting AFF3 at the methylated ICRs, including the intergenic-differentially methylated region (IG-DMR) within the imprinted Dlk1-Dio3 locus. The depletion of DNA methylation or H3K9me3 at IG-DMR after ZFP57 removal leads to the relocation of AFF3 to the nearby Meg3 enhancer region to activate the Meg3 polycistron expression (16). However, how AFF3 functions at the enhancer region remains unknown.
The Krüppel-like zinc finger transcription factor ZFP281 has previously been identified as a major regulator of stem cell pluripotency (17,18). In this paper, through a de novo motif search, we identified a consensus sequence of the Krüppel-like zinc finger transcription factor ZFP281 at the AFF3-occupied enhancer regions in mouse ES cells. ZFP281 functionally interacts with AFF3 at the Meg3 enhancer, but not IG-DMR, to ensure proper expression of the Meg3 polycistron at the transcriptional elongation level. Our results indicate that AFF3 could coordinate with different zinc finger proteins, e.g. ZFP57 and ZFP281, at different genomic loci to maintain allele-specific expression of imprinted genes.
MATERIALS AND METHODS
ES cell culture
Mouse embryonic stem cells (V6.5 mouse ES cell, ZFP57 wild-type mouse ES cell and ZFP57 knockout mouse ES cell) were cultured on irradiated mouse embryonic fibroblast (iMEF) feeder layers in 0.1% gelatin-coated tissue culture flask. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) (D6546, Sigma) supplemented with 15% ES-certified fetal bovine serum (Hyclone), 2 mM L-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol and recombinant LIF (Millipore). For ChIP and RNA analyses, cells were grown for one passage off feeders on tissue culture plates for 30 min.
Lentivirus mediated RNAi
Mouse ZFP281 shRNA constructs were cloned into the pLKO.1 vector (Addgene #10878). The Non-targeting shRNA construct (SHC002) was purchased from Sigma. Lentiviral particle preparation and infection were performed as previously described (19). Briefly, around 70% confluent 293T cells in 150 mm tissue culture plate were co-transfected with 8 μg of the shRNA construct or Non-targeting control shRNA, 6 μg of psPAX2 packaging plasmids and 2 μg of pMD2.G envelope plasmids using X-tremeGENE 9 (Roche). The media were replaced with fresh DMEM supplemented with 10% fetal bovine serum after 16 h of transfection. The lentiviral supernatants were collected 48 and 72 h after the transfection, filtered through 0.45 μm filters and concentrated at 18K rpm for 2 h. The V6.5 ES cells were infected with concentrated lentiviral particles with polybrene (Sigma) at the concentration of 8 μg/ml. Twenty-four hours after infection, the ES cells were subjected to selection with 2 ug/ml of puromycin for an additional 48 h.
Antibodies
Antibody for RNA Pol II was purchased from Santa Cruz (N-20). Antibody for AFF3 was described previously (16). A fragment of human ZFP281 (aa 413–573) was expressed as a His-tag fusion protein in pET-16b, purified on NTA-agarose according to Qiagen's protocol and sent to Pocono Rabbit Farm and Labs for immunization into rabbits.
Immunofluorescence staining
293T cells were rinsed with phosphate buffered saline (PBS) and fixed with 4% of paraformaldehyde for 15 min at room temperature, followed by 3 times of PBS washes. Cells were then permeabilized with 0.2% Triton X-100 for 10 min, washed with PBS, blocked with 5% bovine serum albumin in PBS with Tween 20 (PBST) for 1 h and stained with primary antibodies (anti-ZFP281, 1:1600) at 4°C overnight. After 3 times of 5-min washes with PBST, cells were then incubated with secondary antibody (anti-rabbit IgG Alexa fluor 488, 1:5000) for 1 h, followed by 3 times of 5-min washes with PBST. The cells were mounted in Vectashield mounting medium and then imaged.
Immunoprecipitations and Western blot
Mouse ES cells V6.5 were washed with cold PBS once and lysed in a high-salt lysis buffer (20 mM HEPES [pH 7.4], 10% glycerol, 350 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 mM DTT) containing proteinase inhibitors (Sigma) for 30 min at 4°C. After centrifugation at 13 400 × g for 40 min, the balance buffer (20 mM HEPES [pH 7.4], 1 mM MgCl2, 10 mM KCl) was added to the supernatant to make the final NaCl concentration 300 mM. The lysate was then incubated with antibodies and protein A beads (GE Healthcare) for 4 h at 4°C with gentle rotation. The beads were spun down and washed three times with wash buffer (10 mM HEPES [pH 7.4], 1 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100) before boiling in Sodium dodecyl sulphate loading buffer. Proteins were resolved by sodium dodecylsulphate-polyacrylamide gel electrophoresis gel and developed with Luminata Forte Western horseradish peroxidase (HRP) substrate. Primary antibodies used were AFF3 (1:2000) and ZFP281 (1:7000). HRP-conjugated secondary antibodies from Sigma were used at a dilution of 1:5000.
Bisulfite sequencing analyses
Following ChIP using ZFP281 antibody, DNA methylation of the Meg3 enhancer, was analyzed by bisulfite sequencing. Whole cell extract was also analyzed as a control. Briefly, the DNA samples were subjected to bisulfite treatment and purification using the EpiTect bisulfite kit (Qiagen). The purified DNA samples were then amplified using the primers corresponding to the Meg3 enhancer (16). The PCR products were cloned into the pGEM-T easy vector (Promega). The clones were subjected to Sanger sequencing.
Quantitative RT-PCR and total RNA-seq library preparation
Mouse ES cells V6.5 were infected with lentivirus carrying either Non-targeting shRNA or ZFP281 shRNA in the presence of 8 ug/ml of polybrene (Sigma). Twenty-four hours later, ES cells were selected with 2 ug/ml of puromycin for additional 48 h and then were grown one passage off feeders for 30 min before harvest. Total RNA was isolated with the RNeasy (Qiagen) kit, treated with DNase I (NEB) and re-purified with RNeasy. cDNAs were synthesized with High Capacity RNA-to-cDNA Kit from Applied Biosystems. The expression levels were measured with Fast SYBR Green Master mix from Thermo Fisher on StepOnePlus (Applied Biosystems). Relative expression to housekeeping genes was calculated. For RNA-seq, total RNA was depleted of rRNA using Ribozero (Illumina) before library preparation using Tru-seq (Illumina).
ChIP and ChIP-seq library preparation
A total of 5 × 107 cells were used per ChIP assay according to the previously described protocol. Briefly, cells were cross-linked with 1% paraformaldehyde for 10 min at room temperature; cross-linking was quenched by glycine. Fixed chromatin was sonicated and immunoprecipitated with specific antibody. Libraries were prepared with NEBNext sample prep kit for the further next generation sequencing.
RNA-seq analyses
Total RNA-seq reads were aligned to the mouse genome (UCSC mm9) using the Tophat aligner version 2.0.10 (20) with gene annotations from Ensembl 67, allowing only uniquely mapping reads. For track files, coverage was calculated across the genome and normalized to total reads per million (r.p.m). Cuffdiff (2.2.1) (21) were used to quantify expression and detect differential expressed genes with gene annotation GTF file downloaded from Ensembl (version GRCm38.79). Genes with fold change at least 1.5-fold and q-value < 0.05 were selected as significantly differential expressed genes.
ChIP-seq analysis
ChIP-seq reads were aligned to the mouse genome (UCSC mm9) using Bowtie version 1.0.0 (22). Only uniquely mapping reads with up to two mismatches within the entire length of the read were considered for further analysis. Reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million; RPM). External sequencing data were acquired from GEO as raw reads and aligned in the same way as internally sequenced samples. Peak detection was done using MACS v2.1.1 (23) using default parameters. The motif analysis was performed by script ‘findMotifsGenome.pl’ from Homer (v4.8) with parameters ‘mm9 -size 200 -len 8 -S 10’.
RESULTS AND DISCUSSION
Widespread presence of ZFP281 binding motif at AFF3-bound enhancers
The AFF protein family members (AFF1-4) are scaffolds for the SEC family complexes (SEC, SEC-L2 and SEC-L3), each of which controls the expression of a specific group of genes (10–12). SEC, containing AFF4, plays important roles in the rapid transcriptional induction of genes in response to external stimuli (13). AFF3 occupies both IG-DMR and the Meg3 enhancer within the imprinted Dlk1-Dio3 locus to ensure the proper expression status of Meg3 in mouse embryonic stem cells (16).
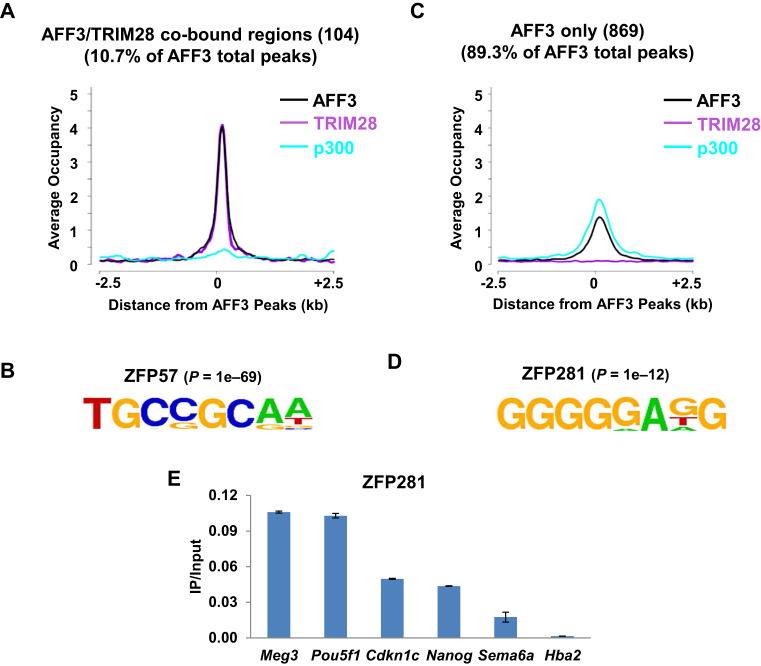
To identify factors required for the association of AFF3 with distinct chromatin regions, here we analyzed the AFF3 ChIP-seq data in mouse ES cells (16). On a genome-wide scale, 104 of AFF3 peaks, including the known ICRs, are also bound by TRIM28 but exhibit relatively low level of the transcriptional coactivator p300 (Figure 1A). We have previously showed that the KRAB domain containing ZFP57 is responsible for the recruitment of AFF3 to ICRs (16). Consistently, we found here by de novo motif search (24) that a specific DNA sequence recognized by ZFP57, 5΄- TGCCGCAA-3΄, is highly enriched in the AFF3 and TRIM28 co-bound regions (Figure 1B).
Figure 1.
ZFP281 binding motif is highly enriched at AFF3 occupied enhancer regions in mouse ES cells. (A) AFF3 and TRIM28 co-occupy 104 genomic loci, including the imprinting control regions (ICRs) of known imprinted loci. The enrichment of the enhancer mark p300 is relatively low at the 104 loci. An average occupancy plot for AFF3, TRIM28 and p300 is shown. (B) ZFP57 DNA-binding motif is the most significantly overrepresented motif among AFF3 and TRIM28 co-bound sites. Statistical significance (P-value) of the over-representation of the ZFP57 binding motif is shown. (C) p300 is relatively high-enriched at the rest of AFF3 binding sites, where the level of TRIM28 is low. An average occupancy plot for AFF3, TRIM28 and p300 is shown. (D) ZFP281 DNA-binding motif is the most significantly overrepresented motif among AFF3 and TRIM28 non-overlapped sites. Statistical significance (P-value) of the over-representation of the ZFP281 binding motif is shown. (E) ZFP281 is enriched at AFF3 bound regions. ChIP of ZFP281 at five randomly selected AFF3 bound regions. The Hba2 gene serves as a negative control for ChIP-qPCR. Error bars represent the standard deviation of two independent measurements.
Besides the presence of AFF3 in the Meg3 enhancer, we also noticed that a large portion of AFF3 peaks are not overlapping with TRIM28-occupied regions, but with high occupancy of p300 (Figure 1C). To gain insight into AFF3 function at the AFF3 and TRIM28 non-overlapping regions, including enhancers, and define how AFF3 might be recruited to these sites, a de novo motif search under these sites was performed (24). We identified the ZFP281 recognition sequence 5΄-GGGGGAGG-3΄ (25) as a highly overrepresented motif at the AFF3 and TRIM28 non-overlapping regions (Figure 1D and Supplementary Figure S1). To confirm the co-occupancy of AFF3 and ZFP281 on chromatin, we generated specific antibody against ZFP281 (Supplementary Figure S2) and interrogated AFF3 bound regions for their association with ZFP281 by performing ZFP281 ChIP assay. We detected significant occupancy of ZFP281 at the Meg3 enhancer and several other randomly selected AFF3 bound sites bearing the ZFP281 recognition sequence in ES cells (Figure 1E).
ZFP281 is required for AFF3΄s enhancer function within the Dlk1-Dio3 locus
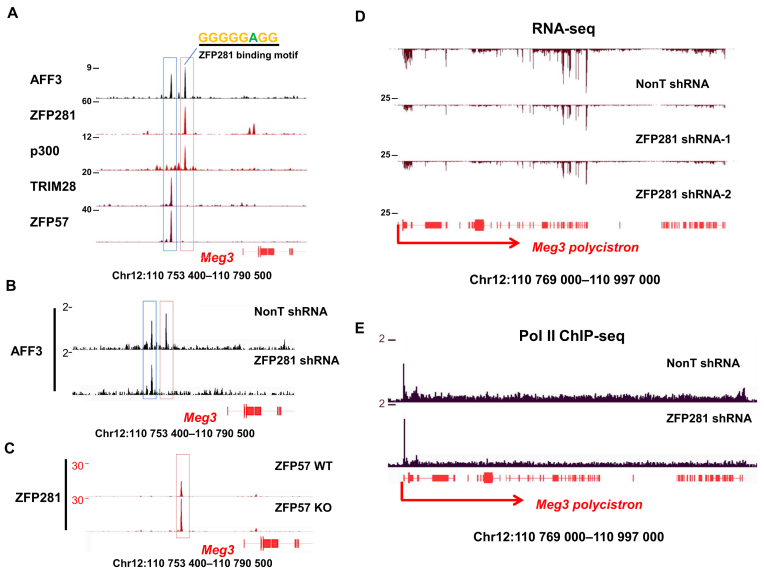
It has been demonstrated previously that AFF3 is a major regulator through its enhancer function in controlling the maternal expression of the Meg3 polycistron within the imprinted Dlk1-Dio3 locus, where the two AFF3-binding sites, located upstream the transcription start site of the Meg3 polycistron, are present. The distal IG-DMR AFF3 peak is co-bound by ZFP57 and its cofactor TRIM28 on the paternal allele (Figure 2A, blue box). AFF3 also binds an enhancer sequence located between the IG-DMR and the Meg3 promoter (Figure 2A, red box) (16). To visualize the occupancy of ZFP281 within the Dlk1-Dio3 locus, we performed ChIP-seq studies using the ZFP281 specific antibody in mouse ES cells. Only the Meg3 enhancer AFF3 peak, but not the IG-DMR peak, is co-bound by ZFP281 within this locus (Figure 2A). It has been previously demonstrated that AFF3 was exclusively recruited to the Meg3 enhancer at the unmethylated maternal allele (16). Here, we also observed the maternal allelic binding of ZFP281 to the Meg3 enhancer in wild type mouse ES cells (Supplementary Figure S3).
Figure 2.
ZFP281 is required for the localization of AFF3 to the Meg3 upstream enhancer within the imprinted Dlk1-Dio3 locus and the transcriptional elongation of the maternally expressed Meg3 polycistron. (A) AFF3 and ZFP281 are co-localized at the Meg3 upstream enhancer within the Dlk1-Dio3 locus. Shown are ChIP-seq binding profiles of AFF3, ZFP281, enhancer mark p300 and ICR marks TRIM28 and ZFP57 at the genomic region upstream of the Meg3 promoter. The intergenic-differentially methylated region (IG-DMR) is highlighted with a blue bar, and Meg3 upstream enhancer region is highlighted with a pink bar. (B) ZFP281 is required for the recruitment of AFF3 to the Meg3 upstream enhancer. Shown is ChIP-seq binding profile of AFF3 at the genomic region upstream of the Meg3 promoter in both control and ZFP281 knockdown mouse ES cells. (C) The occupancy of ZFP281 at the Meg3 upstream enhancer is increased about 2-fold in ZFP57-null cells, where the loss of heterochromatin occurs at the nearby IG-DMR. Shown is ChIP-seq binding profile of ZFP281 at the genomic region upstream of the Meg3 promoter in both ZFP57 wild-type and knockout ES cells. (D) The expression of the maternally expressed Meg3 polycistron is regulated by ZFP281. Shown are RNA-seq track files of the Meg3 polycistron in mouse ES cells bearing control or either of the two independent ZFP281 shRNAs. (E) The Meg3 polycistron is regulated by ZFP281 at the transcriptional elongation stage. ZFP281 knockdown results in reduced occupancy of Pol II in the gene body of the Meg3 polycistron in mouse ES cells. Shown are Pol II ChIP-seq track files at the Meg3 polycistron region in mouse ES cells bearing either control or ZFP281 shRNA.
The Meg3 enhancer occupied by ZFP281 contains the ZFP281 recognition sequence (Figure 2A and Supplementary Figure S2D). As ZFP281 is able to bind DNA directly, we suspect that ZFP281 might function as a recruiter for AFF3΄s localization at chromatin. To further examine whether ZFP281 is required for the localization of AFF3 to the Meg3 enhancer, we depleted ZFP281 by lentivirus-mediated shRNA in mouse ES cells and performed AFF3 ChIP-seq analyses. The results indicated that the occupancy of AFF3 at the Meg3 upstream enhancer was abolished, while AFF3΄s binding at IG-DMR remained unchanged after ZFP281 knockdown (Figure 2B), which indicate that ZFP281 is specifically required for the recruitment of AFF3 to the Meg3 upstream enhancer where ZFP281 and AFF3 are co-localized.
The removal of heterochromatic environment from the methylated IG-DMR by depleting ZFP57 leads to an almost 2-fold increased binding of AFF3 at the Meg3 upstream enhancer and bi-allelic expression of the Meg3 polycistron (16). Consistently, without ZFP57, the occupancy of ZFP281 at this region also nearly doubles (Figure 2C), further substantiating that ZFP281 is a prerequisite for AFF3΄s binding to the Meg3 upstream enhancer.
ZFP281 regulates Meg3 expression at the transcriptional level
To further investigate whether ZFP281 is required for the proper expression of the Meg3 polycistron, we performed total RNA-sequencing analysis in ZFP281-depleted mouse ES cells. Similar to the effects of the AFF3 knockdown (16), Meg3 expression was also down-regulated in ZFP281-depleted cells (Figure 2D). In addition, we also observed a significant reduction of the elongating Pol II at the Meg3 polycistron after the depletion of ZFP281 (Figure 2E). AFF3, the central factor of the elongation complex SEC-L3, regulates the expression of the Meg3 polycistron at the transcriptional elongation level (16). Thus, it is very likely that ZFP281 regulates the expression of Meg3 partially through AFF3.
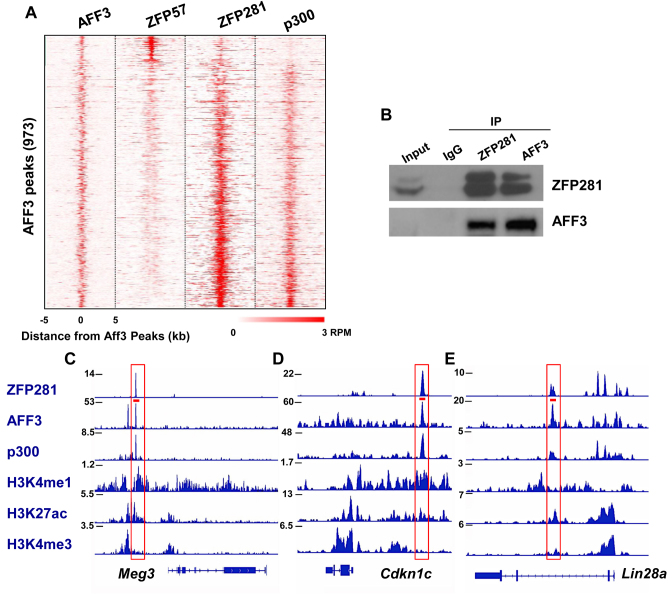
ZFP281 is generally required for the association of AFF3 with many enhancers
To explore whether ZFP281 is generally associated with AFF3 on chromatin, we analyzed the genome-wide occupancies of AFF3 and ZFP281 in mouse ES cells. In line with the motif analysis, a small subset of AFF3 bound sites are also bound by ZFP57, while the most majority of the AFF3 sites tend to be associated with ZFP281 and p300 (Figure 3A). We also examined the potential interaction of ZFP281 with AFF3 in mouse ES cells. Endogenous immunoprecipitation experiments did show an interaction between these two proteins (Figure 3B). The co-localization of AFF3 and ZFP281 is not limited to the imprinted regions, such as the Meg3 and Cdkn1c enhancers (Figure 3C and D). AFF3 and ZFP281 also co-bind the non-imprinted regions, including the Lin28a enhancer (Figure 3E).
Figure 3.
ZFP281 co-localizes with AFF3 in mouse ES cells. (A) Heat maps of binding profiles in mouse ES cell for AFF3, ZFP57, ZFP281 and p300 are shown within 5 kb of the center of AFF3 peaks. (B) Endogenous immunoprecipitations (IP) followed by Western blotting show the interaction between AFF3 and ZFP281. (C–E) Genome browser track examples for the binding profiles of ZFP281, p300 and AFF3, and histone markers H3K4me1, H3K4me3 and H3K27ac. ZFP281 and AFF3 co-bind at the enhancers of the (C) Meg3, (D) Cdkn1c and (E) Lin28a genes. The red bar indicated the primer pair amplified regions in Figure 1E.
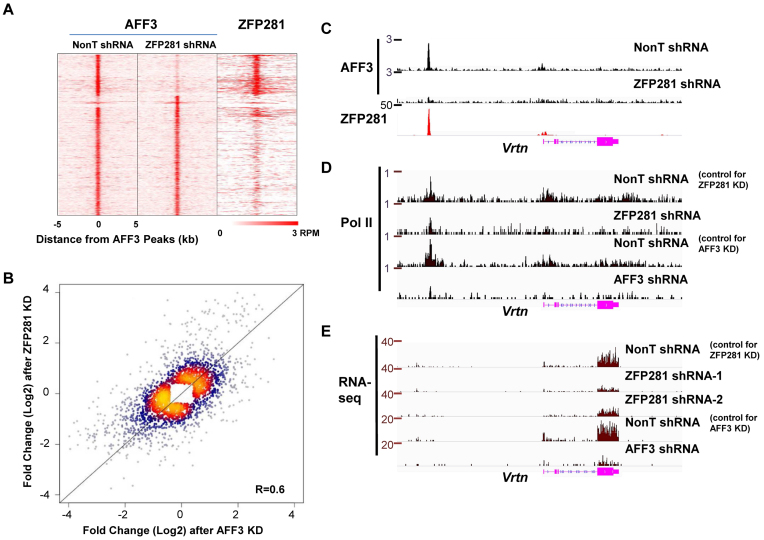
We then examined whether the presence of ZFP281 is generally required for the association of AFF3 with chromatin. The global occupancy of AFF3 was mapped in control and ZFP281-depleted ES cells, respectively. Upon depletion of ZFP281, AFF3 occupancy was reduced at a subset of ZFP281 and AFF3 co-occupied regions (Figure 4A). Since ZFP281 is largely dispensable for the self-renewal of ES cells (26), the reduction in AFF3 binding is not due to a change in cell type.
Figure 4.
ZFP281 is a major cofactor of AFF3 in regulating gene expression in mouse ES cells. (A) Heat maps of binding profiles in mouse ES cell are shown, within 5 kb of the center of AFF3 peaks, to compare the occupancy of AFF3 in control and ZFP281 knockdown ES cells. The binding profile of ZFP281 is also shown. The loss of AFF3 occupancy occurs at the regions where ZFP281 and AFF3 are co-localized. (B) Scatter plot demonstrating differentially expressed genes, assessed by RNA-seq, in ZFP281 knockdown versus AFF3 knockdown. Correlation coefficient is shown. (C) ZFP281 is required for the recruitment of AFF3 to region upstream of the Vrtn gene. Shown is ChIP-seq binding profile of AFF3 at the Vrtn gene locus in both control and ZFP281 knockdown mouse ES cells. (D) Pol II occupancies at the Vrtn gene locus is reduced after either ZFP281 or AFF3 depletion. Shown are Pol II ChIP-seq track files at the Vrtn gene locus in mouse ES cells bearing control, ZFP281 or AFF3 shRNA. (E) The expression of the Vrtn gene is regulated by both ZFP281 and AFF3. Shown are RNA-seq track files of the Vrtn gene in mouse ES cells bearing control, ZFP281 or AFF3 shRNA. RNA-seq and Pol II ChIP-seq after AFF3 knockdown were downloaded from Luo 2016.
To further investigate the functional link between ZFP281 and AFF3, we analyzed differential gene expression using RNA-seq data after ZFP281 and AFF3 knockdown. The correlation in gene expression change between the ZFP281 knockdown and the AFF3 knockdown is about 0.6 (Figure 4B), suggesting that these two factors regulate similar transcription program in mouse ES cells. The functional gene ontology term analysis indicated that molecular pathways including positive regulation of mesenchymal cell proliferation, cell adhesion, angiogenesis, cartilage development, lens development in camera-type eye are significantly enriched at those down-regulated genes after the depletion of AFF3 and ZFP281 (Supplementary Table S1). Many of these down-regulated genes are co-occupied by AFF3 and ZFP281 (Supplementary Table S2). For examples, both AFF3 and ZFP281 co-occupy the upstream intergenic regions of the Vrtn, Lin28a and Cdkn1c genes (Figures 3D, E and 4C). The recruitment of AFF3 to these regions is dependent on ZFP281 (Figure 4C and Supplementary Figure S4A, S4B and S4C). Depletion of either AFF3 or ZFP281 leads to their down-regulation, as reflected by both Pol II occupancy and RNA levels are reduced after ZFP281 and AFF3 knockdown (Figure 4D and E and Supplementary Figures S4A, S4B and S4D). Thus, these data reveal that a large part of gene expression regulation attributed to AFF3 in ES cells is dependent on ZFP281.
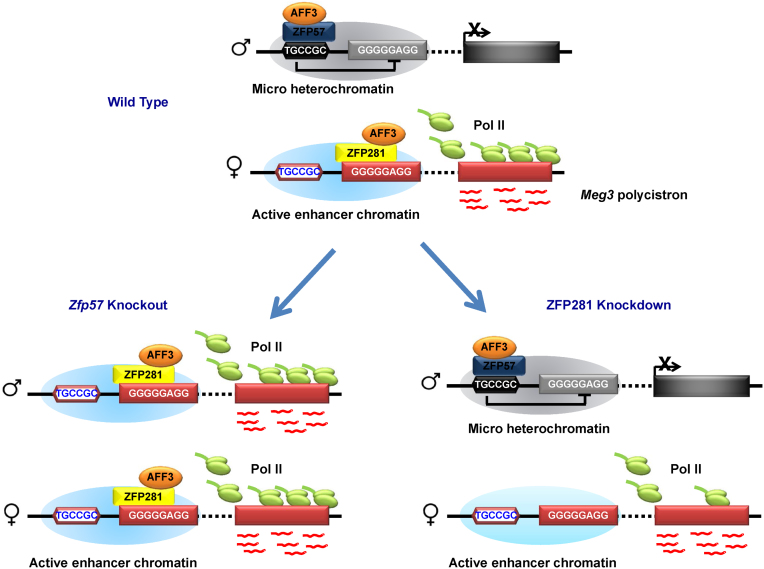
In summary, we have shown that AFF3 binds to both silent and active chromatin regions within the imprinted Dlk1-Dio3 locus through associating with different zinc finger proteins to fine-tune the expression of imprinted genes. Our work represents a significant advance in understanding how AFF3 functions in regulating the dosage-critical, allele-specific expression of imprinted genes. In the imprinted allele, the KRAB domain containing zinc finger protein ZFP57 and the heterochromatic environment that ZFP57 entails mediate the recruitment of AFF3 to the methylated ICR, at which the gene activating function of AFF3 is restricted (Figure 5). The kinase module of SEC-L3, CDK9, is not co-localized with AFF3 at the ZFP57-bound methylated DMRs. It is possible that the formation of SEC-L3 is inhibited by ZFP57 and its related heterochromatin environment. The exact function of AFF3 at methylated DMRs needs to be further investigated. In the active allele, while, AFF3 is recruited by ZFP281 to the active Meg3 enhancer, promoting transcriptional elongation of the downstream Meg3 polycistron (Figure 5).
Figure 5.
Cartoon model illustrates the functional mode of ZFP281 and AFF3 in regulating imprinted gene expression. AFF3 is localized to the methylated IG-DMR on paternal allele and the unmethylated Meg3 upstream enhancer on maternal allele, respectively. The recruitment of AFF3 to methylated IG-DMR at the paternal allele is mediated by ZFP57 and/or the heterochromatic environment that ZFP57 maintains. ZFP281 is required for the loading of AFF3 to the Meg3 upstream enhancer of the maternal allele. When ZFP281 is depleted by shRNA mediated RNAi, the recruitment of AFF3 to the Meg3 upstream enhancer is affected and the expression of the Meg3 polycistron is down-regulated. In the condition of ZFP57 knockout, where heterochromatin is lost at the IG-DMR on the paternal allele, ZFP281 is able to bind to corresponding Meg3 enhancer on the paternal allele and recruit AFF3 to activate the expression of the downstream Meg3 polycistron, leading the bi-allelic expression of the Meg3 polycistron.
The ZFP281-dependent AFF3 regulation is not limited to the imprinted loci. Many developmentally regulated genes, including genes involved in mesoderm and ectoderm development (Supplementary Table S1), are also co-regulated by ZFP281 and AFF3. These findings might provide a possible explanation as to why the depletion of ZFP281 or AFF3 has no obvious effect on the stem cell self-renewal, but impairs their differentiation potentials. It has been reported that ZFP281 can directly bind to the GC-rich DNA sequences (27). ZFP281 can also associate with c-MYC, subunits of nucleosome remodeling and histone deacetylation (NURD) complex, and also the DNA demethylase TET1 in modulating local chromatin environment, thus regulating gene expression and involved in the maintenance of ES cell pluripotency (25,28–29). Thus, it is possible that ZFP281 could facilitate the recruitment of AFF3 to enhancer regions through altering the accessibility of the local chromatin.
The SEC family members play versatile roles in transcription through regulating transcriptional elongation during development and disease pathogenesis (12–13,30–31). However, whether DNA-binding factors are required to set a permissive binding state for SEC family members to active gene expression is still undefined. Here, we report that different zinc finger proteins control AFF3΄s association with distinct chromatin states, thus maintaining proper gene activities. The role of ZFP281 in recruiting AFF3 to its chromatin targets provides a novel mechanism for the regulation of transcriptional elongation by DNA binding factor. In addition, we have previously reported the target gene specificity of SEC family members (11). It is likely that different functions of SEC members could be mediated by their differential recruitment to different genomic loci by different transcription factors, which could also explain the linkage between different SEC family members and different human diseases.
ACCESSION NUMBERS
ChIP-seq and RNA-seq data have been deposited at GEO under the accession number GSE77115. Other data sets come from previously published studies. RNA-seq data after AFF3 depletion in mouse ES cells are from GEO accession number GSE64489 (16); p300 ChIP-seq data in mouse ES cells are from GEO accession number GSE24164 (32); TRIM28 ChIP-seq data in mouse ES cells are from GEO accession number GSE41903 (33); ZFP57 ChIP-seq data in mouse ES cells are from GEO accession number GSE55382 (34).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Prof. Xiajun Li for providing ZFP57 wild type and knockout cell lines for this study. The authors thank Toby Wai Kiat Chin, Joel Celio Francisco, and Zhiqun Tang for the technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Thousand Young Talents Plan of China [to C.L.]; National Natural Science Foundation of China [31671343 to C.L.]; Natural Science Foundation of Jiangsu Province of China [BK20160026 to C. L. and BK20160666 to Z. L.]; Fundamental Research Funds for the Central Universities [2242015K42024 to C.L. and 20155101 to W.X.]. Funding for open access charge: Thousand Young Talents Plan of China [to C.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartolomei M.S., Ferguson-Smith A.C.. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011; 3:a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow D.P., Bartolomei M.S.. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014; 6:a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 2014; 15:517–530. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.T., Bartolomei M.S.. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013; 152:1308–1323. [DOI] [PubMed] [Google Scholar]
- 5.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A. et al. . In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011; 44:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A.C.. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008; 15:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung D., Du T., Wagner U., Xie W., Lee A.Y., Goyal P., Li Y., Szulwach K.E., Jin P., Lorincz M.C. et al. . Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6690–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H.. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004; 429:900–903. [DOI] [PubMed] [Google Scholar]
- 9.Alexander K.A., Wang X., Shibata M., Clark A.G., Garcia-Garcia M.J.. TRIM28 Controls Genomic Imprinting through Distinct Mechanisms during and after Early Genome-wide Reprogramming. Cell Rep. 2015; 13:1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C., Smith E.R., Takahashi H., Lai K.C., Martin-Brown S., Florens L., Washburn M.P., Conaway J.W., Conaway R.C., Shilatifard A.. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010; 37:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z., Lin C., Guest E., Garrett A.S., Mohaghegh N., Swanson S., Marshall S., Florens L., Washburn M.P., Shilatifard A.. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol. Cell. Biol. 2012; 32:2608–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Z., Lin C., Shilatifard A.. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012; 13:543–547. [DOI] [PubMed] [Google Scholar]
- 13.Lin C., Garrett A.S., De Kumar B., Smith E.R., Gogol M., Seidel C., Krumlauf R., Shilatifard A.. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011; 25:1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobhian B., Laguette N., Yatim A., Nakamura M., Levy Y., Kiernan R., Benkirane M.. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010; 38:439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N.J., Alber T., Zhou Q.. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010; 38:428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Z., Lin C., Woodfin A.R., Bartom E.T., Gao X., Smith E.R., Shilatifard A.. Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 2016; 30:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidalgo M., Shekar P.C., Ang Y.S., Fujiwara Y., Orkin S.H., Wang J.. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011; 29:1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z.X., Teh C.H., Chan C.M., Chu C., Rossbach M., Kunarso G., Allapitchay T.B., Wong K.Y., Stanton L.W.. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells. 2008; 26:2791–2799. [DOI] [PubMed] [Google Scholar]
- 19.Lin C., Garruss A.S., Luo Z., Guo F., Shilatifard A.. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013; 152:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fidalgo M., Huang X., Guallar D., Sanchez-Priego C., Valdes V.J., Saunders A., Ding J., Wu W.S., Clavel C., Wang J.. Zfp281 Coordinates Opposing Functions of Tet1 and Tet2 in Pluripotent States. Cell Stem Cell. 2016; 19:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn S., Hermeking H.. ZNF281/ZBP-99: a new player in epithelial-mesenchymal transition, stemness, and cancer. J. Mol. Med. (Berl). 2014; 92:571–581. [DOI] [PubMed] [Google Scholar]
- 27.Lisowsky T., Polosa P.L., Sagliano A., Roberti M., Gadaleta M.N., Cantatore P.. Identification of human GC-box-binding zinc finger protein, a new Kruppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 1999; 453:369–374. [DOI] [PubMed] [Google Scholar]
- 28.Fidalgo M., Faiola F., Pereira C.F., Ding J., Saunders A., Gingold J., Schaniel C., Lemischka I.R., Silva J.C., Wang J.. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16202–16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch H.B., Zhang R., Verdoodt B., Bailey A., Zhang C.D., Yates J.R. 3rd, Menssen A., Hermeking H.. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007; 6:205–217. [DOI] [PubMed] [Google Scholar]
- 30.Luo Z., Lin C.. Enhancer, epigenetics, and human disease. Curr. Opin. Genet. Dev. 2016; 36:27–33. [DOI] [PubMed] [Google Scholar]
- 31.Izumi K., Nakato R., Zhang Z., Edmondson A.C., Noon S., Dulik M.C., Rajagopalan R., Venditti C.P., Gripp K., Samanich J. et al. . Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 2015; 47:338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. . Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe H.M., Kapopoulou A., Corsinotti A., Fasching L., Macfarlan T.S., Tarabay Y., Viville S., Jakobsson J., Pfaff S.L., Trono D.. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013; 23:452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strogantsev R., Krueger F., Yamazawa K., Shi H., Gould P., Goldman-Roberts M., McEwen K., Sun B., Pedersen R., Ferguson-Smith A.C.. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015; 16:112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.