Abstract
Spirochetes of the genus Borrelia possess unusual genomes harboring multiple linear and circular replicons. The linear replicons are terminated by covalently closed hairpin (hp) telomeres. Hairpin telomeres are formed from replicated intermediates by the telomere resolvase, ResT, in a phosphoryl transfer reaction with mechanistic similarities to those promoted by type 1B topoisomerases and tyrosine recombinases. There is growing evidence that ResT is multifunctional. Upon ResT depletion DNA replication unexpectedly ceases. Additionally, ResT possesses RecO-like biochemical activities being able to promote single-strand annealing on both free ssDNA and ssDNA complexed with cognate single-stranded DNA binding protein. We report here that ResT possesses DNA-dependent ATPase activity that promotes DNA unwinding with a 3΄-5΄ polarity. ResT can unwind a variety of substrates including synthetic replication forks and D-loops. We demonstrate that ResT's twin activities of DNA unwinding and annealing can drive regression of a model replication fork. These properties are similar to those of the RecQ helicase of the RecF pathway involved in DNA gap repair. We propose that ResT's combination of activities implicates it in replication and recombination processes operating on the linear chromosome and plasmids of Borrelia burgdorferi.
INTRODUCTION
Borrelia species possess unusual genomes composed of multiple circular and linear replicons. The prototype Borrelia burgdorferi B31 genome has a linear chromosome and 23 plasmids that are a mix of circular and linear replicons (1–3). The linear replicons are terminated by structures called hairpin (hp) telomeres (4–6). The hp telomeres help overcome the dual end-replication and end-protection problems faced by linear DNAs. The linear replicons are copied by DNA replication that initiates at an internal oriC. DNA replication proceeds bidirectionally toward the hp telomeres (7,8). Replisomes that successfully round the hp telomeres would produce inverted repeat chromosome or plasmid dimers joined by replicated telomere (rTel) junctions. The resulting intermediate has been replicated, but cannot be segregated to daughter cells, until a specialized DNA breakage and rejoining reaction, referred to as telomere resolution, occurs at the rTel junctions. Telomere resolution generates a pair of linear replicons terminated by hp telomeres. rTel junctions are converted in vivo and in vitro into hp telomeres (9,10). The essential telomere resolvase that performs this reaction for Borrelia is known as ResT (11,12). A similar replication strategy has been demonstrated for the lysogen of the N15 bacteriophage; the N15 prophage exists as a linear plasmid terminated by hairpin telomeres (13–15). ResT resolves rTel junctions by a 2-step transesterification reaction with similarity to that promoted by type 1B topoisomerases and tyrosine recombinases (9,16–20). Briefly, the reaction is characterized by ResT forming/stabilizing an underwound conformation of the rTel, which is then cleaved 6 bp apart, on the opposing strands around the symmetry axis. The cleaved strands are refolded into a hairpin conformation and the second pair of transesterifications reseals the DNA backbone, generating the hp telomere products (16,20,21).
There is growing evidence that ResT is multifunctional. The phenotype of ResT depletion in a strain with conditional expression of ResT was unexpectedly complex. DNA replication was predicted to generate the dimer intermediates with rTel junctions that would then persist causing the cells to filament as cell division was inhibited. Upon ResT depletion unit sized linear forms did disappear, becoming progressively more complex forms that had an equal mix of hp telomeres and unresolved rTel junctions (12). However, ResT-depleted cells did not filament, and, unexpectedly, DNA replication ceased, as if ResT was required for continued replication (12).
ResT possesses unexpected biochemical properties. ResT has been found to promote single-strand annealing reactions (22). This ability to promote annealing of complementary ssDNA was found to extend over plasmid-length molecules and was operative even with ssDNA sequestered by B. burgdorferi's single-stranded DNA binding protein (SSB). The annealing of SSB-complexed ssDNA was found to be promoted by ResT–SSB interactions that involve the conserved C-terminal tail of SSB (SSB-Ct; (23)). These characteristics are reminiscent of those of the recombinational mediator protein RecO of the RecF pathway (24). These unexpected biochemical properties prompted us to propose that ResT may play a role in recombinational repair of daughter strand gaps, a role normally played by the highly conserved RecQFOR proteins, all of which lack homologues in the B. burgdorferi genome (1,23).
In this report, we extend these findings by describing the unexpected ability of ResT to promote ATP-dependent DNA unwinding. We find that ResT possesses DNA-dependent ATPase activity preferentially stimulated by ssDNA, that ATP binding promotes single-strand annealing but that the presence of ATP does not affect the ability of ResT to catalyze telomere resolution. Additionally, we show that ResT possesses ATP-dependent DNA unwinding activity operative on a variety of substrates including branched substrates that have three DNA arms such as synthetic replication fork and D-loop mimics. Finally, we show that ResT promotes regression of a partially mobile model replication fork. The T4 UvsW helicase and many members of the RecQ family of helicases share this unusual combination of DNA annealing and DNA unwinding activities found within the same polypeptide (25,26). We propose that ResT's combination of activities implicates it in replication and recombination processes operating on the linear chromosome and plasmids of Borrelia.
MATERIALS AND METHODS
DNAs
ϕX174 virion and RFI DNAs were purchased from New England Biolabs (NEB). All oligonucleotides were purchased from Integrated DNA Technologies (IDT). Details of the oligonucleotides and substrate annealings used in this study are presented in the Supplementary Materials and Methods section (Supplementary Table S1).
Proteins
ResT was purified as reported in (16,22), ResT (1–163) and ResT (164–449) were purified as reported in (27). ResT (Q181A), ResT (F93AW94A), ResT (F92AF93AW94A) and ResT (F92AF93AW94AQ181A) were generated by site-directed mutagenesis using the mutagenic oligonucleotides detailed in Supplementary Table S1. Borrelia burgdorferi SSB (locus BB_0114) was purified as reported in (23).
ATP photoaffinity binding assay
The ability to bind ATP was assayed by incubating 575 nM ResT, ResT (1–163), ResT (164–449) or SSB in reaction buffer containing 25 mM HEPES (pH 8.2), 2 mM EDTA (pH 8.0), 1 mM DTT, 50 mM NaCl, 25 μM ATP and 66 nM [γ32P]ATP in a total volume of 60 μl. After incubation on ice for 5 min the 1.5 ml reaction tubes were placed on a transilluminator and subjected to UV irradiation (312 nm) for 2 min. The samples were prepared for gel loading by addition of 5× SDS-load dye to a 1× concentration, followed by denaturation of the samples at 95°C for 6 min. After the gel run the gel was washed for 30 min in several changes of water then Coomassie stained to visualize the position of the bands. The gel was then wrapped in plastic and exposed to a phosphorimaging screen to detect protein that had become crosslinked to [γ32P]ATP.
ATPase assays
ATPase activity was determined by incubating ResT, at the concentration indicated in the figure legend, in buffer containing 25 mM HEPES (pH 8.2), 2 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin, 50 mM NaCl, 100 μM ATP and 66 nM [γ32P]ATP in a total volume of 30 μl. After incubation at 37°C for the times indicated in the figure legend, an aliquot of the reaction was spotted onto a polyethyleneimine thin-layer chromatography plate that was developed with 1 M formic acid and 0.5 M LiCl. The plate was air dried and then exposed to a phosphorimaging screen for determination of the free phosphate to total ATP ratio. When present, the indicated DNA effectors were present at 10 μg/ml.
DNA unwinding assays
DNA unwinding assays were performed by incubation at 37°C in buffer containing 25 mM HEPES (pH 8.2), 2 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin, 50 mM NaCl and 2 mM ATP or ATP-γ-S. The 5΄ 32P-endlabeled DNA substrates were present at 15 nM and ResT was used at the concentration indicated in the figure legends. Reactions were terminated by addition of reaction aliquots into SDS load dye to 1× a final concentration. 1× SDS load dye contains 20 mM EDTA, 3.2% glycerol, 0.1% SDS and 0.0024% bromophenol blue. The products of DNA unwinding were visualized by application of the samples to 20 cm × 20 cm 8% PAGE 1× Tris–acetate EDTA (TAE)/0.1% SDS gels electrophoresed at 13 V/cm for 2 h. The gels were dried and exposed to phosphorimager screens and developed on a BioRad FX phosphorimaging machine.
DNA annealing assays
DNA annealing assays were performed by incubation at 37°C in buffer containing 25 mM HEPES (pH 8.2), 2 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin, 50 mM NaCl and 2 mM ATP or ATP-γ-S. The 5΄ 32P-endlabeled substrates DNAs were present at 15 nM and ResT was used at 37 nM. Additional details are available in the Supplementary Methods and Materials section.
Telomere resolution assays with ATP
Telomere resolution timecourses were performed in 120 μl reactions incubated at 37°C. 18 μl aliquots were removed at the timepoints indicated in the legend and the reaction was terminated by addition of SDS load dye to a 1× final concentration (1× SDS load dye contains 20 mM EDTA, 3.2% glycerol, 0.1% SDS and 0.0024% bromophenol blue). Reactions contained 5.25 nM of 5΄-32P radiolabeled rTel substrate (constructed using oligos OGCB127/128) and 74 nM ResT. Reaction buffer contained 25 HEPES (pH 8.2), 2 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin and 50 mM NaCl. When present, ATP or ATP-γ-S were added to 2 mM.
Standard telomere resolution assays
Telomere resolution assays in Figure 4 were performed in 60 μl reactions incubated at 30°C for 15 min. Reactions contained 5.25 nM of 5΄-32P radiolabeled rTel substrate and the concentration of ResT indicated in the figure legend. Reaction buffer contained 25 mM Tris–Cl (pH 8.5), 1 mM EDTA (pH 8.0), 100 μg/ml bovine serum albumin and 100 mM NaCl.
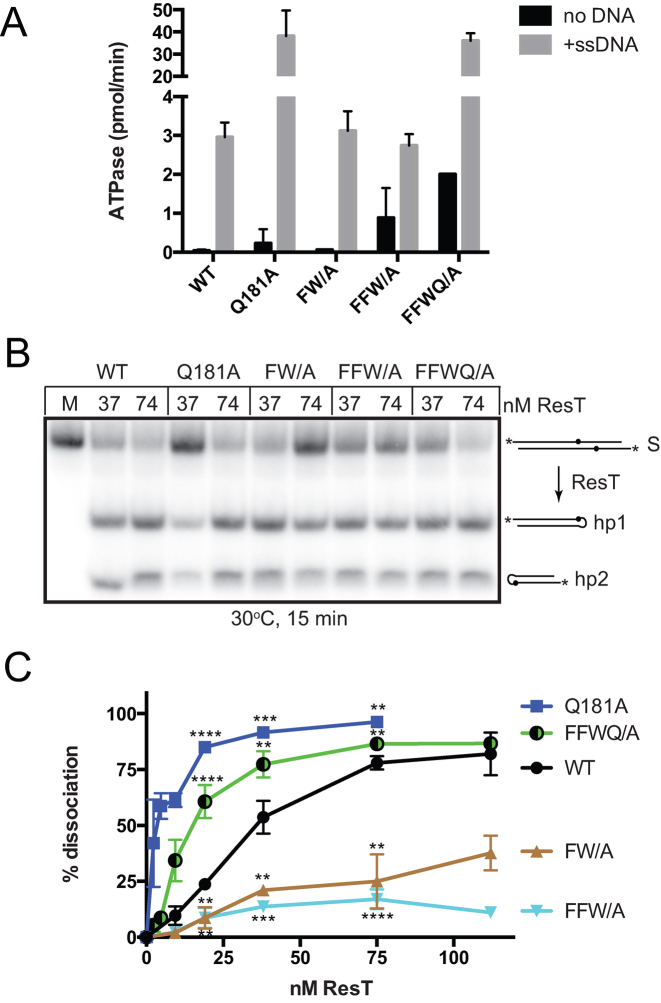
Figure 4.
ResT mutations that differentially affect telomere resolution and DNA unwinding. (A) Summary of ATPase results of the indicated ResT variants, −/+ 10 μg/mL ϕX174 virion. ATPase assays containing 74 nM ResT were incubated at 37°C for 30 min (Q181A) or at 37°C for 120 min for wild type ResT (WT), F93AW94A (FW/A), F92AF93AW94A (FFW/A), or F92AF93AW94AQ181A (FFWQ/A). The mean and standard deviation of at least three independent experiments is shown. (B) 8% PAGE 1× TAE/0.1%SDS gel analysis of telomere resolution assays conducted at 30°C for 15 min. The schematic to the right of the gel indicates the structure of the substrate (S) and of the resulting hp telomere products (hp1 and hp2). The asterisk denotes 5΄-endlabels and the dots the position of the scissile phosphates. (C) % Strand dissociation vs. ResT concentration curves of the indicated ResT variants. The helicase assays were performed by 60 min incubation at 37°C, with 15 nM substrate. Reactions were performed in triplicate and the mean and standard deviation are shown. Differencesbetween each mutant and wild type ResT in reaction conditions containing 37 nM ResT were examined using unpaired t-tests. ** P ≤ 0.01 and *** P ≤ 0.001.
RESULTS
We have previously reported that ResT, besides forming hp telomeres, has RecO-like properties including the ability to promote single-strand annealing of both free ssDNA and ssDNA bound by cognate SSB (22,23). Borrelia burgdorferi lacks the highly conserved RecF pathway that includes the RecFOR proteins that act as mediators that load RecA onto gapped DNA, and the RecQ helicase (1,28). Mechanistic studies of the telomere resolution reaction suggest that telomere resolution involves ResT-mediated formation/stabilization of an underwound pre-cleavage intermediate (16,20). This activity was reminiscent of part of the essential description of DNA helicase reactions which are characterized by DNA translocation and unwinding steps. We investigated the possibility that ResT may have properties in common with helicases despite lacking signature helicase motifs in its primary sequence.
ResT binds ATP and possesses DNA-dependent ATPase activity
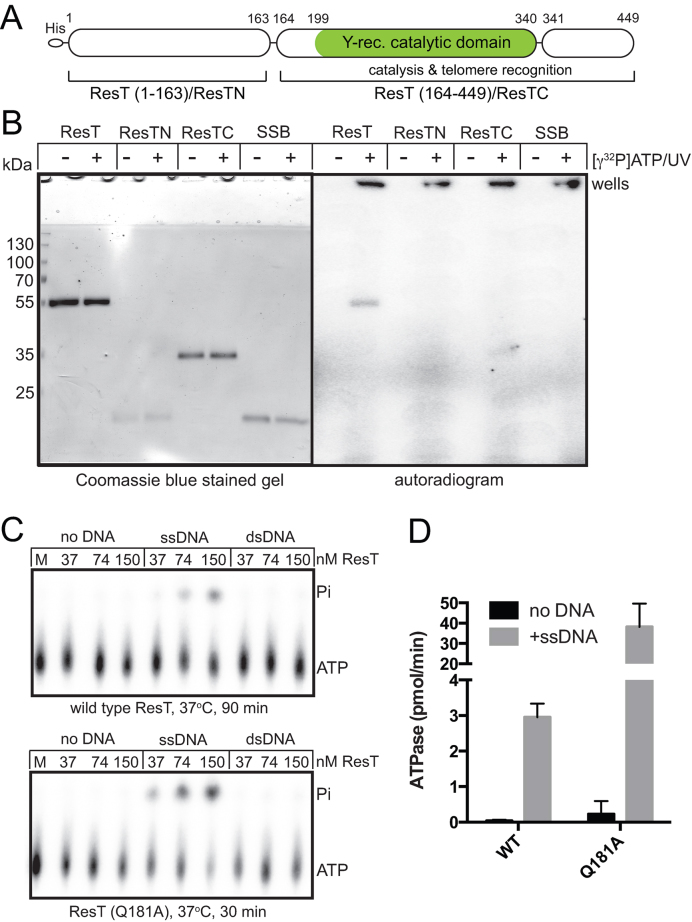
We investigated whether ResT can bind ATP. To directly assay for the ability of ResT to bind to ATP we employed an ATP photoaffinity crosslinking approach. [γ32P]ATP was incubated with ResT and its individual domains, ATP binding was captured by crosslinking the [γ32P]ATP to the protein by UV irradiation (Figure 1B). ATP binding was readily detectable with full-length ResT but not with the individual sub-domains or with an irrelevant negative control (B. burgdorferi SSB). An unusual property of the observed ATP binding was its Mg2+-independence (Figure 1B and Materials and Methods). Most nucleotide binding proteins and NTPase's bind nucleotides with Mg2+ but there are several examples of Mg2+-independent binding (29–31). The observation that ResT could bind ATP prompted us to ask if ResT might possess ATPase activity. We examined wild type ResT, along with mutants created for other studies, in ATPase assays. ResT was found to possess DNA-dependent ATPase activity that was preferentially stimulated by ssDNA (Figure 1C and D). One of the ResT mutants tested (Q181A) showed a 14-fold stimulation of the DNA-dependent ATPase activity relative to wild type (Figure 1D). In contrast, to the ATP binding, the ATPase activity demonstrated the expected dependence upon the presence of Mg2+ (data not shown). The observed ATPase activity is weak having a specific activity range of 1.35–18 pmol ATP/min/pmol ResT. HrpA, the only biochemically characterized helicase from B. burgdorferi has been reported to have a similar weak RNA-dependent ATPase specific acitivity of 7 pmol ATP/min/pmol HrpA (32). By comparison, a strong DNA helicase such as Escherichia coli UvrD has a DNA-dependent ATPase specific activity of ∼1.9 nmol ATP/min/pmol UvrD (33).
Figure 1.
ResT binds ATP and possesses DNA-dependent ATPase activity. (A) ResT's domain structure. The N-terminal His-tag is shown and amino acid numbering shown starts at the first ResT-derived residue. Shaded/green is the region of ResT with similarity to the catalytic domain of tyrosine recombinases (Y-rec. catalytic domain). For simplicity ResT (1–163) is referred to as ResTN while ResT (164–449) is referred to as ResTC. (B) 12% SDS-PAGE analysis of ATP photoaffinity binding assays of ResT and its sub-domains. SSB was included as a negative control. (C) Polyethyleneimine thin-layer chromatography ATPase assay results with ResT and ResT (Q181A) −/+ 10 μg/ml ϕX174 virion and ϕX174 RF1 DNA. (D) Summary of ATPase assay results with ResT and ResT (Q181A) −/+ 10 μg/ml ϕX174 virion. ATPase assays containing 74 nM ResT were incubated at 37°C for 30 min (Q181A) or at 37°C for 120 min for ResT (WT). The mean and standard deviation of at least three independent experiments is shown.
Telomere resolution is unaffected by ATP
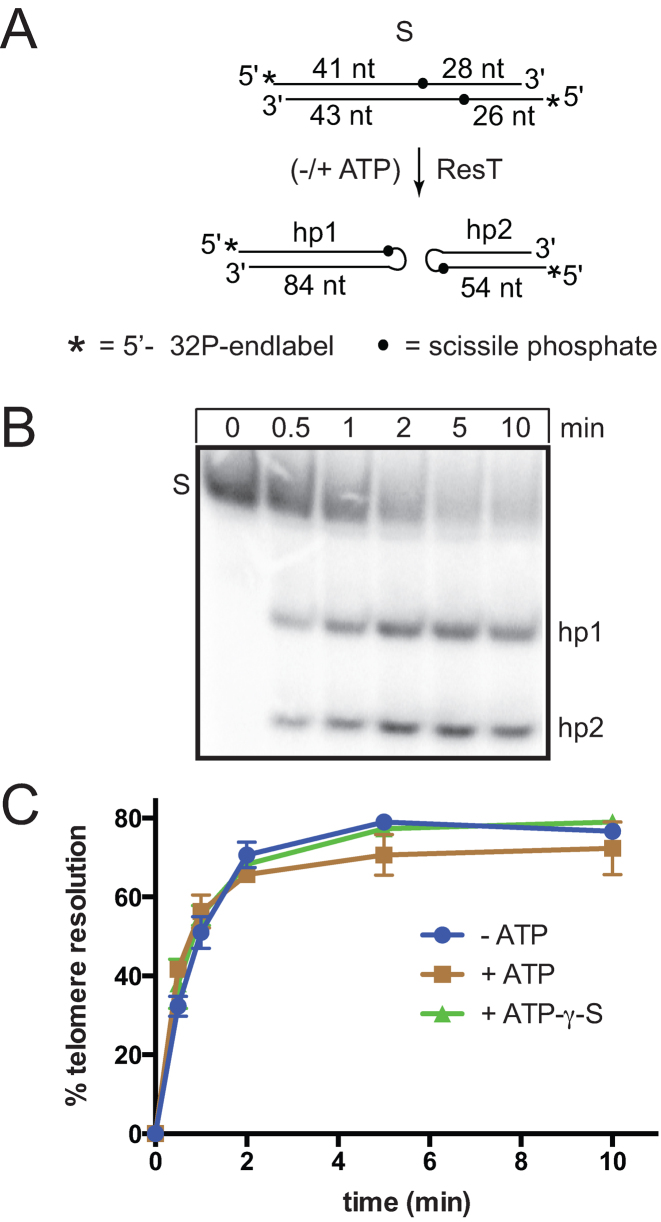
Telomere resolution catalyzed by ResT does not require divalent metal ions or high-energy cofactors like ATP, so our standard reaction buffer has not normally included Mg2+ or ATP (9). We assessed whether ATP had a detectable effect on telomere resolution reactions (Figure 2). Telomere resolution reactions were performed in a buffer containing MgCl2 (needed for ATP hydrolysis) with and without ATP or ATP-γ-S addition. ATP is not required for telomere resolution and its inclusion has no effect on the reaction (Figure 2C). The strong bias for an ssDNA effector to activate the ATPase activity helps rationalize the lack of effect of ATP on telomere resolution, a reaction with a double-stranded substrate.
Figure 2.
Telomere resolution is unaffected by ATP. (A) Schematic representation of the replicated telomere (rTel) substrate and the resultant hp telomere products of telomere resolution. The asterisks denote 5΄ 32P-endlabels; S, substrate; hp1 and hp2, denote the hp telomere products; dots, the position of the scissile phosphates. The strand lengths in the rTel are from the scissile phosphates to the end of the DNA strand, the strand lengths in the hp telomeres indicate the total strand length of the products. (B) Representative 8% PAGE 1× TAE/0.1%SDS gel panel of a telomere resolution timecourse reaction performed in ATPase assay buffer at 37°C with 5.25 nM rTel and 74 nM ResT. The example shown is a reaction without ATP supplementation. (C) % Telomere resolution vs. time plots of telomere resolution assays performed without ATP addition, with ATP or with ATP-γ-S addition. Reactions were performed in triplicate and the mean and standard deviation are shown.
ResT unwinds DNA with a 3΄-5΄ polarity
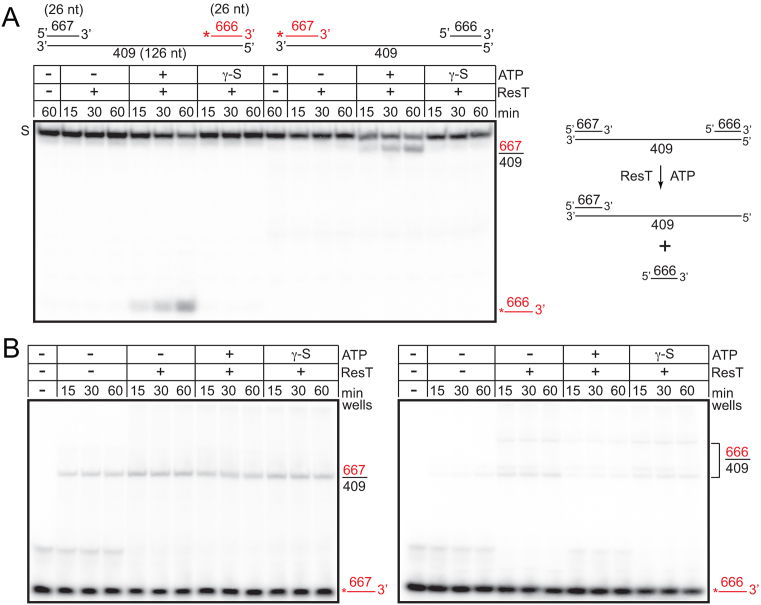
ResT's possession of DNA-dependent ATPase activity preferentially stimulated by ssDNA suggested that it may possess helicase activity. We designed an oligonucleotide substrate with two 26 bp duplex regions separated by a 74 nt single-stranded gap to test for DNA unwinding activity and to determine the polarity of any observed unwinding (Figure 3). ResT supported unwinding of only one of the duplex regions. The observed unwinding was indicative of a 3΄-5΄ DNA reaction polarity, with respect to the ssDNA in the gapped substrate. The observed activity was dependent upon ATP hydrolysis, as use of the poorly hydrolysable ATP analogue, ATP-γ-S, did not support the reaction (Figure 3A). The observed DNA unwinding activity could use ATP or dATP and Mg2+ or Mn2+ but not other triphosphate nucleotides or Ca2+ (data not shown). ResT is a potent single-strand annealing protein. It was unclear, therefore, why ResT did not simply reanneal the unwound strand obscuring the unwinding activity. We addressed the issue of whether the DNA strand displaced in the unwinding assay documented in Figure 3A could be (re)-annealed by ResT under the reaction conditions used. The 26 nt strands used to construct the unwinding substrate were very poorly annealed by ResT. The presence of hydrolysable ATP in annealing reactions with the strand that could be unwound by ResT (666) suppressed the slow annealing seen in other conditions while its presence did not affect annealing of the strand that cannot be displaced (667) since there was no competing unwinding reaction (Figure 3B). ResT has minimum strand length requirements for the annealing reaction that depends upon the GC-content of the DNA to be annealed (22). We infer that we can readily detect DNA unwinding with this substrate because the 26 nt displaced strand is inefficiently reannealed.
Figure 3.
ResT has 3΄-5΄ DNA unwinding activity. (A) 8% PAGE 1× TAE/0.1%SDS gel analysis of DNA unwinding polarity. The red/shaded asterisk indicates a 5΄-endlabel and the red/shaded line indicates DNA strands that are radiolabeled, unlabeled strands are shown with a black line. The name of the oligonucleotides used to assemble the substrate and strand length is indicated in the first schematic. The gel migration position of displaced 666 strand and the partial duplex product 667/409 are shown to the right of the gel. S, indicates the gel migration position of the substrate DNA. The substrate and ResT were present at 15 and 37 nM, respectively. An interpretive schematic of the unwinding reaction promoted by ResT with hydrolysable ATP is presented to the right of the gel. (B) 8% PAGE 1× TAE/0.1%SDS gel analyses of DNA annealing reactions. The gel migration positions of labeled 666, 667 strands and the partial duplex products (666/409 and 667/409) of annealing are shown to the right of the gels. The substrate DNA strands and ResT were present at 15 and 37 nM, respectively.
The effect of ATP on previously characterized ResT-promoted annealing reactions was assayed using a pair of 35 nt complementary oligonucleotides that anneal into a 35 bp blunt-ended duplex product ((23); Supplementary Figure S1). Inclusion of ATP or ATP-γ-S had a mild, but significant, stimulatory effect on annealing (Supplementary Figure S1B). ResT is unable to unwind the 35 bp duplex DNA product of annealing, so the competing unwinding reaction does not contribute to the results (data not shown).
The discovery of helicase activity for ResT was unexpected. We ensured that the observed helicase activity was not due to a contaminant from E. coli by assessing the protein purity using sensitive protein staining, western blotting and mass spectroscopy analysis (Supplementary Figure S2). Wild type ResT has a small amount of ResT cleavage product present. ResT (Q181A) also has proteolytic fragments of ResT present (∼5%) and some contaminating E. coli proteins present at a low level. Mass spectroscopy analyses showed that there were no DNA-dependent ATPase's or helicases present in the purifications (Supplementary Figure S2B).
ResT mutations that differentially affect DNA unwinding vs. telomere resolution
Among the ResT mutants examined for ATPase and helicase activity were mutations of a FFW motif in the N-terminal domain conserved in all Borrelial ResT's that changed the aromatic sidechains to alanines. The F93AW94A (FW/A) and F92AF93AW94A (FFW/A) mutants are unaffected for the DNA-dependent ATPase activity and are active for telomere resolution (Figure 4A and B). However, these mutants were found to be inefficient in the DNA unwinding assay (Figure 4C). ResT (Q181A), in contrast, displayed hyperactivated ATPase and helicase activities (Figures 2 and 4). This mutant was not similarly hyperactive for telomere resolution, instead it showed a slightly compromised ability to make hp telomeres relative to wild type (Figure 4B). Combining the Q181A mutation with the F92AF93AW94A mutations produced a ResT variant with an intermediate phenotype, having stimulated ATPase activity, like that seen with the Q181A mutant, but with attenuated unwinding activity compared to the Q181A mutant. At present, the molecular basis for the hyperactivation of the ATPase/helicase activities conferred by the Q181A mutation is not understood. However, ResT is known to also be subject to autoinhibition for replicated telomere recognition and resolution (21,27).
Telomere resolution proceeds via a phosphoryl transfer mechanism similar to that of type IB topoisomerases and tyrosine recombinases (21). These enzyme families have been reported to use, under certain conditions, alternatives to the active site tyrosine and 5΄-hydroxyl containing cleaved strands as nucleophiles producing hydrolytic reactions of DNA, RNA and the transient phosphotyrosine intermediate (34–37). Therefore, we investigated the possibility that the ATPase activity of ResT may represent an unusual repurposing of the telomere resolvase active site for cleavage of the phosphoanhydride bond between the β and γ phosphates in ATP (Supplementary Figure S3). However, ResT mutants in the catalytic residues required for telomere resolution all showed activity in the ATPase and helicase assays, effectively ruling out this possibility (Supplementary Figure S3).
ResT unwinds replication fork and displacement loop mimics
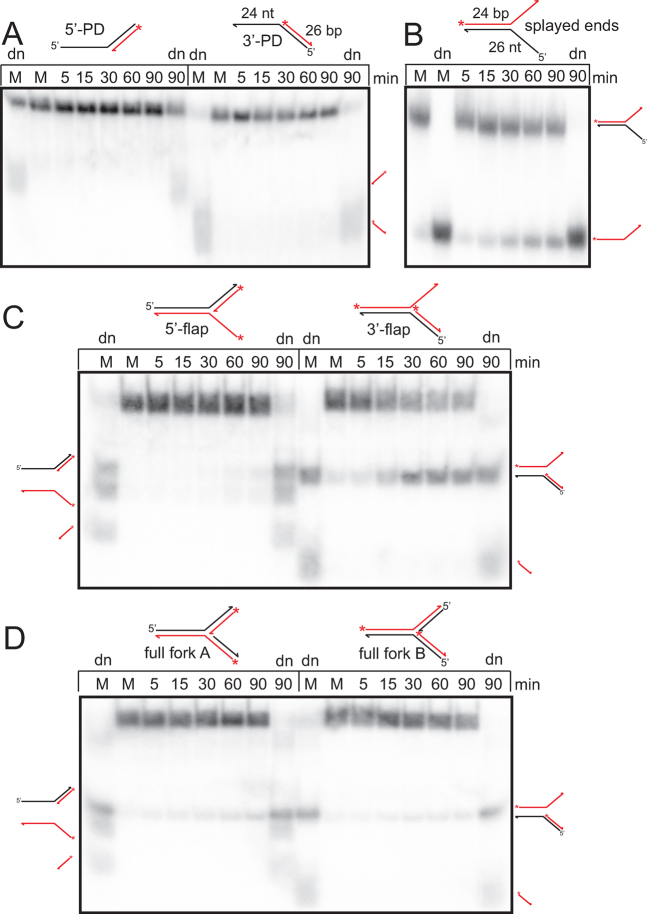
Several DNA helicases, including many RecQ family members and T4 UvsW, share with ResT the property of possessing DNA annealing and unwinding activities in the same polypeptide. Many of these enzymes have been reported to promote branch migration of DNA structures like Holliday junctions (HJ) and/or replications forks (25,26,38,39). ResT was tested in DNA unwinding assays with various partial and full assemblies of a synthetic replication fork mimic (Figure 5). Wild type ResT was found to promote unwinding of the replication fork mimic lacking both nascent strands (splayed end; Figure 5B) and the fork mimic with a missing nascent leading strand (3΄-flap; Figure 5C) consistent with its ssDNA-stimulated ATPase activity and 3΄-5΄ polarity bias. A more limited activity was seen with the 5΄-flap and full fork mimics (Figure 5C and D). The hyperactive Q181A mutant was found to be able to unwind all these substrates except the partial duplex with a 5΄-tail (5΄-PD; Supplementary Figure S4). We assessed whether the differential activity of ResT on the various fork assemblies was due to differences in the ability of ResT to bind the different substrates. ResT displayed binding KD values ranging from 50 to 120 nM for the 5΄, 3΄-flap and full fork versions of the fork (Supplementary Figure S5). Despite showing low activity on the 5΄-flap substrate, ResT bound this substrate with an affinity roughly equivalent to that seen with 3΄-flap substrate (Supplementary Figure S5A). Lower affinity binding that produced mostly wellshifted material rather than discrete ResT-DNA complexes was seen with the 5΄ and 3΄-partial duplex arms (Supplementary Figure S5A). Inclusion of ATP in the binding buffer was accompanied by a ∼2-fold decrease in binding affinity (Supplementary Figure S5B). This is consistent with models of helicase action in which gripping and release of substrate DNA, tied to the ATP binding, hydrolysis and ADP/Pi release facilitates DNA translocation and unwinding (40). Interestingly, the F92AF93AW94A mutant had a 4-fold higher affinity for the replication fork mimics, relative to wild type, raising the possibility that this mutant has poor unwinding activity because of inefficient translocation due to the enhanced binding affinity (Supplementary Figure S6). ResT was also assayed with a branched substrate with 4 arms, the well-characterized, partially mobile synthetic X26 Holliday junction (41). ResT and ResT (Q181A) were both unreactive with this substrate (data not shown).
Figure 5.
ResT unwinds synthetic replication forks. 8% PAGE 1× TAE/0.1%SDS gel analysis of timecourse reactions incubated at 37°C with various assemblies of a replication fork mimic with heterologous arms. (A) Reactions with partial duplex (PD) substrates (B) with the splayed ends substrate (C) with the flap substrates and (D) with two alternate labeling assemblies of the full fork substrate are shown. The substrates are diagrammed and named above the gel panels. The red/shaded asterisk indicates a 5΄-endlabel and the red/shaded line indicates DNA strands that are radiolabeled, unlabeled strands are shown with a black line. M, denotes mock incubation at 37°C for 90 min without ResT addition; dn, indicates heat denaturation at 95°C for 6 min in SDS-load dye prior to gel loading. The SDS-load dye allows partial renaturation of the heated species, accounting for the multiple species indicated in those lanes. The schematics to the side of the gels indicate the gel migration position of the various products of substrate unwinding and heat denaturation/partial renaturation. Assignment of the gel migration position of the various species was verified on separate gels with alternative substrate labeling configuration and markers. Substrate was present at 15 nM and ResT at 37 nM.
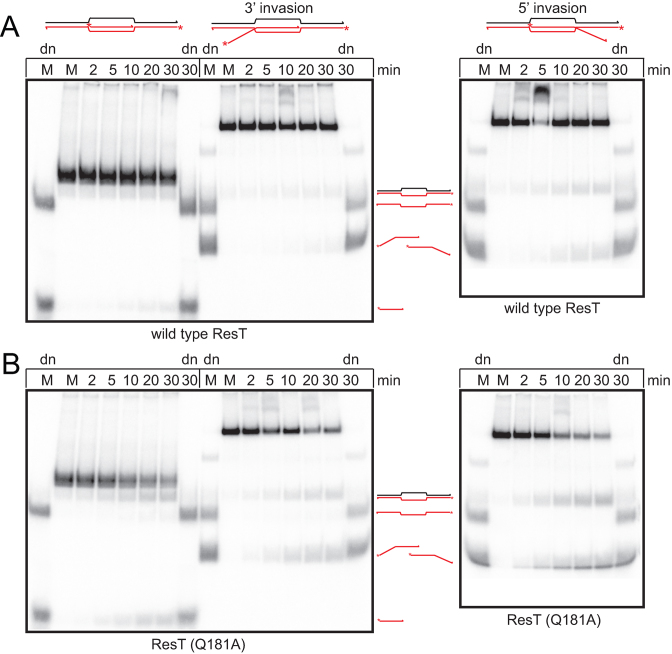
A second class of branched DNA substrate with three arms that RecQ helicases and T4 UvsW can act on is the displacement loop (D-loop) structure formed by strand invasion during homologous recombination (25,26). ResT was also found to promote unwinding of the invading strand in synthetic D-loops. Wild type ResT was active on the D-loop that results from strand invasion using a 5΄ end, consistent with the preference for unwinding substrates with 3΄ ssDNA tails (Figure 6A). ResT (Q181A) was able to displace the invading strand of all three versions of the D-loop (Figure 6B).
Figure 6.
ResT unwinds synthetic D-loops. 8% PAGE 1× TAE gel analysis of (A) wild type ResT and (B) ResT (Q181A) timecourse reactions incubated at 37°C using synthetic D-loop mimics. The red/shaded asterisk indicates a 5΄-endlabel and the red/shaded line indicates DNA strands that are radiolabeled, unlabeled strands are shown as black lines. M, denotes mock incubation at 37°C for 30 min without ResT addition; dn, indicates heat denaturation at 95°C for 6 min in the SDS-load dye prior to gel loading. The schematics to the side of the gels indicate the gel migration position of the various products of substrate unwinding and heat denaturation. Master reactions were incubated at 37°C and aliquots were withdrawn at the times indicated above the gel panels, substrate was present at 15 nM and ResT at 37 nM.
ResT promotes fork regression
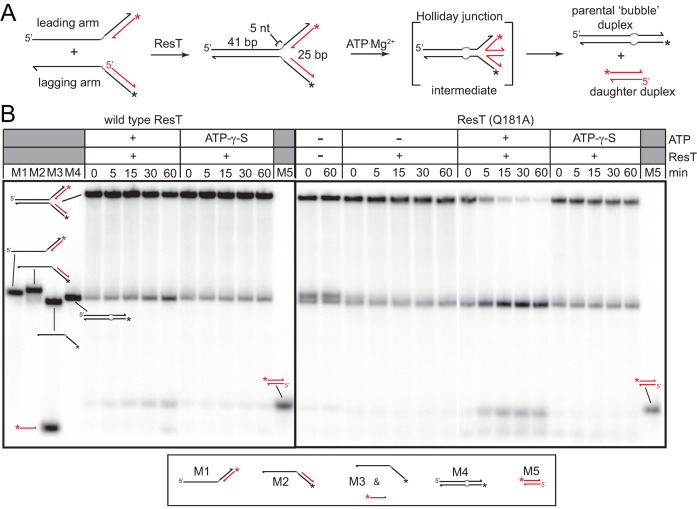
DNA helicases that promote strand annealing and unwinding are often capable of promoting branch migration (42,43). The replication fork mimics used in Figure 5 were designed with heterologous arms to aid substrate annealing into defined structures, thus yielding data that were easier to interpret with respect to ResT's binding and polarity preferences. A more realistic replication fork mimic was designed with homologous arms that would allow branch migration to be observed (Figure 7). Spontaneous branch migration was inhibited by a substrate design that incorporated a 5 nt region of sequence heterology at the ss-dsDNA junction (Figure 7A and ((43)). This partially mobile fork design was tested with ResT in reactions in which the leading and lagging strand partial duplex arms were annealed with the aid of ResT in a buffer lacking ATP.Mg2+. ATP.Mg2+ was then added to induce DNA unwinding (Figure 7A and B). ResT supported an almost complete conversion of the partial duplex arms into the fork structure (Figure 7B, t = 0). Addition of ATP.Mg2+ resulted in the slow accumulation of parental ‘bubble’ duplex and daughter duplex DNA products. Addition of ATP-γ-S.Mg2+ did not result in a similar conversion of the assembled fork into products, indicating that the products in the ATP.Mg2+ conditions are largely the result of ResT-promoted DNA unwinding rather than of spontaneous branch migration. The assignment of the products produced was assessed by gel mobility against all the possible products of unwinding (Figure 7B, markers M1-5) and by the pattern of sensitivity of the products and markers to T7 endonuclease I (data not shown). ResT produces parental duplex (with a T7 endonuclease I-sensitive 5 nt bubble) and daughter duplex in a concerted manner without the appearance of other unwinding products. This strongly suggests that DNA unwinding promotes the fork regression reaction pictured in Figure 7A rather than unwinding of the fork into partial duplex and single-strands that are then specifically reannealed into parental and daughter duplex DNAs. Consistent with the hyperactive phenotype of Q181A on the immobile forks (Supplementary Figure S4) this ResT variant promotes a much faster fork regression reaction with the partially mobile replication fork mimic (Figure 7B). Despite Q181A's hyperactivity the reaction promoted with the partially mobile fork substrate is, nonetheless, specific for fork regression rather than alternative unwinding reactions (Figure 7B).
Figure 7.
ResT promotes fork regression. (A) Fork regression assay using a synthetic replication fork with homologous arms. Leading and lagging arm partial duplexes are annealed with ResT. The resulting replication fork mimic has homologous arms but a 5 nt heterology on each template strand at the ss-dsDNA junction inhibits spontaneous branch migration. ATP.Mg2+ is added to induce ResT's helicase activity. (B) 8% PAGE 1× TAE/0.1%SDS gel analysis of fork regression promoted by wild type ResT and ResT (Q181A). After a 10 min annealing step, fork unwinding was induced by addition of ATP.Mg2+ (or ATP-γ-S.Mg2+). The possible products of fork unwinding were loaded as markers M1-5. The structure of the markers is indicated in the legend. The partial duplex arm substrates were present at 15 nM and ResT at 37 nM.
DISCUSSION
In this report, we have demonstrated that the B. burgdorferi telomere resolvase, ResT, possesses DNA-dependent ATPase activity that powers the unwinding of a variety of branched DNAs typical of replication and recombination intermediates. ResT is known to function as a telomere resolvase to create the hp telomeres of Borrelia through resolution of replicated telomeres (9,12). ResT also has RecO-like properties, in that it can anneal ssDNA both in its free and SSB-complexed forms. The annealing reaction with SSB-bound ssDNA is promoted by ResT–SSB interactions involving the conserved C-terminal tail of SSB (22,23). The discovery of DNA unwinding activity for ResT was unexpected, as ResT does not have obvious homology to characterized ATPase or helicase enzymes and because it has the seemingly contrary activity of single-strand annealing. Nonetheless, we have shown that ResT can bind ATP, that ATP binding modulates single-strand annealing and alters ResT's affinity for branched DNAs. Furthermore, we have characterized ResT mutations that have hyper- or hypoactive ATPase/helicase phenotypes that do not greatly affect the enzyme's ability to catalyze telomere resolution, yielding a preliminary view of the function of invariant residues in Borrelial ResT's not required for telomere resolution.
There have been a couple of indications that ResT may possess a nucleotide-binding pocket prior to this report. A recent study of telomere resolution demonstrated the existence of a pre-cleavage intermediate characterized by an underwound conformation (20). Modification of the substrate with abasic sites between the scissile phosphates alleviated the cold-sensitivity of the resolution. Furthermore, modification with abasic sites conferred site-specific rescue of the telomere resolution defect of certain ResT mutants. As missing base modifications can reduce the need for enzyme stabilization of extrahelical bases, a possible physical basis for these data would be for these nucleotides to be flipped out of the double helix into a nucleotide-binding pocket in ResT (20,44,45). With the new DELTA-BLAST algorithm ResT is found to have weak homology to part of the pterin-binding domain of bacterial methionine synthases, extending in ResT's primary sequence from residues 154–289 (Supplemental Figure S7; (46,47)). The pterin ring system of pterins like methyltetrahydrofolate is derived from the purine nucleotide GTP (48). This homology may indicate that this region of ResT participates in nucleotide binding. Consistent with this view are the observations that the individual ResT domains, ResT (1-163) and ResT (164-449), do not bind ATP presumably because the putative ATP-binding region is split between the two domains (Figure 1B). The hyperactivating Q181A mutation also falls in this region. Residues 154–289 in ResT are part of the conserved telomere resolvase domain (pfam 16684) and can be threaded to the structure of the related telomere resolvase, TelA, from Agrobacterium tumefaciens but poorly the Thermotaga maritima MetH (Supplementary Figure S7B). Methionine synthases have not been reported to have ATPase activity. Examination of the characterized telomere resolvases, TelN and TelK, identify the expected conserved telomere resolvase domain but also reveal weak, partially overlapping homology to P-loop NTPase and UvrD helicase conserved domains, respectively (Supplementary Figure S7A; (49)). These observations suggest the possibility that other telomere resolvases may also possess DNA helicase activity. Experimental mapping of the ATP binding and hydrolysis determinants in ResT and testing other telomere resolvase family members for helicase activity will be required to validate these additional weak domain homologies and to determine the generality of our findings to other hairpin telomere resolvases.
ResT can promote both DNA annealing and unwinding reactions. When ResT anneals strands into a partial duplex with a 3΄ tail, a structure that it can also unwind, the presence of hydrolysable ATP lessens the yield of annealing (Figure 3B). When it anneals strands into a blunt-ended duplex, a structure it cannot unwind, the presence of ATP stimulates the annealing reaction (Supplementary Figure S1). We infer from these data that ATP binding is stimulatory to single-strand annealing and that the two activities are in competition with DNAs that ResT can also unwind. Many helicases that can both anneal and unwind DNA promote branch migration of structures like replication forks or Holliday junctions. Consistent with this idea, we found that ResT can unwind synthetic replication forks and D-loops (Figures 5 and 6) and promote fork regression with a partially mobile, synthetic replication fork (Figure 7).
Potential implications for hp telomere replication and recombination
The current model for replication of the linear DNAs is that that bidirectional replication initiates internally and proceeds to the end of each replicore and rounds the hairpin telomeres to produce the replicated telomere (rTel) junctions that are processed into a pair of hairpin telomeres by ResT. This model is supported by studies that show that both in vivo and in vitro such rTel junctions are resolved into two hairpin telomeres by the resolvase (9,10). Bidirectional replication from an internal origin of replication has been directly demonstrated for B. burgdorferi's chromosome and inferred by bioinformatic studies for all of B. burgdorferi's linear plasmids (7,8).
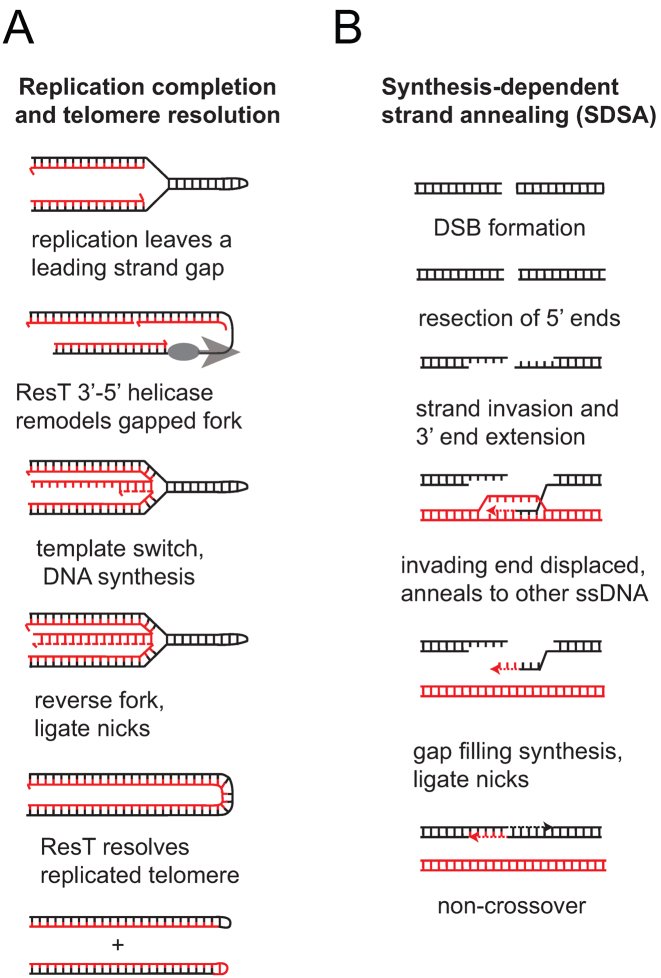
We have pointed out previously that this model assumes that the replisome is able to fully denature the DNA at the hairpin telomere end and that no gaps would be left. The actual behaviour of a replisome approaching a hairpin telomere is not currently known. A conceptual challenge of this model is the issue of the seeming potential for a conflict between the replicative helicase (DnaB) that tracks along the lagging strand template and the leading strand polymerase. Such a conflict could result in a failure by the canonical replication machinery to completely replicate the hp telomere on both strands. The unexpected complexity of the phenotype of ResT depletion (12) and ResT's possession of DNA helicase and RecO-like activities (22,23) suggest that ResT may play a role in replication completion at the hp telomeres, in addition to its well understood role in telomere resolution. We propose a scheme where the replisome leaves a leading strand gap at the hp telomere. This stalled fork structure can be remodeled by ResT-promoted fork regression, allowing a template switch to complete replication across the gap (Figure 8A). Difficulty completing replication near hp telomeres could also, in principle, trigger recombinational attempts to restart or rescue replication. How DNA annealing and unwinding activities could be involved in such repair events is detailed in Figure 8B.
Figure 8.
DNA transactions that require annealing and unwinding activities. (A) Presented is a model in which the replisome leaves a gap on the leading strand at a hairpin telomere due to a hypothesized conflict between the replicative helicase and the leading strand polymerase. ResT is shown as remodeling the fork by unwinding the nascent lagging strand and annealing it to the nascent leading strand. This remodeling is referred to as fork regression and it allows the nascent lagging strand to serve as a template for synthesis to fill in the leading strand gap. Reversal of the fork followed by ligation of the nick produces a replicated telomere junction that can serve as the substrate for ResT to produce the hp telomeres. (B) Presented is a model in which ResT plays a role in double-strand break repair. A DNA break is resected to allow recombinase loading onto the 3΄ tails. One end of the DSB invades an intact duplex at the homologous sequence producing a D-loop, the invading strand is extended by the replication machinery. ResT is hypothesized to unwind the extended D-loop intermediate and to promote annealing of the displaced strand to the complementary sequence on the other side of the DSB via its ability to promote annealing of complementary SSB-coated single-stranded DNAs. Gap filling and nick ligation produces a repaired duplex without causing a crossover between the DNA that suffered the DSB and the donor duplex used to repair the break.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank members of the laboratory for useful discussions and Nicola Schaan for making an observation, in a different context, that lead us to suspect that ResT may bind ATP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) [RGPIN 32 6797-2011]. Funding for open access charge: Internal Publication Grant Fund, University of Saskatchewan and NSERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fraser C.M., Casjens S., Huang W.M., Sutton G.G., Clayton R., Lathigra R., White O., Ketchum K.A., Dodson R., Hickey E.K. et al. . Genomic sequence of a Lyme disease spirochaete Borrelia Burgdorferi. Nature. 1997; 390:580–586. [DOI] [PubMed] [Google Scholar]
- 2.Casjens S., Palmer N., van Vugt R., Huang W.H., Stevenson B., Rosa P., Lathigra R., Sutton G., Peterson J., Dodson R.J. et al. . A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000; 35:490–516. [DOI] [PubMed] [Google Scholar]
- 3.Chaconas G., Kobryn K.. Structure, function, and evolution of linear replicons in Borrelia. Annu. Rev. Microbiol. 2010; 64:185–202. [DOI] [PubMed] [Google Scholar]
- 4.Barbour A.G., Garon C.F.. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987; 237:409–411. [DOI] [PubMed] [Google Scholar]
- 5.Baril C., Richaud C., Baranton G., Saint Girons I.S.. Linear chromosome of Borrelia burgdorferi. Res. Microbiol. 1989; 140:507–516. [DOI] [PubMed] [Google Scholar]
- 6.Ferdows M.S., Barbour A.G.. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:5969–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picardeau M., Lobry J.R., Hinnebusch B.J.. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 1999; 32:437–445. [DOI] [PubMed] [Google Scholar]
- 8.Picardeau M., Lobry J.R., Hinnebusch B.J.. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 2000; 10:1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobryn K., Chaconas G.. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell. 2002; 9:195–201. [DOI] [PubMed] [Google Scholar]
- 10.Chaconas G., Stewart P.E., Tilly K., Bono J.L., Rosa P.. Telomere resolution in the Lyme disease spirochete. EMBO J. 2001; 20:3229–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byram R., Stewart P.E., Rosa P.. The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol. 2004; 186:3561–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandy N.J., Salman-Dilgimen A., Chaconas G.. Construction and characterization of a Borrelia burgdorferi strain with conditional expression of the essential telomere resolvase, ResT. J. Bacteriol. 2014; 196:2396–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deneke J., Ziegelin G., Lurz R., Lanka E.. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7721–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravin N.V., Strakhova T.S., Kuprianov V.V.. The protelomerase of the phage-plasmid N15 is responsible for its maintenance in linear form. J. Mol. Biol. 2001; 312:899–906. [DOI] [PubMed] [Google Scholar]
- 15.Ravin N.V., Kuprianov V.V., Gilcrease E.B., Casjens S.R.. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 2003; 31:6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankhead T., Chaconas G.. Mixing active site components: A recipe for the unique enzymatic activity of a telomere resolvase. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13768–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deneke J., Burgin A.B., Wilson S.L., Chaconas G.. Catalytic residues of the telomere resolvase ResT: a pattern similar to, but distinct from tyrosine recombinases and type IB topoisomerases. J. Biol. Chem. 2004; 279:53699–53706. [DOI] [PubMed] [Google Scholar]
- 18.Kobryn K., Burgin A.B., Chaconas G.. Uncoupling the chemical steps of telomere resolution by ResT. J. Biol. Chem. 2005; 280:26788–26795. [DOI] [PubMed] [Google Scholar]
- 19.Briffotaux J., Kobryn K.. Preventing broken borrelia telomeres: Rest couples dual hairpin telomere formation to product release. J. Biol. Chem. 2010; 285:4100–41018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucyshyn D., Huang S.H., Kobryn K.. Spring loading a pre-cleavage intermediate for hairpin telomere formation. Nucleic Acids Res. 2015; 43:6062–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobryn K., Chaconas G.. Hairpin Telomere Resolvases. Microbiol. Spectrum. 2014; 2, doi:10.1128/microbiolspec.MDNA3-0023-2014. [DOI] [PubMed] [Google Scholar]
- 22.Mir T., Huang S.H., Kobryn K.. The telomere resolvase of the Lyme disease spirochete, Borrelia burgdorferi, promotes DNA single-strand annealing and strand exchange. Nucleic Acids Res. 2013; 41:10438–10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S.H., Kobryn K.. The Borrelia burgdorferi telomere resolvase, ResT, anneals ssDNA complexed with its cognate ssDNA-binding protein. Nucleic Acids Res. 2016; 44:5288–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryzhikov M., Koroleva O., Postnov D., Tran A., Korolev S.. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011; 39:6305–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson S.W., Benkovic S.J.. The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J. Biol. Chem. 2007; 282:407–416. [DOI] [PubMed] [Google Scholar]
- 26.Croteau D.L., Popuri V., Opresko P.L., Bohr V.A.. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014; 83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourand Y., Lee L., Chaconas G.. Telomere resolution by Borrelia burgdorferi ResT through the collaborative efforts of tethered DNA binding domains. Mol. Microbiol. 2007; 64:580–590. [DOI] [PubMed] [Google Scholar]
- 28.Rocha E.P., Cornet E., Michel B.. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005; 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo M., Kioka N., Amachi T., Ueda K.. ATP binding properties of the nucleotide-binding folds of SUR1. J. Biol. Chem. 1999; 274:37479–37482. [DOI] [PubMed] [Google Scholar]
- 30.Guhan N., Muniyappa K.. Mycobacterium tuberculosis RecA intein, a LAGLIDADG homing endonuclease, displays Mn(2+) and DNA-dependent ATPase activity. Nucleic Acids Res. 2003; 31:4184–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Focia P.J., Alam H., Lu T., Ramirez U.D., Freymann D.M.. Novel protein and Mg2+ configurations in the Mg2+GDP complex of the SRP GTPase ffh. Proteins. 2004; 54:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salman-Dilgimen A., Hardy P-O, Radolf J.D, Caimano M.J, Chaconas G.. HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the lyme disease spirochete. PLoS Pathogens. 2013; 9:e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumura K., Sekiguchi M.. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J. Biol. Chem. 1984; 259:1560–1565. [PubMed] [Google Scholar]
- 34.Sekiguchi J., Shuman S.. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997; 1:89–97. [DOI] [PubMed] [Google Scholar]
- 35.Xu C.J., Grainge I., Lee J., Harshey R.M., Jayaram M.. Unveiling two distinct ribonuclease activities and a topoisomerase activity in a site-specific DNA recombinase. Mol. Cell. 1998; 1:729–739. [DOI] [PubMed] [Google Scholar]
- 36.Tian L., Claeboe C.D., Hecht S.M., Shuman S.. Guarding the genome: electrostatic repulsion of water by DNA suppresses a potent nuclease activity of topoisomerase IB. Mol. Cell. 2003; 12:199–208. [DOI] [PubMed] [Google Scholar]
- 37.Ma C.H., Rowley P.A., Macieszak A., Guga P., Jayaram M.. Active site electrostatics protect genome integrity by blocking abortive hydrolysis during DNA recombination. EMBO J. 2009; 28:1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L., Hickson I.D.. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006; 40:279–306. [DOI] [PubMed] [Google Scholar]
- 39.Shereda R.D., Bernstein D.A., Keck J.L.. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 2007; 282:19247–19258. [DOI] [PubMed] [Google Scholar]
- 40.Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 41.Elborough K.M., West S.C.. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990; 9:2931–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P.. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004; 23:2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machwe A., Xiao L., Groden J., Orren D.K.. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006; 45:13939–13946. [DOI] [PubMed] [Google Scholar]
- 44.Li X.Y., McClure W.R.. Stimulation of open complex formation by nicks and apurinic sites suggests a role for nucleation of DNA melting in Escherichia coli promoter function. J. Biol. Chem. 1998; 273:23558–23566. [DOI] [PubMed] [Google Scholar]
- 45.Helmann J.D., deHaseth P.L.. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999; 38:5959–5967. [DOI] [PubMed] [Google Scholar]
- 46.Boratyn G.M., Schaffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L.. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012; 7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A., Derbyshire M.K, Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I. et al. . CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015; 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nar H., Huber R., Meining W., Schmid C., Weinkauf S., Bacher A.. Atomic structure of GTP cyclohydrolase I. Structure. 1995; 3:459–466. [DOI] [PubMed] [Google Scholar]
- 49.Kelley L.M., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.