Abstract
Pervasive transcription is widespread and needs to be controlled in order to avoid interference with gene expression. In Saccharomyces cerevisiae, the highly conserved helicase Sen1 plays a key role in restricting pervasive transcription by eliciting early termination of non-coding transcription. However, many aspects of the mechanism of termination remain unclear. In this study we characterize the biochemical activities of Sen1 and their role in termination. First, we demonstrate that the helicase domain (HD) is sufficient to dissociate the elongation complex (EC) in vitro. Both full-length Sen1 and its HD can translocate along single-stranded RNA and DNA in the 5΄ to 3΄ direction. Surprisingly, however, we show that Sen1 is a relatively poorly processive enzyme, implying that it must be recruited in close proximity to the RNA polymerase II (RNAPII) for efficient termination. We present evidence that Sen1 can promote forward translocation of stalled polymerases by acting on the nascent transcript. In addition, we find that dissociation of the EC by Sen1 is favoured by the reannealing of the DNA upstream of RNAPII. Taken together, our results provide new clues to understand the mechanism of Sen1-dependent transcription termination and a rationale for the kinetic competition between elongation and termination.
INTRODUCTION
Pervasive transcription is a common phenomenon both in eukaryotes and prokaryotes that consists in the massive production of non-coding RNAs (ncRNAs) from non-annotated regions of the genome (1). Pervasive transcription poses a risk that needs to be controlled since it can interfere with normal transcription of canonical genes. In Saccharomyces cerevisiae, a major actor in the control of pervasive transcription generated by the RNA polymerase II (RNAPII) is the Nrd1–Nab3–Sen1 (NNS) complex, which elicits early termination of non-coding transcription and promotes degradation of the RNAs thus produced by the nuclear exosome (2–4). The NNS-complex plays two additional important roles in gene expression. One is through its function in the biogenesis of sn- and snoRNAs, which are important for splicing and rRNA modification, respectively. Both transcription termination and 3΄ end maturation of most sn- and snoRNAs by the exosome depend on the NNS-complex (5). In addition, in a growing number of cases the NNS-complex mediates regulation of gene expression by a mechanism of premature termination or attenuation (for review, see 6).
The NNS-complex is composed of three essential proteins: the sequence-specific RNA-binding proteins Nrd1 and Nab3 and the RNA and DNA helicase Sen1 (7,8). Nrd1 and Nab3 recognize specific motifs on the target ncRNAs, GUAA/G and UCUUG being the optimal respective binding sites (9–12). In addition, Nrd1 interacts with the C-terminal domain (CTD) of the largest subunit of RNAPII (13,14). Both the recognition of the ncRNA and the interaction with the RNAPII CTD are necessary for the recruitment of the NNS-complex to the elongation complex (EC) and for efficient transcription termination (3,15–17).
Sen1 is the only subunit of the NNS-complex that possesses a catalytic activity. It contains a conserved central domain with homology to the Superfamily 1 (SF1) helicases, flanked by N-terminal and C-terminal extensions involved in protein–protein interactions. Specifically, the N-terminal domain has been proposed to mediate the interaction with RNAPII, while the C-terminal domain contains sequences important for the nuclear localisation of Sen1 and for the interaction with the phosphatase Glc7 and Nab3 (7,18,19). Deletion of the N-terminal domain or mutation of the helicase domain provoke transcription termination defects in vivo (19–21). Sen1 is also the only protein of the NNS-complex that is conserved in most eukaryotes. Its human ortholog, senataxin, has also been shown to play a role in transcription termination at several model genes (22–25) and mutations in the most conserved regions of senataxin, the N-terminal and the helicase domains, are linked to amyotrophic lateral sclerosis type 4 (ALS4) and ataxia-ocular apraxia type 2 (AOA2) (26). In addition, introducing AOA2 mutations in the equivalent residues of Sen1 provokes growth defects (19) suggesting that these neurological disorders could be due to termination defects.
Transcription termination can be envisioned as a multi-step process involving the recruitment of termination factors, pausing of the transcribing polymerases and finally the action of the termination factors on the nucleic acid and/or the protein components of the EC to elicit its dissociation (for review, see 6). In a previous study, we have used a highly purified in vitro transcription termination (IVTer) system to address the role of Sen1 in transcription termination. More precisely, we analysed the capacity of Sen1 to achieve the final steps of termination (i.e. dissociation of the EC). Importantly, we have found that Sen1 alone can dismantle a paused EC in vitro. In addition, we have observed that, similarly to the bacterial termination factor Rho, Sen1 needs to interact with the nascent RNA and hydrolyse ATP to promote termination (27). However, many details of the termination reaction remain to be elucidated. For instance, it is unclear whether termination requires the translocation of Sen1 along the RNA, as it is the case for its bacterial counterpart. In addition, although the integrity of the helicase domain is essential for termination, the possible contribution of additional domains of Sen1 to the step of dissociation of the EC has so far not been studied. In this work we employ a variety of in vitro approaches to explore these aspects. First, we have performed a functional dissection of Sen1 protein to understand the role of the different domains of Sen1 in the final step of termination. Importantly, we have found that the helicase domain is sufficient to dissociate the EC and that the presence of the N-terminal domain partially inhibits termination in vitro, suggesting a possible function for this domain in modulating the termination activity of Sen1. Second, we have found that both full-length Sen1 and its helicase domain can translocate along single-strand DNA and RNA with 5΄ to 3΄ polarity. Our data indicate that Sen1 is a low-processivity enzyme that displays a significant preference for DNA over RNA for translocation. However, IVTer assays performed under particular conditions indicate that dissociation of the EC does not involve the interaction of Sen1 with the DNA component of the EC. Nevertheless, substituting the nascent RNA by ssDNA in IVTer reactions substantially increases the efficiency of EC dismantling. Because Sen1 translocation seems significantly more processive on ssDNA than on ssRNA, this result strongly suggests that termination requires Sen1 translocation on the RNA. Furthermore, we provide evidence that Sen1 can promote forward translocation of stalled RNAPII, similar to the bacterial termination factor Rho. Taken together, our data provide considerable advances in the understanding of the mechanisms of action of Sen1 in non-coding transcription termination.
MATERIALS AND METHODS
Strain and plasmid construction
Oligonucleotides used for the constructs are detailed in Supplementary Table S1 and plasmids are described in Supplementary Table S2. Strain YDL2556 for overexpression of wild-type SEN1 at the endogenous locus was obtained by inserting the GAL1 promoter just upstream of SEN1 in a protease-deficient strain (BJ2168, kind gift of B. Seraphin) using standard procedures (28,29). Plasmids for overexpression of SEN1 variants are derived from pCM185 and were constructed by homologous recombination in yeast using previously described methods (28).
Protein purification
Saccharomyces cerevisiae RNAPII was purified from strain BJ5464 (30) by Nickel-affinity chromatography followed by anion exchange essentially as previously described (31), except for a few modifications. For this study, the ammonium sulfate precipitation step was omitted and all the buffers used in Ni2+-affinity chromatography were supplemented with 5 mM MgCl2 and 2 mM β-mercaptoethanol. Recombinant His6-tagged Rpb4/7 heterodimer was purified from Escherichia coli by Ni2+-affinity chromatography followed by gel filtration as previously described (31).
N-terminally TAP-tagged Sen1 proteins were purified from yeast, either from strain YDL2556, in the case of wild-type Sen1, or from strain BJ2168 harboring the appropriate plasmid (see Supplementary Table S2), in the case of Sen1 variants. Overexpression was induced by growing cells in YPA (for wild-type Sen1) or minimal synthetic media (for the mutant versions) containing 20 g/l of galactose for 14–24 h at 30°C. Proteins were purified using a standard TAP protocol as described before (31), with the following modifications: the concentration of NaCl in elution buffers was elevated to 500 mM to increase the elution yield and a treatment with 20 μg/ml of RNase A was included during elution from the IgG-beads at 4°C overnight. The purity of the protein preparation was monitored by SDS-PAGE followed by silver staining and by mass spectrometry (MS) analyses. No traces of Nrd1 and Nab3 or any known partners were detected and only minor amounts of unspecific contaminants (e.g. ribosomal proteins) were revealed by MS (see Supplementary Table S3). To accurately quantify the different Sen1 preparations, we loaded 5–10 μl of each preparation onto a protein gel next to four different concentrations of highly pure BSA (typically 50,100, 200, 500 and 1000 ng). We stained the gel with SYPRO Ruby (Thermo Scientific), scanned it with a Typhoon scanner (GE Healthcare) and quantified the different protein bands using the Image Quant software (GE Healthcare). The concentration of Sen1 proteins was calculated by comparison to the BSA standard curve (signal versus the protein concentration), correcting the values according to the molecular weight of the different proteins.
ATP hydrolysis assay
ATPase assays were performed in 10 μl reactions containing 10 nM of purified Sen1 proteins in 10 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM MgCl2, 1 μM ZnCl2, 1 mM DTT and 15% glycerol in the presence of 50 ng/μl of polyU. The reaction was initiated by the addition of an ATP solution (250 μM of cold ATP and 0.25 μM of 800 Ci/mmol α32P-ATP as the final concentrations in the reaction) and allowed to proceed for a total of 12 min at 28°C. Aliquots (1.5 μl) were removed at various times and mixed with 1.5 μl of quench buffer (10 mM EDTA, 0.5% SDS). The hydrolysis products were separated by thin layer chromatography on PEI cellulose plates (Merck) in 0.35 M potassium phosphate (pH 7.5) and analysed by phosphorimaging using a Typhoon scanner (GE healthcare).
In vitro termination (IVTer) assay
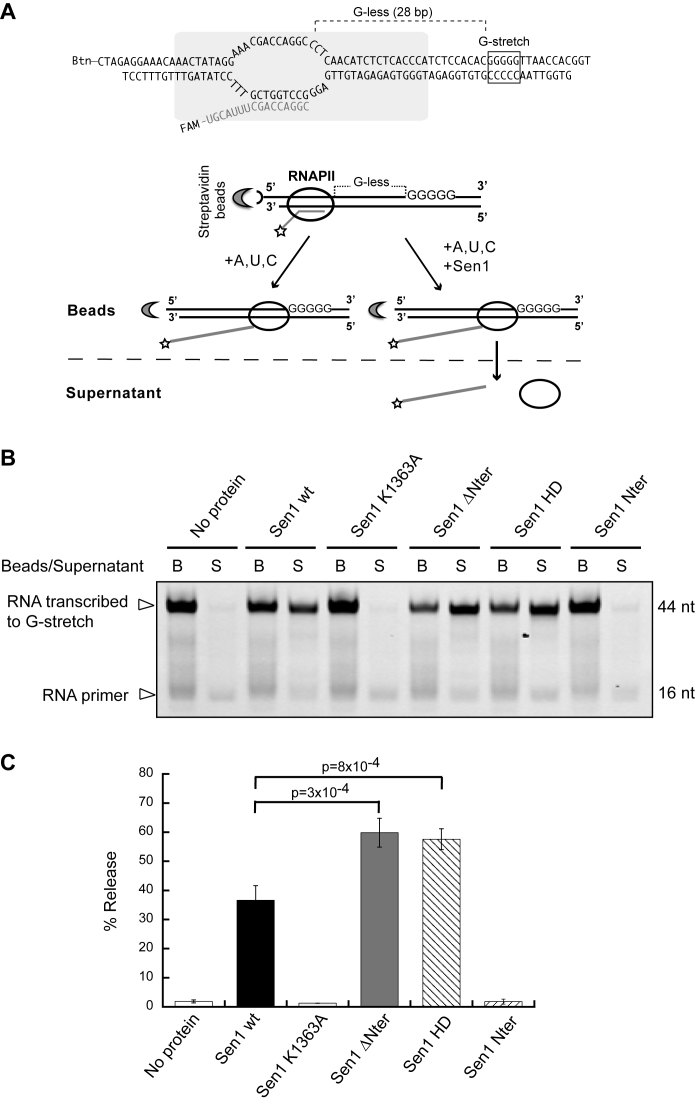
Termination assays were performed basically as previously described (31). Briefly, ternary ECs were assembled in a promoter-independent manner by first annealing a fluorescently labeled RNA with the template DNA and subsequently incubating the RNA:DNA hybrid with purified RNAPII. Next, the non-template strand and recombinant Rpb4/7 heterodimer were sequentially added to the mixture. The ternary ECs were then immobilized on streptavidin beads (Dynabeads MyOne Streptavidin T1 from Invitrogen) and washed with transcription buffer (TB) containing 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 8 mM MgCl2, 10 μM ZnCl2, 10% glycerol and 2 mM DTT; then with TB/0.1% Triton, TB/0.5 M NaCl and finally TB. The termination reactions were performed in TB in the presence of RNase inhibitors in a final volume of 20 μl. The amount of EC-beads used for each assay was optimized experimentally, depending on the efficiency of EC assembly for each template. For standard IVTer assays as presented in Figure 2, transcription was initiated after addition of a mixture of ATP, UTP and CTP at 1 mM each as the final concentration in the reaction, to allow transcription through the G-less cassette up to the first G of a G-stretch in the non-template strand. For assays performed in the absence of transcription as in Figures 5 and 6, the ECs contained a sufficiently long RNA to allow Sen1 loading (i.e. 44-nt long) and termination was assessed in the presence of 1 mM ATP. The reactions were incubated for 15 min at 28°C and then stopped by the addition of 1 μl of 0.5 M EDTA and the mixtures were separated into beads and supernatant fractions. The bead fractions were resuspended in 8 μl of loading buffer (1× Tris–borate–EDTA, 8 M urea) and boiled for 5 min at 95°C and RNAs in the supernatant fractions were ethanol-precipitated and resuspended in 8 μl of loading buffer. Transcripts were subjected to 10% (w/v) denaturing PAGE (8 M urea) and the gels were scanned using with a Typhoon scanner. The efficiency of termination by the different Sen1 variants was inferred from the % of nascent RNA released into the supernatant.
Figure 2.
Analysis of the capability of Sen1 variants to terminate transcription in vitro. (A) Scheme of an in vitro transcription termination assay. A schematic of a ternary EC based on previous biochemical and structural analyses (47,48) is shown on the top. Promoter-independent assembly of ECs is performed using a 9 nt RNA:DNA hybrid that occupies the RNAPII catalytic center. Ternary ECs are attached to streptavidin beads via the 5΄ biotin of the non-template strand allowing subsequent separation of bead-associated (B) and supernatant (S) fractions. The RNA is fluorescently labeled with FAM at the 5΄-end. The transcription template contains a G-less cassette followed by a G-stretch in the non-template strand. After adding an ATP, UTP, CTP mix, the RNAPII transcribes until it encounters the G-rich sequence. Sen1 provokes dissociation of ECs paused at the G-rich stretch and therefore the release of RNAPII and associated transcripts to the supernatant. (B) Denaturing PAGE analysis of RNAs from a representative IVTer assay in the absence and in the presence of Sen1 proteins. (C) Quantification of the fraction of transcripts released from ECs stalled at the G-stretch as a measure of the termination efficiency. Values represent the average and standard deviation of three independent experiments. The p-value associated with a t-test (p) is indicated.
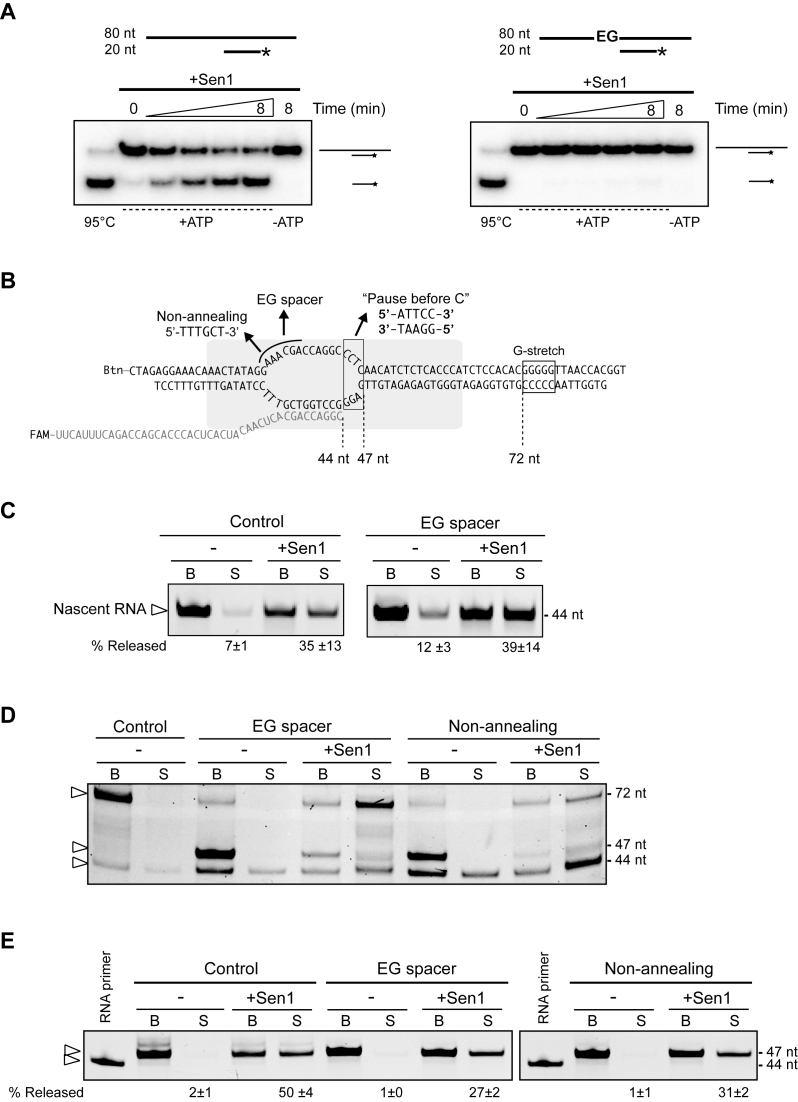
Figure 5.
Analysis of the role of the ssDNA in the transcription bubble in Sen1-dependent termination. (A) Unwinding assays showing that Sen1 cannot translocate through ethyleneglycol (EG) spacers. Top: Scheme of the substrates used for the unwinding assays. The duplex consists in the non-template strand from the IVTer assays annealed to a 20-nt DNA oligo, leaving a 34-nt single-strand overhang at the 5΄ end, either in the presence or in the absence of two hexa-ethyleneglycol spacers in tandem that replace 6 nt of the non-template strand. An asterisk denotes the radioactive label at the 5΄ end of the short DNA oligonucleotide. Bottom: Representative gels of unwinding assays. The experiments were performed twice, with the same results. (B) Schematic of the different ternary ECs used as termination substrates in the experiments shown in panels (C), (D) and (E). (C) IVTer assays comparing the efficiency of EC dissociation in the absence and in the presence of the EG. ECs were assembled on both types of templates using a 44-mer RNA, so that the nascent RNA is sufficiently long to support termination in the absence of transcription. Values represent the average and SD from two independent experiments. (D) IVTer assays performed in the presence of ATP, CTP and UTP to allow transcription up to the G-stretch. The ECs harboured either a normal complementary (control), an EG-containing or a non-annealing portion of non-template DNA. Sen1 can rescue stalled ECs but can only induce EC dissociation when transcription is prevented by the absence of an incoming nucleotide (i.e. at the G-stretch). The major RNA species detected are indicated by an arrowhead. (E) Experiments performed with ECs containing a different sequence at the downstream portion of the transcription bubble as indicated in panel (B) as ‘Pause before C’ templates. Assays were conducted in the presence of ATP and UTP to induce RNAPII pausing after addition of 3 nt. The 44-mer RNA primer used for the assembly of ECs was loaded next to the IVTer reactions. Values correspond to the release of the 47 nt RNA species (average and SD of two independent experiments).
Figure 6.
Analysis of the role of the nascent RNA in Sen1-mediated termination. (A) IVTer assays showing that a single-strand non-template DNA cannot replace the nascent RNA for termination. A schematic of the modified EC is shown on the top. The ECs were assembled using a shortened template strand so that the non-template strand upstream of the transcription bubble is single-stranded. A 15-bp region of upstream duplex DNA was maintained to avoid perturbations of the RNAPII-DNA interaction network that might lead to destabilization of the EC. (B) IVTer assays showing that substituting the nascent RNA by ssDNA increases the efficiency of termination. ECs were assembled using either a 44-mer RNA, as in Figure 5, or a chimeric DNA-RNA molecule in which the 16 nt at the 3΄end (embedded in RNAPII) are RNA and the additional 28 nt (exposed for Sen1 interaction) are DNA (indicated by a dotted line). The sequence of the DNA templates and the structure of the EC is the same as in Figure 2A. Values represent the average and SD from three independent experiments. The P-value for the comparison of the ssRNA and the ssDNA in termination is 0.03.
Duplex unwinding assay
The DNA:DNA and RNA:DNA substrates for the unwinding assays were formed by annealing a 5΄-end radiolabeled short DNA oligonucleotide to either the 5΄ or 3΄ end of a longer DNA or RNA oligonucleotide (Supplementary Table S1). The DNA oligonucleotides were purchased from Sigma and PAGE-purified before use. The RNA was obtained by in vitro transcription with the appropriate templates (see Supplementary Table S1) using the MEGAshortscript T7 kit (Ambion). Unwinding assays were performed in unwinding buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 7.5 μM ZnCl2, 0.5 mM DTT, 10% glycerol, 0.1 mg/ml BSA) in 20 μl reactions. Sen1 proteins were preincubated with the corresponding duplex substrate and the reaction was initiated by adding a mixture containing ATP and MgCl2 at 2 mM as the final concentrations, and an excess of unlabeled competitor oligonucleotide (0.1 μM) to trap the unwound unlabeled oligonucleotide. Aliquots were withdrawn at the indicated times and mixed with 1 volume of stop/loading buffer containing 50 mM EDTA, 1% SDS and 20% glycerol. The samples were separated by electrophoresis on a 20% native PAGE. Gels were directly exposed on phosphorimager screens overnight at -80°C and subsequently analysed using a Typhoon scanner.
Processivity assays were performed on a substrate composed of a 76-nt 5΄ radiolabeled DNA oligonucleotide annealed to two tandem shorter DNA oligos (20-nt and 19-nt long, respectively, see Supplementary Table S1). A 1.5-fold excess of the short oligonucleotides was used to ensure complete substrate formation. Briefly, the tandem duplex substrate (0.25 nM final concentration) was mixed with an excess of Sen1 (30 nM) in unwinding buffer and incubated for 10 min at 28°C. The reaction was initiated by adding a mixture containing ATP and MgCl2 (2 mM final concentrations), an excess of competitor oligonucleotides (25 nM final concentration) to trap the unwound unlabeled oligonucleotides and heparin (2.5 μg/ml final concentration) to trap free/released Sen1 molecules. Reaction aliquots were withdrawn at various times and mixed with 1 volume of stop/loading buffer. Samples were separated by electrophoresis on a native 8% PAGE. Gels were dried and analyzed by phosphorimaging as before.
RESULTS
The helicase domain of Sen1 is sufficient for transcription termination in vitro
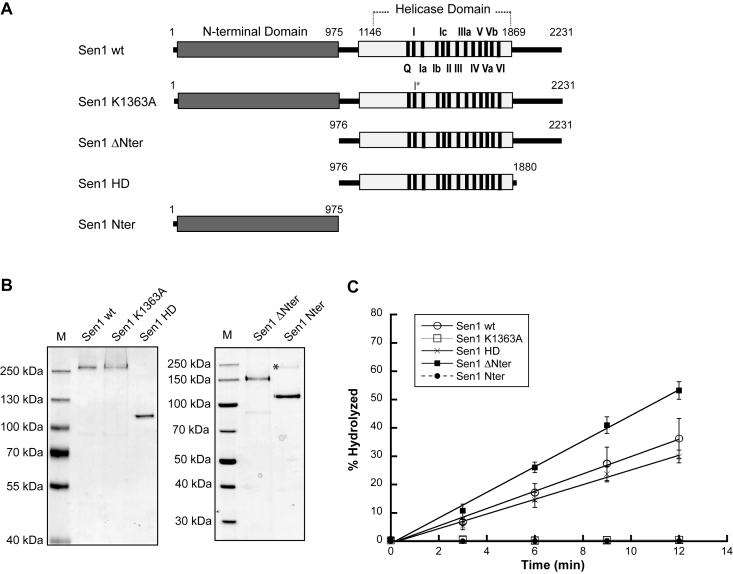
Sen1 contains three different structural modules: a conserved helicase domain and two adjacent regions with proposed roles in protein–protein interactions. In order to understand which domains of Sen1 are actually involved in the step of dissociation of the EC, we purified several truncated versions of Sen1 from yeast and we assessed their capacity to terminate transcription in vitro (Figures 1 and 2). As a control we produced a catalytic mutant (Sen1 K1363A) harboring a substitution in an essential lysine at the conserved motif I (also known as Walker A motif, Figure 1A). Because Sen1-mediated transcription termination strictly requires ATP hydrolysis both in vivo and in vitro (21,27), we first monitored the ATPase activity of the different Sen1 variants (Figure 1). As expected, the Sen1 K1363A mutant and the Sen1 N-terminal domain did not display any detectable ATPase activity. Deletion of the N-terminal domain alone or together with the C-terminal region did not significantly affect the efficiency of ATP hydrolysis, suggesting that these domains do not substantially contribute to or regulate Sen1 ATPase activity. We next analysed the capability of the different Sen1 versions to terminate transcription using a highly purified in vitro transcription–termination system containing solely purified RNAPII and Sen1 proteins (27 and Figure 2A). Briefly, in this system we assemble, in a promoter-independent manner, ternary ECs composed of RNAPII (12 subunits), the transcription templates and a short nascent RNA. ECs are immobilized on streptavidin beads via a biotin moiety present at the 5΄-end of the non-template strand. The transcription templates contain a G-less cassette followed by a G-stretch in the non-template strand so that upon addition of a nucleotide mix devoid of GTP, the RNAPII transcribes until the G-stretch and is retained in the beads. The capacity of the different Sen1 variants to elicit EC dissociation is inferred from the fraction of nascent RNA released into the supernatant. Consistent with the notion that transcription termination is ATP-dependent, the Sen1 K1363A mutant and the Sen1 N-terminal domain failed to terminate transcription in our IVTer assay (Figure 2B). Importantly, the helicase domain alone was sufficient to terminate transcription in vitro, indicating that this region retains all the activities and properties that are required for dissociation of the EC. Both Sen1 ΔNter and HD actually exhibited a somewhat more efficient termination activity, suggesting that the N-terminal region might play an inhibitory role in termination. This inhibition was only observed in cis, since addition of increasing concentrations of purified N-terminal domain did not affect the efficiency of termination by Sen1 ΔNter or HD in vitro (Supplementary Figure S1).
Figure 1.
Analysis of the ATPase activity of different Sen1 variants. (A) Schematic of the Sen1 proteins analyzed. The two regions predicted to form globular domains (the N-terminal and the helicase domains) are indicated by gray bars. A black line represents the regions that are predicted to be disordered. Highly conserved motifs in SF1 helicases are indicated. The K1363A mutant harbouring a mutation at the universally conserved motif I serves as control. (B) Coomassie-stained protein gel showing the Sen1 variants purified from yeast (M, molecular size marker). An asterisk denotes a fraction of Sen1 Nter that exhibits anomalous migration on a SDS-PAGE (as identified by MS). (C) Graphical representation of the ATP hydrolysed by the different Sen1 variants as a function of time. The values correspond to the average and standard deviation (SD) of three independent experiments.
Sen1 can translocate on both ssRNA and ssDNA in the 5΄ to 3΄ direction
Our previous observation that Sen1-dependent termination in vitro requires both the interaction of Sen1 with the nascent RNA and ATP hydrolysis (27) suggests that, akin to Rho-dependent termination in bacteria, disruption of the EC by Sen1 might require Sen1 translocation along the RNA. To investigate this possibility, we decided to analyze the capacity of Sen1 proteins to unwind different nucleic acid duplexes, which is an indirect way to monitor translocation. We tested both DNA:DNA and RNA:DNA duplexes containing a single-strand overhang at either the 5΄ or the 3΄ end (Figure 3). Similarly to its Schizosaccharomyces pombe ortholog, both full-length Sen1 and the helicase domain could dissociate DNA:DNA and RNA:DNA duplexes containing a 51–55 nt single-strand 5΄ overhang but were essentially inactive on duplexes with a 3΄ overhang, suggesting that these proteins can translocate on ssDNA or ssRNA in the 5΄-3΄ direction (Figure 3A and B). We tested several lengths of overhang and we found that a 5΄ overhang as short as 5 nt can actually support unwinding (although less efficiently, data not shown). However, the full-length protein and the helicase domain exhibited a somewhat different behaviour. Whereas the helicase domain was slightly more efficient for DNA:DNA duplex unwinding, it was significantly less active than the full-length protein for RNA:DNA duplex unwinding, suggesting that additional regions in the full-length protein might be important for optimal activity on RNA:DNA duplexes. The Sen1 ΔNter variant exhibited levels of RNA:DNA duplex unwinding activity that were similar to those of full-length Sen1, or even higher, suggesting that the C-terminal domain contains sequences that improve the activity on RNA:DNA duplexes (Supplementary Figure S2). Strikingly, both the full-length protein and the helicase domain worked much more efficiently on a DNA:DNA duplex than on an RNA:DNA duplex, since both the amplitude and the rate of the unwinding reaction were substantially higher for the former compared to the latter duplex (Figure 3C). Because we observed a similar affinity for the ssDNA and for the ssRNA or even a higher affinity for the RNA, (Supplementary Figure S3), these results suggest that Sen1 is significantly more processive on ssDNA than on ssRNA and/or very sensitive to the stability of the duplex (i.e. unwinding is less efficient for more stable duplexes). While this work was in progress, an independent study using the recombinant Sen1 helicase domain obtained similar results regarding the preference of Sen1 for ssDNA over ssRNA (32). The authors of that study performed complementary analyses and concluded that the stability of the duplex only partially accounts for the differences in unwinding efficiency, strongly suggesting that Sen1 is substantially more processive on ssDNA than on ssRNA.
Figure 3.
Nucleic acid duplex unwinding assays with full-length Sen1 (wt) and its helicase domain. (A) Representative gels showing ATP-dependent unwinding of DNA:DNA versus RNA:DNA duplexes containing a 5΄ single stranded overhang by Sen1 variants. The RNA molecule is shown in grey. An asterisk indicates the radioactive label at the 5΄ end of the short DNA molecule. The first lanes correspond to heat-denatured (95°C) samples and the last lanes are control reactions incubated with Sen1 proteins in the absence of ATP. DNA:DNA duplex unwinding reactions were performed with 0.5 nM of the substrate and 3 nM of Sen1 proteins whereas RNA:DNA duplex reactions contained 0.5 nM of the substrate and 30 nM of Sen1 proteins. Sen1 K1363A was used as a negative control. (B) Same assays as in (A) but using duplexes containing a 3΄ single stranded overhang. (C) Graphs showing the fraction of duplex unwound by Sen1 proteins as a function of time. Values represent the average and SD from three independent experiments. Data were fitted with Kaleidagraph to the Michaelis–Menten equation.
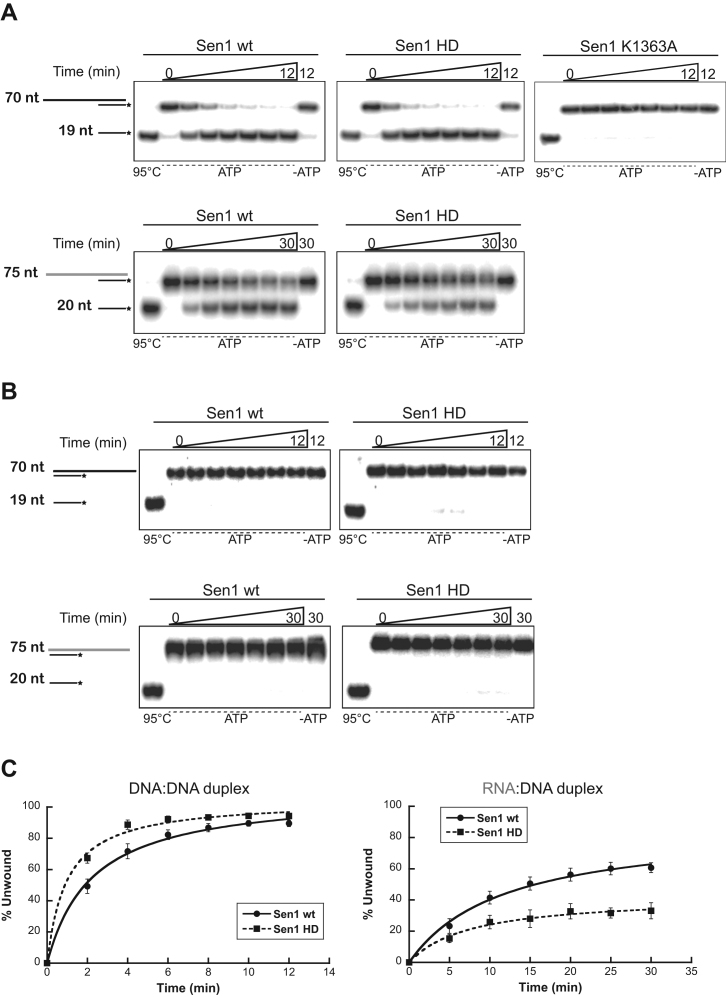
Sen1 is a relatively low-processivity helicase
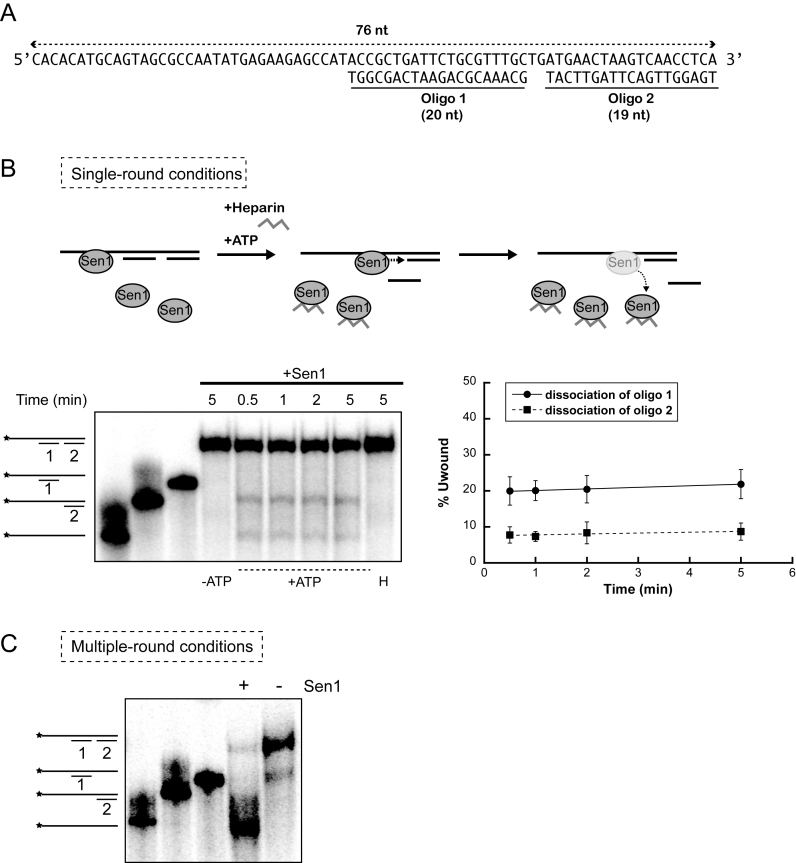
The former unwinding assays do not allow to directly evaluate Sen1 processivity, defined as the average number of nucleotides that Sen1 can translocate before dissociating from the nucleic acid substrate. The reasons are that: (i) these assays were performed in multiple-round conditions, such that the overall level of duplex dissociation might be the result of several proteins acting subsequently on the same substrate and (ii) a single duplex substrate does not allow to detect translocation distances shorter or longer than the length of this duplex. Because the processivity of translocation might be a critical parameter of the termination reaction mediated by Sen1, we set out to assess directly Sen1 processivity on nucleic acids. To this end, we performed single-round unwinding experiments using a substrate composed of a 76-nt ssDNA molecule annealed to two short oligos (20-nt and 19-nt) in tandem and containing a 35-nt single-strand overhang at the 5΄ end (Figure 4A). Because the second oligo can only be dissociated after translocation of Sen1 throughout the first duplex region, comparing the efficiency of release of the first and the second oligo allows the estimation of the processivity of translocation. In order to perform single-round assays, heparin was added together with ATP to initiate the unwinding reactions so that Sen1 reloading on the substrate was prevented. Heparin effectively prevented Sen1 reassociation with the duplex because when added before Sen1, the reaction was fully inhibited (Figure 4B, control ‘H’). We observed a dramatic decrease of the overall unwinding efficiency in single-round experiments compared to multiple-round assays performed with the same substrate in the absence of heparin (Figure 4C), suggesting that heparin might outcompete the binding of a fraction of Sen1 proteins to the substrate and/or that part of Sen1 molecules do not even translocate long enough to dissociate the first duplex. We found that the reaction is relatively fast, since already after 30 sec the maximal level of unwinding was reached, (for technical reasons we could not monitor the activity at times shorter than 30 s). The fact that the fraction of unwound duplex did not increase after 30 s further validates our conditions to reproduce single-round reactions. Strikingly, we observed a clear accumulation of the product of dissociation of the first oligo and only 30% of the substrates from which the first oligo was released were fully unwound by Sen1. This indicates that >70% of Sen1 molecules dissociate from the substrate DNA after unwinding the first duplex and before reaching the 3΄ end, which implies that Sen1 processivity is ≈ 20–40 nt. We attempted similar experiments using two tandem RNA:DNA duplexes but the resulting unwinding activity was too low to be accurately quantified (data not shown). These results indicate that Sen1 is a relatively poorly-processive translocase, which has important implications for the mechanisms of termination (see Discussion).
Figure 4.
Analysis of the processivity of Sen1 translocation on ssDNA. (A) Sequence and structure of the substrate used in these experiments. (B) Unwinding experiments performed in single-round conditions with Sen1 HD. An experiment performed with full-length Sen1 gave identical results (data not shown). Top: Schematic description of the assay. The presence of heparin in the reaction prevents re-association of Sen1 molecules that have dissociated from the substrate, thus precluding new rounds of unwinding. Bottom-left: Representative gels of unwinding experiments. An asterisk indicates the radioactive label at the 5΄ end of the long DNA molecule. The first three lanes show different nucleic acid molecules that could be detected in these experiments. –ATP, control reaction incubated with Sen1 HD in the absence of ATP. H, control reaction in which heparin was added before challenging the substrates with Sen1 HD. Bottom-right: Graphs showing the fraction of oligonucleotide dissociated by Sen1 HD as a function of time. Values represent the average and SD from three independent experiments. (C) Control multiple-round unwinding assay. The reaction proceeded for 5 min. In the absence of heparin, Sen1 can undertake multiple cycles of association-translocation-dissociation that allow carrying the unwinding reaction to completion (89% of the substrate fully unwound).
Termination does not require the interaction of Sen1 with the ssDNA in the transcription bubble
The striking preference of Sen1 for ssDNA over ssRNA led us to ask whether this property of Sen1 would be part of the mechanism of termination. Because the only portion of ssDNA that is present in our transcription system is the unwound DNA in the transcription bubble, we considered the possibility that, in addition to interacting with the nascent RNA, Sen1 recognizes the single stranded non-template DNA in the transcription bubble. Pulling the non-template DNA by virtue of its helicase activity might help Sen1 destabilize the EC. To explore this possibility, we set out to analyse whether Sen1 can dismantle an EC in which the non-template strand just upstream of stalled RNAPII has been substituted by a non-nucleic acid spacer. Specifically, we used two molecules of hexa-ethyleneglycol (EG) in tandem to replace the 6 nucleotides at the edge of the bubble (Figure 5). We first assessed whether the presence of the EG spacer could block the progression of Sen1 along the DNA by monitoring the capability of Sen1 to dissociate a duplex containing the EG spacer immediately upstream of the double strand region. We tested a duplex composed of the non-template strand containing the spacer annealed to a short complementary DNA oligonucleotide. An identical duplex without the spacer served as control and was efficiently unwound by Sen1 (Figure 5A). However, the presence of the EG spacer completely abolished unwinding, indicating that translocation was effectively prevented. We next assessed whether Sen1 could dissociate a stalled EC containing the EG as described above (Figure 5B–C). In order to place the EG at the desired position, we assembled ECs with a long RNA primer (44-mer) so that 28 nt of nascent transcript are exposed to interact with Sen1 and we assessed termination in the absence of transcription. As shown in Figure 5C, in these conditions Sen1 could induce dissociation of the EC, indicating that termination does not strictly require ongoing transcription. Importantly, the presence of the spacer in the transcription bubble did not significantly affect the efficiency of EC dismantling, strongly suggesting that Sen1 does not interact with or translocate on the non-template DNA to elicit termination.
Sen1 can promote forward translocation of stalled RNAPII
The presence of the EG spacer in the non-template strand might alter the catalytic properties of the EC that, under certain conditions, might respond differently to Sen1. We assessed whether ECs containing the EG spacer were catalytically competent, by monitoring the capacity of RNAPIIs to transcribe in the presence of nucleotides (Figure 5D). Interestingly, we observed that the EG-containing ECs stalled after addition of 3 nt. This was due to the inability of the upstream DNA to rewind, because substituting the same portion of non-template strand by a sequence non-complementary to the template produced the same effect. Strikingly, the addition of Sen1 helped these stalled RNAPIIs to resume elongation and transcribe up to the G-stretch where they stalled because of the absence of GTP. Sen1 required both the nascent RNA and ATP hydrolysis to promote elongation, because neither a short, non-exposed nascent RNA nor the addition of a non-hydrolyzable ATP analogue supported EC rescue (Supplementary Figure S4). This result indicates that, by acting on the nascent RNA, Sen1 can apply a mechanical force on the EC that promotes forward translocation of RNAPII. Importantly, because in these conditions Sen1 promotes elongation rather than dissociation of RNAPII from the templates, these data also suggest that termination requires a persistent paused state of RNAPII or a particular conformation that RNAPII does not adopt when it stalls due to the lack of upstream DNA rewinding. Finally, we performed additional experiments to assess whether Sen1 could induce termination instead of promoting elongation of the former stalled complexes if elongation were precluded. To this end, we assembled ECs with transcription templates containing a sequence that in the absence of cytidine provokes RNAPII stalling after transcribing 3 nt (Figure 5B). We then analysed the capacity of Sen1 to dismantle ECs harboring normal complementary, EG-containing or non-annealing non-template strand in the absence of cytidine (Figure 5E). Interestingly, termination occurred in these conditions, but with reduced efficiency in the presence of the EG spacer or a non-complementary non-template DNA. This result suggests that reannealing of the DNA at the upstream portion of the transcription bubble is necessary for fully efficient termination.
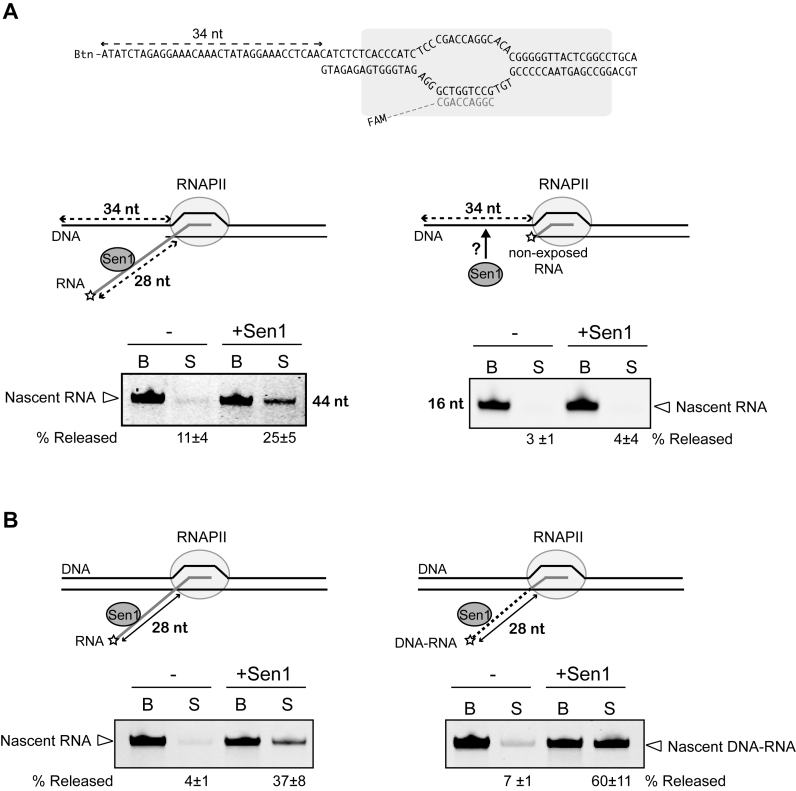
A long portion of ssDNA at either the non-template or the template strand cannot replace the nascent RNA in termination
The above results strongly suggest that the nascent RNA is the only nucleic acid molecule required for Sen1-mediated termination. We decided to explore in more detail the role of the nascent transcript in termination. The RNA could be a mere way to bring Sen1 in contact with specific surfaces of RNAPII. Alternatively, because Sen1 can promote forward translocation of stalled RNAPII, the RNA could support translocation of Sen1 towards the paused polymerase, which in particular conditions might ultimately prompt RNAPII forward movement without nucleotide addition and therefore EC destabilization. This would be analogous to the so-called ‘hypertranslocation’ model proposed for the bacterial termination factor Rho (for review, see 33). If Sen1 employs such a mechanism of termination, given that it can translocate along the ssDNA, it would be conceivable that a sufficiently long portion of ssDNA at the non-template strand upstream of RNAPII could support termination in the absence of exposed nascent RNA. In order to test this possibility, we performed IVTer assays under modified conditions in which most of the template strand upstream of RNAPII was omitted leaving 34 nt of single-strand non-template DNA exposed for the interaction with Sen1. As shown in Figure 6A, in the presence of a long nascent RNA Sen1 could elicit dissociation of the EC, although with somewhat reduced efficiency compared to previous assays under normal conditions. Importantly, in the absence of exposed RNA Sen1 failed to dismantle the EC, indicating that the ssDNA at the non-template strand cannot functionally replace the nascent transcript in the termination reaction. We also tested whether a 25 nt single-strand extension of the template strand downstream of the assembled EC could support termination by Sen1 (Supplementary Figure S5). Again, Sen1 could induce EC dissociation only in the presence of an exposed RNA, indicating that the single-strand template DNA even if it should support the translocation of Sen1 toward the RNAPII, cannot substitute the nascent RNA in termination.
Substitution of the extruded RNA by ssDNA increases the efficiency of termination
The above results indicate that Sen1-mediated termination specifically requires the interaction with the nascent RNA. We envisaged two possible explanations for this. Because the behaviour of Sen1 on DNA and on RNA is different (Figure 3), the first possibility is that it is the nature of the nucleic acid that matters (i.e. only the interaction with RNA promotes the termination activity of Sen1). The second possibility is that the position of the nucleic acid is crucial, for instance because it brings Sen1 to a specific region of RNAPII. In order to distinguish between these two possibilities we performed modified IVTer assays in which we compared the efficiency of termination on normal ECs and on complexes assembled with a chimeric DNA-RNA in which the nucleic acid portion that is exposed for interaction with Sen1 is ssDNA instead of ssRNA (Figure 6B). We found that Sen1 can elicit dissociation of an EC with emerging ssDNA, indicating that Sen1-dependent termination specifically requires the interaction with the nascent RNA due to its position relative to RNAPII as any single-strand nucleic acid at the place of the nascent RNA can support termination. Strikingly, we reproducibly observed substantially higher efficiency of termination on ECs harbouring the chimeric DNA–RNA relative to a normal RNA transcript. Because Sen1 seems more processive on ssDNA than on ssRNA, this result suggests that the translocation processivity is a limiting parameter in the termination reaction. Importantly, this strongly suggests that termination requires the translocation of Sen1 along the RNA.
DISCUSSION
Transcription termination by Sen1 plays several crucial roles in maintaining the integrity of the functional transcriptome, from preventing genome deregulation by uncontrolled pervasive transcription to regulating gene expression by premature termination. The high conservation of Sen1 across species together with evidence linking mutations in the human Sen1 orthologue to several neurological disorders argues for evolutionarily conserved functions of major biological relevance for Sen1 proteins. However, despite intense investigations on the function of Sen1 in vivo during the last few years, little is known about the Sen1 biochemical properties and precise mechanisms of action. The large size of Sen1 (252 kDa) makes its purification very challenging and our attempts to produce a recombinant version of full-length Sen1 using several overexpression systems have failed. Yet, we have succeeded in purifying functional Sen1 from its natural host, S. cerevisiae. In a previous work, we reconstituted Sen1-dependent transcription termination using a minimal in vitro system and thus unveiled some of the key features of Sen1 mechanisms of action. However, many relevant questions about the mechanisms of termination remain unanswered. In this study, we characterize the helicase activity of Sen1 and investigate its role in transcription termination in vitro. Our results lead us to propose a model according to which, akin to Rho-dependent termination in bacteria, translocation of Sen1 on the nascent RNA is a critical step in the termination reaction.
The functional architecture of Sen1
Helicases are enzymes that often have little substrate specificity and that require additional factors to specify their biological function and regulate their activity. In the case of Sen1, previous in vivo studies have assigned functions to both the N-terminal and the C-terminal domain in mediating the interaction with RNAPII and Nab3, respectively (7,18). Because Sen1 itself does not exhibit any clear sequence preference in RNA binding (27,34), these interactions might be critical for Sen1 recruitment to the EC and for loading of Sen1 on non-coding transcripts harbouring the characteristic motifs of NNS-dependent targets (i.e. Nrd1 and Nab3 recognition sequences). However, whether the N- and C-terminal domains of Sen1 participate in the step of EC dissociation or whether they play a direct role in modulating Sen1 activity have remained open questions.
In this study, we have performed a functional dissection of Sen1 to explore these aspects. Importantly, we have found that the helicase domain is sufficient for the step of dissociation of the EC, indicating that this region contains all the properties that are essential for the final step of termination. In a previous report, we showed that the capacity to dismantle an EC is not an unspecific property of any RNA helicase (27). Thus, it would be interesting to undertake structural analyses of Sen1 helicase domain to try to identify the determinants of Sen1 termination activity.
Our results therefore suggest that the N-terminal and C-terminal domains of Sen1 are implicated in earlier steps of the termination process that are only limiting in vivo, possibly related to the timely and specific recruitment of Sen1 to its targets as discussed above. In addition, we have found that in vitro, the presence of the N-terminal domain decreases the termination efficiency. This behaviour is reminiscent of that of the closest homologue of Sen1, the helicase Upf1, a key factor in the nonsense-mediated decay pathway for RNA quality control. In the case of Upf1, intra-molecular interactions mediated by N-terminal and C-terminal extensions of the helicase domain induce autorepression. The autoinhibition exerted by the N-terminal domain, also called CH domain, is relieved upon interaction with the Upf1 partner Upf2 (35,36). It remains to be tested whether the N-terminal domain of Sen1 also mediates inhibitory intra-molecular interactions. Alternatively, its mild inhibitory role might be due to its large size and its position towards the 3΄ end of the RNA, and therefore towards the RNAPII. It is possible that the N-terminal domain partially occludes regions of interaction with RNAPII that might be important for termination.
We have also observed that the presence of the C-terminal domain improves Sen1 helicase activity, especially on RNA:DNA duplexes. Helicases often possess additional nucleic-acid binding domains at the extensions of their core helicase domains (37). It is therefore possible that the C-terminal domain contains an additional RNA-binding domain, a hypothesis that we are currently exploring. Such possible RNA-binding activity could be particularly relevant for termination in vivo, where the presence of a multitude of competing RNA-binding proteins (e.g. RNP factors) might limit the access to the nascent RNA.
An intrinsically low-processivity helicase
In a previous report we did not succeed in detecting Sen1 duplex unwinding activity (27). Later, we discovered that minor contaminations of RNA in our protein preparations prevented Sen1 from exhibiting nucleic-acid binding and unwinding activity (unpublished observations). Thus, we have modified our purification protocol (see the Methods for details) and we have managed to observe that Sen1 can unwind both RNA:DNA and DNA:DNA duplexes in an ATP-dependent manner, but only in the presence of a 5΄ overhang, indicating that Sen1 can translocate along nucleic acids in the 5΄ to 3΄ direction (Figure 3). Surprisingly, our tests using tandem duplexes have revealed that Sen1 is a relatively low-processivity enzyme since more than half of Sen1 molecules dissociate before translocating ≈40 nt (Figure 4). This behaviour is in contrast with that of Upf1, which can translocate over more than 10 kb (38).
This feature of the Sen1 helicase activity could be important for its regulation. Given the specificity of Sen1 function, which contrasts with its low inherent capacity to discriminate between target and non-target RNAs, several mechanisms must exist to ensure timely termination at the appropriate substrates. On one hand, Sen1 protein levels are kept low (around 100 molecules/cell; 39). On the other hand, the interaction of Sen1 with Nrd1 and Nab3, which recognize specific motifs on ncRNAs, likely plays an important role in attracting Sen1 to the right targets. Low processivity might have been selected for Sen1 proteins throughout evolution to provide an additional layer of control as it leaves a relatively narrow window for the action of Sen1 and makes termination highly dependent on slowing down elongation or polymerase pausing, an event that might naturally occur or be induced by specific factors at termination regions.
Indeed, a previous in vivo study has revealed a kinetic competition between Sen1-mediated termination and RNAPII elongation as fast RNAPII mutants exhibit delayed termination (40). Our observation that in vitro Sen1 can only operate on paused polymerases (27; and Figure 5D) together with our results showing that Sen1 is an intrinsically low-processivity enzyme provide mechanistic data to better understand the former in vivo evidences. It remains possible that the processivity of translocation on unpaired RNA is significantly higher than on a duplex, yet in vivo the RNA is unlikely to exist devoid of secondary structures and bound proteins. A more direct assessment of translocation with higher resolution techniques (e.g. single-molecule systems) in the absence and in the presence of a barrier (a nucleic acid duplex or RNA-bound proteins) would be required to fully understand what is the contribution of Sen1 translocation rate to the kinetics of termination and to estimate the impact of competing processes such as elongation and RNP assembly.
Our results demonstrating the relatively low processivity of Sen1 on RNA are also relevant in the light of a previously-proposed role for Sen1 and senataxin in removing R-loops that form during transcription when the nascent RNA hybridizes with the DNA template (22,25,41). In humans R-loops that form in vivo are very long (even more than 1 kb in humans, see 42). Sen1, given its low processivity, could not in principle unwind such long structures. However, it is possible that in yeast R-loops are much shorter. Alternatively, the accumulation of R-loops observed upon Sen1 mutation could be an indirect consequence of impaired transcription termination. Finally, an interesting possibility is that in vivo the processivity of Sen1 is significantly enhanced by the interaction with other factors, for instance with the Sen1 partners Nrd1 and Nab3.
The mechanism of transcription termination
One of the most relevant but also most elusive questions in the field is what is the precise mechanism of termination by Sen1. Our previous results showing that ATP-dependent termination by Sen1 requires its interaction with the nascent RNA together with in vivo evidence of a kinetic competition between termination and transcription elongation suggested a mechanism of termination involving Sen1 translocation on the nascent RNA (27,40). However, a formal proof for this model has been missing. Here, using classical duplex unwinding assays, we have proved that Sen1 can translocate on single stranded nucleic acids. Our observation that Sen1 is at least one order of magnitude more efficient on DNA:DNA than on RNA:DNA duplexes together with data from another group obtained with a recombinant helicase domain strongly suggest that Sen1 is more processive on ssDNA than on ssRNA.
The prominent preference of Sen1 for ssDNA over ssRNA led us to ask whether Sen1 might need to interact with the ssDNA in the transcription bubble to elicit termination. However, the results of our IVTer assays with modified transcription templates strongly suggest that this is not the case (Figure 5). Nevertheless, we have taken advantage of the different behaviour of Sen1 on ssDNA and ssRNA to obtain further insight into the mechanism of termination. We have shown that the substitution of the extruded nascent RNA by ssDNA increases the efficiency of termination in vitro (Figure 6B). Because the affinity of Sen1 for the ssDNA and the ssRNA is similar (Supplementary Figure S3, note that we have measured the affinity for the same sequences used in the experiments in Figure 6B), we exclude the possibility that this enhanced termination is the result of better recruitment to the EC. Instead, because Sen1 seems more processive on ssDNA than ssRNA, the enhancement of termination obtained with ssDNA is most likely due to increased processivity, supporting the idea that Sen1-mediated termination requires Sen1 translocation on the nascent RNA (Figure 7).
Figure 7.
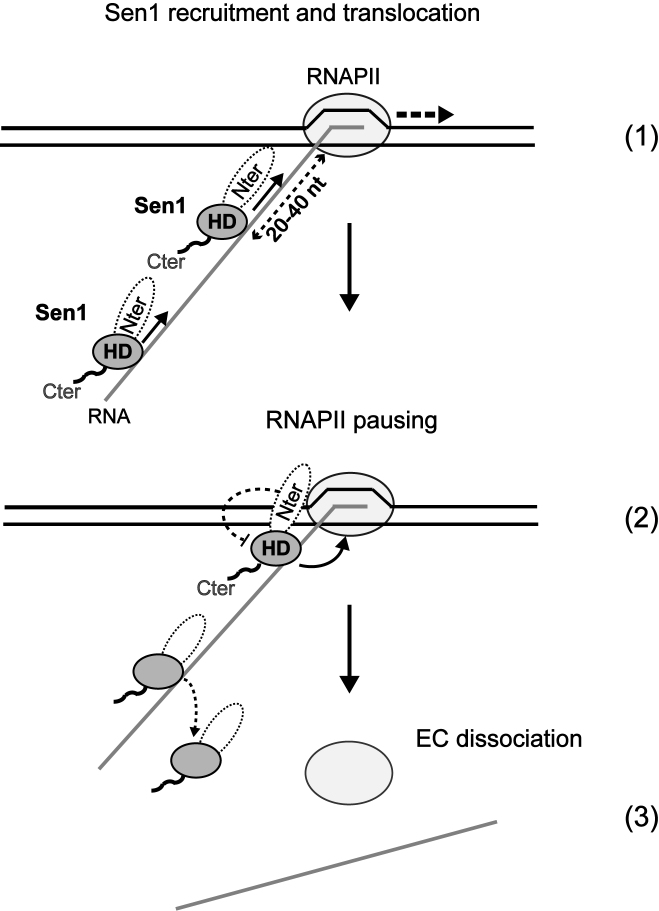
Model for the mechanisms of transcription termination by Sen1. After recruitment to the EC by the other NNS components, Nrd1 and Nab3, and/or the direct interaction with RNAPII, Sen1 must be loaded onto the nascent RNA in close proximity to the RNAPII. Sen1 molecules loaded more than 40 nt upstream of the RNAPII would dissociate before accomplishing termination. Sen1 translocation along the RNA allows dismantling of the EC in a reaction that requires exclusively the action of the helicase domain of Sen1 and likely involves ‘hypertranslocation’ of RNAPII. Termination strictly requires RNAPII pausing. In the absence of additional factors, the Sen1 N-terminal domain partially inhibits the transcription termination activity of the helicase domain.
The efficient activity of Sen1 on ssDNA remains striking and might be relevant for its previously proposed roles in DNA repair and replication. Indeed, the N-terminal domain of Sen1 has been shown to mediate the interaction with the nucleotide excision repair (NER) factor Rad2 (18) and to be critical for efficient transcription coupled repair (TCR), an NER subpathway (43). Our data showing that Sen1 can promote forward translocation of stalled RNAPIIs in vitro (Figure 5D) suggest that, similar to the bacterial transcription-repair coupling factor Mfd (44), in vivo Sen1 might facilitate DNA repair by displacing the lesion-stalled polymerases and promoting the recruitment of the NER machinery. In addition, given the efficient DNA:DNA unwinding activity of Sen1, it is tempting to speculate an additional role for Sen1 in subsequent steps of the DNA repair process involving the exposure of ssDNA regions. Furthermore, a previous report suggested that Sen1 associates with replication forks and promotes their progression through highly transcribed regions (45). The DNA helicase activity of Sen1 might be important for this role in replication.
The behavior of Sen1 is reminiscent of the bacterial terminator factor Rho. Three alternative models have been proposed to explain the mechanism of termination by Rho. The ‘hypertranslocation’ model posits that the powerful helicase activity of Rho would exert a pushing force on the RNAP that would force it to translocate without transcription and this would ultimately provoke EC dissociation. The ‘hybrid shearing’ model postulates that Rho would rather disrupt the hybrid in the catalytic centre by dragging the nascent RNA out of the EC with a similar destabilizing effect as in the previous model. Finally, the allosteric model proposes that the simultaneous interaction of Rho with the RNAP and the nascent transcript allows Rho to induce a conformational change in RNAP that ultimately leads to EC disassembly (33 and references therein).
Our finding that Sen1 can apply a mechanical force on a stalled EC allowing the RNAPII to resume elongation would support a ‘hypertranslocation’ model (Figure 5D). In addition, we have observed that disrupting the base-pairing of the duplex region immediately upstream of the transcription bubble decreases the efficiency of termination (Figure 5E). This suggests that bubble rewinding provides additional energy that assists EC dissociation by Sen1, as previously shown for its bacterial counterpart Rho (46). Yet, given the extensive interactions of RNAPII with the upstream duplex DNA in the EC (47), we cannot exclude the possibility that the modifications introduced in the non-template DNA alter the conformation of the EC making it a less favourable substrate for termination. Finally, additional evidence supports a more complex model for termination with not only mechanical but also allosteric components. Indeed, in our previous study we showed that Upf1 could not terminate RNAPII transcription in our in vitro assay, indicating that a processive RNA translocase activity is not sufficient for termination. In addition, we found that Sen1 cannot terminate transcription by the bacterial RNAP, suggesting that specific recognition of RNAPII might be required for termination (27). Understanding whether Sen1 interacts with specific regions of RNAPII to elicit termination and the nature of the molecular transitions leading to EC dissociation remain a major challenge and is the focus of our future research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karine dos Santos for her attempts in producing recombinant Sen1 and for helpful discussions. We thank the proteomic platform of Institut Jacques Monod for technical assistance. We thank all other members of the Libri lab for their input and Marc Boudvillain, Terence Strick, Anne-Lise Haenni and Bronislava Leonaite for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Agence National pour la Recherche [ANR-08-Blan-0038-01 and ANR-12-BSV8-0014-01 to D.L.]; Fondation pour la Recherche Medicale (programme Equipes 2013 to D.L.); PhD fellowship from the China Scholarship Council (to Z.H.). Funding for open access charge: Agence National pour la Recherche [ANR-12-BSV8-0014-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jensen T.H., Jacquier A., Libri D.. Dealing with pervasive transcription. Mol. Cell. 2013; 52:473–484. [DOI] [PubMed] [Google Scholar]
- 2.Arigo J.T., Eyler D.E., Carroll K.L., Corden J.L.. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006; 23:841–851. [DOI] [PubMed] [Google Scholar]
- 3.Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D.. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006; 23:853–864. [DOI] [PubMed] [Google Scholar]
- 4.Schulz D., Schwalb B., Kiesel A., Baejen C., Torkler P., Gagneur J., Soeding J., Cramer P.. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 2013; 155:1075–1087. [DOI] [PubMed] [Google Scholar]
- 5.Steinmetz E.J., Conrad N.K., Brow D.A., Corden J.L.. RNA-binding protein Nrd1 directs poly (A)-independent 3΄-end formation of RNA polymerase II transcripts. Nature. 2001; 413:327–331. [DOI] [PubMed] [Google Scholar]
- 6.Porrua O., Libri D.. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015; 16:190–202. [DOI] [PubMed] [Google Scholar]
- 7.Nedea E., Nalbant D., Xia D., Theoharis N.T., Suter B., Richardson C.J., Tatchell K., Kislinger T., Greenblatt J.F., Nagy P.L.. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008; 29:577–587. [DOI] [PubMed] [Google Scholar]
- 8.Vasiljeva L., Buratowski S.. Nrd1 interacts with the nuclear exosome for 3΄ processing of RNA polymerase II transcripts. Mol. Cell. 2006; 21:239–248. [DOI] [PubMed] [Google Scholar]
- 9.Hobor F., Pergoli R., Kubicek K., Hrossova D., Bacikova V., Zimmermann M., Pasulka J., Hofr C., Vanacova S., Stefl R.. Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J. Biol. Chem. 2011; 286:3645–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunde B.M., Hörner M., Meinhart A.. Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM. Nucleic Acids Res. 2011; 39:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porrua O., Hobor F., Boulay J., Kubicek K., D'Aubenton-Carafa Y., Gudipati R.K., Stefl R., Libri D.. In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. EMBO J. 2012; 31:3935–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wlotzka W., Kudla G., Granneman S., Tollervey D.. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011; 30:1790–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubicek K., Cerna H., Holub P., Pasulka J., Hrossova D., Loehr F., Hofr C., Vanacova S., Stefl R.. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012; 26:1891–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A.. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008; 15:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinmetz E.J., Ng S.B.H., Cloute J.P., Brow D.A.. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 2006; 26:2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tudek A., Porrua O., Kabzinski T., Lidschreiber M., Kubicek K., Fortova A., Lacroute F., Vanacova S., Cramer P., Stefl R. et al. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol. Cell. 2014; 55:467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudipati R.K., Villa T., Boulay J., Libri D.. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008; 15:786–794. [DOI] [PubMed] [Google Scholar]
- 18.Ursic D., Chinchilla K., Finkel J.S., Culbertson M.R.. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004; 32:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Müller U., Sundling K.E., Brow D.A.. Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human Senataxin that elicit cerebellar ataxia. Genetics. 2014; 198:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkel J.S., Chinchilla K., Ursic D., Culbertson M.R.. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics. 2010; 184:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmetz E.J., Warren C.L., Kuehner J.N., Panbehi B., Ansari A.Z., Brow D.A.. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006; 24:735–746. [DOI] [PubMed] [Google Scholar]
- 22.Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suraweera A., Lim Y., Woods R., Birrell G.W., Nasim T., Becherel O.J., Lavin M.F.. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 2009; 18:3384–3396. [DOI] [PubMed] [Google Scholar]
- 24.Wagschal A., Rousset E., Basavarajaiah P., Contreras X., Harwig A., Laurent-Chabalier S., Nakamura M., Chen X., Zhang K., Meziane O. et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012; 150:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao D.Y., Gish G., Braunschweig U., Li Y., Ni Z., Schmitges F.W., Zhong G., Liu K., Li W., Moffat J. et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016; 529:48–53. [DOI] [PubMed] [Google Scholar]
- 26.Bennett C.L., La Spada A.R.. Unwinding the role of senataxin in neurodegeneration. Discov. Med. 2015; 19:127–136. [PubMed] [Google Scholar]
- 27.Porrua O., Libri D.. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 2013; 20:884–891. [DOI] [PubMed] [Google Scholar]
- 28.Longtine M.S., McKenzie A. 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast Chichester Engl. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 29.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B.. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999; 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- 30.Kireeva M.L., Lubkowska L., Komissarova N., Kashlev M.. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 2003; 370:138–155. [DOI] [PubMed] [Google Scholar]
- 31.Porrua O., Libri D.. Characterization of the mechanisms of transcription termination by the helicase Sen1. Methods Mol. Biol. Clifton NJ. 2015; 1259:313–331. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Tumasz S., Brow D.A.. Saccharomyces cerevisiae Sen1 Helicase Domain Exhibits 5΄- to 3΄-Helicase Activity with a Preference for Translocation on DNA Rather than RNA. J. Biol. Chem. 2015; 290:22880–22889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters J.M., Vangeloff A.D., Landick R.. Bacterial transcription terminators: the RNA 3΄-end chronicles. J. Mol. Biol. 2011; 412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creamer T.J., Darby M.M., Jamonnak N., Schaughency P., Hao H., Wheelan S.J., Corden J.L.. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly (A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011; 7:e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarti S., Jayachandran U., Bonneau F., Fiorini F., Basquin C., Domcke S., Le Hir H., Conti E.. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011; 41:693–703. [DOI] [PubMed] [Google Scholar]
- 36.Fiorini F., Boudvillain M., Le Hir H.. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2013; 41:2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiorini F., Bagchi D., Le Hir H., Croquette V.. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015; 6:7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong Y.T., Koh J.L.Y., Friesen H., Duffy S.K., Duffy K., Cox M.J., Moses A., Moffat J., Boone C., Andrews B.J.. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell. 2015; 161:1413–1424. [DOI] [PubMed] [Google Scholar]
- 40.Hazelbaker D.Z., Marquardt S., Wlotzka W., Buratowski S.. Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Mol. Cell. 2012; 49:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mischo H.E., Gomez-Gonzalez B., Grzechnik P., Rondon A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J.. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011; 41:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu K., Chedin F., Hsieh C.-L., Wilson T.E., Lieber M.R.. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 43.Li W., Selvam K., Rahman S.A., Li S.. Sen1, the yeast homolog of human senataxin, plays a more direct role than Rad26 in transcription coupled DNA repair. Nucleic Acids Res. 2016; 44:6794–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howan K., Smith A.J., Westblade L.F., Joly N., Grange W., Zorman S., Darst S.A., Savery N.J., Strick T.R.. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012; 490:431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alzu A., Bermejo R., Begnis M., Lucca C., Piccini D., Carotenuto W., Saponaro M., Brambati A., Cocito A., Foiani M. et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012; 151:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J.-S., Roberts J.W.. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes C.O., Calero M., Malik I., Graham B.W., Spahr H., Lin G., Cohen A.E., Brown I.S., Zhang Q., Pullara F. et al. Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol. Cell. 2015; 59:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kireeva M.L., Komissarova N., Waugh D.S., Kashlev M.. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000; 275:6530–6536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.