Abstract
The CST complex is a phylogenetically conserved protein complex consisting of CTC1/Cdc13, Stn1 and Ten1 that protects telomeres on linear chromosomes. Deletion of the fission yeast homologs stn1 and ten1 results in complete telomere loss; however, the precise function of Stn1 is still largely unknown. Here, we have isolated a high-temperature sensitive stn1 allele (termed stn1-1). stn1-1 cells abruptly lost telomeric sequence almost completely at the restrictive temperature. The loss of chromosomal DNA happened without gradual telomere shortening, and extended to 30 kb from the ends of chromosomes. We found transient and modest single-stranded G-strand exposure, but did not find any evidence of checkpoint activation in stn1-1 at the restrictive temperature. When we probed neutral-neutral 2D gels for subtelomere regions, we found no Y-arc-shaped replication intermediates in cycling cells. We conclude that the loss of telomere and subtelomere DNAs in stn1-1 cells at the restrictive temperature is caused by very frequent replication fork collapses specifically in subtelomere regions. Furthermore, we identified two independent suppressor mutants of the high-temperature sensitivity of stn1-1: a multi-copy form of pmt3 and a deletion of rif1. Collectively, we propose that fission yeast Stn1 primarily safeguards the semi-conservative DNA replication at telomeres and subtelomeres.
INTRODUCTION
Telomeres at the ends of linear chromosomes are essential for maintenance of chromosomal stability. The native DNA ends need to be distinguished from deleterious DNA breaks, thereby preventing gratuitous activation of the DNA damage checkpoint. Telomeres consist of tandem repeats of short DNA composed of G-rich and complementary C-rich sequences called the G-strand and C-strand, respectively. It has been shown that telomeres are intrinsically difficult to replicate, and can thus be considered a special kind of fragile site (1,2). Because defective replication at telomeres and subtelomeres (regions adjacent to the telomeric repetitive DNA sequences) leads to chromosomal anomalies, telomeres are equipped with both general and specific mechanisms that facilitate their replication (1).
The six-component protein complex, shelterin, is conserved in a wide range of eukaryotic species, including fission yeast and mammals. It is known that a member of shelterin, TRF1 in mammals and Taz1 in fission yeast, is required for efficient replication (2,3). The CST complex is another highly conserved protein complex in budding yeast, fission yeast, plants, and humans. The prototypic CST complex in budding yeast consists of Cdc13, Stn1, and Ten1 (4–6). The three members possess OB-fold (oligonucleotide/oligosaccharide-binding fold) domains, which provide CST with ssDNA-binding activities (7). Budding yeast cells deleted for Cdc13, Stn1 or Ten1 are inviable, and high-temperature sensitive alleles (cdc13-1, stn1-13 and ten1-31) showed extended exposure of single stranded G-strands at the restrictive conditions, leading to a cell cycle arrest at G2/M phase (4–6). It is widely believed that the primary function of budding yeast CST is the protection of telomeres. In this report, hereafter, we define the term ‘telomere protection’ as protecting telomere DNA from exonucleolytic attack, aberrant DNA repair events such as end-to-end fusions, and DNA damage checkpoint activation. In addition, there are multiple lines of evidence suggesting that CST promotes the C-strand fill-in reaction subsequent to G-strand synthesis by either replicative leading strand synthesis or telomerase. Moreover, it is known that budding yeast Stn1 and Cdc13 associate with the B-subunit and the catalytic subunit of DNA polymerase α-primase, respectively (8,9). In vertebrates and plants, the CST complex consists of CTC1, STN1 and TEN1 (10,11). Homology between CTC1 and Cdc13 is limited, albeit both possess OB-fold domains (10). CST partly localizes at telomeres, and knockdown or knockout of any of the subunits results in abnormal telomeres, such as fragile telomeres, an increase in telomere loss, and extended G-tails. These observations are consistent with the notion that CST is involved in the replication of telomere duplex, including the C-strand fill-in step. (10–16). It was also reported that human CST regulates telomerase activity by limiting telomerase processivity and, thereby, telomere length (17). CST plays a role in both telomeric and non-telomeric regions to facilitate restart of arrested DNA replication forks (12).
In fission yeast, although stn1 and ten1 homologs were reported, a Cdc13/CTC1 counterpart has not yet been identified. Whereas most stn1- or ten1-deleted cells exhibit cell cycle arrest and cell elongation, a minor population loses telomeres completely and survives by self-circularizing all three chromosomes (18). However, the precise function of Stn1-Ten1 in telomere maintenance is largely unknown. Recent reports showed that SUMOylation of Tpz1, one of the shelterin components in fission yeast, permits the recruitment of Stn1-Ten1 to the telomeres, and the complex represses telomerase activity to limit telomere length (19,20).
Here, we have isolated a temperature-sensitive stn1 allele in fission yeast (termed stn1-1) to analyze the immediate consequences of Stn1 dysfunction. We found that stn1-1 at the restrictive temperature lost telomeric sequence completely without gradual shortening. The erosion of chromosome ends was not restricted to the telomeres but extended to subtelomeres. We also found evidence of frequent semi-conservative DNA replication fork collapse at subtelomeres that likely lead to DNA double-strand breaks (DSBs) and telomere loss. Furthermore, we screened for genes that suppress stn1-1 temperature sensitivity and identified several stn1-related genes that suppressed the severe telomere loss.
MATERIALS AND METHODS
Strains and media
Schizosaccharomyces pombe strains used in this study are listed in Supplemental Table S1. Basic manipulations and MM media composition are described elsewhere (21). Hydroxyurea (Sigma-Aldrich) was used at 40 mM in YES (Bacto™ Yeast extract (BD) 0.5%, glucose 3%, leucine 200 mg/l, uracil 100 mg/l, adenine 200 mg/l and histidine 200 mg/l) and yeast were incubated for 4 h to elicit Cds1 activation. Bleomycin hydrochloride (Wako) was used at 5 μg/ml in YES and incubated 1 h to induce Chk1 activation. (S)-(+)-Camptothecin (Sigma-Aldrich) was used at 500 nM in SD (Difco™ Yeast Nitrogen Base w/o Amino Acids (BD) 0.67%, glucose 1% and appropriate nutrients) plates to exclude self-circularized chromosome survivors in the screen for stn1-1 suppressor strains.
Random mutagenesis in stn1
Ustn1-stn1-3xflag-LEU2-Dstn1 (where Ustn1 or Dstn1 means approximately 500 nt upstream or downstream region of stn1 ORF, respectively) inserted in the pUC119 plasmid was randomly mutagenized by error-prone PCR with the following conditions: 1xEX Taq buffer, 2 mM MgCl2, 480 μM each dNTP, 2 μM each primer, 25 units EX Taq (TaKaRa) (22). The cycling conditions were: 95°C 5 min, [95°C 30 s, 52°C 1 min, 72°C 6 min] 30 times, 72°C 2 min. Primers used for the error-prone PCR were 5΄-ccggagctcttagaattatccggtatatgaag-3΄ and 5΄-ccggcatgcggcaatcggatgccttaatc-3΄.
Isolation of stn1-1
Wild-type strain (JK317) was transformed with the randomly mutagenized stn1 DNA. The transformed cells were plated onto SD-leucine medium and incubated at 25°C for 1 day and replicated onto the same medium. The original plate was incubated at 25°C and the replicated one was incubated at 37°C. Colonies that appeared at 25°C but not at 37°C were isolated.
Southern hybridization
The extracted genomic DNAs were digested with EcoRI to detect telomeres, telo-5k, telo-9k, telo-15k, telo-33k, control region (genomic location 2232548–2233109 chromosome II, http://www.ncbi.nlm.nih.gov/nuccore/CU329671.1), and his1 (genomic location 4314109–4314600 chromosome I, http://www.ncbi.nlm.nih.gov/nuccore/CU329670.1) fragments. EcoRI-digests were separated by electrophoresis and transferred to Hybond-XL membranes (GE healthcare). The membranes were hybridized with terminally 32P-labeled C-rich oligonucleotide (5΄-tgtaaccgtacccctgtaaccccctgtaacc-3΄) or G-rich oligonucleotide (5΄-ggttacagggggttacaggggtacggttaca-3΄) to detect G-strand or C-strand, respectively. To detect other regions, the membrane was hybridized with internally 32P-labeled PCR products amplified with 5΄-acactcaattcaaatcaacttc-3΄ and 5΄-gtgtttgaaaattgagcttatg-3΄ for telo-5k, 5΄-aaagcatcatttattacttttgc-3΄ and 5΄-atgatcgcttttgaagacatg-3΄ for telo-9k, 5΄-caactggtttacaatggtatc-3΄ and 5΄-aaaattggaaaattgtatacgtc-3΄ for telo-15k, 5΄-gccaatacagaagtaggatg-3΄ and 5΄-ccgagaataacgtctaacag-3΄ for telo-17k, 5΄-aacgtggcacaatttcatttc-3΄ and 5΄-gaacatgactcagctgatgc-3΄ for telo-33k, 5΄-gaggaattcagtggaattctc-3΄ and 5΄-ataactatgaagaatcggcttc-3΄ for control region, 5΄-tcactagttagatgacgagg-3΄ and 5΄-tttcacatctggtcgtactc-3΄ for rDNA, and 5΄-gctcccatacctcaacaagc-3΄ and 5΄-gggtatcatacttggaacagg-3΄ for his1, respectively. Probes were hybridized in hybridization buffer (7% SDS, 500 mM pH7.2 Na phosphate, and 1 mM EDTA) at appropriate temperatures (42°C for G- and C-strand probes and 55–65°C for other probes) and membranes were washed with wash buffer (1% SDS, 40 mM pH7.2 Na phosphate and 1 mM EDTA). The membranes were exposed to imaging plates (FUJIFILM) and signals were detected with Typhoon 9400 or Typhoon FLA 7000 (GE healthcare). In-gel hybridization and signal quantification were performed as described in (23). For in-gel hybridization we used 10 times the amount of gDNA than in Figure 1E because the fission yeast G-tail is known to be very short and difficult to detect.
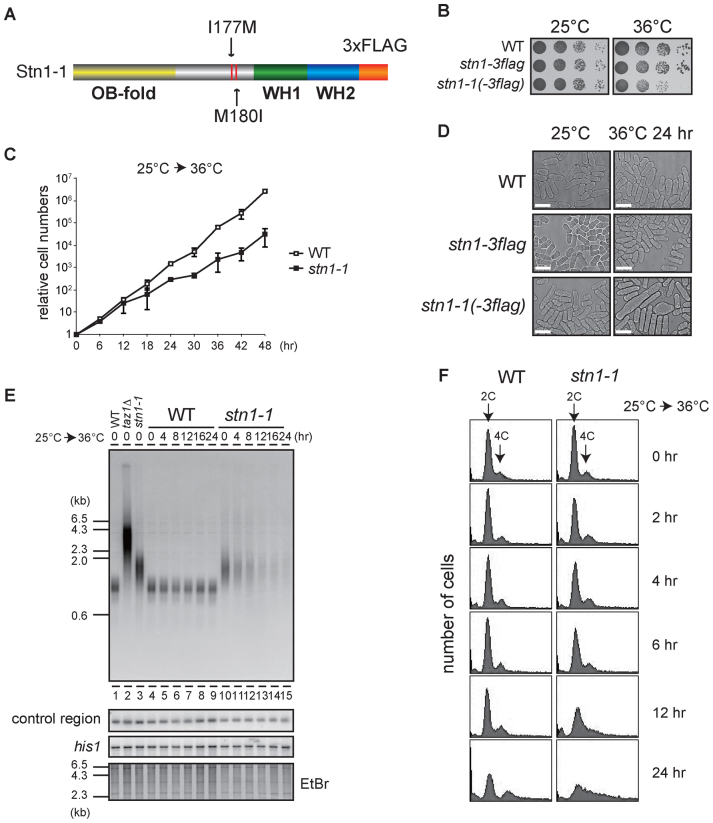
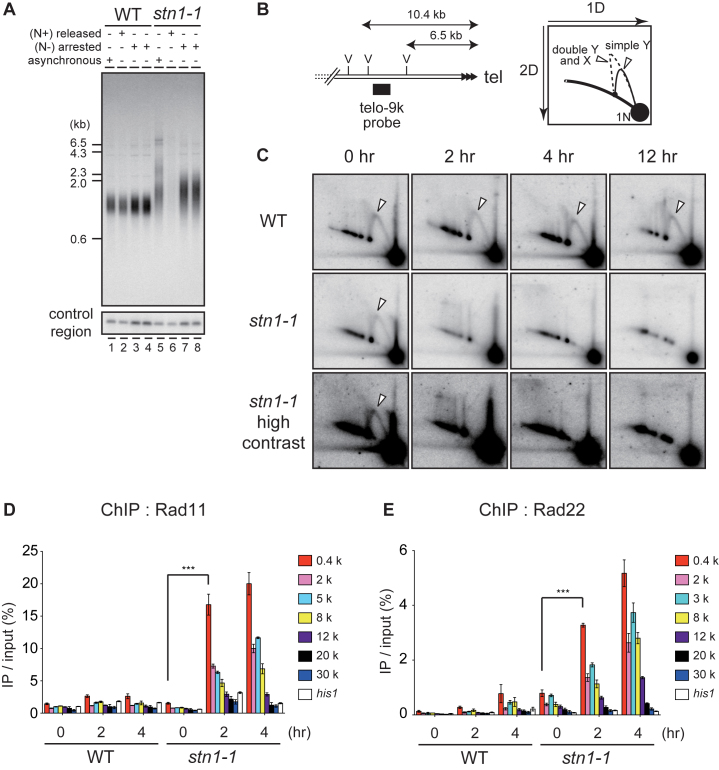
Figure 1.
Loss of telomeres at high temperature in temperature-sensitive stn1-1 mutant yeast. (A) The domain structure of Stn1-1. Positions of I177M and M180I substitutions are indicated. (B) Ten-fold serial dilutions were spotted onto YES plates and incubated at 25°C and 36°C for 3 days. (C) Growth curves of wild-type and stn1-1. Wild-type (JK317) and stn1-1 were cultured at 25°C in liquid YES medium and the temperature was shifted to 36°C for 48 h. Cell numbers were counted every 6 h. (D) Micrographs of WT, stn1-3flag, and stn1-1 obtained with a phase-contrast optical microscope (DeltaVision Elite). The scale bar represents 10 μm. (E) Wild-type, taz1Δ, and stn1-1 were cultured at 25°C (lanes 1–3). Wild-type and stn1-1 were cultured at 36°C for the indicated times (lanes 4–15). EcoRI-digested genomic DNA fragments were analyzed by Southern hybridization with a telomeric probe to detect both G- and C-strand telomere DNAs. The same membrane was re-hybridized with a probe specific to an interior region of chromosome II (control region) and the his1 gene locus (see Materials and Methods). The EtBr-stained gel was photographed before blotting. (F) Wild-type and stn1-1 cultured in YES at 36°C for the indicated times were stained with Propidium Iodide (PI) and analyzed with FACS.
Quantitative PCR (qPCR) at subtelomeric regions
Telomeric and subtelomeric sequences were derived from pNSU70 (telomere) and pT2R1 (right arm of chromosome II). The sequence files are archived in Pombase (http://ebi.edu.au/ftp/databases/pombase/pombe/Archived_directories/Cosmid_sequences/). The extracted genomic DNAs were quantified with a StepOnePlus™ Real-Time PCR System (Applied Biosystems) using the following primers:
5΄-attaattgggtaacggagtaacaatataga-3΄ and 5΄-ctatttctttattcaacttaccgcacttc-3΄ for telo-0.4k; 5΄-acgattactcgccttacgtc-3΄ and 5΄-aattcaaagttcacttagtcag-3΄ for telo-3k; 5΄-acactcaattcaaatcaacttc-3΄ and 5΄-ggatgtagtgtgcatgagtg-3΄ for telo-5k; 5΄-cgaattcaagtattccagcttc-3΄ and 5΄-aacatgatggatgaagcgaatg-3΄ for telo-8k; 5΄-agtagacgtttctcgtaaaac-3΄ and 5΄-agatggttgccagacaaacg-3΄ for telo-12k; 5΄-gtgcctgtagtttcgaaatc-3΄ and 5΄-aaaggccacctaccaatg-3΄ for telo-13k; 5΄-tctcgtcacatcgtttttgc-3΄ and 5΄-tcagggttccattctcgttc-3΄ for telo-20k; 5΄-attcgtcattttataaaccaaac-3΄ and 5΄-cgttataaatgttgtctctcc-3΄ for telo-25k; 5΄-gatgaatgaaatgacaaggtatcg-3΄ and 5΄-tcgtcgtcctattcaaacga-3΄ for telo-30k; 5΄-tcggaagtaccatcggtttc-3΄ and 5΄-atagccctggtgcagatcac-3΄ for telo-40k; 5΄-tagttgttatcctgatggtag-3΄ and 5΄-caataggcccccaaatttcac-3΄ for telo-50k; 5΄-tgtccacctcggaatcactg-3΄ and 5΄-cgaagacgtgcttcagcga-3΄ for his1.
Alkaline, neutral-neutral 2D, and Pulsed-field gel electrophoresis
Alkaline gel electrophoresis and neutral-neutral 2D gel electrophoresis were performed mainly as described in (23) with minor modifications. DNA was separated in a 1% agarose gel in a solution containing 50 mM NaOH and 1mM EDTA for alkaline gel electrophoresis. Agarose-stiffened cell-plugs were prepared for neutral-neutral 2D gel electrophoresis with the same method as pulsed-field gel electrophoresis. Pulsed-field gel electrophoresis was performed essentially as described in (24).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed essentially as described in (25). Sequences of primers used in the ChIP assays were mentioned above, except for telo-2k; 5΄-caattaattatgactgagtgaac-3΄ and 5΄-ggtgcagtaagtagaataaagg-3΄.
Monitoring aberrant mitosis
Cells were stained with Hoechst 33342 and images were captured and analyzed with DeltaVision Elite (GE Healthcare). We counted anaphase to telophase mitotic cells on the basis of the following criteria: two nuclei in one cell; the distance between the separating chromatids was more than 5 μm; and apparent septation was not observed.
Immunoprecipitations and immunoblotting
Immunoprecipitations were performed mainly as described in (25) with minor modifications. Anti-HA 16B12 was used to immunoprecipitate HA-tagged Chk1 and Cds1 and immunoblotting was performed with anti-HA Y11 (Santa Cruz) in Figure 5. Anti-Flag M2-agarose resin (Sigma-Aldrich) was used to immunoprecipitate Flag-tagged Stn1 and immunoblotting was performed with anti-Flag F7425 (Sigma-Aldrich) and anti-HA 16B12 (BAbCO) in Supplementary Figure S2.
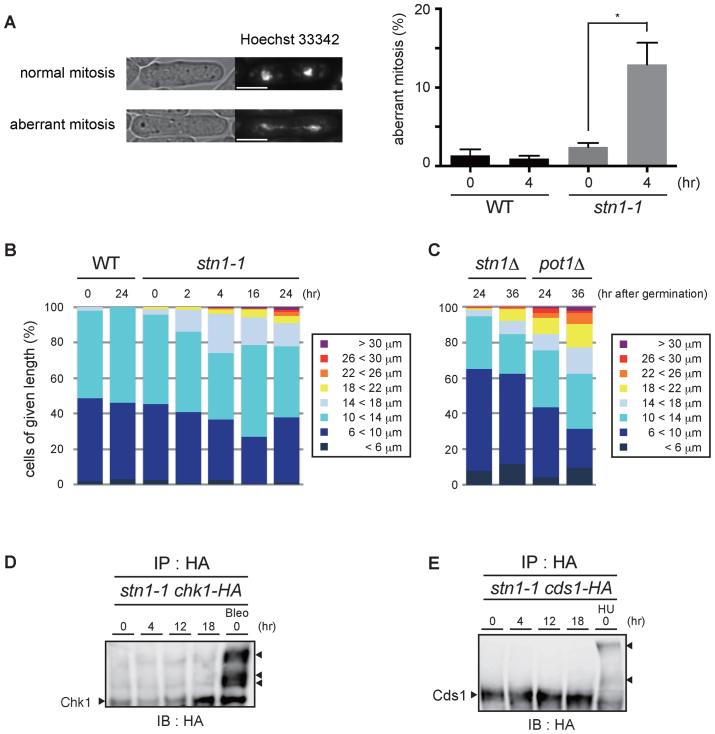
Figure 5.
stn1-1 cells show aberrant mitoses and chromosomal bridges, but the DNA damage checkpoint is not activated at the restrictive temperature. (A) Representative images of normal and aberrant mitotic cells (left). The white scale bar represents 5 μm. The bar graph represents the frequencies of aberrant mitotic cells among total mitotic cells that were scored (right). Error bars show mean values of three independent experiments with SD. * P value was 0.020 calculated with a two-tailed Student's t-test. More than 180 mitotic cells were counted in all experiments. (B) WT and stn1-1 cells were grouped into eight classes according to cell length. More than 200 cells were counted in all conditions. (C) stn1Δ and pot1Δ cells at 24 and 36 h after germination were grouped into eight classes. More than 200 cells were counted in all conditions. (D) C-terminally HA-tagged Chk1-expressing stn1-1 cells were cultured at 36°C for the indicated times. The positive control (Bleo) was treated with bleomycin at 25°C. All lysates were immunoprecipitated with anti-HA antibody, resolved by Phos-tag™ PAGE, and immunoblotted using anti-HA antibody. The right arrowhead indicates activated Chk1. (E) C-terminally HA-tagged Cds1-expressing stn1-1 cells were cultured at 36°C for the indicated times. The positive control (HU) was treated with hydroxyurea at 25°C. All lysates were immunoprecipitated with anti-HA antibody, resolved by Phos-tag™ PAGE, and immunoblotted using anti-HA antibody. The right arrowheads indicate band-shifted Cds1.
Phos-tag™ PAGE
Phos-tag™ PAGE (Wako) was performed according to the manufacturer's instructions with minor modifications. The separating gel consisted of 6% acrylamide, 0.2% N,N’-methylenebisacrylamide, 357 mM pH 6.8 Bis–Tris–HCl, 50 μM Phos-tag™, 100 μM ZnCl2, 0.67% Ammonium Peroxodisulfate, 0.13% N,N,N’,N’-tetramethylethylenediamine.
Isolation of stn1-1 suppressors
stn1-1 was transformed with an S. pombe genomic DNA library (26,27). After the transformed cells were plated on SD medium containing 500 nM camptothecin and incubated for 1 day at 25°C, the cultivation temperature was shifted to 36°C for 2∼3 days.
Scoring cell length after germination
Spores were prepared essentially as described in (28) with minor modifications. Spores were germinated in YES media containing 10 μg/ml G418 disulfate (Nacalai Tesque). We have confirmed that yeast cells lacking the kanMX6 gene do not proliferate in the presence of 10 μg/ml G418 (data not shown). Cell lengths were measured on images captured with DeltaVision Elite (GE Healthcare).
RESULTS
Isolation of a temperature-sensitive stn1 mutant in Schizosaccharomyces pombe
We isolated several high-temperature sensitive mutants by transforming the wild-type fission yeast strain (JK317) with randomly mutated stn1 DNAs generated by error-prone PCR. One isolated strain grew as vigorously as wild-type at 25°C and showed severe growth defects when it was cultivated at 37°C. We found three amino acid substitutions (I177M, M180I and V249A) in the strain. To pinpoint the substitution(s) responsible for temperature sensitivity, we generated stn1 mutants harboring various combinations of the three amino acid substitutions (I177M; M180I; V249A; I177M M180I; I177M V249A; M180I V249A; and I177M M180I V249A). We found that I177M M180I and I177M M180I V249A mutants were sensitive to high-temperature condition (37°C), but the others were not (Supplementary Figure S1A). We therefore concluded that the combined I177M M180I mutations of Stn1 are sufficient to confer high-temperature sensitivity to cells, and named the strain stn1-1 (Figure 1A). While stn1-1 grew normally at the permissive temperature (25°C), the strain showed low viability, retarded growth rates, and aberrant cell morphologies at the restrictive temperature (36°C or higher) (Figure 1B–D). To test whether stn1-1 is recessive or dominant, we generated a stn1+/stn1-1 diploid strain and conducted spot assays (Supplementary Figure S1B). Because the diploid strain grew as well as wild-type cells at 36°C, we concluded that stn1-1 is a recessive mutant.
Stn1 protein is composed of an N-terminal OB-fold domain and two putative human Rpa2-like C-terminal winged helix-turn-helix (WH) motifs (29). The two amino acid substitutions in Stn1-1 were located within the intervening region connecting the two types of domains (Figure 1A). It has been shown that Schizosaccharomyces pombe Stn1 associates with Ten1 through Stn1's OB-fold domain (29), suggesting that the interaction between Stn1-1 and Ten1 is not disrupted in stn1-1. Indeed, immunoprecipitation-immunoblotting experiments showed unaltered association between Stn1-1 and Ten1 at both the permissive and restrictive temperatures (25°C and 36°C, respectively) (Supplementary Figure S2).
We conducted Southern hybridizations using a telomeric probe to observe effects of Stn1 defects at telomeres. DNA samples were obtained from wild-type and stn1-1 cells that were harvested at intervals (0, 4, 8, 12, 16 or 24 h) after shifting from 25°C to 36°C. We found that stn1-1 had slightly longer telomeres than wild-type at 25°C (Figure 1E, lanes 3 and 10). When the stn1-1 cultures were shifted from 25°C to 36°C, however, the intensity of telomere signals in Southern blots was markedly reduced at 4 h and barely visible at 12, 16 and 24 h, whereas signal intensities at the control region (see Materials and Methods) and his1 gene locus were not reduced at all (Figure 1E, lanes 10–15). In contrast, the wild-type cultures did not show any change in signals for the three probes (Figure 1E, lanes 4–9). Interestingly, the wild-type and stn1-1 strains proliferated at similar rates up to 12 h after the temperature shift, by which time stn1-1 had lost most of the telomere signals (Figure 1C). Cell cycle progressions of wild-type and stn1-1 under the restrictive temperature were examined by FACS (fluorescence activated cell sorting) analyses. DNA content histograms of wild-type showed no apparent change at 36°C up to 24 h incubation. stn1-1 shifted to 36°C showed similar histograms from 0 h throughout 6 h, suggesting the cell cycle proceeded normally during that period. However, those of stn1-1 exhibited a single broad peak covering 2C and 4C DNA content at 12 h, and the peak tail spread over 4C and more at 24 h (Figure 1F). These results suggested that Stn1 dysfunction in stn1-1 does not cause immediate effects on the cell cycle progression at least until 6 h after the temperature shift, although telomere DNA was reduced at 4 h and significantly lost at 8 h. In addition, continuous Stn1 dysfunction for >6 h led to abnormal DNA content, suggesting that chromosome instability occurred in this time window. Indeed, we found telomere loss and subsequent chromosome end-to-end fusions at 12 h (Supplementary Figure S3A and B). We also found that the growing stn1-1 cells cultivated at 36°C for several days bypassed the telomere loss via self-circularization of all three chromosomes (Supplementary Figure S3C), comparable to previously reported stn1-deleted cells (18). Importantly, this study has revealed that Stn1 dysfunction results in rapid loss of telomeric repeat DNA. This phenotype is in contrast to that of telomerase-defective cells, which shorten telomere length gradually during ∼10 days, culminating in complete telomere loss and senescence (30). The stn1-1 telomeres vanished abruptly within 12 h of post-temperature shift without noticeable telomere shortening, whereas other chromosome regions were intact (Figure 1E).
Stn1 dysfunction leads to loss of telomeric and subtelomeric regions
To examine to what extent the proximal subtelomeric DNA regions were lost in stn1-1 cells at 36°C, we performed Southern hybridizations with the EcoRI-digests of genomic DNA obtained from wild-type and stn1-1 cells harvested at intervals (0, 4, 8, 12, 16, 20 and 24 h) after shifting from 25°C to 36°C. The black boxes in Figure 2A depict the positions of the probes used in this study: telo-33k, -15k, -9k and -5k probes, which are located approximately 33, 15, 9 and 5 kb centromeric from the telomeric repeat, respectively. The telo-33k probe detected signals of similar intensities in all samples derived from wild-type and stn1-1 cells. However, we found that the signal intensities in stn1-1 observed at telo-15k, -9k, and -5k were significantly reduced at 4 h and that further signal reduction was observed at 12, 16, 20 and 24 h. Incubation at 36°C did not significantly change the signal intensities in wild-type cells (Figure 2B). If severe nucleolytic attack or site-specific DSBs had occurred in the subtelomeric region, there should have been damaged intermediates such as signal tailing or mobility shifts; however, DNA fragments detected with the probes showed no electrophoretic mobility changes in any samples, suggesting that neither frequent nucleolytic attack nor site-specific DSBs occurred.
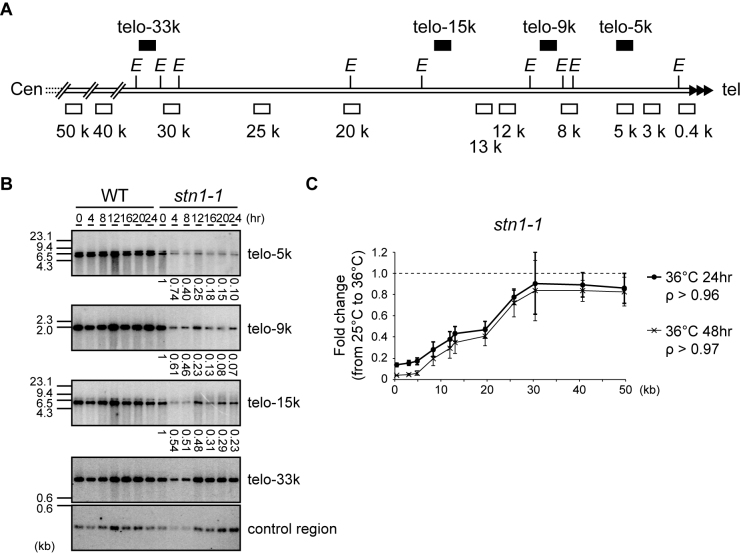
Figure 2.
Stn1 dysfunction leads to subtelomere loss. (A) Southern hybridization probes (upper black boxes) and qPCR primer sets (lower open boxes) were designed for the subtelomeric region of chromosome II right arm (see Materials and Methods for sequences). The letter E refers to EcoRI sites. (B) Wild-type and stn1-1 were incubated at the restrictive temperature for the indicated times. EcoRI-digested genomic DNAs were analyzed by Southern hybridization. Each signal was first quantified and normalized to the signal from the control region; the values under the lanes represent the fold changes relative to the 25°C condition. (C) stn1-1 was cultured at 25°C and 36°C for 24 and 48 h. Fold changes of the abundance of qPCR products at 36°C compared to those at 25°C were calculated for each primer set. Error bars show mean values of three independent experiments with SD. The letter ρ represents the Pearson correlation between the Y-axis and X-axis from 0.4 to 30 kb.
To measure the relative frequency of deletion at individual regions, we performed qPCR using the primer sets depicted in Figure 2A (open boxes). stn1-1 cells were harvested at 25°C and at two time points, 24 and 48 h after shifting cells to 36°C. Note that stn1-1 continued to grow at a retarded rate at 48 h (Figure 1C). We used a primer set located at the his1 gene locus for normalization across every sample because this internal region, on the right arm of chromosome I, was intact even at 36°C (Figure 1E). The PCR products obtained from stn1-1 cells grown at 25°C and 36°C were normalized to his1 and the amount of DNA/genome was estimated. The fold changes between cultures at 36°C and those at 25°C were calculated by dividing the quantified PCR signal at 36°C by the signal at 25°C. We found that subtelomeric regions located 0.4–30 kb from the telomere were significantly lost in stn1-1 cells at 36°C. The signal intensities at 36°C for subtelomeric regions located closer to the telomeric repeat DNA were lower than the signal intensities for more centromeric regions, suggesting that the distal regions of subtelomeres (close to termini) were deleted more severely than the proximal regions (Figure 2C). The quantified signals obtained with the 0.4–30 kb primer sets were linearly correlated with the distance of the primer set from the telomeric repeat DNA. Interestingly, after 24 h the extent of subtelomeric DNA loss at each respective position was approximately the same as at 48 h; that is to say, further deletion, if any, was limited (compare 24 and 48 h in Figure 2C). Taken together, Stn1 dysfunction caused rapid loss of telomere repeats and partial loss of approximately 30-kb of adjacent subtelomeric DNA. The loss occurred significantly by 4 h, and the frequencies of cells that had experienced the telomere loss gradually increased thereafter up to 48 h at 36°C.
Single-strand DNA resection at telomeres does not account for telomere loss
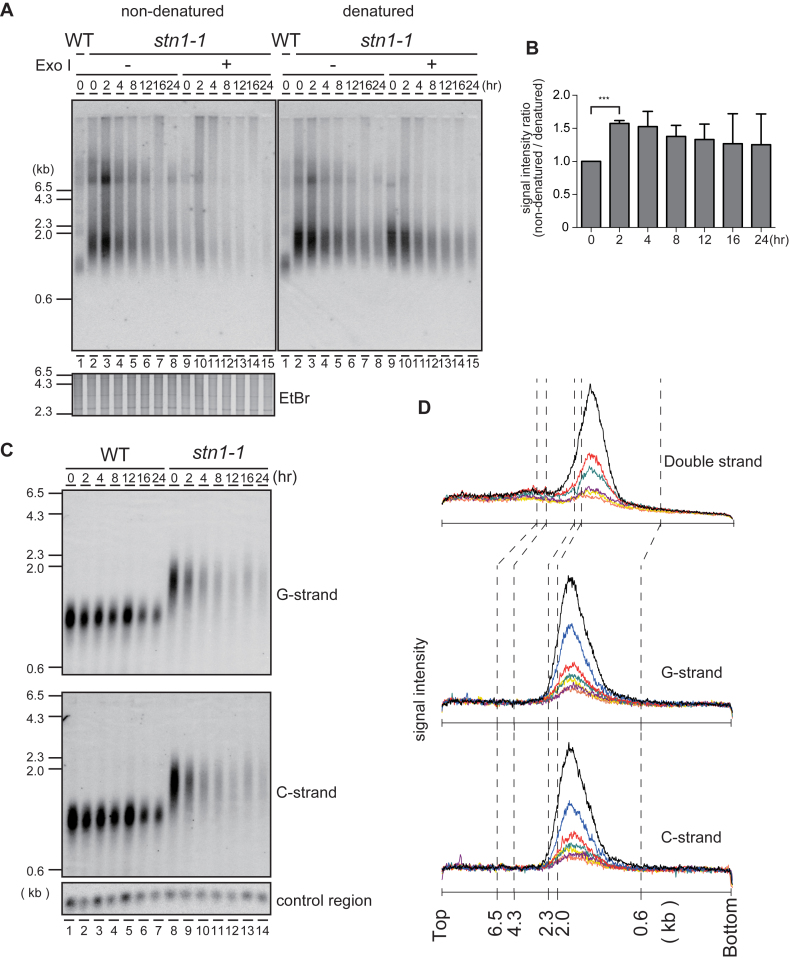
Budding yeast cdc13-1 and stn1-13 temperature-sensitive mutants cultivated at the restrictive temperature show excessive G-strand exposure and activate the G2/M checkpoint pathway (4,5). To test whether fission yeast stn1-1 also shows G-strand exposure similar to the budding yeast mutants, we performed in-gel non-denaturing hybridization with a telomeric C-rich probe to detect telomeric G-strands in EcoRI-digests derived from wild-type and stn1-1 cells harvested at intervals (0 h for wild-type and 0, 2, 4, 8, 12, 16 and 24 h for stn1-1) after shifting the temperature from 25°C to 36°C. After we detected the G-strand signals, the gel was denatured and re-hybridized with the same C-rich probe (Figure 3A). The signal intensity ratio (non-denatured/denatured) reflects the amount of exposed single-stranded G-strand relative to the entire telomeric G-strand. Immediately after the temperature shift, stn1-1 G-strand-specific signals were elevated transiently in the non-denatured gel (at 2 h, Figure 3A, lanes 2–3, Figure 3B). These signals were largely sensitive to Exonuclease I, indicating that they represented mostly G-tail DNA (Figure 3A, lanes 9–15). These results indicate that impaired Stn1 function leads to increased G-strand overhangs. We next investigated whether excessive C-strand resection, which could uncover G-strands, or damaged DNAs, such as nicks or gaps, occurred in stn1-1 prior to telomere disappearance. EcoRI-digested genomic DNA fragments were separated by alkaline gel electrophoresis, blotted onto a membrane, and hybridized with a C-rich probe or a G-rich probe to detect telomeric G-strands or C-strands, respectively (Figure 3C). We quantified telomeric signal distributions from the alkaline Southern blot, along with the signal distributions from the non-denaturing gels (Figure 3D upper; obtained from Figure 1E, lanes 10–15), single-stranded G-strand (Figure 3D middle; obtained from Figure 3C, lanes 8–14 G-strand), and single-stranded C-strand (Figure 3D lower; obtained from Figure 3C, lanes 8–14 C-strand). We did not detect significant size changes between denatured C-strand, denatured G-strand, and non-denatured G-strand, suggesting that there were few, if any, nicks or gaps in telomeric double-stranded DNAs, and no profound single-strand degradations.
Figure 3.
The G-tail is transiently exposed at Stn1-dysfunctional telomeres. (A) An analysis of EcoRI-digested genomic DNA was performed with in-gel non-denaturing hybridization using a C-rich telomeric probe (left). The same gel was denatured and re-hybridized with the same probe (right). The EtBr-stained gel was photographed before hybridization. (B) The signal intensities between 0.6 and 4.3 kb were quantified in Figure 3A denatured and non-denatured lanes 2–8. Ratios of intensities for non-denatured versus denatured signals were calculated for each time point, and the ratios were normalized to the 0 h time point. Error bars show mean values of three independent experiments with SD. *** P value was 0.0020 calculated with a two-tailed Student's t-test. (C) EcoRI-digested genomic DNAs were electrophoresed in an alkaline-denaturing gel, and Southern hybridization was performed with a telomeric C-rich probe (upper, G-strand) and G-rich probe (middle, C-strand). (D) The signal quantifications from the top of the gel to the bottom were obtained from: Figure 1E standard gel electrophoresis (upper plot); Figure 3C, G-strand (middle plot); and Figure 3C, C-strand (lower plot). stn1-1 cultured at 36°C for 0 h (black), 2 h (blue), 4 h (red), 8 h (green), 12 h (purple), 16 h (yellow) and 24 h (orange) were quantified.
If the telomere loss was generated through single-strand DNA resection, exonuclease activities would be indispensable. However, when either Exo1 or Mre11, two exonucleases responsible for telomere single-strand DNA resection (31,32), were deleted, we still observed telomere disappearance (Supplementary Figure S4A). It is known that redundant pathways process telomeres and DSB ends. Recent reports showed that excessive G-strand exposure caused by shut-off of Pot1, a telomere ssDNA-binding protein, was significantly suppressed by simultaneous deletions of exo1 and rqh1 (33,34). However, we still observed severe telomere loss in a stn1-1, exo1Δ, rqh1Δ triple mutant strain at the restrictive temperature (Supplementary Figure S4B). Taken together, we concluded that although stn1-1 at 36°C results in transient G-strand exposure, exonuclease activities that account for C-strand resection do not cause the telomere loss. This is in contrast with the excessive C-strand resection observed with the budding yeast cdc13-1 and stn1-13 cells (4,5). It was shown that a deletion of pot1 results in immediate and complete telomere loss, leading to chromosome circularization (35). The circularization depends on the SSA (single-strand annealing) pathway (36). We examined whether the SSA pathway was involved in the rapid telomere and subtelomere loss in stn1-1. We found that deletion of rad16, a gene encoding the catalytic subunit of ERCC1/XPF endonuclease, which is required for the SSA pathway, did not suppress telomere loss in stn1-1 at the restrictive temperature (Supplementary Figure S5). These results indicate that telomere loss in stn1-1 was not mediated by the SSA pathway.
Stn1-1 abruptly causes replication fork collapse at subtelomeres
As described above, neither DNA exonuclease activities nor gradual telomere attrition accompanied by cell growth in the absence of telomerase are likely to explain the loss of telomeres and subtelomeres in stn1-1 at 36°C. We hypothesized that the replication fork very frequently collapses at subtelomeres in stn1-1, leading to DSB formation at the subtelomeres, followed by the telomere disappearance.
We found that telomere loss was not observed in stn1-1 cells undergoing cell cycle arrest by nitrogen starvation and cultivated at 36°C, suggesting that stn1-1 telomere loss is incurred concomitant with DNA replication in S phase (Figure 4A). Next, we analyzed the replication of subtelomeres by neutral-neutral 2D gel electrophoresis of EcoRV-digests probed with the telo-9k probe, which detected a 3.9-kb long fragment 6.5–10.4 kb distant from telomeric repeat DNA (Figure 4B). We found that replication intermediates primarily consisted of simple Y-arcs in wild-type cultivated at 25°C or 36°C, and stn1-1 cultivated at 25°C (indicated with white triangles, in Figure 4C). In contrast, we did not observe subtelomeric replication intermediates in stn1-1 cultivated at 36°C for 2 h or longer (Figure 4C). We observed similar results at the telo-17k region (Supplementary Figure S6A), where DNA loss was observed in the non-permissive condition (Figure 2B and C). The correlation of loss of replication intermediates in 2D gels with the DNA signal in Southern blotting and PCR experiments further suggest that replication fork collapse caused the subtelomere DNA loss. These results strongly suggest that semi-conservative DNA replication at subtelomeres became defective immediately in stn1-1 cultivated at 36°C. We excluded the possibility that the loss of replication intermediates was a consequence of telomere disappearance, because telomere and subtelomere loss was modest within 4 h incubation at 36°C (Figures 1E and 2B). Since stn1-1 grew logarithmically at 36°C during the initial 12 h (Figure 1C), the replication error was unlikely to be distributed throughout the genome but rather was likely specific to subtelomeres. Indeed, we observed replication intermediates in the rDNA repeat region in stn1-1 at the restrictive temperature, although stn1-1 showed slight reduction in the replication intermediate signals (Supplementary Figure S6B).
Figure 4.
stn1-1 at 36°C frequently arrests DNA replication at subtelomeres, leading to DSBs. (A) Southern hybridization was performed with EcoRI-digested genomic DNA from cells harvested after culture in the following conditions: asynchronously growing cells at 25°C in MM medium (lanes 1 and 5); starved for nitrogen for 16 h at 25°C and released by NH4Cl addition and cultured at 36°C for 24 h (lanes 2 and 6); starved for nitrogen for 16 h at 25°C then shifted to 36°C for 12 h (lanes 3 and 7) and 24 h (lanes 4 and 8), respectively. (B) Positions of the telo-9k probe and EcoRV sites (left). (C) Wild-type and stn1-1 were cultured at 25°C and 36°C for the indicated times. EcoRV-digests were resolved by neutral-neutral 2D gel electrophoresis and probed by Southern hybridization with the telo-9k probe. The white triangles indicate the replication intermediates, simple Y-arcs. (D) Wild-type and stn1-1 cells expressing C-terminally myc-tagged Rad11 were cultured at 36°C for the indicated times and were analyzed by ChIP-qPCR using qPCR primer sets depicted in Figure 2A. Error bars show mean values of three independent experiments with SD. *** P value was 0.0043 calculated with a two-tailed Student's t-test. (E) Wild-type and stn1-1 cells expressing C-terminally myc-tagged Rad22, cultured at 36°C for the indicated times, were analyzed by ChIP-qPCR. Error bars show mean values of three independent experiments with SD. *** P value was 0.0018 calculated with a two-tailed Student's t-test.
When a DNA replication fork is collapsed, the HRR (homologous recombination repair) pathway rescues the arrested replication fork (37,38). Looking for evidence of HRR at subtelomeric regions, we performed ChIP assays using the PCR primers indicated in Figure 2A, and anti-myc antibodies to immunopreciptate myc-tagged Rad11, the largest subunit of the RPA (Replication protein A) complex, as well as a myc-tagged homologous recombination protein, Rad22 (the fission yeast homologue of budding yeast Rad52), in the wild-type and stn1-1 backgrounds. Both proteins were minimally accumulated at subtelomeres and a control locus, his1, in wild-type cells incubated at 25°C or 36°C. In marked contrast, Rad11-myc and Rad22-myc were strongly and specifically accumulated at subtelomeres in the stn1-1 cells cultured at 36°C (Figure 4D and E). Moreover, subtelomeric regions located nearer to the telomere repeat DNA had stronger ChIP signals in a direct relation. These results showed a correlation with the distribution of telomere-subtelomere signal loss observed in Figure 2C, supporting the notion that replication forks are frequently collapsed at Stn1-dysfunctional subtelomeres at the restrictive temperature.
Stn1 dysfunction does not elicit a DNA damage and replication checkpoint, but leads to deprotected chromosome ends
The disappearance of replication forks and the accumulation of Rad11 and Rad22 at restrictive temperatures are consistent with the notion that Stn1 dysfunction leads to replication fork collapse at subtelomeric regions. However, the cell cycle was not arrested at a specific phase for the initial 6 h as expected for cells activating checkpoints (Figure 1C and F). We hypothesized that chromosomes that had not completed subtelomere and telomere replication, and that had not undergone cell cycle arrest would incur chromosome breaks at mitosis. We examined mitotic chromosomes especially from anaphase to telophase (see Materials and Methods). As we expected, aberrant fibrous DNA structures connecting the two segregated nuclei were frequently observed in stn1-1 cells shifted to 36°C for 4 h (Figure 5A). Together with the presence of Rad11 and Rad22, these results imply that stn1-1 accumulates deprotected recombinogenic chromosomal ends immediately after the temperature shift.
Fission yeast Pot1 inactivation causes telomere loss and results in activation of the G2/M checkpoint via the ATR-Chk1 pathway (39). Elongated cell morphology is typically observed in fission yeast cells arrested by a DNA damage checkpoint. As seen in Figure 1D, some stn1-1 cells at 36°C for 24 h exhibited aberrant morphology. We scored the cell length of stn1-1: severe cell elongation (longer than 18 μm) was <10% and a substantial number of stn1-1 cells were not elongated after 24 h at 36°C (Figure 5B). These results raised the possibility that stn1-1 cells at 36°C escape a checkpoint-induced cell cycle arrest, at least temporarily, after the temperature is shifted to 36°C.
To clarify the immediate phenotypes caused by deletions of stn1 and pot1, we measured cell lengths of nascent stn1- or pot1-deleted haploid cells obtained at 24 and 36 h after germination of stn1Δ/stn1+ and pot1Δ /pot1+ diploid cells. The stn1 and pot1 deletion alleles were obtained by substituting the kanMX6 gene for the endogenous wild-type alleles. We obtained stn1Δ and pot1Δ haploid cells en masse by drug selection with G418 (see Materials and Methods). We confirmed that G418-resistant diploid cells constituted <4.4% of the cell population at 36 h after germination induction (data not shown). The resulting stn1Δ and pot1Δ haploid cells grew with similar kinetics after the germination (Supplementary Figure S7A). The nascent pot1Δ cells showed an elongated morphology, as expected (15.1% and 22.8% were longer than 18 μm, at 24 and 36 h after germination, respectively). In contrast, we found that the elongated morphology of the nascent stn1Δ cells was less obvious than for pot1Δ (Figure 5C, Supplementary Figure S7A, B, 1.6% and 7.8% were longer than 18 μm, at 24 and 36 h after germination, respectively). Importantly, cell lengths of the nascent stn1Δ cells were similar to those of stn1-1 at the restrictive temperature cultured for comparable periods (Figure 5B and C). These results support the following points. First, checkpoint-mediated cell cycle arrest in stn1Δ, occurred, if at all, to a lesser extent compared to pot1Δ immediately after inactivation of cognate genes. Second, the behavior of stn1-1 cells at the restrictive temperature, in terms of circumvention of the DNA damage checkpoint, is similar to that for cells immediately after stn1 deletion, although it is difficult to formally prove this premise.
We explored this notion further by examining whether the two major checkpoint transducers Chk1 and Cds1 were activated in stn1-1 at the restrictive temperature. We tested electrophoretic mobility shifts in immunoprecipitation and immunoblotting experiments. Neither Chk1 nor Cds1 band-shifts were observed up to 18 h after the temperature shift (Figure 5D and E). As such, even though stn1-1 cells cultivated at the restrictive temperature lost telomere repeat DNA, the cell cycle checkpoint pathways seemed not to be activated. These results suggest that in the absence of fully functional Stn1, replication fork collapse at subtelomeres does not by itself elicit checkpoint-induced cell cycle arrest, despite frequent chromosome breaks that occur in these regions in stn1-1 cells. A minor fraction of cells showed elongated cell morphology, likely resulting from an indirect effect of stn1-1 inactivation, i.e. chromosome end-to-end fusions between DSBs formed at subtelomeres and consequent chromosomal instability (Supplementary Figure S3B).
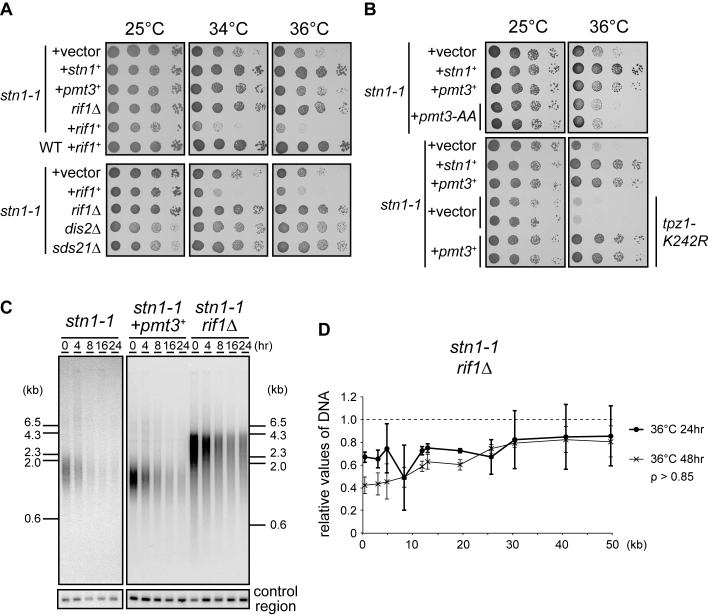
rif1 and pmt3 genetically interact with stn1
To understand how aberrant subtelomeric replication is triggered in stn1-1, we performed a screen for suppressor genes that bypassed the high-temperature sensitivity. We transformed stn1-1 with a plasmid-based S. pombe genomic DNA library and obtained transformants that no longer showed growth defects at 36°C. Temperature-sensitive stn1-1 survived by self-circularizing the chromosomes at the restrictive temperature (Supplementary Figure S3C). We wanted to obtain genuine suppressor genes that bypassed the sensitivity without chromosome self-circularization. It is known that cells harboring circular chromosomes are highly sensitive to DNA-damaging agents (40). We introduced camptothecin, a topoisomerase I inhibitor, to the selective medium to eliminate cells possessing circular chromosomes (see Materials and Methods). We found that one suppressing plasmid contained the 3΄-terminal half of the rif1 gene, comprising an ORF (2769061–2770572 nt of chromosome 1) encoding the C-terminal 503 amino acids out of a total of 1400 amino acids for the full-length Rif1 protein. Although the native rif1+ promoter was deleted in the plasmid, we suspected that somehow the partial rif1 sequence was transcribed to produce an N-terminally deleted Rif1 protein, which produced a dominant negative effect on the endogenous wild-type rif1+. To follow up on this line of reasoning, we deleted the rif1+ gene from stn1-1, and found that the rif1 deletion in the stn1-1 mutant suppressed the high temperature sensitivity associated with stn1-1 (Figure 6A). By contrast, stn1-1 transformed with a high-copy plasmid expressing full-length rif1 showed synthetically deleterious growth (Figure 6A). Recent reports showed that Rif1 protein recruits protein phosphatase 1, Dis2 and Sds21 in fission yeast, to control the timing of replication origin firing (41,42). We generated dis2 and sds21 deletion mutants in stn1-1, and found that they also suppressed the high-temperature sensitivity of stn1-1, suggesting that the Rif1, Dis2 and Sds21 pathway contributes to the high-temperature sensitivity in stn1-1 (Figure 6A bottom).
Figure 6.
stn1 genetically interacts with rif1 and pmt3. (A) Ten-fold serial dilutions of the indicated strains were spotted onto SD plates (without camptothecin) and incubated for 5 days at 25°C, and 3 days at both 34°C and 36°C. (B) Cells were spotted onto SD (top) and SD + camptothecin plates (bottom) and incubated for 4 days at 25°C, and 3 days at 36°C, respectively. (C) stn1-1, stn1-1 + pmt3+, and stn1-1 rif1Δ were incubated at 25°C and shifted to 36°C for the indicated times. EcoRI-digested genomic DNAs were analyzed by Southern hybridization, using a telomere probe, and the same membrane was then stripped and re-hybridized with the control region probe. (D) stn1-1 rif1Δ cells were cultured at 25°C, 36°C for 24 h, and at 36°C for 48 h. qPCR and quantifications were carried out as for Figure 2C.
We found that another independent plasmid contained a full-length pmt3 gene, which encodes a precursor of SUMO. stn1-1 transformed with a high-copy plasmid expressing full-length pmt3 showed normal growth at 36°C, indicating that pmt3 is a suppressor of stn1-1. SUMOylation affects the localization or function of proteins (43). High copy number pmt3 can alter protein SUMOylation levels. Indeed, pmt3-AA, a nonconjugatable form of SUMO, did not suppress the temperature sensitivity of stn1-1 cells (Figure 6B top), suggesting that protein SUMOylation is required for pmt3 overexpression to suppress stn1-1 phenotypes. Because Tpz1-K242 is SUMOylated to regulate telomerase recruitment (19,20), it was possible that enhancement of this SUMOylation reaction was responsible for the suppression. However, pmt3 overexpression in stn1-1 cells with a tpz1-K242R mutation still suppressed the high temperature sensitivity (Figure 6B bottom). Together, these results suggested that pmt3 overexpression suppresses stn1-1 phenotypes through SUMOylating proteins at lysine residues other than Tpz1-K242.
We analyzed the telomeres and subtelomeres of pmt3-overexpressing and rif1-deleted strains by Southern hybridization and qPCR. These assays revealed that overexpression of pmt3 and deletion of rif1 partially suppressed telomere loss (Figure 6C, D, and Supplementary Figure S8). These results indicate that both suppressors did not fully complement the function of Stn1-1 at the restrictive temperature. Indeed, we did not observe any suppression of stn1-1 replication fork collapse (Supplementary Figure S6C).
DISCUSSION
In this study, we isolated a fission yeast stn1 high-temperature sensitive allele, stn1-1. The mutant cells lost telomeres and subtelomeres rapidly during cultivation at 36°C. We also found that the defective Stn1 function in stn1-1 cells led to transient G-tail exposure and frequent replication fork collapse at subtelomeres, without checkpoint activation, which probably led to frequent chromosome breaks during mitosis. These results indicate that fission yeast Stn1 plays crucial roles in proper semi-conservative replication at telomeres and subtelomeres. We also identified that overexpression of pmt3 and deletion of rif1 individually suppressed the lethality at the restrictive temperature. These alleles suppressed telomere loss that would otherwise lead to the stn1-1 lethality.
Stn1 is required for telomere maintenance
In budding yeast, many reports have shown that the CST complex plays telomere protective roles. High-temperature sensitive alleles stn1-13 and stn1-td cause excessive C-strand resection, leading to G-strand exposure and G2/M cell cycle arrest (5,44). Stn1 interacts with DNA polymerase α B subunit Pol12 for completion of lagging strand synthesis (8). It is known that knockdown of human STN1 leads to G-tail extensive exposure (10). Fission yeast Stn1-Ten1 regulates telomerase activity by interacting with SUMOylated Tpz1 (19,20). These results suggest that Stn1 maintains telomeres through G-strand elongation and C-strand fill-in. In this study, stn1-1 showed transient G-tail exposure at the restrictive temperature, suggesting that the C-strand fill-in function was defective. Unlike stn1-13 and stn1-td strains, however, stn1-1 showed complete telomere loss and did not activate the DNA damage checkpoint. Immediately after the temperature shift, stn1-1 showed complete loss of replication intermediates at subtelomeres, suggesting its essential role in replicating telomeres and subtelomeres. We also observed that stn1-1 showed a slight reduction in the replication intermediate signals at rDNA (Supplementary Figure S6B). This result raised the possibility that Stn1 also participates in DNA replication at the rDNA arrays. It is tempting to speculate that Stn1 is required for DNA replication of specific regions with some common features, such as repetitive DNA. Future studies are required to test this hypothesis. It was reported that the Kluyveromyces lactis stn1-M1 mutant showed an ALT-like phenotype (45). Knockdown of hStn1 results in increasing numbers of fragile telomeres (46). These results indicate that Stn1 functions are not completely conserved among species.
Differential functions achieved by fission yeast Stn1 and Pot1
The phenotypes described for stn1-1 cells at a restrictive temperature in this study very closely resemble those of pot1-1, a temperature-sensitive mutant in fission yeast (39). Both cells showed immediate loss of telomere DNA without gradual shortening after temperature shifts. However, while pot1-1 cells underwent extensive C-strand resection, concomitantly with the telomere loss, stn1-1 cells generated only limited and transient G-strand exposure after the temperature shift. pot1-1 activates the G2/M DNA damage checkpoint, as revealed by the appearance of highly elongated cells. These phenotypes are shared by the budding yeast CST temperature-sensitive mutants. In contrast, we have shown that stn1-1 does not display either excessive C-strand resection or DNA damage checkpoint activation, two major phenotypes associated with deprotected telomeres. Instead, we observed that conventional DNA replication is abrogated specifically at subtelomeres in stn1-1 cells at the restrictive temperature. When we arrested the cell cycle, thereby preventing entry into S phase, telomere loss was not observed in stn1-1 cells at the restrictive temperature (Figure 4A). From these criteria, we argue that the primary role of fission yeast Stn1 in telomere maintenance is the facilitation of telomere and subtelomere replication. Its role in telomere protection (protecting telomere DNA from exonucleolytic attack, aberrant DNA repair such as end-to-end fusions, and DNA damage checkpoint activation) is relatively minor (Supplementary Figure S9).
Frequent DNA replication fork collapse should lead to chromosome breaks at subtelomeres, which immediately manifests as telomere loss. It was reported that mouse Stn1 (AAF-44) stimulates the activity of DNA polymerase α (47). Xenopus laevis Stn1 promotes the priming step in DNA replication in egg extracts (48). We therefore hypothesize that fission yeast Stn1 facilitates lagging strand DNA synthesis not only at the very ends of telomeres but also at subtelomeres. Furthermore, we detected DNA polymerase Pol1 (α), Pol2 (ε), and Pol3 (δ) accumulation at subtelomeres at 36°C (Supplementary Figure S10A–C). These results indicate that the DNA replication fork does not completely collapse in the subtelomeres in stn1-1 cells at the restrictive temperature. We favor a hypothesis that the efficiency of lagging strand synthesis is specifically impaired at subtelomeres in stn1-1. Severe ssDNA exposure at lagging strand templates would disrupt the Y-arc structure. The imbalance between processivity of the lagging and leading strand syntheses may lead to frequent replication fork arrests, resulting in the accumulation of RPA and homologous recombination repair factors. Mammalian telomeres are difficult to replicate, and possess properties common to fragile sites (2). Interestingly, it was reported that mouse CTC1 knockout cells showed phenotypes similar to those found in this study with stn1-1 at the restrictive temperature: telomere loss without gradual shortening, frequent chromosome fusion, defects in telomere replication and the presence of t-circles, suggesting elevated homologous recombination at telomeres (14). We propose that fission yeast telomeres and subtelomeres are also difficult to replicate, and Stn1 is required to properly accomplish semi-conservative DNA replication at telomeres and subtelomeres. It was reported that Taz1 is required for proper replication of telomere sequences (3). While deletion of taz1 causes replication fork stalling, taz1Δ cells retain highly elongated telomeres. Simultaneous deletion of taz1 and trt1 results in precipitous telomere loss (3). The phenotypes observed in taz1Δ trt1Δ are close to stn1-1 shown in this study. However, we have not obtained strains deleted for taz1 in the stn1-1 background despite several attempts, leaving a study of this line for the future.
The budding yeast CST complex is required for telomere protection and efficient telomere DNA replication. Given the differential phenotypes observed with the stn1-1 and pot1-1 mutants in fission yeast as summarized above, we propose that the two major roles of the budding yeast CST complex are accomplished individually by Stn1 (replication) and Pot1 (telomere protection).
Dormant DNA damage checkpoint in stn1-1 at restrictive temperatures
Curiously, stn1-1 at the restrictive temperature did not activate replication or DNA damage checkpoints, and continued progressing through the cell cycle while sustaining severe replication fork collapse at subtelomeres and telomere loss, immediately after the temperature shift. It is not unprecedented that the checkpoints do not respond to DNA replication fork arrest in fission yeast, however. Using a strain in which DNA replication forks could be inducibly blocked at a single locus, it was shown that neither the DNA replication checkpoint nor the DNA damage checkpoint were activated, and the cell cycle proceeded in the first cell cycle in the presence of replication fork arrest (49). However, cells subsequently activated the DNA damage checkpoint robustly after passing through the first M phase, presumably due to the occurrence of DNA double-stranded breaks produced through the breakage of the dicentric chromosomes in M phase. The authors argued that the presence of a single replication fork arrest was not enough to activate either checkpoint. These observations are very similar to what we found with stn1-1 cells. Conceivably, replication fork arrests and/or slow-downs at subtelomeres, while other genomic regions are efficiently replicated, are not recognized by the checkpoint mechanism. Alternatively, it is possible that Stn1 is involved in detecting replication fork stalling and/or activating replication checkpoint arrest at telomeres and subtelomeres, which should be tested in the future. By contrast, it is well established that stn1-deleted cells strongly activate the checkpoint as evidenced by the highly elongated cells in micro-colonies (18). However, that report did not describe how cells responded to Stn1 dysfunction immediately after the stn1 deletion. It is possible that the checkpoint activation found in stn1-deleted cells in micro-colonies was actually induced by secondary events produced after rounds of cell cycle progression. Cells initially cycling normally might produce dicentric chromosomes, which in the following M phase physically tear apart, leading to gross chromosomal rearrangements and activation of the DNA damage checkpoint. To address this possibility, we examined the immediate effects of the deletion of stn1 and pot1 (Figure 5C and Supplementary Figure S7). We demonstrated that stn1Δ, as well as stn1-1 at the restrictive temperature, showed elongated morphology only to a less extent, compared to pot1Δ, at the earliest time point we could observe after the gene inactivation. These results suggest that fission yeast Stn1 plays a primary role in DNA replication at subtelomeres, and that Pot1 rather than Stn1 plays a major role in telomere protection. An alternative explanation for the dormant checkpoint activation is that the stn1-1 is a hypomorphic allele that preserves some telomere protection functions.
Suppressors of stn1-1
cdc13-1 is synthetically lethal with deletion of rif1, whereas a high-copy number rif1-expressing plasmid suppressed cdc13-1 lethality (50). Overexpression of Rif1 in budding yeast inhibits RPA and checkpoint protein accumulation on ssDNA that results in suppression of checkpoint activation (50–52). Contrary to these reports in budding yeasts, fission yeast stn1-1 temperature sensitivity was suppressed by deletion of rif1; moreover, stn1-1 carrying a high-copy number plasmid expressing rif1 was synthetically deleterious (Figure 6). This is likely because the stn1-1 lethality was caused by telomere loss; by contrast, the inability of cdc13-1 to form colonies is due to excessive G-strand exposure, which activates the DNA damage checkpoint.
Recent reports showed that Rif1 has several roles, including recruitment of protein phosphatase 1 and regulation of the timing of origin firing (41–42,53). Because Rif1 recruits protein phosphatase 1 and localizes to telomeres, we can assume Rif1 regulates protein phosphorylation levels in association with telomeric chromatin, and that these phosphorylation/dephosphorylation events are integral to telomere maintenance. Protein phosphorylation patterns at telomeres can be disrupted when rif1 is deleted, and conceivably this disruption can suppress Stn1-dysfunction. Note that stn1-1 sds21Δ at 36°C showed slightly healthier colonies than stn1-1 dis2Δ (Figure 6A); this could reflect the fact that Sds21 localizes at telomeres more abundantly than Dis2 (42). Telomere loss could be suppressed due to changes in the timing of replication origin firing at subtelomeres, because disruption of the PP1 interaction domain in Rif1 changes the timing of origin firing (42). Recent reports showed that mammalian RIF1 cooperates with 53BP1 in the choice to repair DSBs by homologous recombination (HR) or non-homologous end joining (NHEJ) (54,55). These reports prompted us to speculate that Rif1 depletion in stn1-1 down-regulates the activity of NHEJ and instead up-regulates HR around telomeres, which might explain the accumulation of Rad22 at subtelomeres at the restrictive temperature. Zaaijer et al. have recently identified that deletion of rif1 suppressed defects in chromosome segregation observed in taz1Δ at the cold temperature condition (56). Similarly, Rog et al. found that mutation of Rqh1 SUMOylation sites rescues anaphase bridges observed in taz1Δ cells (57). The lethality of stn1-1 could also stem from the defects in chromosome segregation at M phase and it can similarly be suppressed by deletion of rif1.
SUMOylation affects the localization or function of proteins (43). High copy number pmt3 can alter protein SUMOylation patterns. Thus, both Rif1 and Pmt3 might indirectly affect the patterns of posttranslational modification of DNA repair factors, such as HR factors; however, replication fork collapse was still observed either in rif1 deletion or pmt3 overexpression (Supplementary Figure S6C). Future studies are necessary for unraveling the molecular mechanisms underlying these genetic interactions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Tamura and Y. Watanabe for technical assistance, and A. Katayama, F. Maekawa, A. Shirabuchi, E. Yamazaki and S. Fukumura for excellent secretarial work and J. Hejna and Y. Nakaseko for discussion.
Footnotes
Present address: Yusuke Tarumoto, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research [22220012]; Research Grants from Princess Takamatsu Cancer Research Fund. Funding for open access charge: JSPS KAKENHI [JP15H02383].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ishikawa F. Portrait of replication stress viewed from telomeres. Cancer Sci. 2013; 104:790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller K.M., Rog O., Cooper J.P.. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006; 440:824–828. [DOI] [PubMed] [Google Scholar]
- 4.Garvik B., Carson M., Hartwell L.. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995; 15:6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandin N., Reed S.I., Charbonneau M.. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997; 11:512–527. [DOI] [PubMed] [Google Scholar]
- 6.Grandin N., Damon C., Charbonneau M.. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001; 20:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitton-Fry R.M., Anderson E.M., Hughes T.R., Lundblad V., Wuttke D.S.. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002; 296:145–147. [DOI] [PubMed] [Google Scholar]
- 8.Grossi S., Puglisi A., Dmitriev P.V., Lopes M., Shore D.. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004; 18:992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi H., Zakian V.A.. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000; 14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F.. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell. 2009; 36:193–206. [DOI] [PubMed] [Google Scholar]
- 11.Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E.. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell. 2009; 36:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D., Wright W.E., Price C.M.. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012; 31:3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasbek C., Wang F., Price C.M.. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 2013; 288:30139–30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu P., Min J.N., Wang Y., Huang C., Peng T., Chai W., Chang S.. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012; 31:2309–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Stewart J.A., Kasbek C., Zhao Y., Wright W.E., Price C.M.. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012; 2:1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Dai X., Chai W.. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012; 22:1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L.Y., Redon S., Lingner J.. The human CST complex is a terminator of telomerase activity. Nature. 2012; 488:540–544. [DOI] [PubMed] [Google Scholar]
- 18.Martín V., Du L.L., Rozenzhak S., Russell P.. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14038–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyagawa K., Low R.S., Santosa V., Tsuji H., Moser B.A., Fujisawa S., Harland J.L., Raguimova O.N., Go A., Ueno M. et al. SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1(Tpp1) to modulate shelterin-Stn1 interaction in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5950–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg M., Gurung R.L., Mansoubi S., Ahmed J.O., Davé A., Watts F.Z., Bianchi A.. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO-dependent Stn1 telomere association. EMBO Rep. 2014; 15:871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfa C., Fantes P., Hyams J., McLeod M., Warbrick E.. Experiments with fission yeast: a laboratory manual. Experiments with Fission Yeast: A Laboratory Course Manual. 1993; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 22.Noguchi C., Noguchi E.. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007; 175:553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nabetani A., Ishikawa F.. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 2009; 29:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi T., Kanoh J., Saito M., Ishikawa F.. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008; 320:1341–1344. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki H., Tarumoto Y., Ishikawa F.. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012; 26:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka K., Yonekawa T., Kawasaki Y., Kai M., Furuya K., Iwasaki M., Murakami H., Yanagida M., Okayama H.. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 2000; 20:3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C.. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1(+)-encoding syntaxin-like protein. Mol. Biol. Cell. 2001; 12:3955–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shikata M., Ishikawa F., Kanoh J.. Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J. Biol. Chem. 2007; 282:5346–5355. [DOI] [PubMed] [Google Scholar]
- 29.Sun J., Yu E.Y., Yang Y., Confer L.A., Sun S.H., Wan K., Lue N.F., Lei M.. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009; 23:2900–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R.. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997; 277:955–959. [DOI] [PubMed] [Google Scholar]
- 31.Maringele L., Lydall D.. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002; 16:1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larrivée M., LeBel C., Wellinger R.J.. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004; 18:1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.M., Liu X.M., Ding Y.H., Xiong L.Y., Ren J.Y., Zhou Z.X., Wang H.T., Zhang M.J., Yu Y., Dong M.Q. et al. Fission yeast Pxd1 promotes proper DNA repair by activating Rad16XPF and inhibiting Dna2. PLoS Biol. 2014; 12:e1001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanbu T., Nguyên L.C., Habib A.G., Hirata N., Ukimori S., Tanaka D., Masuda K., Takahashi K., Yukawa M., Tsuchiya E. et al. Fission Yeast Exo1 and Rqh1-Dna2 Redundantly Contribute to Resection of Uncapped Telomeres. PLoS One. 2015; 10:e0140456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumann P., Cech T.R.. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001; 292:1171–1175. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Baumann P.. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol. Cell. 2008; 31:463–473. [DOI] [PubMed] [Google Scholar]
- 37.Lambert S., Froget B., Carr A.M.. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst.). 2007; 6:1042–1061. [DOI] [PubMed] [Google Scholar]
- 38.Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M.. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005; 121:689–702. [DOI] [PubMed] [Google Scholar]
- 39.Pitt C.W., Cooper J.P.. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res. 2010; 38:6968–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain D., Hebden A.K., Nakamura T.M., Miller K.M., Cooper J.P.. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010; 467:223–227. [DOI] [PubMed] [Google Scholar]
- 41.Hiraga S., Alvino G.M., Chang F., Lian H.Y., Sridhar A., Kubota T., Brewer B.J., Weinreich M., Raghuraman M.K., Donaldson A.D.. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014; 28:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davé A., Cooley C., Garg M., Bianchi A.. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014; 7:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raman N., Nayak A., Muller S.. The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma. 2013; 122:475–485. [DOI] [PubMed] [Google Scholar]
- 44.Vodenicharov M.D., Wellinger R.J.. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006; 24:127–137. [DOI] [PubMed] [Google Scholar]
- 45.Iyer S., Chadha A.D., McEachern M.J.. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 2005; 25:8064–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boccardi V., Razdan N., Kaplunov J., Mundra J.J., Kimura M., Aviv A., Herbig U.. Stn1 is critical for telomere maintenance and long-term viability of somatic human cells. Aging Cell. 2015; 14:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casteel D.E., Zhuang S., Zeng Y., Perrino F.W., Boss G.R., Goulian M., Pilz R.B.. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 2009; 284:5807–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakaoka H., Nishiyama A., Saito M., Ishikawa F.. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 2012; 287:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohebi S., Mizuno K., Watson A., Carr A.M., Murray J.M.. Checkpoints are blind to replication restart and recombination intermediates that result in gross chromosomal rearrangements. Nat. Commun. 2015; 6:6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue Y., Rushton M.D., Maringele L.. A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet. 2011; 7:e1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribeyre C., Shore D.. Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 2012; 19:307–313. [DOI] [PubMed] [Google Scholar]
- 52.Anbalagan S., Bonetti D., Lucchini G., Longhese M.P.. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet. 2011; 7:e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattarocci S., Shyian M., Lemmens L., Damay P., Altintas D.M., Shi T., Bartholomew C.R., Thomä N.H., Hardy C.F., Shore D.. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep. 2014; 7:62–69. [DOI] [PubMed] [Google Scholar]
- 54.Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013; 339:711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T.. 53BP1 regulates DSB repair using Rif1 to control 5΄ end resection. Science. 2013; 339:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaaijer S., Shaikh N., Nageshan R.K., Cooper J.P.. Rif1 Regulates the Fate of DNA Entanglements during Mitosis. Cell Rep. 2016; 16:148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rog O., Miller K.M., Ferreira M.G., Cooper J.P.. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell. 2009; 33:559–569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.