Abstract
Aberrant formation of interstitial telomeric sequences (ITSs) promotes genome instabilities. However, it is unclear how aberrant ITS formation is suppressed in human cells. Here, we report that MLH1, a key protein involved in mismatch repair (MMR), suppresses telomeric sequence insertion (TSI) at intra-chromosomal regions. The frequency of TSI can be elevated by double-strand break (DSB) inducer and abolished by ATM/ATR inhibition. Suppression of TSI requires MLH1 recruitment to DSBs, indicating that MLH1's role in DSB response/repair is important for suppressing TSI. Moreover, TSI requires telomerase activity but is independent of the functional status of p53 and Rb. Lastly, we show that TSI is associated with chromosome instabilities including chromosome loss, micronuclei formation and chromosome breakage that are further elevated by replication stress. Our studies uncover a novel link between MLH1, telomerase, telomere and genome stability.
INTRODUCTION
Telomeres are specialized structures that play an essential role in maintaining genome stability. In humans, telomeric DNA is made up of ∼3–15 kb of (TTAGGG)n tandem repeats oriented 5΄→3΄ toward the end of the chromosome. The six-member protein complex known as shelterin binds to telomeric DNA and promotes the formation of the ‘capping’ structure, preventing chromosome ends from being recognized as damaged DNA (1). Telomere dysfunction, induced either by loss of telomeric DNA or by deficiency in telomere binding proteins, ‘uncaps’ chromosome ends and activates the ATM/ATR DNA damage response pathways, resulting in unwanted DNA repair activities at telomere ends that lead to genome instabilities such as end-to-end chromosome fusions, inappropriate recombination, and exonucleolytic degradation. Therefore, telomere instability is critically linked to cancer development (2).
While canonical telomeric repeats are predominantly found at chromosome termini, interstitial telomeric sequences (ITSs) also exist at intra-chromosomal loci in some vertebrate species including Homo sapiens (3,4). Previous studies have shown that these sequences are prone to chromosome breakage, recombination and rearrangement (5–7). In addition, the presence of TTAGGG repeats at intra-chromosomal regions is linked to genome instabilities involving chromosome recombination, translocation, and rearrangements (8–13). Thus, erroneous intra-chromosomal insertion of telomeric sequences is normally repressed to keep the genome stable. It has been described recently that in telomerase-negative cancer cells that use recombination for telomere maintenance (the so-called ALT cells), telomeric DNA can be added to discrete sites throughout the genome when nuclear receptor NR2C/F is deficient, suggesting that NR2C/F plays an important role in suppressing telomere sequence insertion in ALT cells (13). Interestingly, the NR2C/F-regulated telomere insertion is specific to ALT cells, and NR2C/F deficiency does not appear to induce such insertion in non-ALT cancer cells including telomerase-expressing cancer cells (13). Hence, the mechanism for preventing telomere insertion in non-ALT cells remains unknown.
While we were investigating the function of the MMR protein MLH1 in telomere maintenance, we unexpectedly observed the involvement of MLH1 in suppressing telomeric sequence insertion (TSI) at intra-chromosomal regions in telomerase-positive cells. The MMR system, consisting of multiple homologs of MutS and MutL, EXOI, PCNA, RPA, DNA polymerase δ and DNA ligase I, is an essential pathway in maintaining genome stability (14). It corrects single nucleotide mismatches or small insertion/deletion loops that often arise during DNA replication. Defects in MMR proteins increase spontaneous mutation rate and lead to microsatellite instability, giving rise to cancer (14,15). In fact, deficiency in MMR genes is the leading cause of Lynch syndrome, also known as hereditary non-polyposis colorectal cancer (HNPCC). Aside from their role in repairing DNA base pairing errors, MMR proteins participate in several MMR-unrelated DNA metabolism pathways that are important for safeguarding genome integrity. First, various MMR proteins mediate DNA damage response and are involved in signaling checkpoint activation. They promote cell cycle arrest and/or programmed cell death in response to certain types of DNA damage. Deficiency in MMR genes often leads to the development of anti-cancer drug resistance (16–21). Second, MMR proteins inhibit homeologous recombination (recombination between related but non-identical DNA sequences) (22–26), although the mechanism is unclear. In cells with MMR defects, the rate of recombination between diverged sequences at double-stranded breaks (DSBs) increases dramatically, leading to gene conversions at recombined sites. Such error-prone recombination events further contribute to genome instability and tumorigenesis. Third, several reports implicate a role of MMR proteins especially MLH1 in modulating non-homologous end joining (NHEJ). Upon irradiation or exposure to radiomimetic chemicals, MLH1, MSH2, MSH3, MSH6 and PMS2 are recruited to damaged sites (27,28), although the function of these proteins in modulating NHEJ remains unclear (29). Finally, MLH1 has specific roles in meiotic recombination (30,31), class switching, and somatic hypermutation (32,33).
Here, we report that MLH1 deficiency induces TSI in multiple telomerase-expressing cancer cell lines and telomerase-immortalized fibroblasts. The frequency of TSI is further elevated by a DSB inducer, but abrogated by ATM or ATR inhibition, suggesting that TSI is likely under the control of the ATM/ATR damage response pathway. Using domain-specific mutants, we find that MLH1 recruitment to DSBs is needed for suppressing TSI, supporting the idea that MLH1 suppresses TSI in response to DNA damage. Importantly, analysis of TSI with telomerase inhibition and in cells deficient in p53 or Rb reveals that TSI is dependent on telomerase activity but not affected by the functional status of p53 or Rb. Our data also suggest that the newly-formed ITSs are unstable, and there is perhaps an equilibrium between TSI formation and TSI loss. Additionally, we show that TSI correlates with genome instabilities including chromosome loss, micronuclei formation, and chromosome breakage that are further elevated by replication stress. Despite MLH1's role in TSI suppression, MLH1 deficiency induces no obvious telomere dysfunction in telomerase-positive cells under unchallenged condition, suggesting that MLH1 plays a minimal role in telomere maintenance. Our findings suggest that MLH1 suppresses the formation of intra-chromosomal telomeric sequences that likely originates from aberrant addition of telomeric repeats by telomerase, and provide new insights into the potential mechanism of carcinogenesis associated with MLH1 deficiency.
MATERIALS AND METHODS
Plasmids
pBabe-hygro-FLAG-MLH1 was constructed by PCR amplifying FLAG-MLH1 cDNA from pcDNA6-FLAG-MLH1 (21), and then cloning into the EcoRI site of pBabe-hygro. QuikChange Site-directed Mutagenesis kit (Agilent) was used to introduce mutations at shRNA targeting region in pBabe-hygro-FLAG-MLH1 to create RNAi-resistant MLH1. The resulting pBabe-hygro-FLAG-MLH1 was then used as a template to construct pBabe-hygro-FLAG-MLH1 N-terminus mutant (2–389 amino acids) and pBabe-hygro-FLAG-MLH1 C-terminus mutant (390–756 amino acids) using the QuikChange Site-directed Mutagenesis kit (Agilent). All constructs were sequenced to ensure sequence accuracy.
RNA interference
MLH1 shRNA sequences targeting GGGGGAAGTTATCCAGCGG (sequence 1) and TTGGATGTGAGGATAAAAC (sequence 2) were used. The corresponding shRNA sequence was cloned into pSIREN-retro-puro (Clontech). Infection and selection were performed following the manufacturer's protocol.
Cell culture
HeLa, BJ/E6E7, BJ/E6E7/hTERT and BJ/hTERT cells were passaged in DMEM supplemented with 10% cosmic calf serum (Hyclone) at 37°C containing 5% CO2. HCT116 and HCT116MLH1 were cultured in McCOY'S media supplemented with 10% fetal bovine serum. HCT116MLH1 was cultured in the presence of G418 (0.3 mg/ml) to maintain stable expression of MLH1. HeLa cells stably expressing full length or truncated FLAG-MLH1 were generated by retroviral transduction followed by hygromycin selection.
Materials
The following agents were used: etoposide (Sigma), aphidicolin (Sigma), ATM inhibitor KU55933 (Sigma), ATR inhibitor VE-821 (EMD Millipore), BIBR1532 ((E)-2-(3-(naphthalen-2-yl)but-2-enamido)benzoic acid, Shelleckchem) and MNNG (N-methyl N'-nitro-N-nitrosoguanidine, Toronto Research Chemicals). All agents were dissolved in DMSO and stored under sterile conditions at −20°C in dark.
Antibodies
The following primary antibodies were used: monoclonal anti-MLH1 (ThermoFisher), rabbit polyclonal anti-FLAG (Cell Signaling), monoclonal mouse anti-actin (BD Biosciences), rabbit polyclonal anti-53BP1 (Novus), rabbit anti-phospho-Chk1 (Ser317) (Cell Signaling), rabbit anti-phospho-Chk2 (Thr68) (Cell Signaling), rabbit anti-Chk1(Cell Signaling), rabbit anti-Chk2 (Cell Signaling), mouse anti-retinoblastoma (Rb) (BD Pharmingen), mouse anti-p53 (Santa Cruz). Secondary antibodies were horseradish peroxidase conjugated anti-mouse IgG and anti-rabbit IgG (BD Biosciences) for western blotting, Dylight 488-anti-mouse IgG (ThermoFisher) and Dylight 549-anti-rabbit IgG (ThermoFisher) for immunofluorescence.
Immunofluorescence (IF)
Cells were grown in chamber slides and fixed with 4% paraformaldehyde, permeabilized with 0.15% Triton X-100, blocked with 3% BSA at 37°C for 1 h in humidified chamber, incubated with primary antibodies for overnight at 4°C, washed with PBS three times, and then incubated with secondary antibody at 37°C for 1 h. Z-stack images were taken at a 0.275 μM thickness per slice under Zeiss AxioImager M2 epifluorescence microscope with a 100× oil objective.
MTT assay
HeLa-shLUC, HeLa-shMLH1, HCT116 and HCT116MLH1 cells (2500 cells per ml) were seeded in 96-well plates for 24 h. Fresh media containing MNNG at indicated concentrations were added. After 20 h of treatment, MTT (5 mg/ml) was added into each well. Following 4 h of incubation, media was removed, and 150 μl of DMSO was added to dissolve the formazan granules generated by live cells. The plate was then shaken for 10 min at r.t., and the optical densities at 490 nm was measured by Synergy H1 microplate reader (BioTek).
Terminal restriction fragment (TRF) analysis
Telomere length was determined by TRF analysis as described (34). Briefly, genomic DNA was isolated using DNeasy kit (Qiagen), digested with a mixture of four enzymes (RsaI, CfoI, AluI, HinfI) at 37°C overnight, and resolved on a 0.7% 1× TAE agarose gel (20 cm × 24cm) at 65 V (15 h). The gel was then denatured for 30 min in 1.5 M NaCl/0.5 M NaOH, neutralized for 15 min in 1.5 M NaCl/0.5 M Tris–HCl pH8.0, and dried at 55°C for 1.5 h. The gel was then hybridized to a 32P-labeled (TTAGGG)3 probe at 42°C overnight, washed twice in 0.1% SDS/0.1× SSC for 15 min each time, and radioactive signals from telomeric repeats were acquired with PhosporImager (GE Healthcare).
FISH and quantitative-FISH (Q-FISH)
Metaphase cell collection and FISH were performed as previously described (35). For shRNA expressing cells, cells were collected 7–8 days after puromycin selection for metaphase preparation. For HeLa, HCT116 and HCT116MLH1, cells were treated with 3 μM etoposide, recovered for 24 h, treated with colcemid (0.1 μg/ml), and metaphase cells were collected. For BJ/E6E7, BJ/hTERT and BJ/E6E7/hTERT, cells were treated with 2 μM etoposide for 2 h, followed by 24 h recovery prior to preparation of metaphase spreads. Images were acquired with a Zeiss AxioImager M2 epifluorescence microscope with a 100× oil objective. During image analysis, unprocessed monochrome images were examined to locate all TSIs, since black/white offers the greatest contrast for human eyes to identify inserted telomeric sequences. Q-FISH was performed as described (36,37) by using the linear relationship between mean telomere fluorescence from ∼4000 telomere ends and distribution of telomere lengths of HCT116 cells as established by TRF.
Conventional chromosome analyses (karyotyping)
Metaphase chromosomes were obtained from cultured BJ/hTERT shLuc and shMLH1 cells were stained with G-bands by trypsin using Giemsa with a standard protocol (38). Metaphase images were captured with Zeiss microscope AxioImager Z2 with a PA 63×/1.4 oil objective. Slides were scanned with a horizontal movement of the microscope stage, overlapping the scanning paths by 10%. Two people independently analyzed all images to ensure data accuracy. The karyograms were prepared using the Metasystems Ikaros Imaging computer-based imaging system.
Statistical analysis
Means were compared using a Student's t-test. Proportions were compared using a binomial Z-statistic. A Holm–Bonferroni correction was used on all multiple pairwise comparisons to control for familywise-error.
RESULTS
MLH1 deficiency has little, if any, effect on telomere maintenance
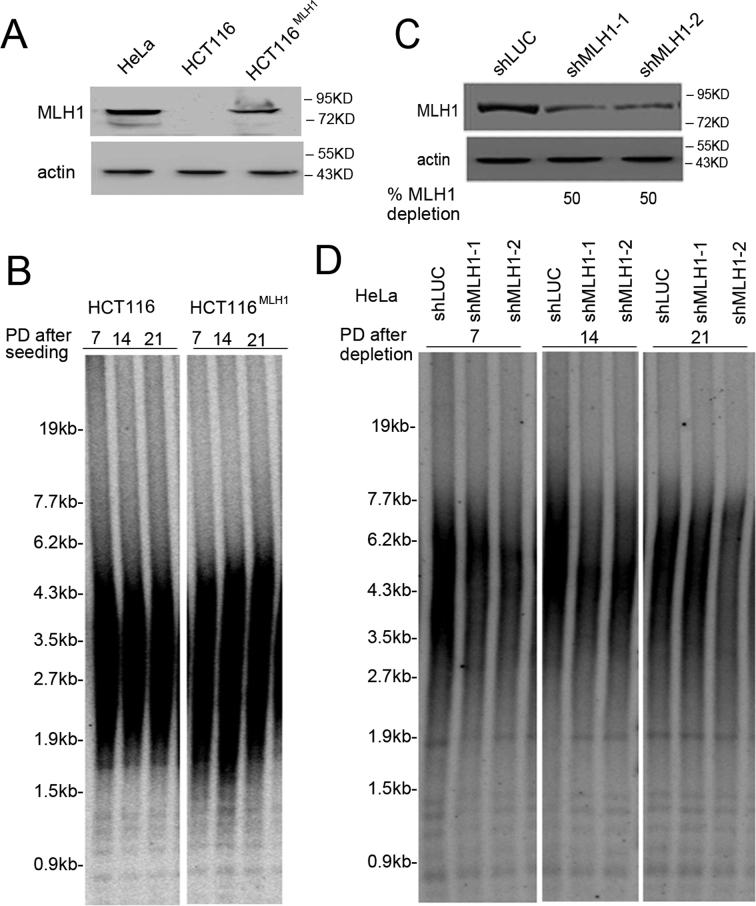
To determine whether MLH1 deficiency impacted telomere maintenance, we first examined telomere length with the TRF assay using two cell lines from different origins: first, the MLH1-deficient human colorectal tumor cell line HCT116 and its isogenic cell line complemented with wild-type MLH1, HCT116MLH1 (39) and second, HeLa depleted of MLH1 with two shRNA sequences. Western blotting confirmed the absence of MLH1 expression in HCT116 and MLH1 knockdown in HeLa (Figure 1A and C). Due to the partial depletion of MLH1 in HeLa cells, we then used an independent assay to test MLH1 deficiency. It is well established that MLH1 deficiency leads to resistance against the cytotoxicity induced by MNNG, an alkylating agent that induces DNA damage (40–42). As shown in Supplementary Figure S1, both HCT116 and HeLa shMLH1 knockdown cells displayed resistance to MNNG, further confirming MLH1 deficiency. TRF analysis showed no significant difference in telomere length between HCT116 and HCT116MLH1 (Figure 1B), or between HeLa control and MLH1 knockdown cells (Figure 1D).
Figure 1.
MLH1 deficiency has little effect on telomere maintenance. (A) Immunoblotting shows the absence of MLH1 in HCT116 and the restore of MLH1 expression in HCT116MLH1. (B) Telomere length measured by TRF analysis in HCT116 and HCT116MLH1. (C) Immunoblotting shows MLH1 knockdown in HeLa cells. Two shRNA sequences were used to deplete endogenous MLH1. Cells were collected at 7 days after puromycin selection. (D) Telomere length measured by TRF analysis in HeLa expressing MLH1 shRNA .
We also performed telomere FISH to analyze various telomere abnormalities including end-to-end fusions, fragile telomeres, and telomere sister chromatid exchanges (T-SCE). No significant change was observed between control and MLH1 deficient cells in either HCT116 or HeLa cell lines (Supplementary Figure S2). Taken together, we conclude that MLH1 deficiency has little effect on telomere homeostasis in telomerase-positive cancer cells. Our results are consistent with previous reports showing that MLH1 deficiency yields little effect on global telomere length or T-SCE in colon cancer cells (43), and that telomere shortening rate was not significantly affected by MLH1 deficiency in leukocytes derived from Lynch syndrome patients (44).
MLH1 deficiency increases TSI
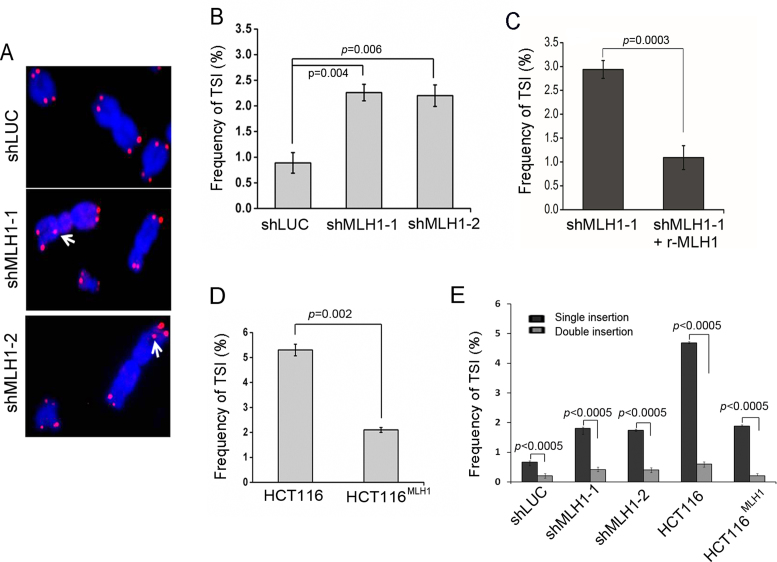
While analyzing metaphase chromosomes, we observed that silencing MLH1 by two different shRNA sequences increased the formation of ITSs (Figure 2A and B). In this experiment, double-blinded FISH analysis was performed to eliminate possible human bias. Expressing RNAi-resistant MLH1 in knockdown cells restored TSI suppression (Figure 2C), suggesting that TSI was specifically induced by MLH1 deficiency.
Figure 2.
MLH1 suppression induces TSI at non-telomeric sites. (A) Metaphase chromosomes from MLH1 suppressed HeLa cells (8 days after puromycin selection) were hybridized by telomeric probe. Representative FISH images are shown. White arrow points to inserted telomeric sequences at intra-chromosomal loci. (B) Frequency of TSI (expressed as % chromosomes with inserted telomere sequences) in HeLa shMLH1 and control shLUC cells. Two tailed t-test was used to calculate statistical significance in all data presented in this manuscript unless state otherwise. Error bars are SEM in all experiments shown in this manuscript. (C) Frequency of TSI after rescuing MLH1 knockdown. RNAi-resistant wild-type MLH1 was expressed in HeLa cells, followed by shRNA expression to knockdown endogenous MLH1. TSI was detected by FISH. (D) Frequency of TSI in HCT116 cells and HCT116MLH1. (E) Percentage of TSI events appearing on a single sister chromatid (single insertion) vs TSI in two sister chromatids (double insertion). In each experiment, >1500 chromosomes from each sample were analyzed and each experiment was repeated with three independent replicates.
To test whether this phenotype was cell line specific, we performed telomere–FISH in HCT116 and it isogeneic MLH1-complemented cell line HCT116MLH1. Similar to HeLa cells, HCT116 cells showed elevated TSI frequency, and complementing MLH1 in HCT116 reduced the frequency of TSI (Figure 2D). Therefore, we conclude that MLH1 deficiency induces TSI.
For easy visualization to readers, only images containing strongly stained TSI signal are presented in main figures. However, it is important to point out that the majority of TSI signals were weak (<500 bp) (Supplementary Figure S3), which was within the range of telomeric repeats that telomerase can add in a single cell cycle (45,46). It is possible that longer insertions may have arisen from expansion of telomeric repeats at preexisting ITSs.
Although the frequency of TSI observed in our study was low, with ∼2-5% of chromosomes containing TSI in MLH1-deficient cells, it is highly plausible that a portion of TSI events might have fallen below the threshold required for FISH detection, and therefore, the actual frequency of TSI may be higher than that detected by telomere–FISH in this study. In addition, we noticed that the appearance of TSI was asymmetrical, with TSI being predominantly found on one sister chromatid (single insertion) rather than both sister chromatids of one chromosome (double insertion) in both HeLa and HCT116 cell lines (Figure 2E), indicating that the majority of TSIs are not stably inherited in cell proliferation.
TSI is elevated by DSB inducer and can be abolished by ATM or ATR inhibition
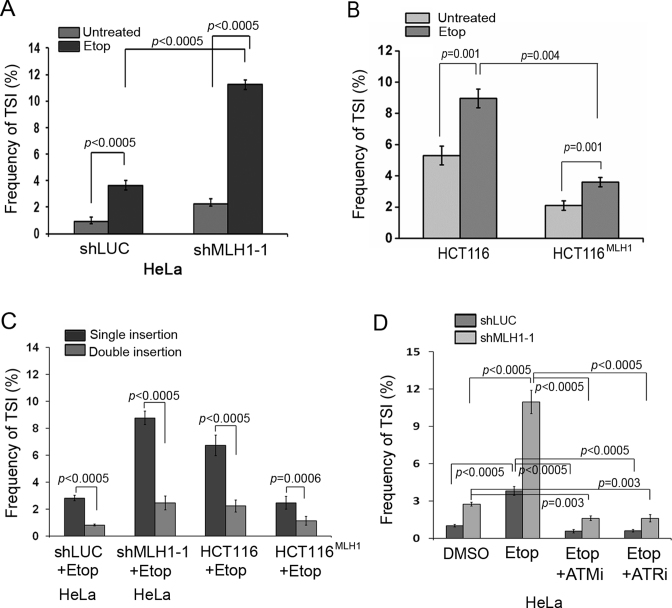
The incorporation of telomeric sequences at interstitial chromosome loci is reminiscent of DNA break rejoining. If TSI takes place at DNA breaks, we would expect that generation of more breaks would increase the frequency of TSI. To test this possibility, we treated both MLH1-proficient and -deficient cells with DSB inducer etoposide (3 μM) for 2 h. Cells were then allowed to repair the induced damage after etoposide removal prior to metaphase collection. Etoposide treatment induced a marked increase in TSI frequency in HeLa with MLH1 knockdown as well as in the MLH1-deficient HCT116 (Figure 3A and B), indicating that TSI might occur at or near DSBs. Similar to untreated cells, the majority of TSI events occurred on one sister chromatid (Figure 3C).
Figure 3.
MLH1 deficiency-induced TSI is elevated by etoposide and is abolished by ATM or ATR inhibition. HeLa expressing shMLH1 or control shLUC, HCT116, and HCT116MLH1 were treated with or without etoposide (3 μM) for 2 h and recovered for 24 h after drug removal. Metaphase cells from drug treated and untreated samples were analyzed by telomere FISH. (A) Frequency of TSI measured in HeLa expressing shLUC and shMLH1 following etoposide treatment. (B) Frequency of TSI measured in HCT116 cells and HCT116MLH1 following etoposide treatment. (C) Percentage of TSI events on a single sister chromatid (single insertion) versus TSI events on two sister chromatids (double insertion) following etoposide treatment. (D) Frequency of TSI measured in HeLa-shLUC and HeLa-shMLH1-1 treated with 10 μM ATM inhibitor (ATMi) or 10 μM ATR inhibitor (ATRi). In each experiment, >1500 chromosomes from each sample were analyzed and each experiment was repeated with three independent replicates.
The key regulators for sensing and repairing DNA damage in mammalian cells are ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) protein kinases. Since a DSB inducer increased the frequency of TSI, we then tested whether TSI was controlled by the ATM or ATR pathway. HeLa MLH1 knockdown cells were concurrently treated with etoposide and a specific inhibitor of ATM (KU55933) (47) or ATR (VE-821) (48). Inhibition of the ATM or ATR activity was confirmed by examining the abolishment of Chk2 and Chk1 phosphorylation (Supplementary Figure S4). As shown in Figure 3D, ATM or ATR inhibition abolished etoposide-induced TSI, suggesting that the ATM/ATR damage response pathway regulates TSI. We also noticed that ATM and ATR inhibition reduced TSI in untreated control cells, suggesting that ATM/ATR inhibition may also suppress TSI caused by spontaneous DBS breaks (49).
The N-terminal, but not the C-terminal domain of MLH1 is required for TSI suppression
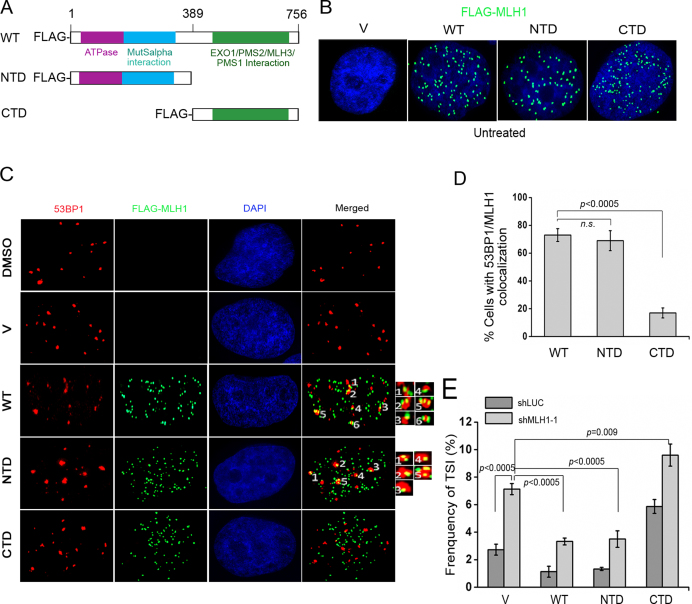
Given that MLH1 has been implicated in signaling DNA damage (27,29,50,51), and inhibition of ATM/ATR abolished TSI (Figure 3D), we reasoned that the damage signaling function of MLH1 might play a role in suppressing TSI. We then attempted to use domain-specific MLH1 mutants to test this hypothesis. Crystal structures of MLH1 orthologues reveal that MLH1 consists of a conserved N-terminal domain (NTD) and a C-terminal domain (CTD) that are connected with a linker region (52–54) (Figure 4A). The NTD of MLH1 possesses an ATPase activity that is required for its MMR function and also orchestrates DNA DSBs end processing (55). The CTD is necessary for heterodimerization with PMS2 and MLH3 (56) and is also responsible for interacting with other proteins including EXO1 or BLM (57), and therefore is needed for mismatch recognition and excision during the MMR process. It has been reported that the NTD of human MLH1 localizes at DSBs, while the CTD alone fails to do so (27). Thus, in this set of experiments, we utilized the RNAi-resistant full-length MLH1 and two domain-specific truncations, the NTD (containing amino acids 2–389) and CTD (containing amino acids 390–756) to determine whether the damage signaling function of MLH1 played a role in suppressing TSI. The WT and two mutants were stably expressed in HeLa cells using retroviral induction. Immunofluorescence staining showed that both truncated proteins predominantly localized in nucleus with expression comparable to WT MLH1 (Figure 4B). These cells were then treated with etoposide, fixed, and subjected to co-immunostaining with FLAG and 53BP1 antibodies. Both the wild-type MLH1 and the NTD were recruited to damage sites, as shown by colocalization of wild-type MLH1 or NTD with 53BP1 foci (Figure 4C and D). In contrast, the majority of CTD failed to co-localize with 53BP1 (Figure 4C and D). Although it appeared visually that colocalization might have arisen from random overlaps of numerous FLAG-MLH1 foci with 53BP1, all images were taken in Z-stacks and only colocalizations present in multiple Z-stacks were considered during image analysis. Furthermore, CTD staining was equally abundant as WT and NTD, yet little random overlap existed between CTD and 53BP1 (Figure 4C and D). We therefore considered the observed MLH1/53BP1 colocalization to be valid. Our results suggest that deletion of the N-terminus of MLH1 abrogates MLH1's recruitment to DSBs, consistent with previous report that the NTD but not the CTD is required for MLH1 localization to DSBs induced by laser micro-irradiation (27).
Figure 4.
The NTD but not the CTD of MLH1 rescues TSI caused by MLH1 deficiency. (A) Schematic representation of functional domains in MLH1 and its deletion mutants. Purple box: ATPase domain. Blue box: MutSα-interacting domain. Green box: EXO1/PMS2/MLH3-binding domain. (B) Truncated FLAG-MLH1 proteins localize in nucleus in untreated cells. WT and truncated FLAG-MLH1 were stably expressed with retroviral transduction. FLAG antibody (green) was used for IF to detect FLAG-tagged WT MLH1 and mutants. (C) Recruitment of full-length MLH1 (WT) and NTD but not CTD of MLH1 to DNA damage sites induced by etoposide treatment (0.3 μM, 2 h). After etoposide treatment, IF was performed with anti-53BP1 (red) and anti-FLAG (green) in cells expressing WT or mutated FLAG-MLH1. Representative co-localizations of 53BP1 and FLAG-MLH1 were labeled with numbers and then enlarged in insets. (D) Percentage of cells with MLH1/53BP1 co-localization (>5 colocalizations per cell). (E) Frequency of TSI measured in HeLa shLUC and shMLH1 cells with concurrent expression of vector (v), WT-MLH1, NTD, or CTD. Cells were treated with 3 μM etoposide for 2 h and then recovered for 24 h prior to FISH. In each experiment, >1500 chromosomes from each sample were analyzed and each experiment was repeated with three independent replicates.
Next, we knocked down endogenous MLH1 in cells stably expressing the RNAi-resistant WT, NTD or CTD (Supplementary Figure S5). As shown in Figure 4E, expression of either WT MLH1 or the NTD reduced TSI to the level comparable to that in MLH1-proficient shLUC control cells, suggesting that both WT and the NTD were able to suppress TSI. In contrast, expression of CTD alone failed to suppress TSI (Figure 4E). Because CTD was defective in DSB response, and NTD alone was sufficient to recruit MLH1 to DSBs (Figure 4C and D), our data imply that MLH1's role in DNA damage response and/or mediating damage signaling may be involved in TSI suppression. It is noticeable that expression of CTD alone increased TSI frequency in shLUC control cells in the presence of endogenous MLH1, indicating that CTD might be a dominant-negative mutant that interferes with WT MLH1 function.
MLH1 deficiency-induced TSI is independent of p53 and Rb
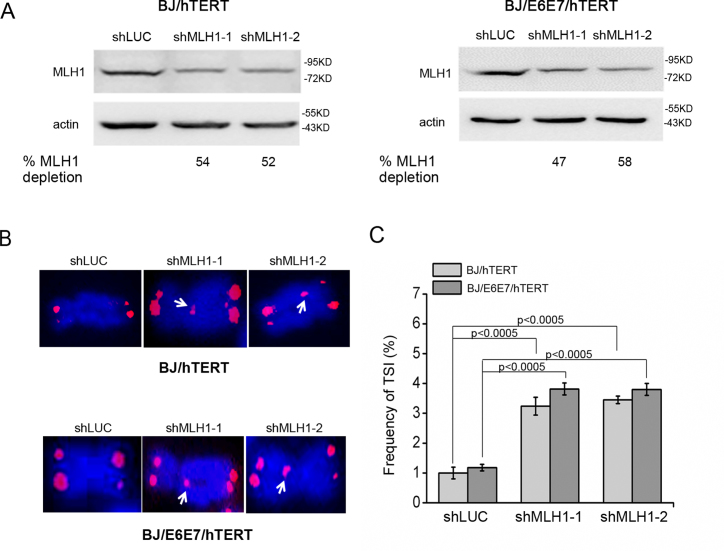
Since MMR proteins modulate the p53-mediated damage signaling pathway and cell cycle arrest in response to both DNA damage and dysfunctional telomeres (29,50,51,58–61), we then determined whether the p53 or Rb pathway was involved in regulating TSI. MLH1 was knocked down in telomerase-immortalized BJ foreskin fibroblasts (BJ/hTERT), which maintain functional p53 and Rb pathways, and BJ/E6E7/hTERT cells (Figure 5A), which ectopically express human papillomavirus E6 and E7 proteins that fully inhibit the functions of p53 and Rb (Supplementary Figure S6). We found that suppression of MLH1 in both cells induced TSI at comparable levels (Figure 5B, C). Thus, the induction of TSI is independent of the functional status of p53 or Rb.
Figure 5.
MLH1 deficiency-induced TSI is independent of p53 or Rb. (A) Western blot of MLH1 knockdown in BJ/hTERT and BJ/E6E7/hTERT cells. Two sequences were used to deplete endogenous MLH1. (B) Telomere-FISH of BJ/hTERT and BJ/E6E7/hTERT cells with MLH1 or control knockdown. White arrows indicate inserted telomeric sequences. (C) Frequency of TSI in BJ/hTERT and BJ/E6E7/hTERT after MLH1 depletion. In each experiment, >1500 chromosomes from each sample were analyzed and each experiment was repeated with three independent replicates.
MLH1 deficiency-induced TSI is dependent on telomerase
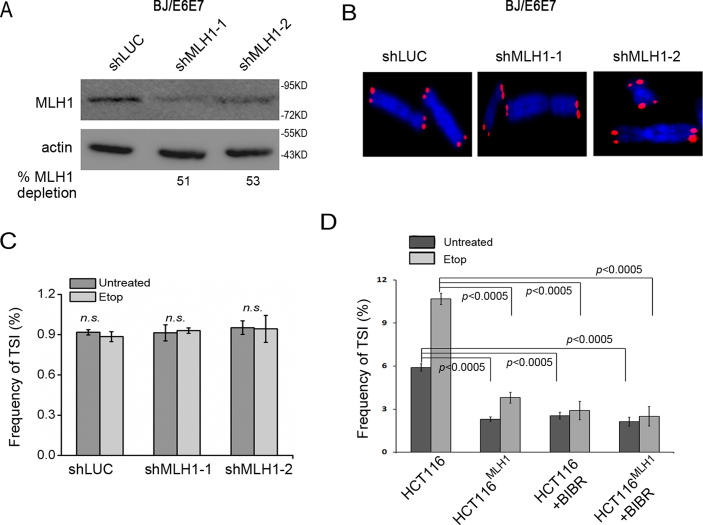
Since telomerase is able to add telomeric repeats to free 3΄ ends at DNA breaks with weak sequence homology to telomerase RNA (62,63), we next set out to determine the potential involvement of telomerase in TSI generation. We depleted MLH1 in telomerase-negative BJ/E6E7 fibroblasts (Figure 6A). Suppression of MLH1 in BJ/E6E7 did not induce TSI in either untreated or etoposide-treated cells (Figure 6B and C). The failure to observe TSI in BJ/E6E7 was unlikely due to incomplete depletion of MLH1, as MLH1 depletion in BJ/E6E7 was comparable to that in other telomerase-expressing cell lines used in this study (Figure 6A).
Figure 6.
MLH1 deficiency-induced TSI is dependent on telomerase. (A) Western blot of MLH1 knockdown in BJ/E6E7 cells. (B) Telomere-FISH of BJ/E6E7 cells with MLH1 or control knockdown (LUC). (C) Frequency of TSI in BJ/E6E7 treated with or without etoposide. (D) Frequency of TSI in HCT116 and HCT116MLH1 treated with or without telomerase inhibitor BIBR1532 (10 μM, 24 h) and etoposide (3 μM, 2 h). In each experiment, >1500 chromosomes from each sample were analyzed and each experiment was repeated with three independent replicates.
To firmly test whether telomerase activity was required for TSI, we inhibited telomerase activity in MLH1-deficient HCT116 cells with a telomerase inhibitor BIBR1532 alone or with BIBR1532 along with etoposide (64). As shown in Figure 6D, telomerase inhibition reduced TSI frequency to baseline regardless of etoposide treatment. Together, our data suggest that TSI requires telomerase activity.
The inserted ITSs are unstable and TSI correlates with the increase of chromosome instabilities
Several observations in our study suggest that the newly-formed ITSs are unstable. First, the majority of ITSs were present at one sister chromatid instead of both chromatids (Figures 2E and 3C). If these sequences were stable, they would be duplicated into sister chromatids in the next cell cycle and appear at both chromatids. Second, restoring MLH1 expression in MLH1-deficient HCT116 reduced the level of TSI that preexisted in HCT116 (Figure 2D). Similarly, telomerase inhibition decreased preexisting TSIs in MLH1-deficient cells (Figure 6D), suggesting that formation of new ITSs was blocked while the preexisting ITSs were lost. These observations were consistent with previous reports that ITSs display higher levels of instability (8–13).
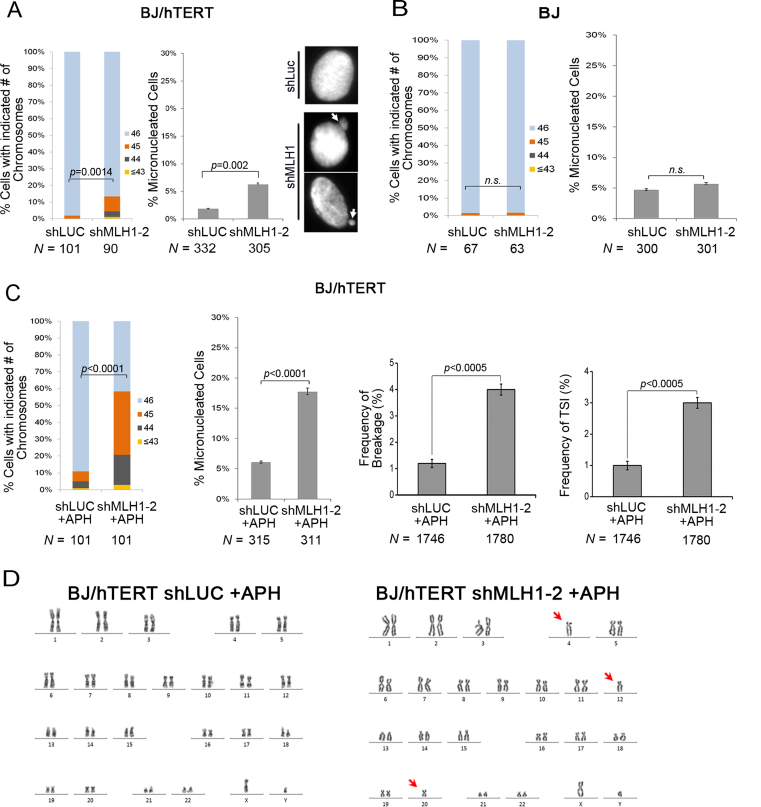
We speculated that elevated levels of TSI could potentially induce chromosome instability. Since cancer cell lines often contain multiple genetic changes, we avoided the MLH1-deficient tumor cell line HCT116. To examine chromosome instabilities after MLH1 suppression, karyotyping analysis was performed in BJ/hTERT, a cell line that is derived from normal primary foreskin fibroblasts by telomerase immortalization and displays normal diploidy with a stable genome. As shown in Figure 7A, MLH1 knockdown led to a statistically significant increase in chromosome loss. In addition, MLH1 deficiency induced a moderate but reproducible increase in micronuclei (MN) formation (Figure 7A), another indicator of chromosome instability (65–68).
Figure 7.
TSI correlates with chromosome instabilities. (A) MLH1 deficiency resulted in an increase in chromosome loss and MN formation in BJ/hTERT. N denotes the number of metaphase spreads (for chromosome loss) or the number of interphase cells (for MN formation) analyzed in each sample. White arrows point to MN formed in shMLH1 expressing cells. (B) Chromosome instabilities and MN formation in BJ cells with MLH1 deficiency. (C) Frequency of chromosome loss, MN formation, chromosome breakage and %TSI in BJ/hTERT control and shMLH1 cells under replication stress (0.3 μM APH, 24 h). (D) An example of karyotypes of BJ/hTERT MLH1 knockdown cells after APH treatment. Arrows point to chromosome loss. Each experiment was repeated with three independent replicates. Chromosome loss was evaluated using a binomial Z-statistic to compare the proportion of intact metaphase spreads (spreads with 46 chromosome).
MMR-deficient cells usually show microsatellite instability but display low levels of genome instabilities (69–71). However, several studies have shown that MLH1-deficient HCT116 cells are heterogeneous, with 5–10% of clones propagated from parental cells containing chromosome gain and losses (69,71–73). In our study, a similar percentage of MLH1 knockdown cells (∼5–10%) displayed chromosome loss and MN (Figure 7A). Therefore, our observation is consistent with chromosome instabilities reported in MLH1-deficient cells.
To determine whether chromosome instabilities observed in BJ/hTERT cells were induced by MLH1 deficiency or by TSI, we measured MN formation and chromosome loss in telomerase-negative BJ cells as well as BJ/hTERT treated with telomerase inhibitor. If chromosome instabilities were caused by MLH1 deficiency per se and irrelevant to TSI, MLH1 depletion would be expected to induce chromosome instabilities regardless of telomerase expression. Conversely, if chromosome instabilities were associated with TSI, then telomerase activity, which is required for TSI (Figure 6), would also be needed for the increase of chromosome instabilities in MLH1 deficient cells. MLH1 knockdown did not induce MN formation or chromosome loss in either telomerase-negative BJ cells or telomerase-inhibited BJ/hTERT cells, as shown in Figure 7B and Supplementary Figure S7. Thus, our results suggest that TSI correlates with chromosome instabilities and MN formation.
Since telomeric repeats are fragile sites and are prone to breakage under replication stress (74), we then performed the above analyses in cells treated with low concentration of aphidicolin (APH), which induces replication stress. APH treatment markedly increased the frequency of chromosome loss and MN formation (Figure 7C, D). Meanwhile, we also observed that MLH1 knockdown further elevated the frequency of chromosome breakage in APH treated cells (Figure 7C). The increase in chromosome instabilities appeared to correlate with the increase in TSI frequency (compare % TSI and % breakage in Figure 7C). Similar correlation was observed in MLH1-deficient BJ/E6E7/hTERT and HeLa cells treated with APH (Supplementary Figures S8B and S8C). Because the majority of TSI signals were weak (Supplementary Figure S3) and the frequency of TSI was low, we failed to find sufficient number of TSI-containing broken chromosomes to determine whether chromosome breakage took place at TSI sites. Nonetheless, such increase was absent in APH-treated telomerase-negative BJ cells (Supplementary Figure S8D), indicating that elevated TSI levels likely contributed to the observed chromosome breakage, in agreement with previous observations that ITSs are prone to APH-induced breakage (75,76). It remains to be determined whether TSI directly causes genome instability or other mechanisms are involved.
DISCUSSION
Using unrelated tumor cell lines and telomerase-immortalized human somatic cells, we unveil a new functional facet of MLH1 in the maintenance of genome stability in that MLH1 suppresses the appearance of telomeric sequences at intra-chromosomal regions. This is the first report that TSI can occur in telomerase-positive cells. Our results suggest that TSI likely requires telomerase, responds to DSB inducer, and is controlled by the ATM/ATR DNA damage response pathway. Furthermore, an MLH1 mutant defective in DSB response fails to suppress TSI, suggesting that MLH1's function in DNA damage response may play an important role in inhibiting TSI. We propose that MLH1 prevents telomerase from erroneously adding telomeric sequences at DSBs in response to DNA damage.
Possible mechanism for TSI in telomerase-positive cells
TSI could potentially be caused by: (a) telomerase directly adding telomeric repeats at DNA breaks during DNA damage repair; (b) translocation of telomeric sequences to interstitial regions; (c) expansion/amplification of preexisting ITSs due to replication slippage in a way similar to trinucleotide expansion; or (d) telomere tethering to intra-chromosomal loci via chromosome looping as described in telomerase-negative myoblast cells (77) and ALT cells (13). While scenario (a) requires telomerase, scenarios (b), (c) and (d) are believed to be independent of telomerase. Our results show that TSI is dependent on telomerase and DSB inducer elevates TSI (Figures 3 and 6), thus favoring the hypothesis that telomerase may be responsible for the addition of telomeric sequences at DNA breaks during the repair process. The proposed involvement of telomerase in TSI is not surprising, since telomerase only needs very minimal sequence homology to recognize the substrate (63) and is able to add telomeric sequences de novo onto non-telomeric DNA breaks (78–84).
Our findings suggest a possible mechanism involving MLH1 in inhibiting TSI in telomerase-positive human cells. While the involvement of MLH1 in suppressing TSI may seem unexpected, it is consistent with the well documented role of MMR proteins in DNA damage response and repair (85). Previous studies have shown that MMR proteins including MLH1 are recruited to damaged sites in response to DSBs (27,28). These proteins participate in DSB repair especially NHEJ and most likely influence the outcome of repair (17,20,25,29,55,86–90). In addition, the MMR proteins promote cell cycle arrest and/or programmed cell death in response to certain types of DNA damage as well as dysfunctional telomeres (16–21,60,61,91,92). Our current study confirms the recruitment of MLH1 to DSBs (Figure 4C). Furthermore, using domain-specific mutants, we have identified that the domain required for MLH1 recruitment of DSBs is sufficient to suppress TSI (Figure 4E). Our results suggest that MLH1 plays important roles in processes distinct from MMR that are implicated in protecting genome stability and preventing tumorigenesis.
Two possible mechanisms may explain how MLH1 suppresses TSI. MLH1 may inhibit telomerase access to DNA breaks during the repair process, thereby preventing telomerase addition of TTAGGG repeats at damaged sites. While this model is appealing, an alternative explanation is that MLH1, likely coordinate with other proteins, may recognize the inserted telomeric sequences following telomerase-mediated insertion and remove these sequences. Our present data do not distinguish these two models, warranting the need for further studies to elucidate the mechanism underlying how MLH1 counteracts telomerase-mediated TSI.
Function of MMR proteins at chromosome ends
Telomeres consist of long tract of tandem TTAGGG repeats that potentially make telomeric DNA highly susceptible to replication errors caused by DNA polymerase slippage as well as G:U mismatches owing to cytosine deamination. Surprisingly, although MMR is expected to be the primary pathway for repairing mismatches and therefore for maintaining the fidelity of telomeric DNA replication, at present little is known about the MMR process at telomeres. Instead, results from yeast, mammalian and human cell systems suggest that defective MMR proteins have diverse effects on telomere maintenance. For instance, in budding yeast, loss of MLH1, MSH2 or PMS1 promotes the recombination-based ALT pathway in telomerase-deficient cells and enhances telomerase-independent cellular proliferation and survival (93). In humans, deficiency in human MSH6 facilitates the engagement of ALT when telomerase is inhibited (43), but it appears that this effect is specific to MSH6 deficiency, since neither loss of MLH1 nor MSH2 deficiency induces ALT (43). This suggests that a subset but not all MMR components may participate in repressing telomere DNA recombination in human cells. In addition, it has been reported that mammalian MMR proteins participate in signaling dysfunctional telomeres. Abrogation of MSH2 or PMS2 attenuates p21 induction in response to short telomeres through the p53 pathway, rescuing aging pathologies associated with short telomeres in telomerase-deficient mice (60,61). Data presented here, together with previous telomere studies on colon cancer cell line and MLH1-deficient leukocytes (43,44), support that MLH1 deficiency has negligible involvement in protecting telomere stability at the majority of chromosome ends under unchallenged condition. Thus, the roles of MMR proteins on telomere maintenance appear to be highly specific to each individual MMR gene, and cannot be generalized to deficiency in MMR. Additionally, studies on telomere maintenance under challenged conditions that elevate DNA mismatches are still lacking. It remains possible that MMR components may be needed for correcting telomeric mismatches and maintaining telomere stability under challenged conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr G. Li (University of Southern California Keck School of Medicine) for HCT116 and HCT116MLH1, and Ms O. Shiva and A. Al Soodani for technical help.
Author contributions: P.J. performed the majority of experiments. M.C. participated in Q-FISH, chromosome loss and micronuclei experiments. Y.Z. performed karyotyping experiments. P.J. and M.C. analyzed data and assembled figures. C.H. and W.C. directed the study and designed experiments. W.C. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM112864] (in part); Ladies Auxiliary VFW Cancer Research Funds (to W.C.). Funding for open access charge: NIH [R01GM112864].
Conflict of interest statement. None declared.
REFERENCES
- 1.Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 2.Longhese M.P. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008; 22:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin K.W., Yan J.. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutat. Res. 2008; 658:95–110. [DOI] [PubMed] [Google Scholar]
- 4.Azzalin C.M., Nergadze S.G., Giulotto E.. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma. 2001; 110:75–82. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni L., Attolini C., Tessera L., Mucciolo E., Giulotto E.. Telomeric and nontelomeric (TTAGGG)n sequences in gene amplification and chromosome stability. Genomics. 1994; 24:53–62. [DOI] [PubMed] [Google Scholar]
- 6.Bouffler S.D., Morgan W.F., Pandita T.K., Slijepcevic P.. The involvement of telomeric sequences in chromosomal aberrations. Mutat. Res. 1996; 366:129–135. [DOI] [PubMed] [Google Scholar]
- 7.Slijepcevic P., Xiao Y., Dominguez I., Natarajan A.T.. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma. 1996; 104:596–604. [DOI] [PubMed] [Google Scholar]
- 8.Kilburn A.E., Shea M.J., Sargent R.G., Wilson J.H.. Insertion of a telomere repeat sequence into a mammalian gene causes chromosome instability. Mol. Cell. Biol. 2001; 21:126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aksenova A.Y., Greenwell P.W., Dominska M., Shishkin A.A., Kim J.C., Petes T.D., Mirkin S.M.. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19866–19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park V.M., Gustashaw K.M., Wathen T.M.. The presence of interstitial telomeric sequences in constitutional chromosome abnormalities. Am. J. Hum. Genet. 1992; 50:914–923. [PMC free article] [PubMed] [Google Scholar]
- 11.Day J.P., Limoli C.L., Morgan W.F.. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis. 1998; 19:259–265. [DOI] [PubMed] [Google Scholar]
- 12.Slijepcevic P., Xiao Y., Natarajan A.T., Bryant P.E.. Instability of CHO chromosomes containing interstitial telomeric sequences originating from Chinese hamster chromosome 10. Cytogenet. Cell Genet. 1997; 76:58–60. [DOI] [PubMed] [Google Scholar]
- 13.Marzec P., Armenise C., Perot G., Roumelioti F.M., Basyuk E., Gagos S., Chibon F., Dejardin J.. Nuclear-Receptor-Mediated Telomere Insertion Leads to Genome Instability in ALT Cancers. Cell. 2015; 160:913–927. [DOI] [PubMed] [Google Scholar]
- 14.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008; 18:85–98. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh P., Yamane K.. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008; 129:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond J.T., Anthoney A., Brown R., Modrich P.. Cisplatin and adriamycin resistance are associated with MutLalpha and mismatch repair deficiency in an ovarian tumor cell line. J. Biol. Chem. 1996; 271:19645–19648. [DOI] [PubMed] [Google Scholar]
- 17.Siehler S.Y., Schrauder M., Gerischer U., Cantor S., Marra G., Wiesmuller L.. Human MutL-complexes monitor homologous recombination independently of mismatch repair. DNA Repair (Amst.). 2009; 8:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q., Vasquez K.M.. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008; 4:e1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyers M., Wagner M.W., Mazurek A., Schmutte C., Fishel R., Boothman D.A.. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J. Biol. Chem. 2005; 280:5516–5526. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y., Lin F.T., Lin W.C.. ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol. Cell. Biol. 2004; 24:6430–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao N., Zhu F., Yuan F., Haick A.K., Fukushige S., Gu L., Her C.. The interplay between hMLH1 and hMRE11: role in MMR and the effect of hMLH1 mutations. Biochem. Biophys. Res. Commun. 2008; 370:338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worth L. Jr, Clark S., Radman M., Modrich P.. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tham K.C., Hermans N., Winterwerp H.H., Cox M.M., Wyman C., Kanaar R., Lebbink J.H.. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Mol. Cell. 2013; 51:326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rayssiguier C., Thaler D.S., Radman M.. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989; 342:396–401. [DOI] [PubMed] [Google Scholar]
- 25.Elliott B., Jasin M.. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 2001; 21:2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villemure J.F., Abaji C., Cousineau I., Belmaaza A.. MSH2-deficient human cells exhibit a defect in the accurate termination of homology-directed repair of DNA double-strand breaks. Cancer Res. 2003; 63:3334–3339. [PubMed] [Google Scholar]
- 27.Hong Z., Jiang J., Hashiguchi K., Hoshi M., Lan L., Yasui A.. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J. Cell Sci. 2008; 121:3146–3154. [DOI] [PubMed] [Google Scholar]
- 28.Shahi A., Lee J.H., Kang Y., Lee S.H., Hyun J.W., Chang I.Y., Jun J.Y., You H.J.. Mismatch-repair protein MSH6 is associated with Ku70 and regulates DNA double-strand break repair. Nucleic Acids Res. 2011; 39:2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bannister L.A., Waldman B.C., Waldman A.S.. Modulation of error-prone double-strand break repair in mammalian chromosomes by DNA mismatch repair protein Mlh1. DNA Repair (Amst.). 2004; 3:465–474. [DOI] [PubMed] [Google Scholar]
- 30.Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996; 13:336–342. [DOI] [PubMed] [Google Scholar]
- 31.Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R. et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996; 85:1125–1134. [DOI] [PubMed] [Google Scholar]
- 32.Wu X., Tsai C.Y., Patam M.B., Zan H., Chen J.P., Lipkin S.M., Casali P.. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J. Immunol. 2006; 176:5426–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casali P., Pal Z., Xu Z., Zan H.. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006; 27:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbert B., Shay J.W., Wright W.E.. Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM. Current Protocols in Cell Biology. 2003; John Wiley & Sons, Inc; Unit 18.16. [Google Scholar]
- 35.Huang C., Dai X., Chai W.. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012; 22:1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lansdorp P.M., Verwoerd N.P., van de Rijke F.M., Dragowska V., Little M.T., Dirks R.W., Raap A.K., Tanke H.J.. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996; 5:685–691. [DOI] [PubMed] [Google Scholar]
- 37.Ji G., Liu K., Okuka M., Liu N., Liu L.. Association of telomere instability with senescence of porcine cells. BMC Cell Biol. 2012; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barch M.J., Knutsen T., Spurbeck J.L.. The AGT Cytogenetics Laboratory Manual. 1997; 3rd edn, Lippincott-Raven Publishers; 269–280. [Google Scholar]
- 39.Koi M., Umar A., Chauhan D.P., Cherian S.P., Carethers J.M., Kunkel T.A., Boland C.R.. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N'-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994; 54:4308–4312. [PubMed] [Google Scholar]
- 40.Kat A., Thilly W.G., Fang W.H., Longley M.J., Li G.M., Modrich P.. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branch P., Aquilina G., Bignami M., Karran P.. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993; 362:652–654. [DOI] [PubMed] [Google Scholar]
- 42.Carethers J.M., Hawn M.T., Chauhan D.P., Luce M.C., Marra G., Koi M., Boland C.R.. Competency in mismatch repair prohibits clonal expansion of cancer cells treated with N-methyl-N'-nitro-N-nitrosoguanidine. J. Clin. Invest. 1996; 98:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W.. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004; 64:3444–3451. [DOI] [PubMed] [Google Scholar]
- 44.Bozzao C., Lastella P., Ponz de Leon M., Pedroni M., Di Gregorio C., D'Ovidio F.D., Resta N., Prete F., Guanti G., Stella A.. Analysis of telomere dynamics in peripheral blood cells from patients with Lynch syndrome. Cancer. 2011; 117:4325–4335. [DOI] [PubMed] [Google Scholar]
- 45.Greider C.W. Telomerase is processive. Mol. Cell. Biol. 1991; 11:4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lue N.F. Adding to the ends: what makes telomerase processive and how important is it. Bioessays. 2004; 26:955–962. [DOI] [PubMed] [Google Scholar]
- 47.Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- 48.Toledo L.I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J.R., Fernandez-Capetillo O.. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 2011; 18:721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marechal A., Zou L.. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Rohde L.H., Emami K., Hammond D., Casey R., Mehta S.K., Jeevarajan A.S., Pierson D.L., Wu H.. Suppressed expression of non-DSB repair genes inhibits gamma-radiation-induced cytogenetic repair and cell cycle arrest. DNA Repair (Amst.). 2008; 7:1835–1845. [DOI] [PubMed] [Google Scholar]
- 51.Li H.R., Shagisultanova E.I., Yamashita K., Piao Z., Perucho M., Malkhosyan S.R.. Hypersensitivity of tumor cell lines with microsatellite instability to DNA double strand break producing chemotherapeutic agent bleomycin. Cancer Res. 2004; 64:4760–4767. [DOI] [PubMed] [Google Scholar]
- 52.Gueneau E., Dherin C., Legrand P., Tellier-Lebegue C., Gilquin B., Bonnesoeur P., Londino F., Quemener C., Le Du M.H., Marquez J.A. et al. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 2013; 20:461–468. [DOI] [PubMed] [Google Scholar]
- 53.Ban C., Yang W.. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998; 95:541–552. [DOI] [PubMed] [Google Scholar]
- 54.Wu H., Zeng H., Lam R., Tempel W., Kerr I.D., Min J.. Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome. Acta Crystallogr. F, Struct. Biol. Commun. 2015; 71:981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chahwan R., van Oers J.M., Avdievich E., Zhao C., Edelmann W., Scharff M.D., Roa S.. The ATPase activity of MLH1 is required to orchestrate DNA double-strand breaks and end processing during class switch recombination. J. Exp. Med. 2012; 209:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang Q., Prolla T.A., Liskay R.M.. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol. 1997; 17:4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dherin C., Gueneau E., Francin M., Nunez M., Miron S., Liberti S.E., Rasmussen L.J., Zinn-Justin S., Gilquin B., Charbonnier J.B. et al. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 2009; 29:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duckett D.R., Bronstein S.M., Taya Y., Modrich P.. hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:12384–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters A.C., Young L.C., Maeda T., Tron V.A., Andrew S.E.. Mammalian DNA mismatch repair protects cells from UVB-induced DNA damage by facilitating apoptosis and p53 activation. DNA Repair (Amst.). 2003; 2:427–435. [DOI] [PubMed] [Google Scholar]
- 60.Martinez P., Siegl-Cachedenier I., Flores J.M., Blasco M.A.. MSH2 deficiency abolishes the anticancer and pro-aging activity of short telomeres. Aging Cell. 2009; 8:2–17. [DOI] [PubMed] [Google Scholar]
- 61.Siegl-Cachedenier I., Munoz P., Flores J.M., Klatt P., Blasco M.A.. Deficient mismatch repair improves organismal fitness and survival of mice with dysfunctional telomeres. Genes Dev. 2007; 21:2234–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morin G.B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991; 353:454–456. [DOI] [PubMed] [Google Scholar]
- 63.Gavory G., Farrow M., Balasubramanian S.. Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vitro. Nucleic Acids Res. 2002; 30:4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pascolo E., Wenz C., Lingner J., Hauel N., Priepke H., Kauffmann I., Garin-Chesa P., Rettig W.J., Damm K., Schnapp A.. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002; 277:15566–15572. [DOI] [PubMed] [Google Scholar]
- 65.Fenech M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., Thomas P.. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011; 26:125–132. [DOI] [PubMed] [Google Scholar]
- 66.Lahdetie J., Keiski A., Suutari A., Toppari J.. Etoposide (VP-16) is a potent inducer of micronuclei in male rat meiosis: spermatid micronucleus test and DNA flow cytometry after etoposide treatment. Environ. Mol. Mutag. 1994; 24:192–202. [DOI] [PubMed] [Google Scholar]
- 67.Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S.W., Meyerson M., Pellman D.. Chromothripsis from DNA damage in micronuclei. Nature. 2015; 522:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leibowitz M.L., Zhang C.Z., Pellman D.. Chromothripsis: a new mechanism for rapid karyotype evolution. Annu. Rev. Genet. 2015; 49:183–211. [DOI] [PubMed] [Google Scholar]
- 69.Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eshleman J.R., Casey G., Kochera M.E., Sedwick W.D., Swinler S.E., Veigl M.L., Willson J.K., Schwartz S., Markowitz S.D.. Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53. Oncogene. 1998; 17:719–725. [DOI] [PubMed] [Google Scholar]
- 71.Lengauer C., Kinzler K.W., Vogelstein B.. Genetic instability in colorectal cancers. Nature. 1997; 386:623–627. [DOI] [PubMed] [Google Scholar]
- 72.Roschke A.V., Stover K., Tonon G., Schaffer A.A., Kirsch I.R.. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002; 4:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Masramon L., Ribas M., Cifuentes P., Arribas R., Garcia F., Egozcue J., Peinado M.A., Miro R.. Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet. Cytogenet. 2000; 121:17–21. [DOI] [PubMed] [Google Scholar]
- 74.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Musio A., Rainaldi G., Sbrana I.. Spontaneous and aphidicolin-sensitive fragile site 3cen co-localizes with the (TTAGGG)n telomeric sequence in Chinese hamster cells. Cytogenet. Cell Genet. 1996; 75:159–163. [DOI] [PubMed] [Google Scholar]
- 76.Bosco N., de Lange T.. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma. 2012; 121:465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robin J.D., Ludlow A.T., Batten K., Magdinier F., Stadler G., Wagner K.R., Shay J.W., Wright W.E.. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014; 28:2464–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flint J., Craddock C.F., Villegas A., Bentley D.P., Williams H.J., Galanello R., Cao A., Wood W.G., Ayyub H., Higgs D.R.. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 1994; 55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 79.Sprung C.N., Reynolds G.E., Jasin M., Murnane J.P.. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao Q., Reynolds G.E., Wilcox A., Miller D., Cheung P., Artandi S.E., Murnane J.P.. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair (Amst.). 2008; 7:1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muller F., Wicky C., Spicher A., Tobler H.. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell. 1991; 67:815–822. [DOI] [PubMed] [Google Scholar]
- 82.Bottius E., Bakhsis N., Scherf A.. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol. 1998; 18:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu G.L., Blackburn E.H.. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991; 67:823–832. [DOI] [PubMed] [Google Scholar]
- 84.Kramer K.M., Haber J.E.. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993; 7:2345–2356. [DOI] [PubMed] [Google Scholar]
- 85.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell. Biol. 2006; 7:335–346. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y., Rohde L.H., Wu H.. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr. Genomics. 2009; 10:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morales M.E., White T.B., Streva V.A., DeFreece C.B., Hedges D.J., Deininger P.L.. The contribution of alu elements to mutagenic DNA double-strand break repair. PLoS Genet. 2015; 11:e1005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu K., Wu X., Tompkins J.D., Her C.. Assessment of anti-recombination and double-strand break-induced gene conversion in human cells by a chromosomal reporter. J. Biol. Chem. 2012; 287:29543–29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao C., Deng L., Chen Y., Kucherlapati R., Stambrook P.J., Tischfield J.A.. Mlh1 mediates tissue-specific regulation of mitotic recombination. Oncogene. 2004; 23:9017–9024. [DOI] [PubMed] [Google Scholar]
- 90.Wang Q., Ponomareva O.N., Lasarev M., Turker M.S.. High frequency induction of mitotic recombination by ionizing radiation in Mlh1 null mouse cells. Mutat. Res. 2006; 594:189–198. [DOI] [PubMed] [Google Scholar]
- 91.Stojic L., Brun R., Jiricny J.. Mismatch repair and DNA damage signalling. DNA Repair (Amst.). 2004; 3:1091–1101. [DOI] [PubMed] [Google Scholar]
- 92.Cejka P., Stojic L., Mojas N., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J.. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003; 22:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rizki A., Lundblad V.. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001; 411:713–716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.