Abstract
Genetic designs can consist of dozens of genes and hundreds of genetic parts. After evaluating a design, it is desirable to implement changes without the cost and burden of starting the construction process from scratch. Here, we report a two-step process where a large design space is divided into deep pools of composite parts, from which individuals are retrieved and assembled to build a final construct. The pools are built via multiplexed assembly and sequenced using next-generation sequencing. Each pool consists of ∼20 Mb of up to 5000 unique and sequence-verified composite parts that are barcoded for retrieval by PCR. This approach is applied to a 16-gene nitrogen fixation pathway, which is broken into pools containing a total of 55 848 composite parts (71.0 Mb). The pools encompass an enormous design space (1043 possible 23 kb constructs), from which an algorithm-guided 192-member 4.5 Mb library is built. Next, all 1030 possible genetic circuits based on 10 repressors (NOR/NOT gates) are encoded in pools where each repressor is fused to all permutations of input promoters. These demonstrate that multiplexing can be applied to encompass entire design spaces from which individuals can be accessed and evaluated.
INTRODUCTION
Genetic engineers often need to explore a defined design space representing a large set of potential alternatives, as opposed to creating an individual DNA construct. In the simplest form, this could be for the creation of libraries where the expression levels of genes are varied randomly or guided by algorithms and screened to identify the top construct (1–12). The variation could be achieved through part substitution, for example by accessing a complete set from a catalog (e.g. the hundreds of promoters in the iGEM/BioFAB/SynBERC Registries (13,14)), or also encompass bigger structural changes, such as the organization of genes into operons (15). A design space could also be for combinatorial chemistry, where libraries of enzymes are combined in different ways to build chemical diversity (16,17). Or it may need to encompass a functionally complete set, e.g. all the ways that transcription factors could be combined to build a genetic circuit (18,19). In many cases, elements of the design space are unguided, where the designer simply does not know what will work best, but wants to be able to access and try a set of defined possibilities.
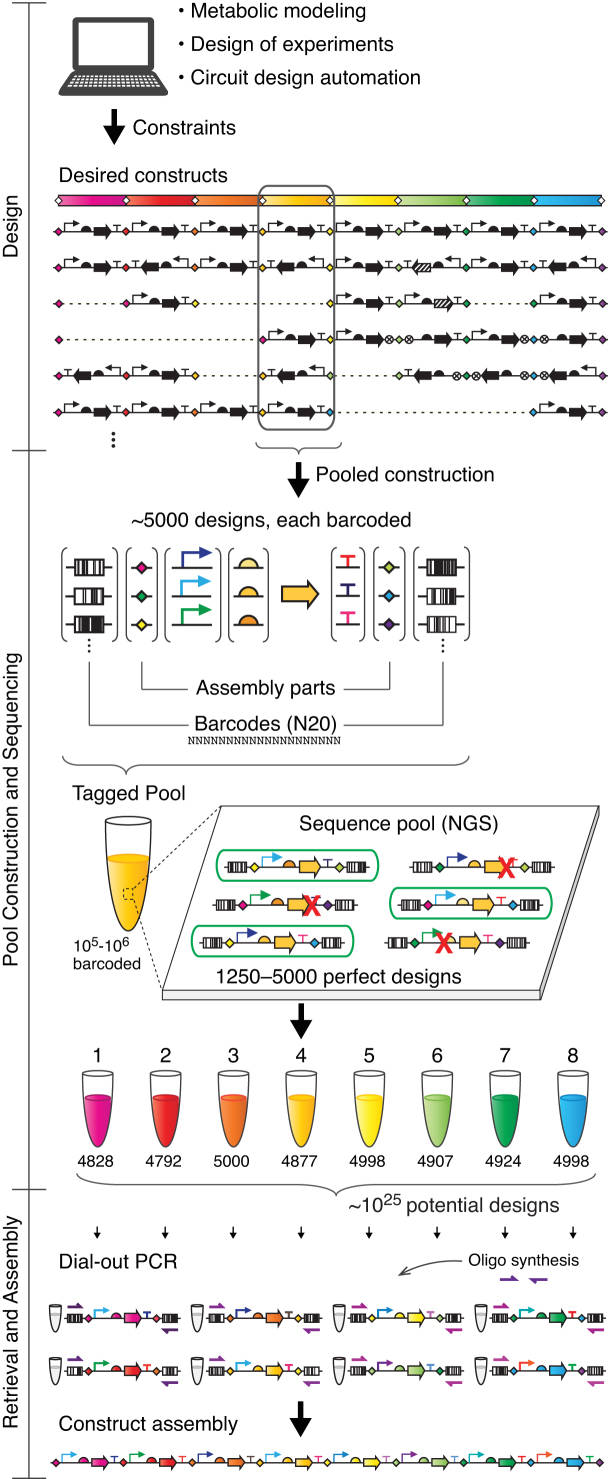
We have developed a method that allows a genetic design space to be captured within a set of multiplexed pools of DNA constructs, from which individual constructs can be retrieved and assembled (Figure 1). Each pool can contain up to 5000 composite parts (combinations of genetic parts) that are sequence-perfect. Creating the pools requires an initial investment in time and cost, but this is readily amortized as new designs can be quickly and repeatedly retrieved from the pools and assembled with high fidelity. Further, the iterative process of creating new designs from the pools is conducive to high-throughput automation, which is increasingly being applied to DNA assembly pipelines (20–23).
Figure 1.
Design and assembly of multi-part constructs from multiplexed, retrievable pools. The input to the pool extraction algorithm is a library of constructs that can be designed computationally or by hand. Dashed lines in the constructs indicate the absence of a gene. The extraction algorithm designs pools of composite parts that encompass the entire design space (combinations of parts). The parts for assembling multi-gene constructs after retrieval are included in the pool design; these can include Type IIS recognition sites and 4 bp linkers (diamonds). Barcode symbol represents the random 20 bp barcodes used for retrieval. Red X's indicate mutations in the composite part construct. Green ovals represent a constructed composite part whose sequenced reads perfectly match a designed composite part in the pool. The pools are colored to correspond to the gene from the desired constructs contained in the composite parts. The numbers below each pool are examples of the number of sequence-verified unique composite parts contained (out of 5000 designs in each pool).
Pool construction required methods to assemble and sequence verify thousands of composite parts, which is referred to as multiplexing when done simultaneously in a single reaction mixture (24,25). The term has been applied to techniques used for next-generation sequencing (NGS), where hundreds of DNA samples can be sequenced as a pool when they are ligated to 8–20 bp barcodes (26,27). Here, multiplexing is applied to assemble parts to create a pool of composite parts ligated to barcodes that are used to retrieve an individual. Previously, various approaches have been applied to the retrieval of DNA from pools (28–30). In this work, we use Dial-out PCR (31–33), where the constructs are barcoded (‘tagged’) with flanking random 20-mer oligonucleotides (1012 possible barcode sequences). The pool is sequenced using NGS, and then a sequence-verified construct can be retrieved by PCR using a primer pair that corresponds to the flanking barcodes. Dial-out PCR has been applied to retrieving long ∼200 nt oligos from pools of chip-synthesized oligonucleotides for gene synthesis (33) and a 11.4 kb penicillin gene cluster (31). The advantages over other retrieval approaches are that it utilizes standard primer synthesis, does not require specialized equipment, is highly scalable and is compatible with any NGS platform. Repurposing Dial-out retrieval for the efficient assembly of large libraries of multi-gene constructs presented several challenges. In particular, the sequencing step did not allow for high-throughput verification of pools of long constructs, e.g. if one of the parts is a full-length gene. This limitation is due to the Illumina platform that, while cost-effective and accurate, generates relatively short reads (up to 2 × 300 bp) (34). Sequencing the pools required a method to fragment the barcoded multi-part constructs and retain the identity of a construct's barcodes on all fragments generated from the pool. For this fragmentation, we adapted PCR-based approaches previously used for genome sequencing (35,36) to sequence targeted regions of composite part constructs in highly multiplexed pools of several million constructs.
Once a particular set of composite parts are retrieved from the pools, they are assembled to create an individual construct. Our approach is compatible with the many DNA assembly methods that have been widely adopted in the field (37–41). Here, we demonstrate the approach using Golden Gate assembly that utilizes Type IIS restriction enzymes (42,43). The recognition sites and overhangs (linkers) are included as parts in the creation of pools and the particular ones retrieved dictate the position, orientation and presence/absence of a gene within the final construct (Figure 1). This allows the pools to be used to create very diverse final designs, as opposed to constraining the designs to a particular gene order or distribution into operons.
In this manuscript, we show the conversion of genetic design spaces into pools of composite parts, from which individual constructs can be retrieved and assembled. First, an algorithm is described that takes a desired set of constructs and determines the pools that need to be built. The set could be generated by one of the computer aided design programs used in synthetic biology and metabolic engineering (19,44–48). Then, the pipeline is described including multiplexed pool assembly, sequence verification, retrieval, large construct assembly and the automation necessary to build libraries. This is applied to two disparate problems in genetic engineering. In the first, a design of experiments algorithm is applied to a 4.5 Mb library of 23 kb constructs to optimize expression levels in a 16-gene pathway (nitrogen fixation). In the second, pools are designed that encompass all possible genetic circuits comprising 10 repressors, including feedback loops. The pools are used to retrieve alternative designs as part of improving a combinational logic circuit. Collectively, these demonstrate the ability for retrievable pools to efficiently capture large design spaces.
MATERIALS AND METHODS
Strains and media
Escherichia coli DH5α was used for routine cloning and propagation of plasmids, except where noted. E. coli MG1655 was used for assaying nitrogenase activity. E. coli NEB 10-beta (New England Biolabs, C3019) was used for assaying genetic circuits. E. coli was cultured in LB Miller medium (Sigma-Aldrich, L3152) for routine cloning and propagation of plasmids. For nif induction, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; ThermoFisher Scientific, 15529019) was added to the medium. Genetic circuits were assayed in M9 minimal media (Sigma-Aldrich, M6030) with supplements (Supplementary Data). Inputs for the sensor promoters were 1 mM IPTG (Sigma-Aldrich, I6758), 2 ng/ml anhydrotetracycline hydrochloride (aTc; Sigma-Aldrich, 37919) and 5 mM L-arabinose (Sigma-Aldrich, A3256).
Pool extraction algorithm
For the automated design of pools, the pool extraction algorithm was implemented as a Java application using the Spring Framework. This pool designer application features a web page interface written in JavaScript and HTML for communication with human users and a representational state transfer application program interface for communication with other applications over the web. The application uses the existing software libraries libSBOLj (49) for importing and exporting construct designs written in SBOL and Neo4j (Neo Technology Inc., San Mateo, CA, USA) for storing and querying pool extraction data as graphs. The source code for the pool designer application is available open source (BSD license) on GitHub at https://github.com/CIDARLAB/poolDesigner/.
Assembly and barcoding of composite part pools
Full method details, including part construction, are available in the Supplementary Data. For multiplex assembly of pools, all part variants in each position were mixed in equimolar proportions. Parts were provided as plasmid constructs or amplified PCR products. For Golden Gate-in vivo-ligation (GVL) composite part GVL construction, multiplex assembly reactions were performed in 5.5 μl with 20 fmol linear backbone PCR product, 40 fmol of mixed parts for each part position, 2.5 U BsaI, 2.5 U T4 DNA ligase (20 U/μl HC; Promega, M1794) in 1X ligase buffer with the protocol: 37°C for 5 h, 10 cycles (37°C for 2 min then 16°C for 5 min), followed by 50°C for 15 min and then 80°C for 20 min. For Stitching-in vivo-Gibson (SVG) and Stitching-in vitro-Gibson (STG) pools, Scarless Stitching was performed in two rounds as previously described (15). For in vivo amplification of SVG pools, transformants of each assembly reaction were plated in ≥10-fold excess of the number of unique designs to harvest pooled plasmid DNA. For in vitro amplification in STG construction, PCR was performed to amplify the composite part constructs using primers that anneal to the backbone. After Scarless Stitching, the assembly parts and homology regions for Gibson assembly were added by emulsion PCR with the Micellula DNA Emulsion and Purification (ePCR) Kit (Chimerx, 3600–02) following the manufacturer's protocol and Gibson isothermal assembly was performed using Gibson Assembly Master Mix (NEB, E2611) according to the manufacturer's recommended protocol. For in vivo amplification, the tagging ratio was applied by performing electroporation using 1 μl of the final assembly reaction and plating the specified number of colonies and harvesting the pooled plasmid DNA after 12–16 h incubation. All part sequences are listed in Supplementary Data and provided in the Supplementary Data.
NGS preparation, sequencing and data processing
To sequence the pool, an aliquot of the purified tagged pool plasmid DNA (∼0.5 μg) was digested with NotI-HF (New England Biolabs, R3189), gel purified, self-circularized using Quick T4 DNA Ligase (New England Biolabs, M2200) and gel purified again to isolate the self-ligated products using E-gel EX 1% agarose gels (Thermo Fisher Scientific, G401001). Next, fragments for Illumina sequencing were generated by inverse PCR performed as an emulsion PCR (ePCR) using an ePCR kit (Chimerx, 3600) according to the manufacturer's protocol. Illumina sequencing adapters were ligated to PCR products by standard A-tailing and enrichment PCR. Prepared samples were sequenced using a MiSeq (Illumina Inc., San Diego, CA, USA) and MiSeq Reagent Kit 600 cycle v3 (Illumina, MS-102-3003). The FASTQ files for each pool were analyzed with custom Python scripts to check each read-pair's barcode identities, perfect sequence match to a composite part design and uniqueness of the barcodes to one design. Full details on sequencing preparation, MiSeq sequencing and data analysis are provided in Supplementary Data. Scripts are available open source (MIT license) on GitHub at https://github.com/VoigtLab/MIT-BroadFoundry/tree/master/dialout-designs/.
Retrieval of composite parts and pool propagation
Retrieval primers were designed using Primer3 (50,51) with a Tm of 63–66°C. For a desired composite part design, the sequence-perfect construct with greatest number of reads and allowable primer designs was retrieved. Retrieval PCRs were performed in 25 μl with Q5 DNA polymerase (New England Biolabs, M0493) with an aliquot of the tagged pool pDNA (0.2 fmol per 1000 constructs) and the protocol according to the manufacturer's recommendations and 25–30 cycles of PCR. Retrieval PCR reactions were DpnI digested and purified by a PCR cleanup using Agencourt AMPure XP magnetic beads (Beckman Coulter, A63881) according to the manufacturer's protocol. Pools have been successfully propagated by re-plating the transformed pool from frozen glycerol stocks (Supplementary Data). The design of retrieval primers for the pool was automated using the Primer3-py wrapper for Primer3 (52) (Supplementary Data). The script is available on GitHub at https://github.com/VoigtLab/MIT-BroadFoundry/tree/master/dialout-designs/.
Type IIS one-pot assembly variations
Optimization of one-pot construct assembly was performed using 16 PCR products (580 bp each) each flanked by BbsI sites and linker sequences as listed in Supplementary Data. Product distribution after assembly was quantified by capillary gel electrophoresis on an Agilent 2100 Bioanalyzer with the 12000 Kit (Agilent, 5067-1508). Original one-pot assembly conditions were based on previous work (43) (Supplementary Data). Engineered conditions increased the BbsI and DNA concentrations, and used the protocol: 100 cycles 37°C for 2 min, 16°C for 5 min followed by 50°C for 15 min and 80°C for 20 min. A custom Python script was written to design orthogonal linker sequences. The rules applied for Designed sets of linkers are the default parameters, and the script is available on GitHub (Supplementary Data).
Combinatorial nif design, nitrogenase assay and statistical modeling
Full method details are provided in the Supplementary Data. The 192-member combinatorial DOE library was designed and analyzed using the JMP Pro 12 software (SAS Institute Inc., Cary, NC, USA). A pool of composite part constructs was built using the SVG method, and composite parts were retrieved by PCR as described above. Hierarchical Type IIS assembly of retrieved composite parts was automated based on MoClo (43) Golden Gate assembly. DNA mixtures containing the retrieved composite part and appropriate destination vector were prepared using an Echo 550 acoustic liquid handling system (Labcyte Inc., Sunnyvale, CA, USA) and the Echo Cherry Pick software (Supplementary Data). Activity of each nitrogenase cluster design was measured using the standard acetylene reduction assay (53) as previously described. Each nif design was assayed in duplicate, and response surface modeling was performed using JMP Pro 12 software (Supplementary Data).
Genetic circuit design, construction and flow cytometry analysis
Circuits were designed and gates were assigned using the Cello software (19) with the default UCF JSON input file (Eco1C1G1T1.UCF.json) and Verilog code for the desired logic function. Pools of gate constructs were assembled by the GVL method described above. For each repressor, a pool of NOT constructs and a pool of NOR constructs was constructed. The pools were sequenced, and composite parts were retrieved by PCR as described above. For one-pot assembly of circuits, the reactions in 5 μl contained 20 fmol of each retrieved composite part, 10 fmol of 3-input circuit backbone, 5 U BbsI, 2.5 U T4 DNA ligase (Promega, M1794) in 1X ligase buffer with the protocol: 37°C for 8 h, followed by 50°C for 15 min and then 80°C for 20 min. The circuit constructs were fully sequenced and co-transformed into E. coli NEB 10-beta with the corresponding output plasmid containing YFP fused to the circuit output promoters (Supplementary Data). Circuits were assayed as previously described (19).
RESULTS
Algorithmic reduction of the design space into pools
The first step is to algorithmically convert a set of desired constructs into the pools necessary to encompass these designs (Figure 1). The set could be the output of any number of design algorithms or could be generated ‘by hand’. For example, when a design space is specified using a set of genetic constraints, then combinatorial design algorithms can create an arbitrary number of constructs that conform to the constraints, and this has been applied to metabolic pathways (1,4,15–16) and genetic circuits using Cello (19). Algorithmic approaches, such as numerical optimization or design of experiments, could also be used to generate the set of constructs (2,5,7,11–12). Once the set is generated, pools then have to be designed that encompass this space. For simple designs, this can be done manually, but we have developed an algorithm that automates the process for large and more complex constructs (Supplementary Data).
The input to the pool extraction algorithm is a library of constructs where parts are defined using the SBOL standard (54,55). Most often, it is desirable to organize the pools around transcription units and this is done automatically based on the SBOL part definitions. Alternatively, the user can optionally specify how the constructs should be broken into constituent pools. Starting from a library of constructs, the software performs three computational steps to generate a pool design. First, the library of constructs is queried based on the default or optionally specified pool structure. Second, the matching lists of parts from each construct are extracted into two pools of forward-oriented and reverse-oriented (i.e. reverse complement) parts. The lists are merged by part order: first with first, second with second and so on. Third, both orientations are aligned and the two pools are merged into the final pool design, where barcodes and restriction enzyme recognition sites are added. In practice, the pool designs for a complex library can be generated quickly (less than 1 min runtime for the libraries described in this work).
Optimization of multiplexed pool assembly
The assembly reaction had to be optimized to create pools that reliably cover the desired design space and produce composite parts with high fidelity. Mixtures of the variable parts are added to multiplex an otherwise standard assembly reaction (56–59). The DNA fragments for pool construction were added to the reaction as either linear double-stranded DNA (PCR products or amplified synthesized oligos) or plasmid DNA. Variable fragments for a given position were mixed in equimolar proportions (Materials and Methods). The constructs in the pool are then barcoded with 20-mer random oligonucleotides before the pool is sequenced.
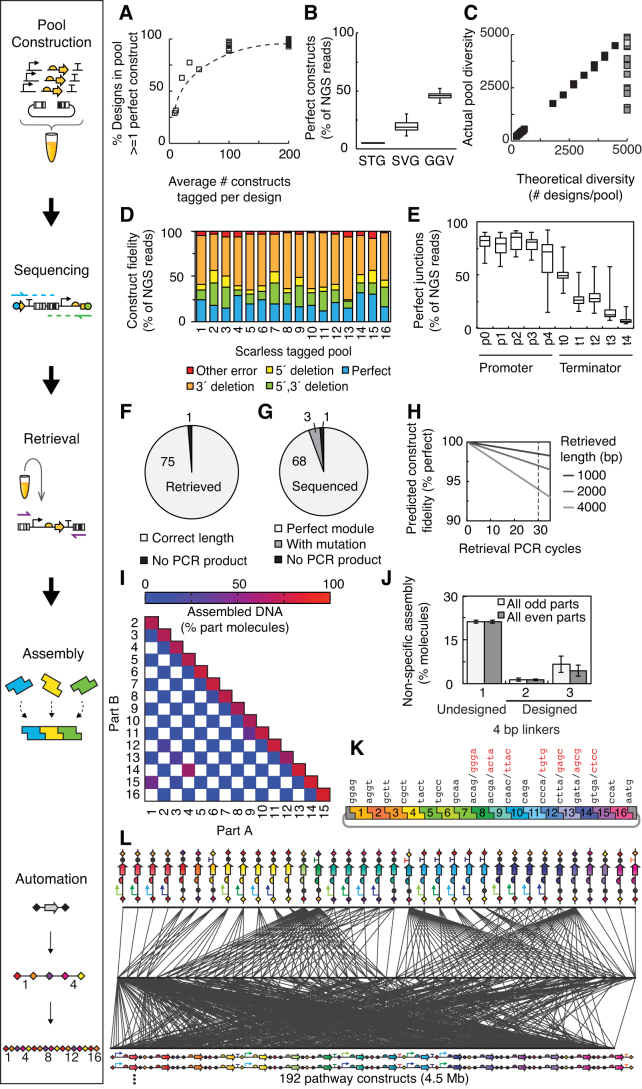
Multiplexing leads to large pools that contain up to 106 constructs of dual-barcoded composite parts. Each of these constructs could be designed to be unique. However, this would lead to many individual designs being absent due to biases, assembly errors and random sampling. Introducing redundancy increases the probability that any given desired composite part is present with a perfect sequence. Because NGS can sequence a few million composite parts (assuming the use of a MiSeq sequencer, see Discussion for the potential of other approaches), we can barcode multiple constructs for each design to introduce redundancy and then select one with a perfect sequence for retrieval. The ‘tagging ratio’ is defined as the average number of barcoded constructs there are in a pool for a given design; e.g. a pool with 5000 composite parts would have 106 barcoded constructs and a tagging ratio of 200 (106/5000 = 200). Pools were constructed that varied the tagging ratio to quantify the trade off between the diversity of the pool and the probability that a perfect construct could be present for all designs (Figure 2A). When the tagging ratio is >100, this yields complete pools with nearly all designs having perfect constructs. In practice, tagging ratios between 100–200 are ideal and this leads to the ability for a pool to reliably contain 5000–10 000 composite parts and have a perfect construct identified for each.
Figure 2.
Optimization of DNA assembly by retrieval. (A) Relationship between depth of sampling and fraction of composite part designs with a sequence-perfect construct. A cutoff of at least one perfect construct for each design of composite parts was used. (B) Accuracy of pools: (i) STG, (ii) SVG and (iii) GVL. Boxes show medians and interquartile ranges, and whiskers denote the maximum and minimum for pools constructed using each method. Accuracy of DNA constructs (% of next-generation sequencing reads) was determined for a paired-read perfectly matching a designed composite part in the pool. A total of 37 pools were constructed (Supplementary Data). (C) Diversity of perfect constructs in a pool compared to the theoretical designed diversity of composite parts. For actual pool diversity, a cutoff of at least one sequence-perfect construct was used. Pools for STG (white), SVG (grey), GVL (black). (D) Analysis of mutations in constructed composite parts for SVG pools (16 pools, 66 million total NGS reads). Imperfect NGS paired-reads were aligned to the closest matching composite part design sequence and deletions at the 5΄ and 3΄ scarless junctions were analyzed. (E) Prevalence of mutations after Scarless Stitching with variable promoter or terminator parts at the scarless junctions for 16 pools (SVG). (F) Accuracy of Dial-out PCR retrievals producing the correct length PCR product. Seventy-six different retrievals (1.2 kb average length) from GVL pools were analyzed by eye and gel electrophoresis (Supplementary Data). (G) Sequence accuracy of 72 different retrieved composite part constructs from SVG pools. PCR products were cloned, and one clone was sequenced for each. (H) Calculated theoretical sequence fidelity of retrieved construct after cycles of retrieval PCR for 5 × 10−7 DNA polymerase fidelity and 1–4 kb PCR retrievals. We used 30 cycles for retrievals (dashed line). (I) Orthogonality of 17 undesigned 4 bp linkers used for Type IIS assembly of 16 parts (undesigned linkers, sequences in red in panel K). Assembly reactions (BbsI) were tested for pairwise combinations of parts (580 bp parts). Assembled DNA was measured by quantitative capillary electrophoresis (71 reactions tested). (J) Non-specific assembly of 4 bp linkers was tested using non-contiguous parts (odd or even positions as shown in panel K). Three sets of linker sequences: (i) undesigned 17 linkers (16 parts, 580 bp each), (ii) designed 17 linkers (16 parts, 580 bp each) and (iii) designed 32 linkers (31 parts, 28 bp each). Reactions were performed in triplicate. (K) Linker sequences tested for 16-part assembly. Set of designed linkers shown in black. Linkers shown in red were used in the undesigned set and changed in the designed set. (L) Example of automated hierarchical assembly after retrieval. Assembly tree shown for a library of 192 designs containing 16 genes that was built using acoustic droplet liquid transfer. Each line represents a liquid transfer step of a DNA construct into an assembly reaction (top to bottom).
The sequencing step of pool construction is critical and requires the ability to provide sequences for multi-part constructs and cover the entire pool while avoiding biases. Previous work with Dial-out PCR was limited to sequencing pools of ∼200-mers (up to 0.5 kb constructs) (31–33). To extend the length, we adapted methods that had been previously applied for targeted sequencing (35,36) and the preparation of self-circularized ‘jumping’ libraries (60) to generate fragments containing targeted regions of the barcoded composite parts (Materials and Methods and Supplementary Data). Within each tagged pool, the barcoded composite parts were constructed on plasmids flanked by NotI restriction sites that are used to digest and then self-circularize the constructs, from which monomers are obtained via a gel extraction (Supplementary Data). Inverse PCR is then performed in emulsion droplets from priming sites internal to the composite part. The PCR products contain the parent construct's barcodes and are sequenced using Illumina paired-end reads (2 × 300 bp). We sequenced up to 7.2 million constructs in one flow cell. Inverse PCR on self-circularized constructs could provide complete sequences for up to 3 kb composite parts. Larger composite parts can be sequenced with these methods, but at most 1.0–1.5 kb (61) at either end is observable. In this paper, we applied this approach to pools of up to 3.8 kb constructs, but in principle larger internal genes are possible (the longest we have constructed is 9.1 kb, not shown).
There are three key choices in designing the reaction to build the pools. The first is the choice of the assembly method. In theory, any could be used but they differ in the biases that they introduce (e.g. due to secondary structure, repeats or high GC content), and some methods introduce scars. Here, we compared two approaches that are good at assembling variable sized DNA parts: Scarless Stitching (15) and Golden Gate assembly (42,43) (when used to build libraries, this leaves a 4 bp scar). The second choice is the method by which the 20 bp random barcodes are added to the composite parts (>1024 unique barcode pairs). This was either performed serially as a separate step using Gibson assembly or in parallel as a ligation done concurrently with the assembly reaction. Finally, the third choice is how the assembled constructs are amplified. This can either be done in vitro (PCR and gel excision) or in vivo (transformation and pooled growth on agar plates).
Three combinations of the above techniques were used to evaluate the above approaches: (i) Stitching-in vitro-Gibson (STG), (ii) Stitching-in vivo-Gibson (SVG) and (iii) Golden Gate-in vivo-ligation (GVL) (Supplementary Data). Scarless Stitching with in vivo amplification was previously used to assemble individual composite parts (15). Here, we sought to develop a multiplexed variation for pool assembly (SVG) and modifications for fully in vitro construction (STG) and improved fidelity of assembly (GVL). A test set of 37 pools was constructed in order to evaluate the three assembly approaches (Materials and Methods). The pools were constructed with an average of 6.8 parts assembled from 4.3 fragments with an average of 5.2 variants at each position (Supplementary Data). Thus, the design space for each pool on average encompasses 3230 potential composite parts. After sequencing, the methods were evaluated two ways. First, we measured the percent of NGS reads that correspond to a perfect sequence from the designed library (Figure 2B). This is an upper bound of errors that are introduced in the assembly or amplification steps as sequencing errors can also contribute. Second, we measured the percent of the potential composite parts in the pool for which perfect sequences can be found (Figure 2C). This is a measure of potential biases. All of the methods are able to cover this diversity well with greater than 90% of the entire space covered. Notably, avoiding in vitro amplification aids assembly fidelity by eliminating multiple PCR steps. Golden Gate assembly is the highest fidelity and could be scarless, but it typically comes at the cost of introducing scars when used to assemble libraries because part plasmids and linkers are reused in different genetic contexts to minimize the number of part plasmids required.
In total, we sequenced 15 million constructs and from this data set the errors that arise from pool construction were determined. The breakdown of errors is shown in Figure 2D for the SVG method. Deletions of 1–3 bp at the part junctions were prevalent in all pools constructed with Scarless Stitching. This effect could be impacted by hairpin formation as significantly more deletions were observed at the junctions with terminators (Figure 2E). For the GVL method, mutations most often occurred when the part was built by long oligo synthesis. In contrast, mutations in previously sequence-verified parts (e.g. genes obtained by DNA synthesis) or due to assembly were rare.
Retrieval and assembly of composite parts
An individual composite part in the pool is obtained by PCR amplification using a pair of primers designed to target the flanking barcodes. We developed computational tools to scan the sequencing data to: (i) check that the barcode pair is unique in the pool, and (ii) design optimal retrieval primers with a Tm of 63–66°C (Materials and Methods). The retrieval PCRs produced a strong band for 99% of the attempts with few off-target products (Figure 2F and Supplementary Data) and 96% were error-free as confirmed by sequencing (Figure 2H). Because mutations arise from PCR, the mutagenesis rate is based on the construct length, polymerase fidelity and number of cycles. Figure 2H shows the predicted probability that the retrieved construct is perfect as a function of these parameters. This calculation guides the number of final constructs that need to be built and screened to identify one that is perfect. For example, the error rate is predicted to be 3% when retrieving 2 kb composite parts using Q5 polymerase (∼5 × 10−7 fidelity) and 30 cycles. When the final assembly requires 20 retrievals, the probability of a final perfect sequence is 54%.
The retrieved composite parts are purified using a bead-based PCR clean-up conducive to automation and are then assembled into larger constructs using Type IIS assembly (Materials and Methods). For constructs involving fewer retrieved fragments, the latter can be done as a single assembly reaction. If the final construct is large, then assembly has to occur via hierarchal steps that build up the construct through intermediate fragments. For example, when we assemble four retrieved fragments that were first individually cloned, they can be combined in a single reaction and 100% of the final constructs (24/24, 5–8 kb) are the correct length (Supplementary Data). We found that up to seven fragments could be assembled in one reaction and then the accuracy decreases with more fragments. Mutations included incomplete assembled constructs, such as short fragments and 1 bp insertions/deletions particularly in poly-T runs of 6–10 bases.
We also improved the design of the 4 bp linker sequences as these form the sticky ends to anneal fragments together. Initially, 17 linker sequences were chosen (11 random and 6 from the literature (43)) and evaluated by assembling 16 fragments. Eight fragments containing a subset of the linkers (non-contiguous fragments in the final construct) were tested for assembly and we found that 17% were able to assemble (Figure 2J). This led to our evaluation of all-possible cross-reactions between linkers within these reactions (Figure 2I). Based on these results, we extracted rules (4 bp length, 25–75% GC and 2 bp max sequence identity to forward or reverse complement) and developed a script to generate orthogonal linkers (Materials and Methods). Using this code, we re-designed the 17 linker sequences that reduced the cross reactions to 1.4% (Figure 2J, ‘Designed’). For 32 linkers, this reduces the probability of cross-reactions to 5.5% (Figure 2J). With the optimized reaction conditions and redesigned linkers, we tested one-pot assembly of 16 fragments (17 linkers, 580 bp parts, Figure 2K) and 31 fragments (32 linkers, 28 bp parts). The former assembled properly 77% of the time (17/22) but no full-length fragments were found for the latter. In practice, to increase the probability of obtaining full-length constructs for complex assemblies, we mix a maximum of 5–7 fragments (depending on length) in a single reaction and then proceed with hierarchal assembly for larger constructs.
We optimized the conditions to increase the number of fragments that can be combined in a single reaction. Assembly reactions were tested using 4, 8, 12 or 16 fragments (each 580 bp PCR products) and the products were analyzed by capillary electrophoresis (Methods). To perform hierarchical assembly of a large library (Figure 2l), we used automated liquid handling that transfers 2.5 nl droplets at a rate of up to 500 Hz using acoustic droplet ejection (Labcyte Echo 550) (Methods). The small transfer volumes allowed us to reduce the volume of the assembly reaction (4-fold to 2.5 μl total) and amount of DNA material required for each reaction. A plate of 96 multi-part assembly reactions was typically transferred in 15–30 min.
Algorithmic exploration of expression space: nitrogen fixation
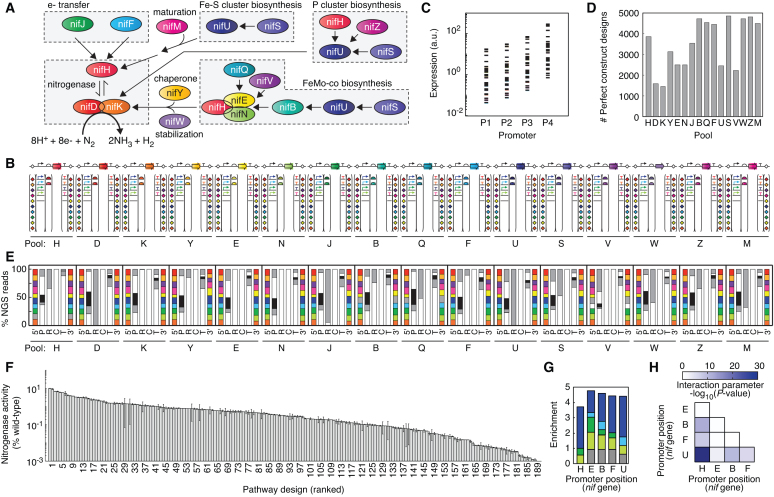
Biological nitrogen fixation is a process carried out by many microorganisms to convert atmospheric N2 into ammonia (62). The pathway from Klebsiella contains 16 genes that are required for this function (Figure 3A). The genes include the nitrogenase enzyme subunits, electron transfer pathway, chaperones and pathways for the biosynthesis and loading of complex metal cofactors (63,64) (Figure 3A). Nitrogenase activity is highly sensitive to changes in the expression levels of these genes. Previously, we re-engineered the Klebsiella pathway to codon optimize the genes, use only synthetic genetic parts and be controlled by T7 RNA polymerase (15,65). Here, we applied the approach to design pools of nif genes that encompass extraordinary potential pathway complexity. This is applied to retrieve and assemble libraries designed to understand how changing the transcription level of operons impacts activity when the pathway is transferred to E. coli.
Figure 3.
Algorithm-guided exploration of genetic design space for a 16-gene nitrogen fixation pathway. (A) An overview of the K. oxytoca pathway (65). (B) Pool designs for composite parts with one nif gene in each pool. Parts were permuted as shown with variants of promoters (5), RBSs (2), terminators (5) and assembly linker (10 × 10) parts in each nif pool. Across the 16 pools, there are 96 part positions in total. Pool names are abbreviated by last letter of gene name. (C) Expression range of promoter-RBS combinations. Expected expression of promoter and RBS part combinations for the design space in the pools. Expression was estimated as the product of promoter strength (REU units) (65) and RBS strength (67,68). (D) Number of sequence-perfect composite part designs in nif pools. A cutoff of at least 1 sequence-perfect construct for each design was applied. (E) Representation of parts in the sequence-perfect constructs (% NGS reads that perfectly match a composite part design) at each position in the 16 pools. Approximately 4 million NGS reads per pool were sequenced and analyzed. Part type abbreviations: 5΄ linker (5΄), promoter (P), RBS (R), CDS (C), terminator (T), 3΄ linker (3΄). Linker positions are colored according to the order listed in Supplementary Data and parts variants at each position (bottom to top) follow the order listed. (F) Assayed activity for a 192-member algorithm-designed library built from the pools. Acetylene substrate was supplied, and ethylene produced was measured by GC analysis. Nitrogenase activity (% wild-type) is calculated using assayed K. oxytoca ethylene production. Error bars show deviation of duplicate assays for each pathway design. (G) Part enrichment for the top 20 pathway designs as compared to the whole 192-member library. Enrichment for each promoter position was calculated based on its frequency (freqTop20/freqLibrary). The nif genes were arranged in the order shown in panel B. The promoter variants include PT7.4 (purple), PT7.3 (blue), PT7.2 (green) and PT7.1 (yellow) and P0 (grey) with sequences listed in Supplementary Data. (H) Significance of pairwise interaction parameters in the fitted response surface model for nitrogenase activity (Methods).
The pools were designed so that the full pathways could contain many different combinations of expression parts for each gene, any operon structure (including monocistronic designs), any orientation, any ordering of the genes and any subset of genes (e.g. to test for essentiality) (Figure 3B). Each pool centers around the control of individual genes. The RBS and promoter combinations were chosen to span a ∼100-fold range in expression level based on individual characterization of promoters (66) and RBS strength predictions (15,67–68) (Figure 3C). We assembled and sequenced 55 848 perfect constructs in the 16 pools (Figure 3D) to generate a highly complex library of pathways that could be accessed (1043 pathway designs). Parts from a set of T7 promoters (66) and strong terminators (69) were included. For each promoter and terminator position, a non-functional spacer part was included to allow for operon structures. The linkers were designed to allow any combination of genes in any order and orientation by including a set of 10 linkers (5 different 4 bp sequences and their reverse complement). The diversity of the perfect constructs in the pools was determined by NGS, and 70% of all possible designs have at least one perfect construct (Figure 3E).
Once constructed, the same pools could be re-used to test any number of hypotheses. Here, we retrieved a subset of pathways to identify the optimal balancing of promoter strengths for a previously identified gene order (65) and similar operon occupancy. In order to quantify the pairwise epistasis, we used a coordinate-exchange algorithm (70) to design constructs. The library encompassed 192 pathways that cover 7.7% of all promoter combinations. The composite parts required to build the pathway designs were retrieved from the pools and assembled hierarchically to generate each of the 23 kb pathway constructs that totaled 4.5 Mb (Figure 2l). The nitrogenase activity of each construct was quantified using an acetylene reduction assay (Figure 3F). The range of activities represented in the library span four orders of magnitude. When the top 20 are compared to the library, the strongest promoter is highly enriched at each position, although many promoters are also observed (Figure 3G). Additional response surface modeling identified significant epistatic effects, where many interaction parameters in the model were found to be significant (5/10 parameters P < 0.001, 7/10 P < 0.05) (Figure 3H and Supplementary Data).
Capturing a complete combinatorial space: genetic circuit design
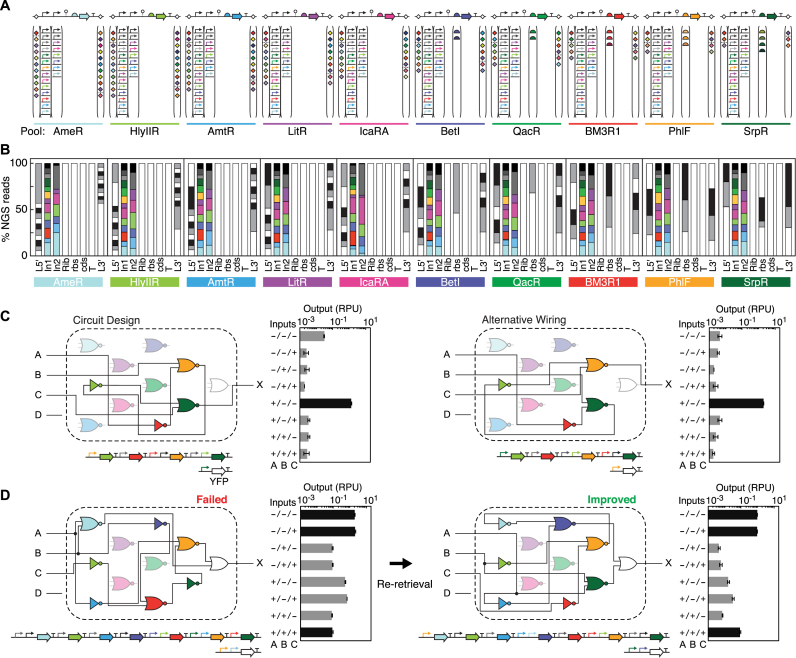
Genetic circuits encompass gene regulatory networks that implement a computational function. They are often composed of transcription factors that regulate each other to produce logic or feedback. Connecting the same transcription factors in different ways can create different circuit behaviors (18). Previously, we characterized a set of repressors (71) that can be used to build NOT gates (a single input promoter) or NOR gates (two input promoters in tandem). The gates can be connected in different ways and layered in order to build larger circuits. Here, we use gates based on a subset of 10 repressors. These can be connected in different permutations to make ∼1030 circuits, including combinational logic (19), analog circuits (72) (all combinations of feedback loops) and dynamic circuits (73). Each of these circuits corresponds with a different pattern of promoters in front of the repressor genes.
We designed pools centered around each of the 10 repressors so that any of the possible circuits could be retrieved and assembled. This involves varying the identities of one promoter (NOT pools) or two promoters (NOR pools) in front of each repressor (Figure 4A and Supplementary Data). The NOT and NOR pools were designed such that the Golden Gate assembly parts (Figure 2K) allowed at each position fix the ordering of the repressors but allow for changes in the orientation (Figure 4A). The 20 pools (10 NOT and 10 NOR) contain 35 746 sequence-perfect gates, which is 98.5% of all possible permutations of promoters and repressors (Supplementary Data). In total there are 3 884 109 sequence-perfect constructs across the 20 pools. On average there are 107 sequence-perfect constructs for each gate design (Supplementary Data). The promoter variability at each position is fairly even, as designed (Figure 4B). The failed 1.5% most often contained the longest promoter (PBAD). Retrievals from these pools were 99% successful (Figure 2F).
Figure 4.
Design space encompassing all possible circuits. (A) Pool designs for NOR gates with one repressor (71) in each pool. A pool of NOT gates (one input promoter) and NOR gates (two tandem input promoters) was constructed for each repressor (20 pools total). Linkers for the forward and reverse orientations were mixed separately in two multiplex PCR reactions; the dashed line represents this split. Colors represent each of the 10 repressors and their corresponding promoters. Assembly parts are colored in the order shown in Supplementary Data. (B) Representation of parts in the sequence-perfect constructs (% NGS reads that perfectly match a composite part design) at each position in the NOR pools. Approximately 5 million NGS reads per pool were sequenced and analyzed. Part type abbreviations: 5΄ linker (L5’), input promoter 1 (In1), input promoter 2 (In2), hammerhead ribozyme (Rib), RBS (rbs), CDS (cds), terminator (T) and 3΄ linker (L3’). Part sequences are listed in Supplementary Data. Input promoter positions are colored according to its corresponding repressor and in the order listed in Supplementary Data (bottom to top). (C) Using the Cello software (19), a circuit was designed for a 3-input logic function (output 00001000) and an alternative gate assignment was subsequently tested. The circuit wiring diagrams are drawn showing all possible gates in a fixed position (10 repressor gates and output OR gate) with lines connecting the gates to show the wiring of input(s) and output for each gate. Gate symbols are colored according to the repressor and lightly shaded if not present in the circuit. For each circuit design, the constructs were built by retrieving the gates from the pools with all-forward orientation and one-pot Type IIS assembly into a backbone with the corresponding 3-input sensor block. Circuits were tested under all 8 input states (presence or absence of 1 mM IPTG, 2 ng/ml aTc and 5 mM L-arabinose) and YFP expression was analyzed by flow cytometry (n = 20 000). Bars are colored to show if predicted output expression is high (black) or low (grey). The average and standard deviation are calculated from triplicate experiments performed on different days. (D) For a 3-input logic function (output 11000001) requiring seven NOT and NOR gates, an initial design failed to generate the desired function. An alternative wiring of gates was tested by re-retrieval from the gate pools.
We then demonstrated the assembly of circuits from gates retrieved from the pools. To simplify the process, we optimized the system for one-pot Type IIS assembly (BbsI) so the circuit could be built in a single reaction after the retrieval of gates (Materials and Methods). We co-transformed the constructed circuit plasmid with the corresponding actuator plasmid containing the output fluorescent reporter (YFP) and measured the circuit output for all input combinations in relative promoter units (19) (Supplementary Data). The first circuit we tested encodes 3-input combinational logic. There are many possible ways that this circuit function could be built from the repressor pools. We assembled two alternative designs, where the gates correspond to different repressors, and both were found to function correctly (Figure 4C).
We next used the same pools to build a much larger circuit, consisting of seven repressors. The first circuit retrieved was found to be non-functional (Figure 4D), so we then retrieved and assembled an alternative design from the pools that performs the same function. An improved circuit was found from this second attempt. This demonstrates how the same pools can be reused to attempt alternative designs to iteratively improve function.
DISCUSSION
In this work, we constructed a total of 109 Mb of unique DNA encoding 91 594 sequence-perfect composite parts and 5 966 773 sequence-perfect constructs (6.57 GB). Physically, these pools of composite parts are stored in a frozen liquid aliquot in a tube (e.g. 37 pools in separate tubes), from which an individual composite part can be retrieved by PCR. This retrieval is possible because each composite part has its own barcode that is identified when the pool is sequenced. By optimizing the protocol for pool construction, we were able to reliably create up to 5000 composite parts per pool that encompass 98.3% ± 1.7% of designs (≥1 sequence-perfect construct for each design). The assembly of the retrieved composite parts into the final 10–25 kb construct was done with an accuracy of 45.8% ± 3.4% (sequence-perfect constructs). The pre-sequencing step of the pools reduced many of the common types of errors observed from assembly reactions. There is an initial investment in time and cost to build and sequence the pools (typically 2–3 weeks). But once they are built, retrieval and assembly is rapid (2 days) and the error rate is lower than would occur from de novo synthesis and assembly. This allows reuse of the pools to rapidly build new designs.
The genetic design space that can be explored by re-retrieval is defined during the initial design of the pools. With up to 5000 composite parts, each pool could be designed to include a broad and weakly constrained design space or preliminary experiments could be performed to determine design constraints that are relevant for the desired function. For example, parts that are held constant across all composite parts (e.g. genes) are typically pre-validated. From an efficiency standpoint, this approach would be most advantageous in terms of time and cost when the pools are reused for many cycles of retrieval and library construction. One way to make the pools more reusable would be to allow for a greater number of composite parts so that they could contain a more expansive design space for iterative investigation.
The primary limitation in the size of the pools is the read depth achievable by NGS. Here, we used a MiSeq to sequence up to 1 million barcoded composite parts in a pool and this allows for confirmation of up to 4876 sequence-perfect constructs. We multiplexed the sequencing step and sequenced up to 20 pools in a single run containing 7.2 million composite part constructs (3.88 million sequence-perfect constructs and 35 746 sequence-perfect designs). With new NGS platforms (e.g. HiSeq 2 × 250 bp) (34,74), there is the potential to sequence up to 180 million composite parts at a time, which is expected to increase the unique retrievable parts to 1.7 million. For composite parts containing variable parts exceeding these read lengths (>500–600 bp), the sequencing methods could be modified to accommodate these pools. For example, the sequencing preparation could be modified to incorporate additional fragmentation, such as multiplexed variations of the inverse PCR. Further improvements in sequencing technologies would allow even greater pool depth and longer composite part constructs.
When considering very large and diverse libraries, the retrieval step can become cost limiting because it requires a pair of primers per retrieval as well as a PCR reaction. There are various ways to reduce this cost, including the application of reusable retrieval primers, combinatorial barcode design, fewer PCR reactions and smaller reaction volumes. Multiplex PCR (75,76) could potentially be used to simultaneously retrieve multiple composite parts at a time. Automation facilitates setting up these reactions may also allow for further miniaturization to reduce the reagents required. In recent work, a set of 4637 reusable barcodes were designed and implemented for Dial-out PCR (33). These designed barcodes could be implemented in place of the degenerate barcodes used in this work in order to have a set of in-house retrieval primers that could be applied across all pools. There are also software platforms that can be applied to reduce the number of oligos and cloning steps needed to build a library (21,77–78).
Intriguingly, the pools potentially offer an efficient way to share materials between labs and provide access to extensive design spaces for typical laboratories without automated liquid-handling capabilities. As pools become deeper, it may be possible to encompass entire ‘registries’ of genetic parts (8,13,69,79–83) in a single aliquot, not just as individual parts but also as useful combinations of retrievable composite parts. A single tube could be sent between labs or from a central repository without the need for the level of maintenance and quality control over individual parts, which does not scale with the rate of data generation in the field. A key challenge in implementing this is being able to amplify a pool efficiently without introducing biases as a means of propagating it and creating the aliquots to be sent. In our hands, in vitro pool amplification, while also technically feasible, has not been successful. However, we have propagated the tagged pools as clonal populations for our own work and retrieved from them successfully without re-sequencing the pool (Materials and Methods).
This work demonstrates the ability to create pools that encompass enormous design spaces and to efficiently retrieve large libraries from these pools. There are many ways that design algorithms can be connected to automate this process. For example, the circuit automation software Cello currently either generates a DNA sequence encoding a circuit or a constraint file that can be used to create diverse designs. The output could easily be the set of oligos required to retrieve a desired circuit from the pools. Similar approaches can be taken for metabolic engineering, where numerical analysis, evolutionary algorithms (84), Bayesian optimization (85), design of experiments (70,86–87) or other approaches could iteratively generate the next round of oligos needed to retrieve the pathways to be tested. Systematically exploring greater regions of design space will ultimately allow genetic designers to access and engineer more complex genetic systems.
Supplementary Material
ACKNOWLEDGEMENTS
L.B.A.W., T.S.M., R.N. and C.A.V. conceived and designed experiments. L.B.A.W. performed experiments. L.B.A.W., T.S.M., T.E.G. performed computational analyses on sequencing data. T.E.G. and D.B.G. wrote scripts to automate analysis of the pools and design linkers. N.R. wrote software to automate the design of pools based on discussions with D.B.G., D.D. and L.B.A.W.. D.D, R.N. and C.A.V. supervised this work. L.B.A.W., T.S.M. and C.A.V. analyzed the data. L.B.A.W. and C.A.V. wrote the manuscript. The authors thank X. Zhang for assistance with next-generation sequencing. The code for the design tools described in this work is available on GitHub (https://github.com/VoigtLab/MIT-BroadFoundry/tree/master/dialout-designs/ and https://github.com/CIDARLAB/poolDesigner/).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Defense Advanced Research Projects Agency's Living Foundries program awards [HR0011-12-C-0067, HR0011-13-1-0001 and HR0011-15-C-0084]; Institute for Collaborative Biotechnologies [W911NF-09-0001 to C.A.V.]; U.S. Army Research Office and by the Office of Naval Research Multidisciplinary University Research Initiative [N00014-13-1-0074 to C.A.V.]; U.S. National Science Foundation's Expeditions in Computing Program [award #1522074 to D.D.]. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. Funding for open access charge: U.S. Defense Advanced Research Projects Agency (DARPA) Living Foundries award [HR0011-15-C-0084].
Conflict of interest statement. L.B.A.W., T.S.M., R.N. and C.A.V. have filed a patent application (U.S. serial no. PCT/US2015/032760) on this work.
REFERENCES
- 1. Pfleger B.F., Pitera D.J., Smolke C.D., Keasling J.D.. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 2006; 24:1027–1032. [DOI] [PubMed] [Google Scholar]
- 2. Ajikumar P.K., Xiao W.H., Tyo K.E.J., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G.. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010; 330:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warner J.R., Reeder P.J., Karimpour-Fard A., Woodruff L.B.A., Gill R.T.. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 2010; 28:856–862. [DOI] [PubMed] [Google Scholar]
- 4. Lee M.E., Aswani A., Han A.S., Tomlin C.J., Dueber J.E.. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013; 41:10668–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farasat I., Kushwaha M., Collens J., Easterbrook M., Guido M., Salis H.M.. Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol. Syst. Biol. 2014; 10:731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cobb R.E., Ning J.C., Zhao H.M.. DNA assembly techniques for next-generation combinatorial biosynthesis of natural products. J. Ind. Microbiol. Biotechnol. 2014; 41:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou H., Vonk B., Roubos J.A., Bovenberg R.A.L., Voigt C.A.. Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor. Nucleic Acids Res. 2015; 43:10560–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E.. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015; 4:975–986. [DOI] [PubMed] [Google Scholar]
- 9. Agmon N., Mitchell L.A., Cai Y.Z., Ikushima S., Chuang J., Zheng A., Choi W.J., Martin J.A., Caravelli K., Stracquadanio G. et al. Yeast Golden Gate (yGG) for the Efficient Assembly of S-cerevisiae Transcription Units. ACS Synth. Biol. 2015; 4:853–859. [DOI] [PubMed] [Google Scholar]
- 10. Nielsen M.T., Madsen K.M., Seppala S., Christensen U., Riisberg L., Harrison S.J., Moller B.L., Norholm M.H.H.. Assembly of highly standardized gene fragments for high-level production of porphyrins in E. coli. ACS Synth. Biol. 2015; 4:274–282. [DOI] [PubMed] [Google Scholar]
- 11. Xu P., Rizzoni E.A., Sul S.-Y., Stephanopoulos G.. Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth. Biol. 2016; doi:10.1021/acssynbio.1026b00187. [DOI] [PubMed] [Google Scholar]
- 12. Jeschek M., Gerngross D., Panke S.. Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat. Commun. 2016; 7:11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 14. Ham T.S., Dmytriv Z., Plahar H., Chen J., Hillson N.J., Keasling J.D.. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012; 40:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smanski M.J., Bhatia S., Zhao D., Park Y., Woodruff L.B.A., Giannoukos G., Ciulla D., Busby M., Calderon J., Nicol R. et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014; 32:1241–1249. [DOI] [PubMed] [Google Scholar]
- 16. Sanchez C., Zhu L., Brana A.F., Salas A.P., Rohr J., Mendez C., Salas J.A.. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narcross L., Bourgeois L., Fossati E., Burton E., Martin V.J.J.. Mining enzyme diversity of transcriptome libraries through DNA synthesis for benzylisoquinoline alkaloid pathway optimization in yeast. ACS Synth. Biol. 2016; 5:1505–1518. [DOI] [PubMed] [Google Scholar]
- 18. Guet C.C., Elowitz M.B., Hsing W.H., Leibler S.. Combinatorial synthesis of genetic networks. Science. 2002; 296:1466–1470. [DOI] [PubMed] [Google Scholar]
- 19. Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A.. Genetic circuit design automation. Science. 2016; 352:aac7341. [DOI] [PubMed] [Google Scholar]
- 20. Ellis T., Adie T., Baldwin G.S.. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 2011; 3:109–118. [DOI] [PubMed] [Google Scholar]
- 21. Beal J., Weiss R., Densmore D., Adler A., Appleton E., Babb J., Bhatia S., Davidsohn N., Haddock T., Loyall J. et al. An end-to-end workflow for engineering of biological networks from high-level specifications. ACS Synth. Biol. 2012; 1:317–331. [DOI] [PubMed] [Google Scholar]
- 22. Dharmadi Y., Patel K., Shapland E., Hollis D., Slaby T., Klinkner N., Dean J., Chandran S.S.. High-throughput, cost-effective verification of structural DNA assembly. Nucleic Acids Res. 2014; 42:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chao R., Yuan Y.B., Zhao H.M.. Recent advances in DNA assembly technologies. FEMS Yeast Res. 2015; 15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian J., Gong H., Sheng N., Zhou X., Gulari E., Gao X., Church G.. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004; 432:1050–1054. [DOI] [PubMed] [Google Scholar]
- 25. Kosuri S., Eroshenko N., Leproust E.M., Super M., Way J., Li J.B., Church G.M.. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010; 28:1295–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melnikov A., Rogov P., Wang L., Gnirke A., Mikkelsen T.S.. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014; 42:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kitzman J.O., Starita L.M., Lo R.S., Fields S., Shendure J.. Massively parallel single-amino-acid mutagenesis. Nat. Methods. 2015; 12:203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Church G.M., Kieffer-Higgins S.. Multiplex DNA sequencing. Science. 1988; 240:185–188. [DOI] [PubMed] [Google Scholar]
- 29. Smith A.M., Heisler L.E., St Onge R.P., Farias-Hesson E., Wallace I.M., Bodeau J., Harris A.N., Perry K.M., Giaever G., Pourmand N. et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010; 38:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quail M.A., Smith M., Jackson D., Leonard S., Skelly T., Swerdlow H.P., Gu Y., Ellis P.. SASI-Seq: sample assurance Spike-Ins, and highly differentiating 384 barcoding for Illumina sequencing. BMC Genomics. 2014; 15:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwartz J.J., Lee C., Shendure J.. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nat. Methods. 2012; 9:913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H., Han H., Ahn J., Lee J., Cho N., Jang H., Kim H., Kwon S., Bang D.. 'Shotgun DNA synthesis' for the high-throughput construction of large DNA molecules. Nucleic Acids Res. 2012; 40:e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein J.C., Lajoie M.J., Schwartz J.J., Strauch E.M., Nelson J., Baker D., Shendure J.. Multiplex pairwise assembly of array-derived DNA oligonucleotides. Nucleic Acids Res. 2016; 44:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hiatt J.B., Patwardhan R.P., Turner E.H., Lee C., Shendure J.. Parallel, tag-directed assembly of locally derived short sequence reads. Nat. Methods. 2010; 7:119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lundin S., Gruselius J., Nystedt B., Lexow P., Kaller M., Lundeberg J.. Hierarchical molecular tagging to resolve long continuous sequences by massively parallel sequencing. Sci. Rep. 2013; 3:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pachuk C.J., Samuel M., Zurawski J.A., Snyder L., Phillips P., Satishchandran C.. Chain reaction cloning: a one-step method for directional ligation of multiple DNA fragments. Gene. 2000; 243:19–25. [DOI] [PubMed] [Google Scholar]
- 38. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 39. Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011; 498:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Kok S., Stanton L.H., Slaby T., Durot M., Holmes V.F., Patel K.G., Platt D., Shapland E.B., Serber Z., Dean J. et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth. Biol. 2014; 3:97–106. [DOI] [PubMed] [Google Scholar]
- 41. Trubitsyna M., Michlewski G., Cai Y., Elfick A., French C.E.. PaperClip: rapid multi-part DNA assembly from existing libraries. Nucleic Acids Res. 2014; 42:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S.. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011; 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hillson N.J., Rosengarten R.D., Keasling J.D.. j5 DNA assembly design automation software. ACS Synth. Biol. 2012; 1:14–21. [DOI] [PubMed] [Google Scholar]
- 45. Lux M.W., Bramlett B.W., Ball D.A., Peccoud J.. Genetic design automation: engineering fantasy or scientific renewal?. Trends Biotechnol. 2012; 30:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blakes J., Raz O., Feige U., Bacardit J., Widera P., Ben-Yehezkel T., Shapiro E., Krasnogor N.. Heuristic for maximizing DNA reuse in synthetic DNA library assembly. ACS Synth. Biol. 2014; 3:529–542. [DOI] [PubMed] [Google Scholar]
- 47. Wilson E.H., Sagawa S., Weis J.W., Schubert M.G., Bissell M., Hawthorne B., Reeves C.D., Dean J., Platt D.. Genotype specification language. ACS Synth. Biol. 2016; 5:471–478. [DOI] [PubMed] [Google Scholar]
- 48. Roehner N., Young E.M., Voigt C.A., Gordon D.B., Densmore D.. Double Dutch: A Tool for Designing Combinatorial Libraries of Biological Systems. ACS Synth. Biol. 2016; 5:507–517. [DOI] [PubMed] [Google Scholar]
- 49. Zhang Z., Nguyen T., Roehner N., Misirli G., Pocock M., Oberortner E., Samineni M., Zundel Z., Beal J., Clancy K. et al. libSBOLj 2.0: a java library to support SBOL 2.0. IEEE Life Sci. Lett. 2015; 1:34–37. [Google Scholar]
- 50. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rozen S., Skaletsky H.. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000; 132:365–386. [DOI] [PubMed] [Google Scholar]
- 52. Koressaar T., Remm M.. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007; 23:1289–1291. [DOI] [PubMed] [Google Scholar]
- 53. Stewart W.D., Fitzgerald G.P., Burris R.H.. In situ studies on nitrogen fixation with the acetylene reduction technique. Proc. Natl. Acad. Sci. U.S.A. 1967; 58:2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Galdzicki M., Clancy K.P., Oberortner E., Pocock M., Quinn J.Y., Rodriguez C.A., Roehner N., Wilson M.L., Adam L., Anderson J.C. et al. The synthetic biology open language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat. Biotechnol. 2014; 32:545–550. [DOI] [PubMed] [Google Scholar]
- 55. Roehner N., Beal J., Clancy K., Bartley B., Misirli G., Grunberg R., Oberortner E., Pocock M., Bissell M., Madsen C. et al. Sharing structure and function in biological design with SBOL 2.0. ACS Synth. Biol. 2016; 5:498–506. [DOI] [PubMed] [Google Scholar]
- 56. Engler C., Gruetzner R., Kandzia R., Marillonnet S.. Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS One. 2009; 4:e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Farrow M.F., Arnold F.H.. Combinatorial recombination of gene fragments to construct a library of chimeras. Curr. Protoc. Protein Sci. 2010; doi:10.1002/0471140864.ps2602s44. [DOI] [PubMed] [Google Scholar]
- 58. Sleight S.C., Sauro H.M.. Randomized BioBrick assembly: a novel DNA assembly method for randomizing and optimizing genetic circuits and metabolic pathways. ACS Synth. Biol. 2013; 2:506–518. [DOI] [PubMed] [Google Scholar]
- 59. Iverson S.V., Haddock T.L., Beal J., Densmore D.M.. CIDAR MoClo: improved MoClo assembly standard and new E-coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth. Biol. 2016; 5:99–103. [DOI] [PubMed] [Google Scholar]
- 60. Kandpal R.P., Kandpal G., Weissman S.M.. Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Head S.R., Komori H.K., LaMere S.A., Whisenant T., Van Nieuwerburgh F., Salomon D.R., Ordoukhanian P.. Library construction for next-generation sequencing: overviews and challenges. BioTechniques. 2014; 56:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Geddes B.A., Ryu M.H., Mus F., Costas A.G., Peters J.W., Voigt C.A., Poole P.. Use of plant colonizing bacteria as chassis for transfer of N-2-fixation to cereals. Curr. Opin. Biotechnol. 2015; 32:216–222. [DOI] [PubMed] [Google Scholar]
- 63. Rubio L.M., Ludden P.W.. Maturation of nitrogenase: a biochemical puzzle. J. Bacteriol. 2005; 187:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rubio L.M., Hernandez J.A., Soboh B., Zhao D., Igarashi R.Y., Curatti L., Ludden P.W.. The role of Nif proteins in nitrogenase maturation. Biol. Nitrogen Fixation. 2008; 42:325–328. [Google Scholar]
- 65. Temme K., Zhao D.H., Voigt C.A.. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:7085–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Temme K., Hill R., Segall-Shapiro T.H., Moser F., Voigt C.A.. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012; 40:8773–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Borujeni A.E., Channarasappa A.S., Salis H.M.. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014; 42:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen Y.J., Liu P., Nielsen A.A.K., Brophy J.A.N., Clancy K., Peterson T., Voigt C.A.. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013; 10:659–664. [DOI] [PubMed] [Google Scholar]
- 70. Meyer R.K., Nachtsheim C.J.. The Coordinate-Exchange Algorithm for Constructing Exact Optimal Experimental-Designs. Technometrics. 1995; 37:60–69. [Google Scholar]
- 71. Stanton B.C., Nielsen A.A., Tamsir A., Clancy K., Peterson T., Voigt C.A.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014; 10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Daniel R., Rubens J.R., Sarpeshkar R., Lu T.K.. Synthetic analog computation in living cells. Nature. 2013; 497:619–623. [DOI] [PubMed] [Google Scholar]
- 73. Elowitz M.B., Leibler S.. A synthetic oscillatory network of transcriptional regulators. Nature. 2000; 403:335–338. [DOI] [PubMed] [Google Scholar]
- 74. Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H.. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009; 4:265–270. [DOI] [PubMed] [Google Scholar]
- 75. Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J.. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010; 7:111–118. [DOI] [PubMed] [Google Scholar]
- 76. Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 2014; 20:1479–1484. [DOI] [PubMed] [Google Scholar]
- 77. Densmore D., Hsiau T.H.C., Kittleson J.T., DeLoache W., Batten C., Anderson J.C.. Algorithms for automated DNA assembly. Nucleic Acids Res. 2010; 38:2607–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Appleton E., Tao J.H., Haddock T., Densmore D.. Interactive assembly algorithms for molecular cloning. Nat. Methods. 2014; 11:657–662. [DOI] [PubMed] [Google Scholar]
- 79. Canton B., Labno A., Endy D.. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008; 26:787–793. [DOI] [PubMed] [Google Scholar]
- 80. Shetty R.P., Endy D., Knight T.F. Jr. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008; 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ham T.S., Dmytriv Z., Plahar H., Chen J., Hillson N.J., Keasling J.D.. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012; 40:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M.. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Curran K.A., Karim A.S., Gupta A., Alper H.S.. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013; 19:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Srinivas M., Patnaik L.M.. Adaptive probabilities of crossover and mutation in genetic algorithms. IEEE Trans. Syst. Man Cybern. 1994; 24:656–667. [Google Scholar]
- 85. Jones D.R. A taxonomy of global optimization methods based on response surfaces. J. Global Optimization. 2001; 21:345–383. [Google Scholar]
- 86. Kushner H.J. A new method of locating the maximum point of an arbitrary multipeak curve in the presence of noise. J. Basic Eng. 1964; 86:97–106. [Google Scholar]
- 87. Močkus J. Marchuk GI. Optimization Techniques IFIP Technical Conference Novosibirsk, July 1–7, 1974. 1975; Berlin, Heidelberg: Springer Berlin Heidelberg; 400–404. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.