Abstract
Both Mcm10 and HP1a are known to be required for DNA replication. However, underlying mechanism is not clarified yet especially for HP1. Knockdown of both HP1a and Mcm10 genes inhibited the progression of S phase in Drosophila eye imaginal discs. Proximity Ligation Assay (PLA) demonstrated that HP1a is in close proximity to DNA replication proteins including Mcm10, RFC140 and DNA polymerase ε 255 kDa subunit in S-phase. This was further confirmed by co-immunoprecipitation assay. The PLA signals between Mcm10 and HP1a are specifically observed in the mitotic cycling cells, but not in the endocycling cells. Interestingly, many cells in the posterior regions of eye imaginal discs carrying a double knockdown of Mcm10 and HP1a induced ectopic DNA synthesis and DNA damage without much of ectopic apoptosis. Therefore, the G1-S checkpoint may be affected by knockdown of both proteins. This event was also the case with other HP family proteins such as HP4 and HP6. In addition, both Mcm10 and HP1a are required for differentiation of photoreceptor cells R1, R6 and R7. Further analyses on several developmental genes involved in the photoreceptor cell differentiation suggest that a role of both proteins is mediated by regulation of the lozenge gene.
INTRODUCTION
Chromatin modification is essential for the regulation of gene expression, and therefore it is also important in cell fate determination and differentiation. Analysis of the proteins involved in this process and how they interact with each other is essential for understanding of development. Heterochromatin is important for the maintenance of genome stability and regulation of gene expression; yet our knowledge of heterochromatin structure and function is incomplete. Heterochromatin protein 1a (HP1a) was originally found in flies as a protein functioning in heterochromatin-mediated gene silencing. In Drosophila, there are at least five HP1 family proteins (1). Among of them, HP1a, HP1b and HP1c (2,3) are well-studied somatic family members. Of these HP1a has been most intensively studied. It is thought to affect chromatin structure through its interactions with other proteins in heterochromatin such as Su(var)3-9, and H3K9me (4–10). HP1a has diverse roles in the nucleus, including the regulation of euchromatic genes. In Drosophila, HP1a interacts directly with HP3, HP4, HP5 and HP6 to recruit them to heterochromatic regions where they can induce heterochromatic gene silencing (11–14). HP1a also acts as a universal docking platform for many mediator proteins that are essential for heterochromatin function.
Roles of HP1a in DNA replication are reported in the following studies. Suppressor of Under-Replication (SUUR) protein is responsible for late replication and for underreplication of intercalary and pericentric heterochromatin in Drosophila (15). Analyses of interaction between SUUR and HP1a suggested that the interaction with HP1a is important for the association of SUUR with chromatin (15). In mouse cells, it is reported that p150 subunit of chromatin assembly factor 1 (CAF-1) plays a key role in the replication of pericentric heterochromatin and S-phase progression and this function is also linked to its ability to interact with HP1a (16). Genome wide mapping of replication timing in HP1a-depleted Drosophila cells revealed that in addition to the repressive role of HP1a for late replication of centromeric DNA, HP1a is required for early replication of euchromatic regions with high levels of repeat sequences, suggesting that of the HP1a-mediated replication complex loading on the chromosome is required for proper activation of these early replication origins (17). However, it is not known yet which replication factor(s) actually interacts with HP1a in replication complex loading. In addition, recent studies have also revealed the possible role of HP1a protein in the DNA Damage Response (DDR) (18–20), although the mechanism regulating the association and dissociation of HP1a with chromatin in response to DNA damage remains unclear.
Minichromosome maintenance protein 10 (Mcm10) is a replication factor required for proper assembly of the eukaryotic replication fork (21–28). Although a number of previous studies demonstrated the role of Mcm10 in initiation of DNA replication, only a few studies have reported the involvement of Mcm10 in regulation of chromatin structure. Recent studies in S. cerevisiae implicate Mcm10 in transcriptional repression of the mating type loci, linking DNA replication proteins to heterochromatin formation (29–31). The depletion of Mcm10 in Drosophila cultured cells leads to under-condensed metaphase chromosomes (32). Additionally, analyses of a hypomorphic mutant of Drosophila Mcm10 demonstrate that the protein has a role in heterochromatic silencing and chromosome condensation, while those with a C-terminal truncation allele of Mcm10 indicate that the CTD of Mcm10 is important for DNA replication (33). These in vivo studies with Drosophila have been performed in limited tissues such as the salivary glands and wing discs (33). In our previous study, we characterised Mcm10 during compound eye development and found that Mcm10 is involved in the differentiation of photoreceptor R7 (34). However, the underlying mechanisms involved are not known yet.
Here, we show that HP1a plays an important role in S-phase progression of eye imaginal disc cells. Proximity Ligation Assay (PLA) suggested that the function of HP1a in S-phase is mediated by its interaction with some DNA replication proteins. Interestingly, many cells in the posterior regions of eye imaginal discs carrying a double knockdown of Mcm10 and HP1a continue to carry out DNA synthesis even in the presence of high levels of DNA damage without inducing much ectopic apoptosis. This event was also the case with other HP family proteins such as HP4 and HP6. These results suggest that Mcm10 and HP proteins play roles in genome maintenance and cell cycle checkpoint. Furthermore, the severely damaged eye phenotypes in these flies are associated with melanotic dots, likely precursors for melanotic tumours, implicating a role for the Mcm10 and HP proteins in tumour development. In addition, we found that HP1a and Mcm10 play roles in photoreceptor cell differentiation. Further analyses suggested that Mcm10 and HP1a are important for the expression of Lozenge and Prospero, but not of Scabrous and Rough.
MATERIALS AND METHODS
Fly stocks
Fly strains were maintained at 25°C on standard food. The UAS-Mcm10IR, UAS-HP1a73-181IR (targeting the region of dHP1a from aa73 to aa181), UAS-HP1a119-206IR (targeting the region of dHP1a from aa119 to aa206), UAS-HP4IR, UAS-HP6IR, UAS-RFC140IR, UAS-dpolε255IR fly lines was obtained from Vienna Drosophila Resource Center. The fly lines HP4 mutant (w; PGSV1-HP4s-147.1), HP6 mutant (w1118; net1 PGT1-HP6BG01429 dpBG01429/ln(2LR)Gla, wgGla-1 PPO1Bc), and all other stocks used in this study were obtained from the Bloomington stock centre (Indiana) and Drosophila Genetic Resource Center (Kyoto).
Scanning electron microscopy
Adult flies were anesthetized, mounted on stages and observed with a VE-7800 (Keyence Inc.) scanning electron microscope. In every experiment, at least five adult flies of each line were chosen for scanning electron microscopy observation to assess the eye phenotype, and these experiments were repeated three times. In the experiments, no significant variation in eye phenotype among the five individuals was observed.
Flip-out experiments
RNAi clones in eye imaginal discs were generated with a flip-out system. Female flies with hs-flp; Act5C>FRT y FRT>GAL4, UAS-GFP were crossed with either UAS-Mcm10IR or UAS-HP1aIR RNAi lines and clones were identified by the presence of green fluorescent protein (GFP) expressed under control of the Act5C promoter. In the control, the female flies with hs-flp; Act5C>FRT y FRT>GAL4, UAS-GFP were crossed with Canton S. Flip-out was induced by heat shock (60 min at 37°C) at 24–48 h after laying the eggs
Plasmid construction for S30-T7 High-Yield Protein Expression System
To construct the plasmids pET16b-fullMCM10-His and pET16b-fullHP1a-His, the full length MCM10 and HP1a cDNAs were amplified by RT-PCR with total RNAs from adult flies and the following combinations of primers:
pET16b-XhoI-Mcm10: 5΄- CGTCATATGCTCGAGATGGGTCCTGCTCAGAAAT
pET16b-Mcm10-EcoRI: 5΄- CGTCTTCAAGAATTCTCACTCTTCATCGGGTACCA
pET16b-XhoI-HP1a: 5΄- CGTCATATGCTCGAGATGGGCAAGAAAATCGACAA
pET16b-HP1a-EcoRI: 5΄- CGTCTTCAAGAATTCTTAATCTTCATTATCAGAGTACCAG
Then, each PCR product was cloned into XhoI- and EcoRI-digested pET-16b (Novagen) by using In-Fusion HD Cloning Kit (Clontech) according to the manufacturer's protocol.
Construction of full length and truncated forms of MCM10 and HP1a in Flexi Vector system
To construct the plasmids: pFN18A-MCM10-Halo, pFN18A-MCM10-N-term-Halo, pFN18A-MCM10-Linker1-Halo, pFN18A-MCM10-ID-term-Halo, pFN18A-MCM10-Linker2-Halo, pFN18A-MCM10-C-term-Halo, pFN18A-HP1a-Halo, pFN18A-HP1a-Chromo-Halo, pFN18A-HP1a-Shadow-Halo, the full length MCM10 and HP1a and their truncated forms were amplified by using PCR with Drosophila cDNAs as templates and the following combinations of primers:
pFN18A-MCM10-Halo-F: 5΄- GACCGCGATCGCCGGTCCTGCTCAGAAATCCGGAACA
pFN18A-MCM10-Halo-R: 5΄- AAGCGTTTAAACCTCTTCATCGGGTACCAGCAAGT
pFN18A-MCM10-N-term-Halo-F: 5΄- TAAAGCGATCGCCGGTCCTGCTCAGAAATCCGGAA
pFN18A-MCM10-N-term-Halo-R: 5΄- CGAGGTTTAAACCTGATCCACCTCCCTCGCCA
pFN18A-MCM10-Linker1-Halo-F: 5΄- GATGGCGATCGCCGAGCTTAAGAAGTCTATCCACGTGGTTACATCC
pFN18A-MCM10-Linker1-Halo-R: 5΄- GCGAGTTTAAACCAGCGCGCCAGCAATGACCC
pFN18A-MCM10-ID-term-Halo-F: 5΄- AGGAGCGATCGCCGTTTCCAAAAATCCTGTAAAAAACAC
pFN18A-MCM10-ID-term-Halo-R: 5΄- TGTCGTTTAAACGGCGAAGGGGGAAACATCAT
pFN18A-MCM10-Linker2-Halo-F: 5΄- GGCTGCGATCGCCCATACCGCTAACCACACCTC
pFN18A-MCM10-Linker2-Halo-R: 5΄- GTCGGTTTAAACTGTGCCTGCTTCAGATTCTT
pFN18A-MCM10-C-term-Halo-F: 5΄- GACCGCGATCGCCCCCGCTGTCAGTATGCCCACTA
pFN18A-MCM10-C-term-Halo-R: 5΄- TTCTGTTTAAACCTCTTCATCGGGTACCAGCAAGT
pFN18A-HP1a-Halo-F: 5΄- GACCGCGATCGCCGGCAAGAAAATCGACAACCC
pFN18A-HP1a-Halo-R: 5΄- GGCAGTTTAAACATCTTCATTATCAGAGTACCAGGATAG
pFN18A-HP1a-Chromo-Halo-F: 5΄- AGCTGCGATCGCCTACGCCGTGGAAAAGATCAT
pFN18A-HP1a-Chromo-Halo-R: 5΄- CGGCGTTTAAACCGCCTCGTACTGCTGGATAAG
pFN18A-HP1a-Shadow-Halo-F: 5΄- AGGAGCGATCGCCCGCGGCCTGGAGGCCGAAAA
pFN18A-HP1a-Shadow-Halo-R: 5΄-TGTCGTTTAAACGGATAGGCGCTCTTCGTAGAAGTGGATTACCAT
Then, each PCR product was cloned into SgfI- and PmeI-digested N-terminal pFN18A Flexi Vector (Promega).
Over-expression of recombinant MCM10 and HP1a proteins in E. coli
The recombinant MCM10 or HP1a full length protein was over-expressed in E. coli BL21 (DE3) (Promega) transfected with the plasmids pFN18A-MCM10-Halo, pET16b-fullMCM10-His, and pET16b-fullHP1a-His. The full length and truncated forms of MCM10 and HP1a proteins were over-expressed in E. coli BL21(DE3)pLysS (Promega) transfected with the plasmids pFN18A-MCM10-N-term-Halo, pFN18A-MCM10-Linker1-Halo, pFN18A-MCM10-ID-term-Halo, pFN18A-MCM10-Linker2-Halo, pFN18A-MCM10-C-term-Halo, pFN18A-HP1a-Halo, pFN18A-HP1a-Chromo-Halo and pFN18A-HP1a-Shadow-Halo. The bacteria containing the Halo-fusion proteins or His-fusion proteins were induced with 1mM IPTG at either 30°C or 37°C for 4 h.
Halotag Pull-down assay
Immobilization of bait proteins (Halo-fusion)
Cell lysis: Cells were harvested from 2 ml of bacterial cultures described above. Then the cells were lysed by freeze-thawing with liquid nitrogen several times prior to the addition of 300 μl Cell Lysis Reagent (Promega) and 6 μl Protease Inhibitor Cocktail (Promega) and 2 μl RQ1 RNase-Free DNase (Promega). Cell lysates were prepared by incubating at 25°C for 30 min with rotation.
Particle equilibration: 100 μl Halolink Resin Particles (Promega) were washed four times with 300 μl Wash Buffer (90 μl NP-40 and 18 ml TBS) and then were re-suspended in 300 μl Wash Buffer.
Binding: Cell lysates were diluted three times in TBS 1× with addition of 1% BSA. Then cell lysates were added to the equilibrated Halolink Resin Particles and incubated for 30 min at 25°C on a rotating platform.
Washing: The binding particles were washed with 1ml Wash Buffer five times at 25°C. The particles were then re-suspended in 20 μl Wash Buffer.
Synthesis of Prey Proteins (His-fusion)
One microgram each of the purified pET16b-fullMCM10-His and pET16b-fullHP1a-His plasmids was used for in vitro transcription/translation using the S30 T7 High-Yield Protein Expression System (Promega) by following the manufacture's instruction.
Capture of Prey Proteins
To one volume aliquot of particles carrying Halo-fusion protein, four volumes of the S30-T7 transcription/translation reaction were added, then, the samples were diluted with Wash Buffer containing 1% BSA. The samples were then incubated for 1 h at 25°C. After washing for five times, the resins were incubated with 50 μl SDS Elution Buffer (Promega) for 30 min at 25°C. The eluted proteins were subjected to western blotting analysis.
Co-immunoprecipitation
Preparation of crude extracts
Drosophila S2 DRSC cells were lysed in NP40 Cell Lysis Buffer (150 mM NaCl, 1% NP40, 50 mM Tris–HCl pH 8.0, RQ1 RNase Free DNase (Promega) and Complete Mini-EDTA free Protease Inhibitors (16)). Cell lysates were prepared by incubating cells at 25°C for 30 min with rotation and then were centrifuged for 5 min at 13 000 g at 4°C. The supernatant was collected for immunoprecipitation.
BS3 cross-linking of the antibodies
Mouse anti-HP1a antibody was cross-linked with Dynabeads Protein G (Invitrogen) by using BS3 (Thermo Scientific), following the manufactures instruction.
Immunoprecipitation from the crude extracts
The HP1a antibody beads were incubated with the extracts at 25°C for 1 h. The bead complex was washed five times with PBS containing 0.1% Tween 20. The immunoprecipitated proteins were then eluted by using 50 mM glycine pH 2.8 at 25°C for 30 min. The eluted proteins were used for western blotting analysis.
Chromatin immuneprecipitation (ChIP)
ChIP assays were performed using ChIP Assay Kit (Millipore) following the manufactures’ instructions. Approximately 1 × 107 S2 DRSC cells were fixed in 1% formaldehyde (Thermo Scientific) at 37°C for 10 min and then quenched in 125 mM Glycine for 5 min at 25°C. Cells were washed twice in PBS containing Complete Mini EDTA-free (16) and lysed in 1 ml of SDS lysis buffer. Lysates were sonicated to shear DNA to lengths between 200 and 1000 base pairs and centrifuged at 15 300 g for 10 min at 4°C. The sonicated cell supernatants were diluted 10-fold in Chip Dilution Buffer and precleared with 75 μl Salmon Sperm DNA/Protein Agarose 50% slurry for 30 min at 4°C. After a brief centrifugation, each supernatant was incubated with 1 μg of normal rabbit IgG or anti-Mcm10 IgG antibody for 16 h at 4°C. Salmon Sperm DNA/Protein A agarose–50% slurry was added, followed by incubation for 1 h at 4°C. After several washings, immunoprecipitated DNA was eluted with elution buffer containing 1% SDS and 0.1 M NaHCO3. Then the protein–DNA crosslinks were reversed by heating at 65°C for 4 h. After deproteinisation with proteinase K (Nacalaitesque). DNA was recovered by phenol-chloroform extraction and ethanol precipitation.
Immunoprecipitated DNA fragments were detected by q-PCR using SYBR Premix ExTag (Takara) and the CFX96 Touch Real-time PCR Detection System (BioRad).
The following primers were used for the Quantitative RT-PCR of ChIP assays:
ChIPLozenge Forward: 5΄-GTGTGTGCCCTCACTGTTC
ChIPLozengeReverse: 5΄-TGTACTGCCTGTGTAGCTTAA
ChIPProsperoForward: 5΄-TATGTTATGAATTTGAATCCC
ChIPProsperoReverse: 5΄-TTTTTGAACATTTTTCGTGC
ChIPScabrousForward: 5΄-TGTATATTTATGGATGGAACA
ChIPScabrousReverse: 5΄-CATGGCTCCAAAAAAAAAT
ChIPRoughForward: 5΄-CACGAAAAATGTTCAAAAA
ChIPRoughReverse: 5΄-CAATGCAAAAATATTTTGTTA
Immunostaining
For third instar larval eye imaginal discs were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 20 min at 25°C. After washing with PBS containing 3% Triton X-100 (PBST), samples were blocked with PBS containing 1.5% Triton X-100 and 10% normal goat serum for 20 min at 25°C and incubated with diluted primary antibodies in PBS containing 0.15% Triton X-100 and 10% normal goat serum for 16 h at 4°C. The following antibodies were used as primary antibodies: rabbit anti-Mcm10 polyclonal (diluted 1:100 (34)), mouse anti-human Mcm10 polyclonal (Abnova) (1:100), mouse anti-dpolα polyclonal (1:100 (35)), rabbit anti-dpolε255 polyclonal (1:100 (36)), rabbit anti-dRFC140 polyclonal (1:200 (37)), mouse anti-HP1a monoclonal (1:200, DSHB) rabbit anti-cleaved Caspase-3 (1:100, BD Biosciences), mouse anti-Prospero (1:200, DSHB), mouse anti-Lozenge (1:100, DSHB), mouse anti-Scabrous (1:100, DSHB), rabbit anti-Ser10 phosphorylated Histone H3 (PH3, 1:200, Cell Signaling Technology), rabbit anti-phospho H2AvD (1:200, Rockland), mouse anti-cyclin E (8B10) (1:100, kindly supplied from Helena Richardson). After extensive washing with PBST, samples were incubated with secondary antibodies labeled with either Alexa488 or Alexa594 (1:400, Invitrogen) for 3 h at 25°C. After further washing with PBST, samples were stained with DAPI and then were mounted in Vectashield Mounting Medium (Vector laboratories) and inspected with a confocal laser-scanning microscope (Olympus FLUOVIEW FV10i and FV1000).
For S2 DRSC cells, the cells were placed in BD Falcon Culture Slides (Fisher Scientific) for 20 min and then fixed in 4% paraformaldehyde and 0.2 M sucrose in PBS (pH 7.5) for 15 min at 25°C. After washing with PBS containing 0.1% Triton X-100 (PBST), samples were blocked with PBST containing 20% normal goat serum for 20 min at 25°C and incubated with diluted primary antibodies in PBS containing 0.1% Triton X-100 and 20% normal goat serum for 16 h at 4°C. The washing steps and secondary antibodies incubation were performed as described in the immunostaining section for eye imaginal discs.
Proximity ligation assay (PLA)
To demonstrate the close distance (<40 nm) between two different proteins in Drosophila tissues and cells, PLA was performed by using Duo-link In Situ-Fluorescence kits from Sigma (Detection Reagents Green 488 or Detection Reagents Red 594) according to the manufacturer's instructions. The eye imaginal discs or S2 cells were treated in immunostaining section until finishing two primary antibodies incubation. The two primary antibodies must have been raised in different species. Then they were washed intensively with PBST and incubated with mixture containing PLA probe Minus and PLA probe Plus for 1 h at 37°C. After washing with PBS containing 0.1% Triton X100, samples were incubated with ligation–ligase solution for 1 h at 37°C. Then the samples were washed with PBS and continued with amplification-polymerase solution incubation for 100 min at 37°C. The samples were finally washed with PBS and mounted in Duolink In Situ Mounting Medium with DAPI (Sigma) and observed with a confocal laser-scanning microscope (Olympus FLUOVIEW FV10i and FV1000).
Western immunoblot analysis
2 × 107 S2 DRSC cells were washed in PBS and lysed in NP40 Cell Lysis Buffer (150 mM sodium chloride, 1% NP40, 50 mM Tris pH 8.0, RQ1 RNase Free DNase (Promega) and Complete Mini-EDTA free Protease Inhibitors (16)). Cell lysates were centrifuged for 20 min at 12 000 g at 4°C and supernatant was electrophoretically separated on 10% polyacrylamide gel containing 10% SDS and transferred to Polyvinylidene difluoride (PVDF) membrane (BIORAD) using Trans-Blot Turbo Transfer System (BIORAD). The blotted membranes were blocked in Tris Buffer Saline Tween 20 (TBST) pH 7.6 (10 mM Tris–HCl, 150 mM NaCl and 0.1% Tween20) containing 5% non-fat milk for 1h at 25°C and then incubated with rabbit anti-Mcm10 polyclonal (diluted 1:1000 (34)), rabbit anti-dpolε255 polyclonal (1:5000 (36)), rabbit anti-RFC140 polyclonal (1:1000 (37)), mouse anti-PCNA monoclonal (1:5000, Abcam), mouse anti-HP1a monoclonal (1:10 000, DSHB), mouse anti-Halotag monoclonal (1:5000, Promega), mouse anti-His6 monoclonal (1:1000, Roche) in TBST containing 1% BSA for 16h at 4°C. After washing with PBST, the blots were incubated with either Stabilized Peroxidase Conjugated Goat anti-mouse IgG (H+L) (1:10 000, Thermo Scientific) or Stabilized Peroxidase Conjugated Goat anti-rabbit IgG (H+L) (1:4000, Thermo Scientific) for 1h at 25°C. Detection was performed with ECL Select Western Blotting Detection Reagent (GE Healthcare) with CS Analyzer version 3.0 and Image Saver 6 for AE-9300H Ez-Capture MG (38).
5΄-Ethynyl-2΄-deoxyuridine (EdU) labeling
Detection of cells in S phase was performed using EdU-labeling kits from Invitrogen (Click-iT EdU Alexa Flour 488 and 594 Imaging Kit). Third instar larvae were dissected in PBS and the imaginal discs were suspended in Grace's insect medium in the presence of 10 mM EdU for 60 min at 25°C. The samples then were fixed with 3.7% paraformaldehyde in PBS adjusting to pH 7.4 for 20 min at 25°C. After fixing, samples were washed with 3% BSA in PBS and were permeabilised in 0.5% Triton X-100 in PBS for 20 min at 25°C. Then, samples were washed with 3% BSA in PBS and incubated with Click-iT reaction cocktails for 30 min at 25°C (following the manufactures’ instructions). After further washing with 3% BSA in PBS and PBS, samples were stained with Hoechst 33342 (Invitrogen) for labeling DNA, and finally samples were mounted and observed as described in immunostaining section.
Quantitative RT-PCR
Sixty eye imaginal discs were dissected and were snap freezing in liquid Nitrogen. Total RNA was isolated from cells using Trizol Reagent (Invitrogen) and aliquots were reverse transcribed with an oligo(dT) primer using Prime Script RT Reagent Kit (Perfect Real Time-Takara). Real-time PCR was performed with SYBR Premix ExTag (Takara) and the CFX96 Touch Real-time PCR Detection System (BIORAD). Glucose-6-phosphate dehydrogenase G6PD was chosen as an endogenous reference gene. To carry out quantitative real-time PCR, the following PCR primers were chemically synthesized:
Lozenge RTPCR Forward: 5΄-TGGCAACCTACGCCAAA
Lozenge RTPCR Reverse: 5΄-GGGAAGCCATCGATGTAGG
Prospero RTPCR Forward: 5΄-AGATCCTCGACCGGAAGTC
Prospero RTPCR Reverse: 5΄-CCGGATTCATGCCCTGTG
HP1a RTPCR Forward: 5΄-CACAGCAAGCAAGCGAAAG
HP1a RTPCR Reverse: 5΄-GGTAGATCCTGAAACGGGAATG
Mcm10 RTPCR Forward: 5΄-CAGGTCGTGGTATCAATGAACTAA
Mcm10 RTPCR Reverse: 5΄-CGATCACGTTCCTTGGTGATTA
G6PD RTPCR Forward: 5΄-GAACAAGAACAAGGCCAACC
G6PD RTPCR Reverse: 5΄-AGGCTTCTCGATAATCACGC
Quantification and statistical analysis
EdU signals, phospho-H2AvD signals, Caspase-3 signals, phospho-Histone H3 signals, the Prospero signals, Lozenge signals, Rough signals, Scabrous signals, and other immunostaining signals in the region posterior to the MF were counted and their signal intensities were measured from 10 independent eye imaginal discs using MetaMorph software (Molecular Devices). The experiments were repeated at least three times. Then, statistical analysis was conducted, as indicated in the figure legends, using GraphPad Prism 6. The grouped data were analysed with 2-way ANOVA, Turkey's multiple comparisons test. Significance is described in the figure legends as *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
RESULTS
Both HP1a and Mcm10 are required for the progression of S phase in eye imaginal disc cells
Drosophila eye imaginal discs have been widely used for the study of DNA replication due to the highly synchronized mitotic waves that pass across the disc. In third instar larvae, the morphogenetic furrow (MF) appears at the posterior end of the eye imaginal discs and slowly moves to the anterior. Cells in front of the MF proliferate asynchronously, while those on the MF are arrested synchronously in the G1 phase. Cells behind the MF either leave the cell cycle and differentiate into the photoreceptors of adult ommatidium, or undergo one more cell division. This cell cycle is a final synchronous round and produces an S-phase and M-phase bands (the second mitotic wave-SMW), after which these cells form a reservoir of cells for subsequent differentiation events. Therefore, eye imaginal discs are a good system to investigate role of any genes and proteins in DNA replication and cell cycle progression.
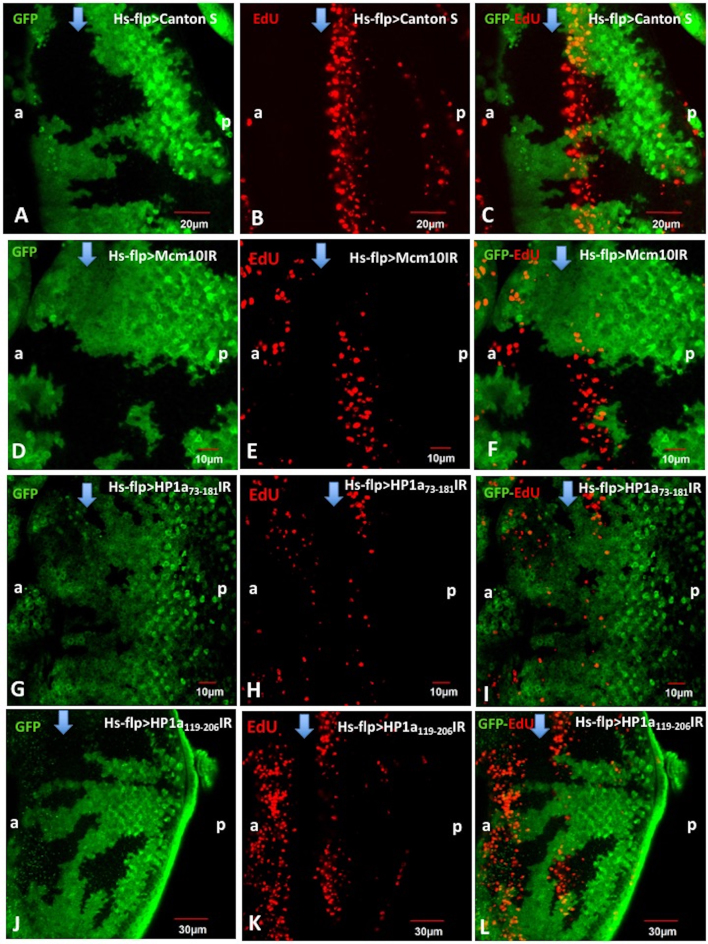
We firstly examined the roles of Mcm10 and HP1a in S-phase progression by flip-out experiments, combined with EdU assays, to determine how the knockdown of these genes affects on S phase progression. Effective knockdown of HP1a or Mcm10 in each flip-out experiment was confirmed by quantifying signal intensities of immunostaining with anti-HP1a antibody (Supplementary Figure S1A–H) or anti-Mcm10 antibody (Supplementary Figure S1 I-P). In the flip-out experiments with the Mcm10 RNAi line, the EdU signals in the S phase zone were decreased in the Mcm10 knockdown area (green), confirming that Mcm10 is required for initiation of DNA replication (Figure 1D–F). These results are consistent with our previous reports and those by other groups (21–28,39,40). In the control experiments expressing GAL4 alone, no difference in the S-phase zone between GFP clones and non-GFP clones was observed (Figure 1A–C). Similar with Mcm10 knockdown data, the analyses with HP1a73-181 RNAi line revealed that in the HP1a knockdown area (green), the number of EdU-positive cells was significantly reduced in compared to the non-knockdown area (Figure 1G–I). A few EdU positive cells were still detected in GFP positive knockdown area (Figure 1). These cells likely represent ineffectively knocked down cells, since we observed a few Mcm10 or HP1a positive cells in GFP positive knockdown area (Supplementary Figure S1). To further confirm this, we performed again with another RNAi line HP1a119-206IR and obtained essentially the same results (Figure 1J–L). All together, we conclude that HP1a is required for the progression of the S phase. This is also consistent with the previous report indicating that HP1a plays a positive role in S-phase progression (17).
Figure 1.
Mcm10 and HP1a are important for initiation and/or progression of the S-phase in eye imaginal discs. Eye imaginal discs from flip-out experiments with Canton S (A–C), UAS-Mcm10IR (D–F), UAS-HP1a73-181 IR (G–I), UAS-HP1a119-206 IR (J–L) flies were performed with EdU labeling assays (Red). The cells expressing Mcm10dsRNA, dHP1a73-181dsRNA, and dHP1a119-206dsRNA are marked with GFP (green). In the control (Canton S), EdU signals in the S phase zone were not changed between GFP and non-GFP clones (A–C). However, we observed the reduction of EdU signals in the GFP clones of the Mcm10 RNAi line (D–F) and in two independent HP1a RNAi lines (G–I and J–L). These data suggest that both Mcm10 and HP1a play important roles for DNA replication in S-phase cells. The flies were reared at 28°C. Scale bars indicate 10, 20 or 30 μm. (a) indicates anterior, (p) indicates posterior. Blue arrows indicate morphogenetic furrow (MF).
HP1a is in close proximity to Mcm10, RFC and DNA polymerase ε 255 kDa subunit (dpolε255) in eye imaginal disc cells
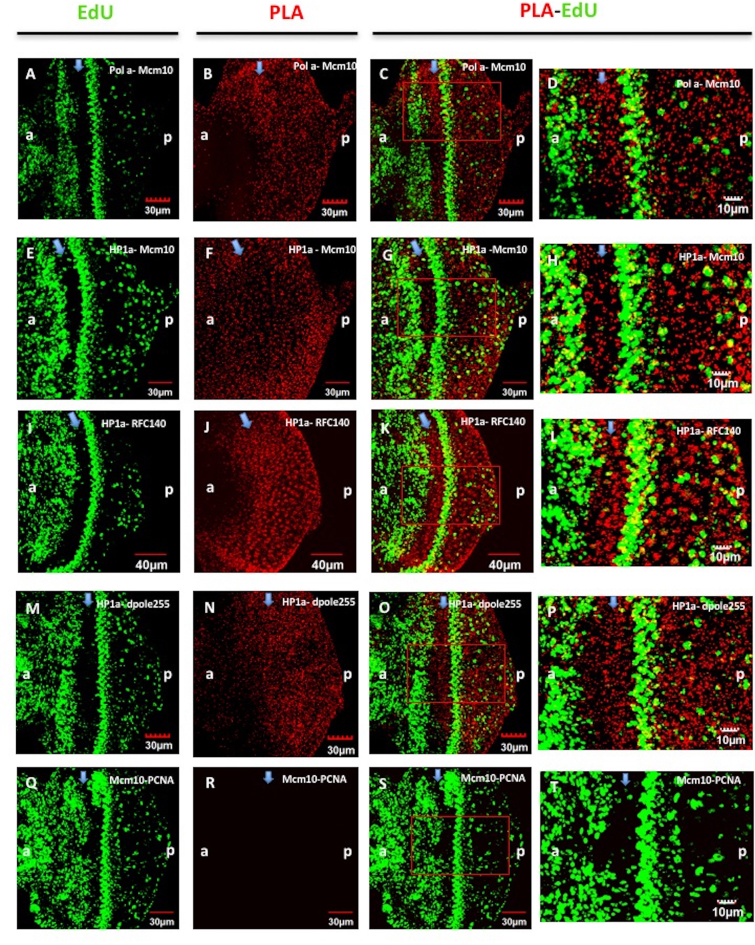
Previous study suggested that HP1a-mediated replication complex loading on the chromosome is required for proper activation of early replication origins in Drosophila (17). In the present study, simultaneous analyses with immunostaining and EdU assays showed that Mcm10 and HP1a distribute ubiquitously in the entire eye imaginal discs including the MF to S phase zone (Supplementary Figure S2A–J). RFC140 and dpolε255 also distribute ubiquitously in the entire eye imaginal discs with slightly higher expression in the S phase zone (Supplementary Figure S2K–T) that merged with EdU signals (Supplementary Figure S2M–O and R–T). These data on DNA replication proteins are consistent with previous reports (34,36,37). Specificities of the antibodies were confirmed by Western immunoblot analyses with anti-HP1a, anti-dpolε255, anti-Mcm10 and anti-PCNA antibodies (Supplementary Figure S3). Based on these observations, we hypothesized that HP1a might interact with some of these replication proteins. We therefore performed PLA assays in eye imaginal discs from Canton S third instar larvae to examine distance between HP1a and each of these replication proteins. The PLA assays have been successfully used to demonstrate close proximity between PCNA and chromatin modifying enzymes at the replication fork (41).
Firstly, we performed PLA with PCNA and RFC140, since both proteins have been demonstrated to interact with each other (42). As expected, strong PLA signals were detected between these two proteins in eye imaginal discs and some of them merged with EdU signals (Supplementary Figure S4). The PLA signals in cytoplasms of non-replicating eye imaginal disc cells may represent a non-replication cytoplasmic scaffold role of PCNA as suggested with human neutrophil (43–45). Secondly, we detected strong PLA signals representing close proximity between Mcm10 and DNA polymerase α (dpolα) (Figure 2A–D). This is consistent with previous studies with mammalian cells, indicating that Mcm10 binds to the catalytic subunit of polα required for chromatin association (46). We could also detect close proximity between HP1a and Mcm10 (Figure 2E–H), HP1a and RFC140 (Figure 2I–L), HP1a and dpolε255 (Figure 2M–P) ubiquitously in the eye imaginal discs, from the MF to the posterior region, and throughout G2, S, G1 and M phases. The interactions between those pairs of proteins are detectable throughout the eye imaginal discs including the EdU positive S phase zone (Figure 2C, G, K and O). In contrast, no PLA signal was detected between Mcm10 and PCNA (Figure 2Q–T). Close proximity between HP1a and each of Mcm10, RFC140 and dpolε255 was also confirmed in Drosophila cultured cells by PLA (Supplementary Figure S5).
Figure 2.
Close proximity of HP1a with Mcm10, RFC140 or dpolε255 in eye imaginal discs. Eye imaginal discs of third instar larvae from Canton S were labeled with EdU (green) (A, E, I, M and Q) and performed with PLA assays to examine close proximity between two target proteins (B, F, J, N and R). PLA examines close proximity between dpolα and Mcm10 (C and D), HP1a and Mcm10 (G and H), HP1a and RFC140 (K and L), HP1a and dpolε255 (O and P) in the S-phase cells labeled with EdU and in the posterior regions to MF where photoreceptor cells differentiate. No PLA signal was detected between PCNA and Mcm10 in eye imaginal discs (Q–T). Enlarged images of the indicated regions (red rectangles) in C, G, K, O and S are shown in D, H, L, P and T, respectively. Scale bars show 10, 30 or 40 μm. (a) indicates anterior, (p) indicates posterior. Blue arrows indicate morphogenetic furrow (MF).
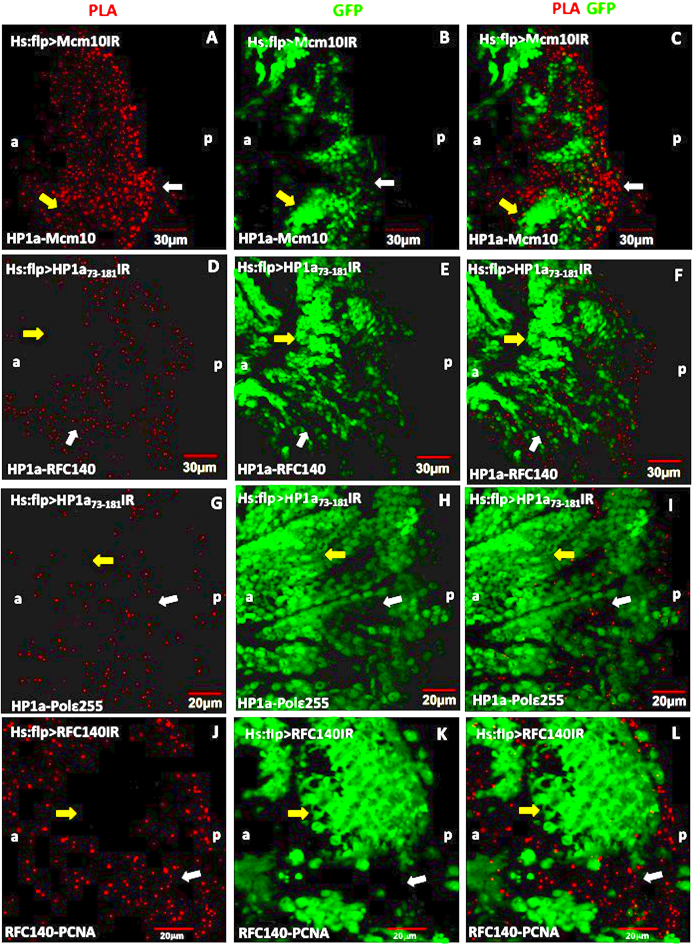
Since we unexpectedly observed PLA signals in non-replicating cells in eye imaginal discs, we examined specificity of the PLA by flip-out experiments in eye imaginal discs (Figure 3). The PLA signals between HP1a and Mcm10 (Figure 3A–C), HP1a and RFC140 (Figure 3D–F), and HP1a and dpolε255 (Figure 3G–I) were significantly reduced in the HP1a knockdown areas (GFP-positive clones, yellow arrows), whereas the PLA signals were mainly detected in the non-HP1a knockdown areas (non-GFP clones, white arrows) (Figure 3C, F, I and L). In the control experiments, we also observed the same phenomenon where the PLA signals between RFC140 and PCNA were significantly reduced in the RFC140 knockdown areas (GFP-positive clones, yellow arrows) (Figure 3J–L). Therefore, again the control data strongly supported the validity of our PLA results in the eye imaginal discs.
Figure 3.
Flip-out experiments combined with the PLA assays showing the interactions between HP1a and Mcm10, HP1a and RFC140, HP1a and dpolε255. Eye imaginal discs from flip-out experiments with UAS-Mcm10IR (A–C), UAS-HP1a73-181IR (D–F), UAS-HP1a119-206IR (G–I), UAS-RFC140IR (J–L) flies were performed PLA (red). (A–C) PLA between HP1a and Mcm10, (D–F) PLA between HP1a and RFC140, (G–I) PLA assays between HP1a and dpolε255. The cells expressing Mcm10 dsRNA (B), HP1a73-181 dsRNA (E and H) and RFC140 dsRNA (K) are marked with GFP (green). The PLA signals were mostly detected in non-GFP clones (yellow arrows), while in GFP clones, the PLA signals were significantly reduced (white arrows). Scale bars indicate 20 or 30 μm. (a) indicates anterior, (p) indicates posterior.
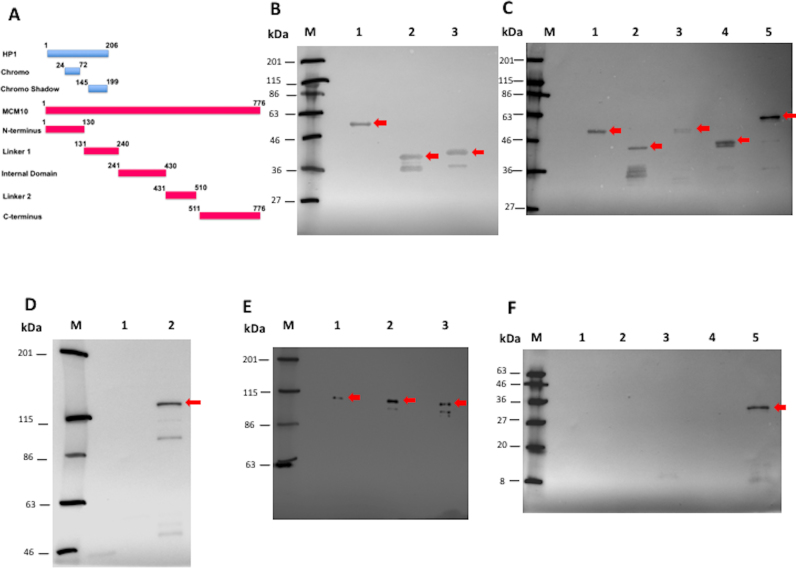
In order to determine the domain interaction between HP1a and Mcm10, we developed the S30-T7 in vitro transcription/translation system together with Halo-tag pull down assays in E. coli. The full-length and truncated forms of Mcm10 and HP1a were expressed in E. coli. Expression of each form of HP1a-Halo-tag was confirmed by western blot analysis with anti-Halo-tag monoclonal antibody (Figure 4B, red arrows). We detected the bands corresponding to the molecular weights estimated from amino acid sequences of full length HP1a with Halo-tag (58 kDa, lane 1), Chromo domain with Halo-tag (40 kDa, lane 2) and Chromo Shadow domain with Halo-tag (41 kDa, lane 3). Expression the full-length HP1a-His tag was confirmed by Western blot analysis with anti-HP1a antibody (Supplementary Figure S6B). The HP1a-His tag protein was detected at the position corresponding to 35 kDa in the blot (Supplementary Figure S6B, lane 2). This band was not detectable with E. coli extracts without IPTG induction (Supplementary Figure S6B, lane 1). The same size of HP1a-His tag protein produced by S30-T7 in vitro transcription/ translation system was also detected in the western blot with mouse anti-His6 monoclonal antibody (Supplementary Figure S6C, lane 1, red arrow). This band was not detectable in the mock control with pFN16 (HQ) containing the Renilla reniformis luciferase gene with humanized codon usage (hRluc) (Supplementary Figure S6B, lane 2) or negative control without template plasmid (Supplementary Figure S6B, lane 3).
Figure 4.
Domain interaction between HP1a and Mcm10 in vitro. (A) Diagram describes the domain constructions of HP1a (Chromo domain and Chromo Shadow domain) and of Mcm10 (N terminus, Linker 1, Internal domain, Linker 2 and C terminus). Numbers of amino acid positions are given in the diagram. (B) Western blots using mouse anti-Halo-tag monoclonal antibody to detect Halo-tagged full length (58 kDa, lane 1), Chromo domain (40 kDa, lane 2), and Chromo Shadow domain (41 kDa, lane 3) of Drosophila HP1a produced in E. coli. (C) Western blots using mouse anti-Halo-tag monoclonal antibody to detect the Halo-tagged N terminal (49 kDa, lane 1), Linker 1 (43 kDa, lane 2), Internal domain (56 kDa, lane 3), Linker 2 (44 kDa, lane 4) and C terminal (64 kDa, lane 5) fragments of Drosophila Mcm10 produced in E. coli. (D) Western blots using mouse anti-Halotag monoclonal antibody to detect the Halo-tagged full length Mcm10 produced in E. coli. The Halo-tagged full length Mcm10 protein was detected at the position corresponding to 121 kDa in E. coli extracts induced with IPTG (lane 2), but not detectable without IPTG (lane 1). (E) Western blot in the S30-T7 and Halotag Pull-Down experiments using mouse anti-His6 monoclonal antibody showed that Mcm10 interacts with HP1a (lane 1) and both two domains: Chromo domain (lane 2) and Chromo Shadow domain (lane 3). (F) Western blot in the S30-T7 and Halo-tag Pull-Down experiments using mouse anti-His6 monoclonal antibody showed that HP1a only binds to the C-terminus of Mcm10 (lane 5). We could not detect the same binding signal to the N-terminus (lane 1), Linker 1 (lane 2), Internal domain (lane 3), and Linker 2 (lane 4). The samples were loaded into SDS-PAGE containing 10% acrylamide and then were transferred to the PVDF membranes (B, C, E and F). The samples were loaded into SDS-PAGE containing 6% acrylamide and then were transferred to the PVDF membranes (D). M indicates protein markers.
Expression of full length and truncated forms of Mcm10-Halo-tag was confirmed by western blot analysis with anti-Halo-tag monoclonal antibody (Figure 4C and D, red arrows). The full length Mcm10-Halo-tag band was detected at the position corresponding to 121 kDa in the blot (Figure 4D, lane 2). This band was not detectable with E. coli extracts without IPTG induction (Figure 4D, lane 1). The bands corresponding to the molecular weights estimated from amino acid sequences of N-terminus region of Mcm10 with Halo-tag (49 kDa, lane 1), Linker 1 region with Halo-tag (43 kDa, lane 2), Internal domain region with Halo-tag (56 kDa, lane 3), Linker 2 region with Halo-tag (44 kDa, lane 4) and C-terminus region with Halo-tag (64 kDa, lane 5) were also detected in the western blots (Figure 4C). In addition, the full length Mcm10-His-tag protein was detected at the position corresponding to 110 kDa in the blot with rabbit anti-Mcm10 antibody (Supplementary Figure S6A, lanes 2 and 3). This band was not detectable with E. coli extracts without IPTG induction (Supplementary Figure S6A, lane 1).
The full length Mcm10-His-tag protein was produced by S30-T7 in vitro transcription/ translation system and then incubated with the full length and truncated forms of HP1a with Halo-tag. The Halo-tag pull down assay showed that Mcm10 binds to full length HP1a (Figure 4E, lane 1) and to both Chromo domain (Figure 4E, lane 2) and Chromo Shadow domain (Figure 4E, lane 3) of HP1a. The results indicate that Mcm10 binds to both Chromo domain and Chromo Shadow domain of HP1a. Besides, the full length HP1a-His-tag protein was also produced by S30-T7 in vitro transcription/ translation system and then mixed with the full length and various truncated forms of Mcm10 with Halo-tag. The pull-down analyses showed that HP1a specifically binds to the C-terminus region of Mcm10 (Figure 4F, lane 5). We could not detect the binding signals to other domains of Mcm10 (Figure 4F, lanes 1–4). These results are consistent with the previous report by yeast two-hybrid system indicating that Drosophila HP1a interacts with C-terminus region of Mcm10 (33). Last but not least, the co-immunoprecipitation of HP1a protein results in Drosophila S2 DRSC cells strongly proved that HP1a directly interacts with Mcm10 (Supplementary Figure S6 D-F).
The close proximity between Mcm10 and HP1a is specifically observed in the mitotic cycle, but not in the endocycle
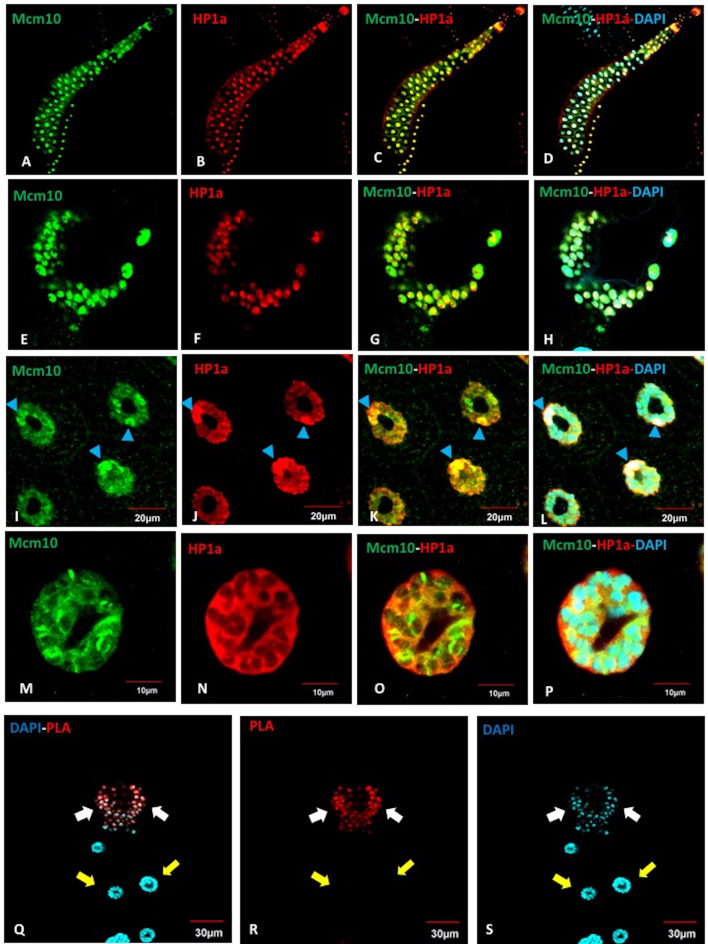
In Drosophila, cells in different regions of tissues undergo either mitotic cycles or endocycles. It is therefore interesting to determine whether the interaction between Mcm10 and HP1a occurs in both types of cycle or not. Salivary gland cells in Drosophila proliferate by repeated rounds of endoreplication, consisting of only S- and G-phase, to form a giant polytene chromosome. Thus, salivary gland cells provide a unique model to visualize the specific chromatin localization patterns of individual proteins. The adult salivary glands develop from imaginal rings located at the proximal end of each larval salivary gland. The imaginal ring cells, a group of diploid cells localizing between the gland and duct, resume mitosis during molting from second to third instar larvae. Immunostaining of the salivary glands from Canton S flies with anti-Mcm10 and anti-HP1a antibodies showed that both Mcm10 and HP1a mostly localize in the nuclei in both imaginal rings (Figure 5E–H) and in whole salivary glands including proximal and distal regions (Figure 5A–D). The nuclei located relatively close to the proximal region show strong HP1 signals in heterochromatic chromocenters near to the nuclear membrane (Figure 5I–L). However, in the nuclei located in the distal region, both Mcm10 and HP1 mainly colocalize in nucleoplasm, although some of them are still on the chromosome (Figure 5M–P). However, surprisingly, we could only detect PLA signals between Mcm10 and HP1a in the imaginal rings where cells undergo mitotic cycle (Figure 5Q–S, white arrows), but no PLA signal is detected in the polytenizing cells of the salivary glands (Figure 5Q–S, yellow arrows).
Figure 5.
Close proximity between Mcm10 and HP1a is specifically observed in mitotic cycle, but not in endocycle. (A–P) Salivary glands of Canton S were immunostained with anti-Mcm10 (A, E, I, M) (Green) and anti-HP1a (B, F, J, N) (Red) antibodies, then with DAPI (Blue) for DNA staining. The Mcm10 and HP1a distribute in the whole salivary glands from the imaginal rings (E–H) to the polytenizing cells (I–P). (I–L) Polytenizing cells in proximal region are shown. (M–P) Polytenizing cells in distal region are shown. (Q–S) Salivary glands of Canton S were subjected to PLA assays (red) and then stained with DAPI (blue). The interactions between Mcm10 and HP1a were only found in the imaginal rings where cells undergo mitotic cycles (Q–S, white arrows), whereas these interactions cannot be detected in the rest part of the glands (Q–S, yellow arrows). (Q) Merged image between PLA signals (R) and DAPI signals (S). The flies were reared at 25°C. Blue arrowheads show chromocenters. Scale bars indicate 10, 20 or 30 μm.
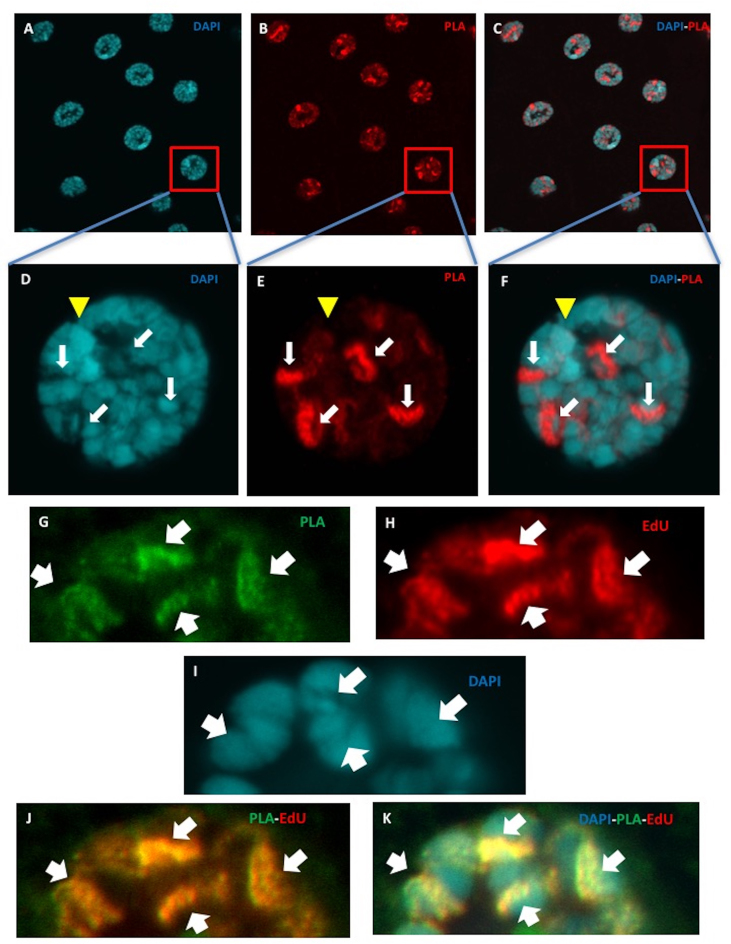
In contrast, we observed the in situ PLA signals between Mcm10 and dpolα in salivary glands in both imaginal rings and either parts of salivary glands (Figure 6A–C). In higher magnification images of the nucleus in Canton S salivary glands, we more clearly observed that the PLA signals between Mcm10 and dpolα were excluded from the nucleolus and heterochromatic regions (Figure 6D–F). Furthermore, these PLA signals coincide with EdU signals, suggesting that Mcm10 and dpolα are in close proximity at the replication forks (Figure 6G–K). The specificity of PLA was further confirmed by flip-out experiments with the Mcm10 RNAi line (Supplementary Figure S7). We could detect the PLA signals in both imaginal ring (Supplementary Figure S7, white arrows) and polytenizing cells in the non-GFP clone (Supplementary Figure S7, yellow arrows). In contrast, no PLA signal was detected in the GFP areas (Supplementary Figure S7, pink arrows).
Figure 6.
Visualisation of in situ interaction between Drosophila Mcm10 and DNA polymerase α at replication fork in endocycle in salivary glands of wild type Canton S. (A–F) Salivary glands of Canton S were subjected to PLA assays (red) (B and E) and then were stained with DAPI (blue) (A and D). (C and F) Merged images between PLA and DAPI signals. (D–F) Higher magnification images of an indicated single nucleus. The bright red PLA signals represent the direct interaction between Mcm10 and DNA polymerase α in the nucleus (B and E, white arrows) and these interactions are excluded from the DAPI dense regions (C and F). Yellow arrowheads indicate heterochromatic chromocenters. (G–K) Salivary glands were labeled with EdU (Red), performed with PLA assays (green) and stained with DAPI (blue). The PLA signals fully coincide with EdU signals showing that Mcm10 and DNA polymerase α are in close proximity at the replication forks where DNA replication occurs. The flies were reared at 25°C.
In addition, the PLA signals between Mcm10 and PCNA were detected in wild type salivary glands in both imaginal rings and whole salivary glands (Supplementary Figure S8E–J), but not detectable in eye imaginal discs (Figure 2Q–T and Supplementary Figure S8A–D). In summary, the data indicate that the close proximity between Mcm10 and HP1a is only observed in the mitotic cycle, but not in endocycling cells. In contrast, close proximity between Mcm10 and dpolα is observed in both mitotic and endocycling cells. Close proximity between Mcm10 and PCNA was only detectable in the salivary glands irrespective to mitotic cycle and endocycle.
Mcm10 and HP1a, HP4, HP6 play a role in genome stability and cell cycle checkpoint in the Drosophila compound eyes
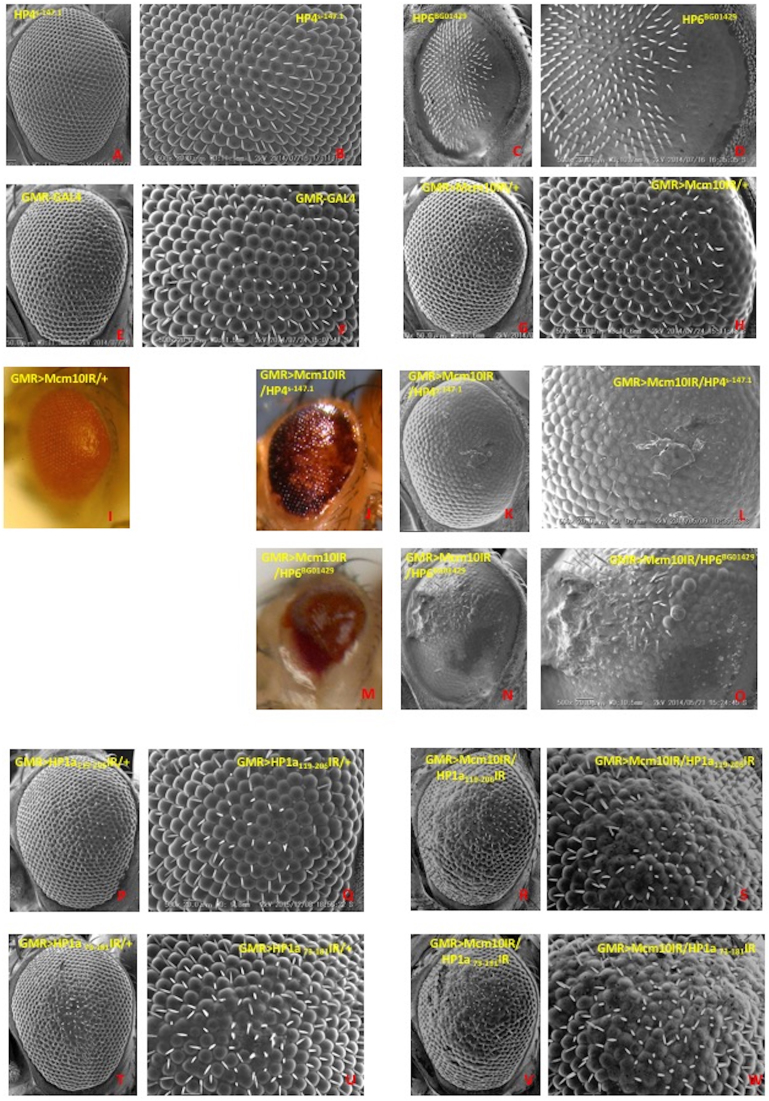
DNA damage induces several cellular responses, including checkpoint activation, DNA repair, and the triggering of apoptotic pathways for irreparable DNA damage. Previous studies indicated that Mcm10 plays roles not only in DNA replication but also in genome stability and checkpoints. The present study with PLA shows the close proximity between Mcm10 and HP1a. This raises the possibility that there may be a link for Mcm10 and other HP family proteins involving in genome maintenance and checkpoint activities. Therefore, we examined genetic interactions between Mcm10 and other HP family genes such as HP2, HP3, HP4, HP5 and HP6. We saw that Mcm10 genetically interacts with HP4 and HP6, since the Mcm10 knockdown-induced rough eye phenotype was enhanced by crossing with either HP4 mutants (Figure 7 J-L) or HP6 mutants (Figure 7 M-O) in compared to the Mcm10 knockdown alone (Figure 7G–I). In the double knockdown of Mcm10 with HP1a (Figure 7R–S and V–W), we noticed again an increase of roughness in the eyes compared to single knockdown flies (Figure 7G–H, P–Q and T–U). Furthermore, the severely damaged eye phenotypes in these flies are associated with melanotic dots, likely precursors for melanotic tumors (47) (Figure 7J–L and M–O).
Figure 7.
Scanning electron micrographs and color photo images of adult compound eyes. Posterior is to the right and dorsal is to the top. The flies were reared at 28°C. Panels I, J and M show color photo images and other panels show SEM images. (A and B) w; +; PGSV1-HP4s-147.1, (C and D) w1118; net1 PGT1-HP6BG01429 dpBG01429/ln(2LR)Gla, wgGla-1PPO1Bc, (E and F) GMR-GAL4/+; +; +, (G–I) GMR-GAL4/+; UAS-Mcm10IR/+; +, (J–L) GMR-GAL4/+; UAS-Mcm10IR/+; PGSV1-HP4s-147.1/+, (M–O) w1118; PGT1-HP6BG01429/In(2LR)Gla; +, (P and Q) GMR-GAL4/+; UAS-HP1a119-206IR/+; +, (R and S) GMR-GAL4/+; UAS-HP1a119-206IR/+; +, (T and U) GMR-GAL4/+; +; UAS-HP1a73-181IR/+, (V and W) GMR-GAL4/+; UAS-Mcm10IR/+; UAS-HP1a73-181IR/+. Panels B, D, F, H, L, O, Q, S, U, W (Scale bars indicate 50μm) are higher magnification images of Figure A, C, E, G, K, N, P, R, T, V (scale bars indicate 20 μm), respectively.
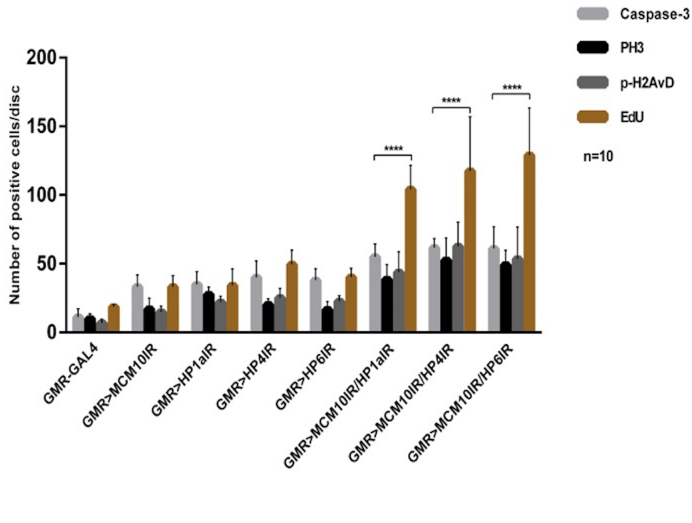
We further investigated the functions of Mcm10 together with HP1a, HP4 and HP6 in genome maintenance in the eye imaginal discs. In single and double knockdown flies for these genes and control driver flies, we performed EdU assays and immunostaining with anti-phospho-H2AvD, anti-phospho-Histone H3 (PH3), anti-Caspase-3 antibodies to detect DNA damage, cell proliferation and cell death. In single-knockdown experiments with Mcm10, HP1a, HP4 or HP6, we observed an increased numbers of cells positive for EdU, phospho-H2AvD, phospho-Histone H3 and Caspase-3 in the posterior regions compared to the control driver alone (Figure 8). Moreover, a partial rescue of the rough eye phenotype induced by knockdown of Mcm10, or HP1a, or HP4, or HP6 was found in flies co-expressing P35 (Supplementary Figure S11G–H, O–P, C–D and K–L, respectively) in compared to the single knockdown of each gene (Supplementary Figure S11A–B, E–F, I–J and M–N). Co-expression of P35 also suppressed the ectopic induction of apoptosis signals (Supplementary Figure S9A’–D’). These rescue effects confirm the apoptosis induction of knockdown Mcm10 or HP1a, HP4 and HP6. Taken together, these data indicate that Mcm10 and those HP proteins are involved in genome stability. Furthermore, double knockdown of Mcm10 with each of HP proteins shows the significant increase in the cell proliferation marked with EdU and phospho-H2AvD signals in the posterior regions (Figure 8 and Supplementary Figure S9 J, P and V) in compared to the single knockdown flies (Figure 8 and Supplementary Figure S9D, G, M, and S). Some of the EdU positive cells merged with phospho-H2AvD positive cells representing cells undergoing DNA replication and DNA damage simultaneously (Supplementary Figure S9J, P and V, blue arrows and Supplementary Figure S10). We also observed a slightly increased number of apoptotic cells in the posterior regions (Figure 8 and Supplementary Figure S9K, Q and W), but they were not comparable to the number of EdU positive cells (Figure 8).
Figure 8.
Mcm10 and HP1a, HP4, HP6 play roles in genome maintenance in Drosophila eye imaginal discs. Quantification of EdU positive cells in posterior regions per disc from three independent EdU labeling and signals of immunostaining assays with indicated antibodies. Each assay was tested with 10 eye imaginal discs (mean SD, ****P< 0.0001, two-way ANOVA and Turkey's multiple comparisons test). Double knockdown of Mcm10 and HP1a, Mcm10 and HP4, Mcm10 and HP6 show significant increase in EdU, phospho-H2AvD (p-H2AvD), PH3 and activated Caspase-3 (Caspase-3) signals in compared to single knockdown of Mcm10.
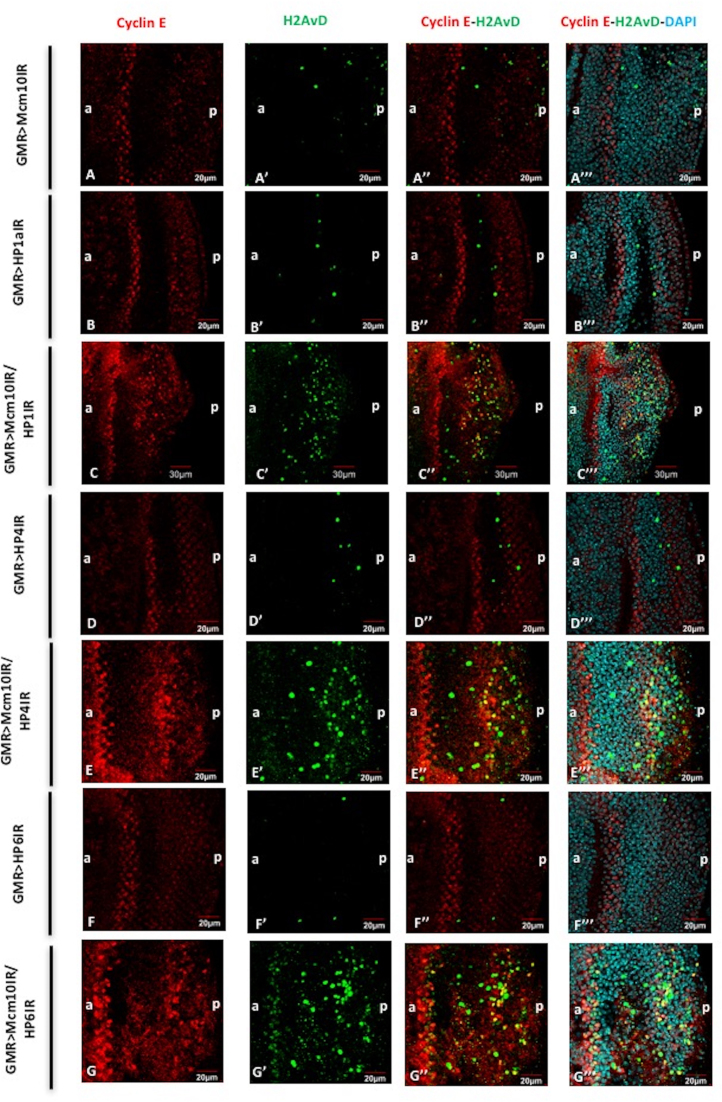
Furthermore, we examined the checkpoint activities in those double knockdown flies. Knockdown of both Mcm10 and HP1a or HP4 or HP6 results in enhanced signal of Cycle E in the eye imaginal discs (Figure 9C–C’’’, E–E’’’ and G–G’’’, respectively). However, we did not detect any effect of Mcm10, HP1a, HP4 or HP6 knockdown alone on the expression level of Cyclin E (Figure 9A–A’’’, B–B’’’, D–D’’’ and F–F’’’, respectively). In addition, the co-immunostaining of phospho-H2AvD and Cyclin E showed that some cells carrying double-strand breaks co-localised with Cyclin E-positive cells (Figure 9C–C’’’, E–E’’’ and G–G’’’). Collectively, the results suggest that the Mcm10 cooperates with each of HP proteins (HP1a, HP4 and HP6) and plays important roles both in cell cycle checkpoint activity and genome maintenance. Double knockdown of Mcm10 and HP1a shows a significant increase of cell proliferation, which partly does not go into apoptosis. Those cells that fail to be arrested by checkpoint may therefore contribute to the formation of tumour precursors in posterior regions of the eye imaginal discs (Figure 7 J–L and M–O).
Figure 9.
Mcm10 and HP1a, HP4, HP6 play roles in G1-S cell cycle checkpoint. Eye imaginal discs from GMR-GAL4/+; UAS-Mcm10IR/+; + (A–A’’’), GMR-GAL4/+; +; UAS-HP1a73-181IR/+ (B–B’’’), GMR-GAL4/+; UAS-Mcm10IR/+; UAS-HP1a73-181IR/+ (C–C’’’), GMR-GAL4/+; +; UAS-HP4IR/+ (D–D’’’); GMR-GAL4/+; UAS-Mcm10IR/+; UAS-HP4IR/+ (E–E’’’), GMR-GAL4/+; +; UAS-HP6IR/+ (F–F’’’), GMR-GAL4/+; UAS-Mcm10IR/+; UAS-HP6IR/+ (G–G’’’) flies were immunostained with anti-Cyclin E (red) and anti-H2AvD (green). All of the discs were stained with DAPI for DNA staining (blue). (A–A’’’) the number of Cyclin E-positive cells was not changed in the eye imaginal discs of Mcm10 RNAi lines. However, eye imaginal discs from double knockdown of Mcm10 and HP1a (C–C’’’), Mcm10 and HP4 (E–E’’’), and Mcm10 and HP6 (G-G’’’) show significantly increased both Cyclin E-positive and phospho-H2AvD-positive cells in posterior regions. Furthermore, some of the phospho-H2AvD-positive cells were merged with Cyclin E-positive cells (C’’, E’’ and G’’) likely representing for cells arrested in G1-S checkpoint. In contrast, we could not find the same phenotypes in the single knockdown of these genes (A’’, B’’, D’’ and F’’). The flies were reared at 28°C. Scale bars indicate 20 or 30 μm. (a) Indicates anterior, (p) indicates posterior.
Mcm10 and HP1a play a role for the differentiation of photoreceptor cells R1, R6 and R7
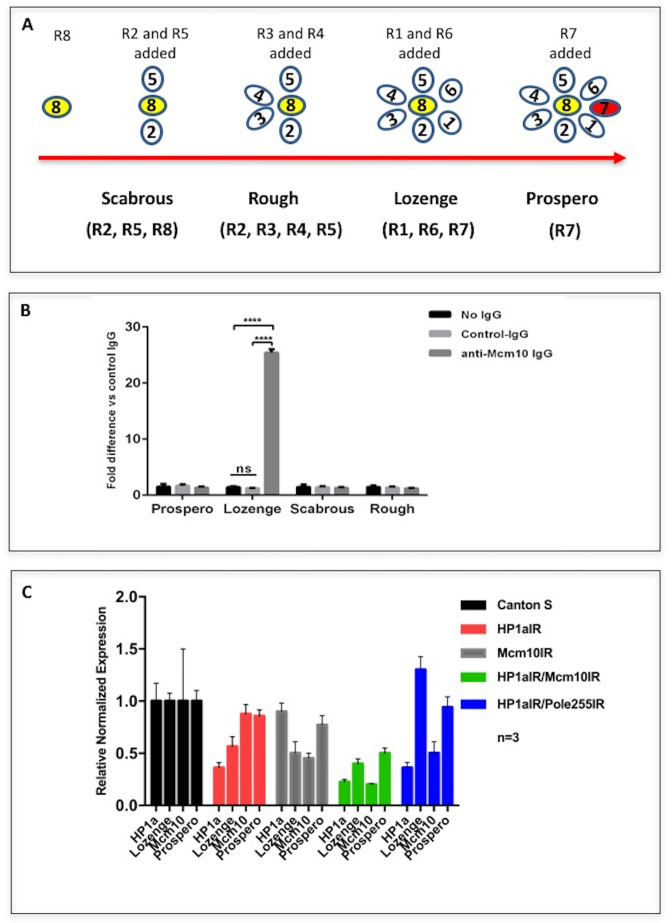
Previously we reported that Mcm10 is required for R7 photoreceptor cell differentiation in eye imaginal discs (34). The present study with PLA shows the close proximity between Mcm10 and HP1a in the posterior region where photoreceptor cells are undergoing differentiation. This raises the possibility that not only Mcm10 but also HP1a may be involved in the differentiation of photoreceptor cells. The photoreceptors (R cells) are divided into three different types depending on their genetic and morphogenetic function. The outer (R1–R6) lies in a ring surrounding two central receptors; R7, the distal, or outer, central cells; and R8, the proximal, or inner, central cells. R cells have been found to be generated sequentially: R8 is generated first, with movement posterior from the MF, then cells are added pair wise (R2 and R5, R3 and R4, and R1 and R6) and R7 is the last photoreceptor to be added to the precluster (48) (Figure 10A).
Figure 10.
(A) A diagram of the differentiation of eight photoreceptor cells in eye imaginal discs and the markers used to mark the relevant photoreceptors in the experiments. Anti-Scabrous antibody was used to mark R2, R5 and R8, anti-Rough antibody was used to mark R2, R3, R4 and R5, anti-Lozenge antibody was used to mark R1, R6 and R7, and anti-Prospero antibody was used to mark R7. (B) Binding of Mcm10 to upstream region of the lozenge gene. Cross-linked chromatin of S2 DRSC cells was immunoprecipitated with anti-Mcm10 IgG, normal rabbit IgG (control IgG) or no IgG. The genomic regions from nucleotide position −1459 to −1300 of the lozenge gene were amplified by real time PCR and compared with the amplification products from the immunoprecipitates with the control IgG. No amplification was observed with 5΄-flanking regions of prospero, scabrous, and rough genes. The statistical analysis was conducted using GraphPad Prism 6. ****P< 0.0001. (C) Endogenous mRNA levels of Lozenge and Prospero were reduced in double knockdown of Mcm10 and HP1 in eye imaginal discs. Sixty eye imaginal discs from Canton S with single knockdown or double knockdown of Mcm10 and HP1a were dissected and mRNAs were extracted for RT-PCR analysis. The signals were normalized relative to the internal control G6PD mRNA. The experiments were repeated three times. Averaged values are shown with standard deviations.
Firstly, we performed double immunostaining with the anti-Prospero antibody (green) and anti-Mcm10 antibody (red) in the wild type Canton S eye imaginal discs to observe the distribution of Mcm10 and Prospero. The results showed that Mcm10 signals not only co-localise with the Prospero signals in the nuclei of R7 cells but also can be detected in other photoreceptor cells (Supplementary Figure S12A–E). We therefore examined the effects of Mcm10 knockdown on other photoreceptors (R1–R6 and R8). We performed the flip-out experiment in either Mcm10 knockdown or HP1a knockdown flies and tested with several markers to follow the fates of individual cells (Figure 11). One of the markers, Prospero is expressed in the R7 equivalence group (the R7 and cone cells) during early stages of their differentiation (49). The level of Prospero expression is initially equivalent, but gradually increases in the R7 cell. The other marker, Lozenge activates Bar gene expression in the R1/R6 pair; an essential function required for specification of the fate of that pair (50). Lozenge is also active in R7 and cone cells (51). The third marker Rough is initially expressed in clusters of cells in the morphogenetic furrow (MF), but at the precluster stage, it is expressed specifically in R2, R3, R4 and R5 (52). The last marker Scabrous is initially expressed in clusters of cells in the morphogenetic furrow representing R2, R5 and becomes subsequently restricted to R8 (53).
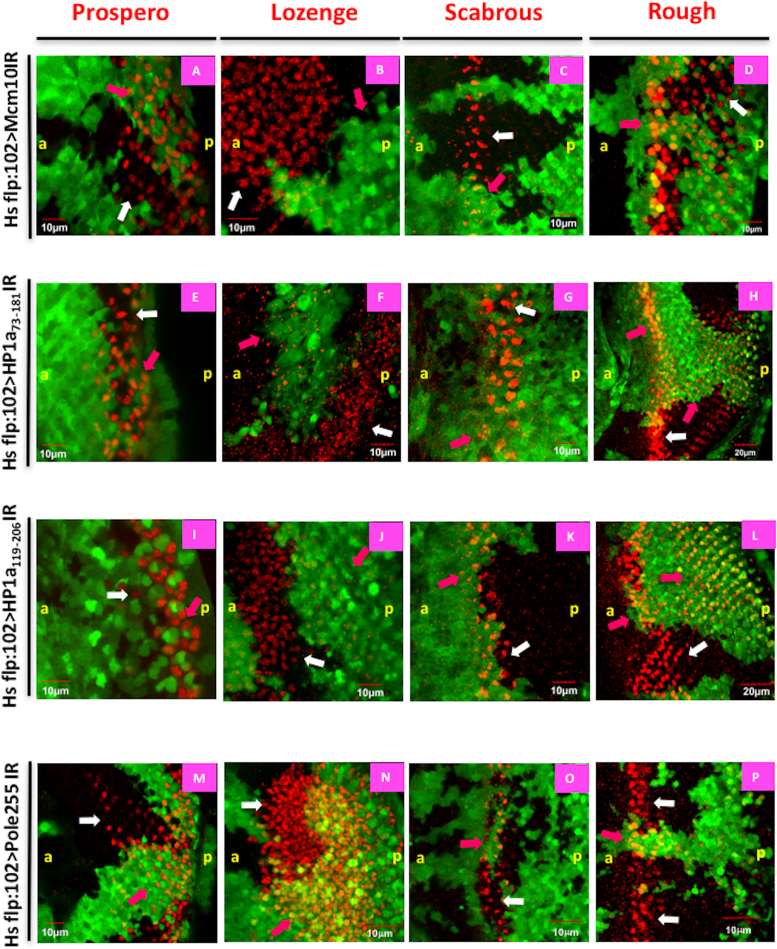
Figure 11.
Both HP1a and Mcm10 involve in the differentiation of photoreceptor cells R1, R6 and R7. Eye imaginal discs from flip-out experiments with UAS-Mcm10IR (A–D), UAS-HP1a73-181IR (E–H), UAS-HP1a119-206IR (I–L) and UAS-dpolε255IR (M–P) flies were immunostained with different developmental markers for photoreceptor cells (red): Prospero (A, E, I and M), Lozenge (B, F, J and N), Scabrous (C, G, K and O), and Rough (D, H, L and P). The cells expressing Mcm10 dsRNA, HP1a73-181dsRNA, HP1a119-206dsRNA, dpolε255 dsRNA are marked with GFP (green). In Mcm10IR, HP1a73-181IR and HP1a119-206IR flies, the expression levels of Lozenge (B, F and J) are significantly reduced in the GFP clones (pink arrows), while Prospero signals (A, E and I) are slightly reduced in knockdown areas (pink arrows) compared to non-knockdown areas (white arrows). In contrast, the signals of Scabrous (C, G and K) and Rough (D, H and L) do not change between knockdown (pink arrows) and non-knockdown (white arrows) areas of these three fly lines. Furthermore, we did not see the difference in the expression pattern of all markers in dpolε255IR flies (M–P). These data indicate that Mcm10 and HP1a are involved in the differentiation of R1, R6 and R7 cells. Scale bars indicate 10 or 20 μm. (a) Indicates anterior, (p) indicates posterior.
We produced flip-out clones in which GFP clones (Figure 11, pink arrows) mark knockdown of target gene and non-GFP clones (Figure 11, white arrows) mark the non-knockdown area. In both Mcm10 knockdown and two different HP1a knockdown flies, a strong reduction of Lozenge (R1, R6, R7 marker) signals (Figure 11 B, F and J) was observed in the GFP clones in the eye imaginal discs. Although there are a few cells expressing both Lozenge and Mcm10 (Supplementary Figure S13D), this is likely because of the inefficient knockdown of Mcm10 even in the GFP positive Mcm10 knockdown area as shown in the Supplementary Figure S1. In addition, DAPI staining shows these three cells still exist in the knockdown area, suggesting that these cells are not lost by apoptosis (Supplementary Figure S13A–E). Under the same conditions, Prospero (R7 marker) (Figure 11A, E and I) signals were slightly reduced. In contrast, the expression levels of Scabrous (R2, R5 and R8 marker) (Figure 11C, G and K) and Rough (R2, R3, R4 and R5 marker) (Figure 11D, H and L) did not change at all in either GFP clones or non-GFP clones. In the flip-out experiments with dpolε255IR, we did not detect any change in the expression levels of any markers (Figure 11M–P), indicating that dpolε255 plays no role in photoreceptor cell differentiation. In addition, we examined Mcm10 signals in HP1a knockdown area of the eye imaginal discs, since it is reported that HP1a can modulate the transcription of some cell-cycle regulators in Drosophila (54). Mcm10 signals were not significantly changed in the HP1a knockdown area in compared to the non-knockdown area, suggesting that HP1a exerts no apparent effect on Mcm10 expression (Supplementary Figure S13F–J).
In order to further confirm these observations, we performed double knockdown experiments on Mcm10 and HP1a. In the double knockdown flies, the Lozenge signals were effectively reduced in the posterior regions of the eye imaginal discs (Supplementary Figure S14A–M). Under the same conditions, the Prospero signals were slightly reduced (Supplementary Figure S12 A-M). However, again neither the Rough (Supplementary Figure S15A–M) nor the Scabrous (Supplementary Figure S16A–M) signal changed in compared to the controls. In addition, the mRNA levels of Lozenge in eye imaginal discs were consistently decreased in the eye imaginal discs of double knockdown of Mcm10 and HP1a in compared to the wild type (Canton S) (Figure 10C). Knockdown of HP1a alone and Mcm10 alone also decreased the mRNA levels of Lozenge in eye imaginal discs by 42% and 50%, respectively (Figure 10C). However, unexpectedly double knockdown of HP1a and dpolε255 marginally increased the mRNA levels of Lozenge, albeit the levels of HP1a and Mcm10 are decreased, suggesting that dpolε may have a negative role for lozenge expression independently to HP1a and Mcm10. It should be noted that dpolε255 genetically interacts with chromatin remodeling factors that could affect expression of some genes through modulation of chromatin structure (36). Further analysis is necessary to clarify this point. In any event, we can here suggest that Mcm10 and HP1a are both required for the differentiation of R1, R6 and R7. In addition, in chromatin immunoprecipitates with an anti-Mcm10 polyclonal antibody, amplification of the lozenge gene upstream region containing binding sites for various insulator associated factors such as MDG4, Su(Hw), CTCF, CP190 and BEAF32 (ModEncode: http://gbrowse.modencode.org/fgb2/gbrowse/fly/?start = 9180673;end = 9182335;ref = X;label = Genes;label = White_INSULATORS_WIG;width = 750#) was 25-fold higher than with control rabbit IgG (Figure 10B). In contrast, no amplification of the upstream regions of prospero, scabrous or rough gene was observed (Figure 10B). These data suggest that Mcm10 binds to the genomic region containing the lozenge gene upstream region. Altogether, both Mcm10 and HP1a are required for differentiation of photoreceptor cells R1, R6 and R7 that is likely mediated by the regulation of lozenge. The mild effect on prospero may be a secondary effect by down regulation of lozenge expression by double knockdown of Mcm10 and HP1a.
DISCUSSION
Mcm10 and HP1a are known to be involved in initiation of DNA replication and genome maintenance in eukaryotic cells. In the present study, we further investigated these roles of Mcm10 and HP1a in Drosophila. Proteomics studies of HP1a-, HP1b- and HP1c-interacting proteins in Drosophila identified 160–310 proteins as candidate interacting partners (55). Some of them are common to these three HP1 families and others are unique to each of them. These candidate proteins can be divided into several groups including chromosome organization, gene expression, cell death and cell cycle. Although Mcm10 was not identified as a HP1a-interacting protein in these analyses, the another group has reported the physical interaction between HP1a and Mcm10 by yeast two hybrid system and genetic interaction in Drosophila wing (33). In this study, we confirmed the physical interaction between these two proteins by co-IP assay, S30 T7 in vitro translation/transcription-Halo-tag pull down for the domain interactions and genetic interaction in the Drosophila compound eye. Furthermore, by using PLA, we visualised close proximity between HP1a and Mcm10, RFC140, and dpolε255 in both Drosophila cultured cells and living flies. Taken together with the previous report (17), HP1a might play a role in loading of replication complex to the chromosomal sites required for DNA replication initiation and/or elongation.
The function of HP1a in replication appears to be specific to the mitotic cell cycle, since the interaction between HP1a and Mcm10 was not detected in endocycling salivary gland cells. In contrast, PLA signals between Mcm10 and DNA polymerase α are detectable in both mitotic cells in the eye imaginal discs and endoreplicating cells in salivary glands. These data suggest that the interaction between different proteins, which even have the same partner, is responsible for different functions. It has been suggested that the origin recognition complex (Orc) is dispensable for endoreplication in salivary gland cells, suggesting differential requirement of Orc subunits for initiation of DNA replication in the mitotic cell cycle and the endocycle (56). Furthermore, N-terminal region of dpolε255 is not required for mitotic replication but essential for endoreplication (36). The present results are therefore consistent with the notion that regulatory mechanism of replication is different between mitotic cells and endocycling cells.
It is reported that loss of Mcm10 engages checkpoint, DNA repair and SUMO-dependent rescue pathways that can counteract replication stress accompanied with chromosome breakage (38,57–66). The present study also suggests that the complex between Mcm10 and HP1a plays an important role in cell cycle progression and genome maintenance. These findings give further insight how Mcm10 protects genome integrity during DNA replication. Notably the single knockdown of Mcm10 did not affect the level of Cyclin E in the eye imaginal discs. However, double knockdown of Mcm10 and HP1a causes increased number of Cyclin E-positive cells in the posterior region of the eye imaginal disc. Generally, one can predict that Cyclin E-positive cells with DNA damage subsequently activate the DNA repair or promote apoptosis to remove these cells with severe DNA damage from the tissues. Surprisingly, however double knockdown of Mcm10 and HP1a shows a significant increase of cell proliferation, which at least partly does not go into apoptosis. Those cells that fail to be arrested by checkpoint may therefore contribute to the formation of tumour precursors in posterior regions of the eye imaginal discs. In fact, the severely damaged eye phenotypes in these flies are associated with melanotic dots, likely precursors for melanotic tumours (47) (Figure 7 J-L and M-O). We can therefore speculate that the knockdown of Mcm10 together with HP proteins could lead to some cancers, while it is only reported that the increased copy number of Mcm10 was observed in some cancer patients (67,68). Although further analyses are necessary, the present study may give a new interpretation for the role of Mcm10 in future cancer studies.
In the present study, we also demonstrated that Mcm10 plays a role in differentiation of R1, R6 and R7 photoreceptor cells. From the results with ChIP assays in S2 cells and quantitative RT-PCR in Mcm10 knockdown flies, Mcm10 appears to be involved in transcriptional regulation of the lozenge gene that is responsible for differentiation of R1, R6 and R7 cells. Although the precise mechanism for the regulation is not known yet, Mcm10 may regulate expression of lozenge by modulating chromatin structure through cooperation or competition with various insulator-associated factors such as MDG4, Su(Hw), CTCF, CP190 and BEAF32. It is well known that Lozenge is specifically expressed in and required for development of the crystal cells, a megakaryocyte-like lineage that participates in clotting (69). The crystal cells secrete components of the phenol oxidase cascade and are involved in the melanisation of invading organisms and in wound repair (69,70). Lozenge interacts and cooperates with another transcription factor, Serpent, to activate the crystal cell differentiation program (71–73). This cooperation is conserved in mammals, where it controls megakaryopoiesis (74–76) and hematopoietic stem cell development (77). It would be therefore interesting to examine roles of Mcm10 and HP1a in the hemocyte system.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly thank to Dr Nguyen Huu Tho for making the fly lines of Mcm10IR in salivary glands, Mr Umegawachi for useful guidance of constructs and primers, Dr Morii and Dr Kotani for kindly using Olympus FLUOVIEW FV1000, Dr Gilles Crevel and Dr Isabelle Crevel for useful advice in in vitro method, Dr Richardson and Dr Burke for good quality Cyclin E antibody, Dr Harada for advice in HP1a structure, Dr Orr-Weaver for contributive comments, and all the lab members of Chromosome Engineering Lab in KIT.
Author contributions: N.V. designed the study, conceived and designed experiments, carried out all experiments, analysed the data and wrote the manuscript. N.Y. performed the immunostaining in salivary glands. N.V. developed the S30-T7 in vitro transcription/translation-Halotag pull down system in E. coli and the co-IP in S2 cells for the revision. N.V. and S.D. performed the revised experiments. M.Y., S.C., H.Y. revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); Japan Society for the Promotion of Science Core-to-Core Program (JSPS), Japan Science Society-Sasakawa Scientific Research Grant and Kyoto Institute of Technology-Venture Laboratory Research Project; Kyoto Institute of Technology Foundation Scholarship Award (to N.V.), Japanese Government (Monbukagakusho) Honors Scholarship for International Students, Kyoto Institute of Technology Promotional Funding for International Exchange; Japanese Government (Monbukagakusho) Honors Scholarship for International Students (to S.D.). Funding for open access charge: Kyoto Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vermaak D., Malik H.S.. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 2009; 43:467–492. [DOI] [PubMed] [Google Scholar]
- 2.James T.C., Elgin S.C.. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986; 6:3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomberk G., Wallrath L., Urrutia R.. The Heterochromatin Protein 1 family. Genome Biol. 2006; 7:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T.. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001; 410:120–124. [DOI] [PubMed] [Google Scholar]
- 5.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T.. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001; 410:116–120. [DOI] [PubMed] [Google Scholar]
- 6.Sims R.J. III, Nishioka K., Reinberg D.. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003; 19:629–639. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs S.A., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S.. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001; 20:5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aagaard L., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B. et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999; 18:1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., Reuter G.. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994; 13:3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zofall M., Grewal S.I.. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:487–496. [DOI] [PubMed] [Google Scholar]
- 11.Greil F., de Wit E., Bussemaker H.J., van Steensel B.. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. EMBO J. 2007; 26:741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joppich C., Scholz S., Korge G., Schwendemann A.. Umbrea, a chromo shadow domain protein in Drosophila melanogaster heterochromatin, interacts with Hip, HP1 and HOAP. Chromosome Res. 2009; 17:19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alekseyenko A.A., Gorchakov A.A., Zee B.M., Fuchs S.M., Kharchenko P.V., Kuroda M.I.. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 2014; 28:1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O. et al. A protein complex network of Drosophila melanogaster. Cell. 2011; 147:690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pindyurin A.V., Boldyreva L.V., Shloma V.V., Kolesnikova T.D., Pokholkova G.V., Andreyeva E.N., Kozhevnikova E.N., Ivanoschuk I.G., Zarutskaya E.A., Demakov S.A. et al. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J. Cell Sci. 2008; 121:1693–1703. [DOI] [PubMed] [Google Scholar]
- 16.Quivy J.P., Gerard A., Cook A.J., Roche D., Almouzni G.. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 2008; 15:972–979. [DOI] [PubMed] [Google Scholar]
- 17.Schwaiger M., Kohler H., Oakeley E.J., Stadler M.B., Schubeler D.. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 2010; 20:771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luijsterburg M.S., Dinant C., Lans H., Stap J., Wiernasz E., Lagerwerf S., Warmerdam D.O., Lindh M., Brink M.C., Dobrucki J.W. et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009; 185:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R.. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008; 453:682–686. [DOI] [PubMed] [Google Scholar]
- 20.Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 21.Wohlschlegel J.A., Dhar S.K., Prokhorova T.A., Dutta A., Walter J.C.. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell. 2002; 9:233–240. [DOI] [PubMed] [Google Scholar]
- 22.Ricke R.M., Bielinsky A.K.. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell. 2004; 16:173–185. [DOI] [PubMed] [Google Scholar]
- 23.Gregan J., Lindner K., Brimage L., Franklin R., Namdar M., Hart E.A., Aves S.J., Kearsey S.E.. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell. 2003; 14:3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller R.C., Kang S., Lam W.M., Chen S., Chan C.S., Bell S.P.. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011; 146:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanke M., Kodama Y., Takahashi T.S., Nakagawa T., Masukata H.. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012; 31:2182–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi M., Yatagai F., Hanaoka F.. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 2004; 279:32569–32577. [DOI] [PubMed] [Google Scholar]
- 27.Yang X., Gregan J., Lindner K., Young H., Kearsey S.E.. Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast. BMC Mol. Biol. 2005; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Deursen F., Sengupta S., De Piccoli G., Sanchez-Diaz A., Labib K.. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012; 31:2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas N.L., Dozier S.K., Donato J.J.. Dual roles for Mcm10 in DNA replication initiation and silencing at the mating-type loci. Mol. Biol. Rep. 2005; 32:197–204. [DOI] [PubMed] [Google Scholar]
- 30.Liachko I., Tye B.K.. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2005; 171:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liachko I., Tye B.K.. Mcm10 mediates the interaction between DNA replication and silencing machineries. Genetics. 2009; 181:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen T.W., Tye B.K.. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell. 2003; 14:2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apger J., Reubens M., Henderson L., Gouge C.A., Ilic N., Zhou H.H., Christensen T.W.. Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics. 2010; 185:1151–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vo N., Taga A., Inaba Y., Yoshida H., Cotterill S., Yamaguchi M.. Drosophila Mcm10 is required for DNA replication and differentiation in the compound eye. PLoS One. 2014; 9:e93450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi M., Hirose F., Nishimoto Y., Naruge T., Ikeda M., Hachiya T., Tamai K., Kuroda K., Matsukage A.. Expression patterns of DNA replication enzymes and the regulatory factor DREF during Drosophila development analyzed with specific antibodies. Biol. Cell. 1995; 85:147–155. [DOI] [PubMed] [Google Scholar]
- 36.Suyari O., Kawai M., Ida H., Yoshida H., Sakaguchi K., Yamaguchi M.. Differential requirement for the N-terminal catalytic domain of the DNA polymerase epsilon p255 subunit in the mitotic cell cycle and the endocycle. Gene. 2012; 495:104–114. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya A., Inoue Y.H., Ida H., Kawase Y., Okudaira K., Ohno K., Yoshida H., Yamaguchi M.. Transcriptional regulation of the Drosophila rfc1 gene by the DRE-DREF pathway. FEBS J. 2007; 274:1818–1832. [DOI] [PubMed] [Google Scholar]
- 38.Raveendranathan M., Chattopadhyay S., Bolon Y.T., Haworth J., Clarke D.J., Bielinsky A.K.. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J. 2006; 25:3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren E.M., Vaithiyalingam S., Haworth J., Greer B., Bielinsky A.K., Chazin W.J., Eichman B.F.. Structural basis for DNA binding by replication initiator Mcm10. Structure. 2008; 16:1892–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du W., Stauffer M.E., Eichman B.F.. Structural biology of replication initiation factor Mcm10. Subcell. Biochem. 2012; 62:197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petruk S., Black K.L., Kovermann S.K., Brock H.W., Mazo A.. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nat. Commun. 2013; 4:2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loor G., Zhang S.J., Zhang P., Toomey N.L., Lee M.Y.. Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997; 25:5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witko-Sarsat V., Pederzoli-Ribeil M., Hirsch E., Sozzani S., Cassatella M.A.. Regulating neutrophil apoptosis: new players enter the game. Trends Immunol. 2011; 32:117–124. [DOI] [PubMed] [Google Scholar]
- 44.Witko-Sarsat V., Mocek J., Bouayad D., Tamassia N., Ribeil J.A., Candalh C., Davezac N., Reuter N., Mouthon L., Hermine O. et al. Proliferating cell nuclear antigen acts as a cytoplasmic platform controlling human neutrophil survival. J. Exp. Med. 2010; 207:2631–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiara A.D., Pederzoli-Ribeil M., Burgel P.R., Danel C., Witko-Sarsat V.. Targeting cytosolic proliferating cell nuclear antigen in neutrophil-dominated inflammation. Front. Immunol. 2012; 3:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chattopadhyay S., Bielinsky A.K.. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell. 2007; 18:4085–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oren-Giladi P., Krieger O., Edgar B.A., Chamovitz D.A., Segal D.. Cop9 signalosome subunit 8 (CSN8) is essential for Drosophila development. Genes Cells. 2008; 13:221–231. [DOI] [PubMed] [Google Scholar]
- 48.Barry D., Ernst H.. Michael B, Alfonso MA. Genetic dissection of eye development in Drosophila. The development of Drosophila melanogater. 1993; II, NY: Cold Spring Harbor Laboratory Press; 1327–1358. [Google Scholar]
- 49.Xu C., Kauffmann R.C., Zhang J., Kladny S., Carthew R.W.. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000; 103:87–97. [DOI] [PubMed] [Google Scholar]
- 50.Daga A., Karlovich C.A., Dumstrei K., Banerjee U.. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996; 10:1194–1205. [DOI] [PubMed] [Google Scholar]
- 51.Flores G.V., Daga A., Kalhor H.R., Banerjee U.. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development. 1998; 125:3681–3687. [DOI] [PubMed] [Google Scholar]
- 52.Tomlinson A., Kimmel B.E., Rubin G.M.. rough, a Drosophila homeobox gene required in photoreceptors R2 and R5 for inductive interactions in the developing eye. Cell. 1988; 55:771–784. [DOI] [PubMed] [Google Scholar]
- 53.Mlodzik M., Baker N.E., Rubin G.M.. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990; 4:1848–1861. [DOI] [PubMed] [Google Scholar]
- 54.De Lucia F., Ni J.Q., Vaillant C., Sun F.L.. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005; 33:2852–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu H.W., Lee D.H., Florens L., Swanson S.K., Washburn M.P., Kwon S.H.. Analysis of the heterochromatin protein 1 (HP1) interactome in Drosophila. J. Proteomics. 2014; 102:137–147. [DOI] [PubMed] [Google Scholar]
- 56.Park S.Y., Asano M.. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:12343–12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee C., Liachko I., Bouten R., Kelman Z., Tye B.K.. Alternative mechanisms for coordinating polymerase alpha and MCM helicase. Mol. Cell. Biol. 2010; 30:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Wu R., Lu Y., Liang C.. Ctf4p facilitates Mcm10p to promote DNA replication in budding yeast. Biochem. Biophys. Res. Commun. 2010; 395:336–341. [DOI] [PubMed] [Google Scholar]
- 59.Haworth J., Alver R.C., Anderson M., Bielinsky A.K.. Ubc4 and Not4 regulate steady-state levels of DNA polymerase-alpha to promote efficient and accurate DNA replication. Mol. Biol. Cell. 2010; 21:3205–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor M., Moore K., Murray J., Aves S.J., Price C.. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1). DNA repair. 2011; 10:1154–1163. [DOI] [PubMed] [Google Scholar]
- 61.Pacek M., Tutter A.V., Kubota Y., Takisawa H., Walter J.C.. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006; 21:581–587. [DOI] [PubMed] [Google Scholar]
- 62.Liang D.T., Forsburg S.L.. Characterization of Schizosaccharomyces pombe mcm7(+) and cdc23(+) (MCM10) and interactions with replication checkpoints. Genetics. 2001; 159:471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009; 35:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grofte M., Chan K.L., Hickson I.D., Bartek J. et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 65.Park J.H., Bang S.W., Kim S.H., Hwang D.S.. Knockdown of human MCM10 activates G2 checkpoint pathway. Biochem. Biophys. Res. Commun. 2008; 365:490–495. [DOI] [PubMed] [Google Scholar]
- 66.Park J.H., Bang S.W., Jeon Y., Kang S., Hwang D.S.. Knockdown of human MCM10 exhibits delayed and incomplete chromosome replication. Biochem. Biophys. Res. Commun. 2008; 365:575–582. [DOI] [PubMed] [Google Scholar]
- 67.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 68.Thu Y.M., Bielinsky A.K.. MCM10: one tool for all-Integrity, maintenance and damage control. Semin. Cell Dev. Biol. 2014; 30:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lebestky T., Chang T., Hartenstein V., Banerjee U.. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000; 288:146–149. [DOI] [PubMed] [Google Scholar]
- 70.Meister M., Lagueux M.. Drosophila blood cells. Cell. Microbiol. 2003; 5:573–580. [DOI] [PubMed] [Google Scholar]
- 71.Waltzer L., Ferjoux G., Bataille L., Haenlin M.. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 2003; 22:6516–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fossett N., Hyman K., Gajewski K., Orkin S.H., Schulz R.A.. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11451–11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferjoux G., Auge B., Boyer K., Haenlin M., Waltzer L.. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev. Biol. 2007; 305:726–734. [DOI] [PubMed] [Google Scholar]
- 74.Elagib K.E., Racke F.K., Mogass M., Khetawat R., Delehanty L.L., Goldfarb A.N.. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003; 101:4333–4341. [DOI] [PubMed] [Google Scholar]
- 75.Pencovich N., Jaschek R., Tanay A., Groner Y.. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011; 117:e1–e14. [DOI] [PubMed] [Google Scholar]
- 76.Tijssen M.R., Cvejic A., Joshi A., Hannah R.L., Ferreira R., Forrai A., Bellissimo D.C., Oram S.H., Smethurst P.A., Wilson N.K. et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell. 2011; 20:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson N.K., Foster S.D., Wang X., Knezevic K., Schutte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E. et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010; 7:532–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.