Abstract
The chief function of the Cpx two-component system is perceiving various cell envelope stresses, but CpxR is also known to regulate the expression of the type III secretion system (TTSS) of Shigella sonnei through transcription of the primary regulator virF. Here, we have isolated novel cpxA mutants that exhibited decreased TTSS expression from Escherichia coli HW1273, which carries the virulence plasmid of S. sonnei. The cpxA deletion strain of HW1273 expressed β-galactosidase activity levels from the virF-lacZ fusion similar to those of HW1273. However, the second regulator InvE (VirB) and the TTSS component IpaB proteins were apparently expressed at a low level. In the cpxA strain, β-galactosidase activity levels from the invE-lacZ transcriptional fusion remained similar to those of HW1273, whereas the β-galactosidase activity level from the translational fusion of invE-lacZ was reduced to 21% of that of HW1273. Therefore, the deletion of the cpxA gene influenced TTSS expression chiefly at the posttranscriptional processing of InvE. In addition, the cpxA deletion strain of S. sonnei showed the same phenotype. These results indicate that the Cpx two-component system is involved in virulence expression through posttranscriptional processing of the regulatory protein InvE, a novel feature of the Cpx two-component system in posttranscriptional processing and virulence expression of Shigella.
Shigella is a pathogen causing bacillary dysentery. Understanding the virulence expression and the development of drugs and vaccines are very important for treatment and control of the disease. Shigella invades epithelial cells lining the intestinal tract, propagates within the epithelium, and provokes inflammation (44, 45, 49). Invasion into the epithelium depends on the type III secretion system (TTSS), which is encoded by the mxi-spa and ipa genes on a virulence plasmid (25, 34, 35, 46). The TTSS of Shigella spp. forms a needle-like structure through which effector molecules are exported into the intracellular milieu of the host cells for bacterial invasion into cells (39).
The expression of TTSS is tightly regulated by the VirF and InvE (VirB) proteins encoded by a virulence plasmid (41, 42, 43) in response to external conditions such as temperature and osmolarity (13, 17, 24, 30). An AraC-type transcriptional regulator VirF, activates transcription of the invE (virB) gene (1, 19, 47). The second regulatory protein InvE activates the transcription of the mxi-spa and ipa genes by replacement of the repressible factor H-NS, a histone-like DNA binding protein (4).
Our previous studies have focused on the pH response of VirF expression. Nakayama and Watanabe isolated an Escherichia coli mutant that exhibited an upregulated β-galactosidase activity from the virF-lacZ fusion plasmid at pH 6.0, with substantial activity at pH 7.4. This mutant had a Tn10 insertion in the cpxA gene, a sensor of the Cpx two-component system (27). Subsequently, a cognate regulator, CpxR (12), was identified as an essential factor for virF expression. Purified CpxR protein binds to the virF promoter, and transcription of virF is activated in vitro by phosphorylated CpxR (28). CpxR has been identified as an important regulator of virulence expression in several gram-negative bacteria, including Legionella pneumophila, an opportunistic pathogen causing respiratory infection (14).
The Cpx two-component system perceives cell envelope stresses such as accumulation of periplasmic proteins (38) and alkaline pH (6). The N-terminal region of CpxA forms a loop structure in the periplasmic space, while the C-terminal region forms kinase and phosphatase domains in the cytosol (31, 48). Various stresses induce phosphorylation of the cognate regulator CpxR by the kinase domain of CpxA (32). Phosphorylated CpxR activates the transcription of various stress response genes such as degP, coding for a periplasmic protease (8), and dsbA, encoding an enzyme that catalyzes disulfide bond formation for maintenance of periplasmic protein folding under stress conditions (7).
In an attempt to isolate the chromosomal factors affecting TTSS expression, we again isolated a cpxA mutant from a screening that selected for decreased expression of the mxi and ipa genes in an E. coli strain that carries the virulence plasmid of Shigella sonnei. The mini-Tn10 insertions were located closer to the 5′ end of the cpxA coding sequence than that of the previously isolated mutant cpxA145::Tn10 (27). In addition, independently constructed cpxA deletions of E. coli and S. sonnei also exhibited decreased TTSS expression. Characterization for deletion of cpxA revealed a new pathway that chiefly contributed to the translational processing of InvE expression. This result indicated novel features of the Cpx two-component system in TTSS expression through posttranscriptional processing.
MATERIALS AND METHODS
Media, reagents, and bacterial strains.
Luria-Bertani (LB) medium (LB Lenox; Difco) and LB plates (containing 1.5% agar) were used for bacterial growth. Ampicillin (50 μg/ml), chloramphenicol (12.5 μg/ml), kanamycin (25 μg/ml), tetracycline (5 μg/ml), rifampin (200 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 100 μg/ml) were added as indicated. All primers were synthesized by Hokkaido System Science. Chemicals were purchased from Sigma. HW1273 is E. coli K-12 strain CC118 (23) carrying the S. sonnei virulence plasmid pSS120::Tn1, which was derived from conjugation of the virulence plasmid of S. sonnei HW383 (19). MS390 is a tetracycline-sensitive derivative of HW383 isolated by fusaric acid selection (22). E. coli S17-1 λ-pir was used for mini-Tn10 mutagenesis (2). The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli K-12 | ||
| CC118 | Δ(ara-leu) araD139 lacX74 galE galK phoA20 rpsE thi-1 argE rpoB recA | 23 |
| HW1273 | CC118 carrying pSS120::Tn1 (Apr Rifr) | 19 |
| ME2523 | HW1273 cpxA63::mini-Tn10 | This study |
| ME2526 | HW1273 cpxA22::mini-Tn10 | This study |
| ME2824 | HW1273 ΔcpxA | This study |
| ME2830 | HW1273 ΔcpxR | This study |
| S17-1 λ-pir | hsdR-M+thi pro recA RP4:2-Tc:Mu:Km:Tn7 λ-pir (Tpr Smr) | 10 |
| S. sonnei | ||
| HW383 | S. sonnei wild-type strain (Tcr) | 45 |
| MS390 | HW383 (Tcs) | This study |
| MS2824 | MS390 ΔcpxA | This study |
| MS2830 | MS390 ΔcpxR | This study |
| Plasmids | ||
| pBSL199 | Mini-Tn10 (Tcr) on R6K suicide vector | 2 |
| pEA225 | GST-InvE fusion on pGEX51 | This study |
| pGEMT-invE | PCR fragment amplified with invE4-invE5 primers on pGEMT-Easy vector | This study |
| pGEMT-virF | PCR fragment amplified with virF1-virF2 primers on pGEMT-Easy vector | This study |
| pHW848 | virF-lacZ fusion on pHSG595 (Cmr) | 27 |
| pSN0329 | E. coli cpxA on pACYC177 (Apr) | 27 |
| pJM0329 | pSN0329 Ω(bla::Tn7npt) (Kmr) | This study |
| pJM1717 | mxiC::Tn3-lac, ipaB-phoA fusion on pTH18kr (Kmr) | This study |
| pJM1718 | mxiC::Tn3-lac, ipaB-phoA fusion on pTH18cr (Cmr) | This study |
| pJM4320 | invE-lacZYA transcriptional fusion on pTH18rrnBT (Cmr) | This study |
| pJM4321 | invE-lacZYA translational fusion on pTH18rrnBT (Cmr) | This study |
| pTH18rrnBT | pTH18cs5 carrying PCR-amplified rrnBT1T2 sequence (Cmr) | This study |
| pTH18cs5 | pSC101 ori, single-copy plasmid (Cmr) | 16 |
Plasmids.
mxiC::Tn3-lac ipaB-phoA plasmid pJM1717 was constructed from pHB74 mxiC::Tn3-lac fusion in our collection (47). A SalI-BamHI fragment containing ipaB≈mxiC::Tn3-lac of pHB74 was cloned into the SalI and BglII sites of pTH18kr-phoA, which carries the PCR-amplified phoA gene from pCB267 (36) at the HindIII and KpnI sites of pTH18kr (16), resulting in pJM1717. Plasmid pJM1718 is a chloramphenicol-resistant version of pJM1717, constructed with the use of pTH18cr (16). The cpxA-encoding plasmid pJM0329 is a kanamycin-resistant version of pSN0329 (27) that carries the npt gene cassette (which confers kanamycin resistance) in the same direction as the β-lactamase gene on pSN0329, inserted by purified Tn7 transposase in vitro (GPS-LS kit; New England Biolabs).
Both transcriptional (pJM4320) and translational (pJM4321) fusions of invE-lacZ plasmids were constructed as follows. The PCR-amplified rrnBT1T2 terminator sequence from pTrc99A (Amersham-Pharmacia) was cloned into the HindIII and PstI sites on single-copy plasmid pTH18cs5 (16) to generate pTH18rrnBT. For transcriptional fusion plasmid pJM4320, the PCR-amplified invE promoter sequence containing bp −260 to −33 (which is the distance upstream from the translation initiation codon ATG; nucleotide A of the start codon is designated +1) was ligated into the BglII and HindIII sites upstream of the Shine-Dalgarno (SD) sequence on promoter cloning vector pCB182 (36) to generate pCB182-invE. The BamHI-EcoRI fragment of pCB182-invE, containing both the invE promoter and the SD sequence, was ligated into the BamHI and EcoRI sites of pFR109 (37) in frame with its lacZYA gene, resulting in pFR109-invETx (transcriptional fusion). The SalI fragment of pFR109-invETx containing invE-lacZYA was recloned into the SalI site downstream of the rrnBT1T2 termination sequence of pTH18rrnBT to generate pJM4320. As a result, the SD region (italic) of pCB182, 5′-AAGCTTCTAGCTAGAGGGTATTAATAATGA-3′, was substituted for that of the invE gene, 5′-AATATAAATCAATATGAATAAACAGGGTGTGATATGG-3′ (the ATG nucleotides of the translation initiation codon are shown in bold).
For the construction of translational fusion plasmid pJM4321, a BglII-BamHI fragment containing the native SD sequence of the invE gene from pEA180 (47) from bp −260 to +118, with nucleotide A of the ATG translation initiation codon designated +1, was ligated into the BamHI site of pFR109 in frame with the lacZYA gene, resulting in pFR109-invETl (translational fusion). A SalI fragment of pFR109-invETl containing invE-lacZYA was recloned into the SalI site downstream of the rrnBT1T2 termination sequence of pTH18rrnBT to generate pJM4321. Since strain HW1273 carrying pJM4320 or pJM4321 showed decreased β-galactosidase activity at 30 or 37°C in LB medium without salt, these constructs retained intact responses to low temperature and low osmolarity (24, 30).
Plasmid pGEMT-Easy was used for construction of the pGEMT-virF and pGEMT-invE vectors after PCR amplification of the virF and invE genes, which generated the RNA probes for the virF and invE genes in Northern blotting. For construction of pEA225, the BamHI fragment of invE from pEA111 (47) was cloned into the BamHI site of pGEX51 (Amersham) in frame with the glutathione S-transferase (GST)-encoding sequence. The integrity of the promoters and the junctions of the above plasmids were checked by DNA sequencing.
The primer pairs for plasmid constructions are as follows. For the construction of pTH18kr-phoA, primers phoA1 (5′-GCTAGAAGCTTCTAGAGATCTTCC-3′) and phoA2 (5′-GGGGTACCACCATCAACACCAGAC-3′) were used; for pTH18rrnBT, rrnBT1 (5′-ATGCAAGCTTGGCTGTTTTGGCG-3′) and rrnBT2 (5′-TTTTCTGCAGGATGGCCTTCTGC-3′); for pJM4320, invE7 (5′-GAAGATCTTATTTCTGTAGTC-3′) and invE8 (5′-CCCAAGCTTGACAAAAGTTAAATGC-3′); for pGEMT-virF, virF3 (5′-CGGAATCCTAAATATAGTTTGGTATATTCTG-3′) and virF4 (5′-ACGCGTCGACCATGGTTAAAATTTTTTATGATATAAG-3′); and for pGEMT-invE, invE5 (5′-CCATGGTGGATTTGTGCAACGACTTG-3′) and invE6 (5′-GTCGACTTATGAAGACGATAGATGGCG-3′).
Isolation of mutants.
Mini-Tn10 was randomly transposed into HW1273 after transconjugation of the mini-Tn10 suicide vector pBSL199 from E. coli strain S17-1 λ-pir as previously described (10). Mini-Tn10 transconjugants grown on LB with ampicillin, tetracycline, and rifampin were transformed with pJM1717. A total of 2 × 104 transformants carrying pJM1717 were spread on LB with tetracycline, ampicillin, kanamycin, and X-Gal, and the mutants that lost β-galactosidase activity from pJM1717 were selected after overnight incubation at 37°C. A total of 20 mutants which appeared to have lost mxiC expression were examined for InvE expression by Western blotting with anti-InvE antiserum. The InvE-reduced mutants were sequenced for the region of mini-Tn10 insertion as follows. The chromosomal DNA was purified with the DNeasy tissue kit (Qiagen), digested with HindIII, and ligated into the HindIII site of pTH18kr. Plasmids carrying the chromosomal DNA with the mini-Tn10 insertion were isolated on LB with tetracycline and kanamycin after transformation of E. coli DH5α and sequenced with an ABI Prism 310 genetic analyzer (Perkin-Elmer) with a Tn10 right-arm primer (5′-ATCATATGACAAGATGTGTATCCACC-3′). The two mutants carried mini-Tn10 insertions at different sites in cpxA. The remaining two mutants had insertions in irrelevant genes which were not found to be involved in TTSS expression, as also determined following construction of their gene deletions.
Construction of cpxA deletion mutant.
For construction of the cpxA and cpxR deletions, a PCR-based gene disruption technique was applied for both E. coli HW1273 and S. sonnei MS390 as described previously (9, 40). For deletion of the cpxA gene of E. coli (ME2824) and S. sonnei (MS2824), the cpxA sequence (extending from the codons for the 18th amino acid residue to the C-terminal end of the CpxA protein) was deleted with a pair of primers, cpxA1 and cpxA2. For deletion of the cpxR gene of E. coli (ME2830) and S. sonnei (MS2830), the cpxR gene (corresponding to the sequence extending from the 17th to the 157th amino acid residues of the CpxR protein) was deleted with a pair of primers, cpxR1 and cpxR2. After construction, the presence of the virulence plasmid that encodes form I antigen was confirmed by agglutination with diagnostic antiserum for S. sonnei (Denka Seiken), and the presence of the invE gene was confirmed by PCR amplification with a pair of primers, invE5 and invE6.
The junctions of deletions were checked by sequencing of PCR products with a pair of primers, cpxA3 and cpxA4 or cpxR3 and cpxR4. At least two independent deletions were constructed for each deletion strain, which showed the same results for all experiments. The primers used are as follows: cpxA1, 5′-TAGGCAGCTTAACCGCGCGCATCTTCGCCATCTTCTGGCTGACGCTGGCGTGTAGGCTGGAGCTGCTTCG-3′; cpxA2, 5′-ATGCCGGATGCGGCGTAAACGCCTTATCCTGCCTGCAAATGCGAAGTTTAATTCCGGGGATCCGTCGACC-3′; cpxA3, 5′-TAAAGATGGTCACCCGTGGTTTAAAACC-3′; cpxA4, 5′-AGCGCCGACTGAGCGGCAAGATCG-3′; cpxR1, 5′-ATGAATAAAATCCTGTTAGTTGATGATGACCGAGAGCTGACTTCCCTATTATTCCGGGGATCCGTCGACC-3′; cpxR2, 5′-CAGATGCTGTGCCAGCAAATAGAGCAGGGTAAACTCAGTACCGGTTAACTTGTAGGCTGGAGCTGCTTCG-3′; cpxR3, 5′-CCCAAGCTTCGCGATTCAACGATAGAGAGTTTACG-3′; and cpxR4, 5′-TTTTAAACCACGGGTGACCATCTTTACG-3′.
Sequencing of cpxRA in S. sonnei.
The cpxRA gene of S. sonnei HW383 was PCR amplified with a pair of primers, cpxA4 and cpxR3, and subsequently, both strands were sequenced with an ABI Prism 310 genetic analyzer (Perkin-Elmer) with the following primers: cpxS1, 5′-GCGTTATCCAGCACTTCACTCC-3′; cpxS2, 5′-ATAAAGACGGTATCACCATTACGG-3′; cpxS3, 5′-GCGGCCATACTTTTTCTTCTGCG-3′; cpxS4, 5′-TTGTGGTTGGCCTGGAGTCTGGC-3′; cpxS5, 5′-GAGCAGGGTAAACTCAGTACCGG-3′; and cpxS6, 5′-GCCGGATCGTAAAGATGGTCACC-3′.
Measurement of β-galactosidase activities and Western blotting for InvE and IpaB.
For Western blotting and measurement of β-galactosidase activities, each strain was inoculated in 2 ml of LB medium with the appropriate antibiotics and grown overnight at 30°C with shaking. The cultures were diluted 100-fold into 5 ml of fresh LB medium in a distilled tube-type optical cell (200-34649; Shimadzu). Each culture was incubated for 3 h at 37°C with shaking, monitored for turbidity at 600 nm with a spectrophotometer (Spectronic 20+; Shimadzu), and harvested at an optical density at 600 nm of 0.8. Although the cpxA deletion strains showed a slightly long doubling time (29), the time of harvesting was delayed by only 10 ± 5 min on average.
β-Galactosidase activity was measured in each of the 50-μl cultures as previously described (26), or 10 μl of whole culture was subjected to Western blotting by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) as previously described (33). The IpaB protein was detected by anti-IpaB monoclonal antibody (18). In this study, the anti-InvE antiserum was prepared with a GST-purified recombinant protein from an E. coli strain carrying pEA225. β-Galactosidase was detected with anti-β-galactosidase antiserum (G4644; Sigma).
Northern blotting and chemical stability of invE mRNA.
For the Northern blotting analysis, total RNA samples were purified from the same cultures that were used for Western blotting. Bacterial cells were quickly trapped on a 0.45-μm filter (HAWP-04700; Millipore) with a vacuum manifold (21003; Millipore). The cells on the filters were immediately mixed and lysed with 1.2 ml of phenol containing RNA extraction reagent (Isogen; Nippon Gene). Total RNA samples were subjected to 1% formaldehyde-agarose gel electrophoreses as previously described (33), and Northern blotting was performed with digoxigenin (DIG EasyHyb, DIG Wash and Block buffer set; Roche) according to the manufacturer's instructions. The digoxigenin-labeled RNA probes were prepared by in vitro transcription from pGEMT-invE or pGEMT-virF with the digoxigenin labeling kit (Roche).
To measure the chemical stability of invE mRNA, rifampin (at a final concentration of 200 μg/ml) was added to the culture at an optical density at 600 nm of 0.8. An aliquot of the cultures (5 ml) was harvested every 1.5 min and subjected to Northern blotting as described above. The chemiluminescent signals were detected with a cold charge-coupled device (CCD) camera (Atto) for 1 h. The intensity of each signal was measured by the CCD processing program (Atto). The relative values of corresponding signals were plotted on a graph. The half-lives of the mRNAs were calculated from the average slope of each graph within the first 3 min for invE mRNA.
All assays were repeated at least three times and yielded comparable results. The statistical significance of the β-galactosidase activities was determined by analysis of variance (P < 0.05, t test) among three independent experiments with duplicate samples.
Nucleotide sequence accession number.
The sequence of the cpxA gene in S. sonnei HW383 was submitted to the DDBJ DNA sequence database with accession number AB161187.
RESULTS
Isolation of mutants.
In an attempt to isolate the chromosomal factor(s) affecting TTSS expression, mini-Tn10 mutants were isolated from E. coli strain HW1273, which carries the virulence plasmid of S. sonnei HW383. Like HW383, HW1273 maintains temperature regulation of the TTSS and contact hemolytic activity for sheep red blood cells (18, 46). Twenty mutants that lost β-galactosidase activity from mxiC::Tn3-lac (mxiC-lacZ) were selected on LB X-gal plates at 37°C from HW1273 carrying pJM1717. To select chromosomal mutations and eliminate mutations in known regulator genes such as virF and invE, all mutants that lost β-galactosidase activity were examined for expression of the InvE protein by Western blotting with anti-InvE antiserum. Sixteen mutants completely lost expression of InvE, probably indicating that the mini-Tn10 insertions were in the virF or invE gene. Four mutants showed decreased expression of InvE. Sequencing of these four mutants indicated that two mutants carried mini-Tn10 insertions in the cpxA gene at different sites. The insertions affected residues 22 (strain ME2526) and 63 (strain ME2523) of the deduced amino acid sequence of CpxA.
In order to confirm the effect of the cpxA mutations, a deletion of the cpxA gene was independently constructed in HW1273 by a PCR-based gene disruption technique (9, 40). The deletion of cpxA (ME2824) also resulted in decreased β-galactosidase activity from mxiC-lacZ plasmid pJM1718 as well as ME2526. The decreased activity was recovered to the level of HW1273 in the presence of plasmid pJM0329, which encodes an intact E. coli cpxA gene (Fig. 1A). IpaB protein, another product of TTSS expression, was detected by Western blotting (Fig. 1B). The expression of IpaB in ME2824 was decreased as well as mxiC-lacZ activity. As examined in the second screening described above, the amount of InvE protein was also low, which was comparable to the expression of IpaB in the same samples (Fig. 1B).
FIG. 1.
(A) β-Galactosidase activities from mxiC-lacZ fusion plasmid pJM1718. Bars indicate strains carrying ME2526 (cpxA::mini-Tn10), ME2824 (cpxA), HW1273 (Wt), ME2824/pJM0329, and CC118 as a negative control (N/C). Error bars indicate standard deviations. (B) Western blotting for InvE and IpaB proteins in E. coli cpxA (ME2824) and wild-type (HW1273) strains. The lowest band is a cross-reacting irrelevant protein detected by the InvE polyclonal antibody as a loading control (L/C).
Transcription of virF in the cpxA strain.
For characterization of the whole cascade of TTSS expression in the cpxA strain, transcription of the virF gene, the primary regulator of TTSS, was examined by the expression of β-galactosidase activity from virF-lacZ fusion plasmid pHW848 (27). The cpxA deletion strain, ME2824, showed β-galactosidase activity levels similar to those of HW1273 (Fig. 2). Consistently, the expression of virF mRNA was almost the same between the cpxA deletion and wild-type strains of S. sonnei (see Fig. 4B). virF expression in the cpxA deletion mutant ME2824 seemed to be sufficient for transcription of the invE gene, because transcription of invE was also maintained at a substantial level in ME2824 (Fig. 3A). On the contrary, virF expression was completely abolished by deletion of the cognate activator cpxR (ME2830), as described previously (Fig. 2) (28).
FIG. 2.
β-Galactosidase activities from the virF-lacZ fusion plasmid pHW848. Bars indicate strains ME2830 (cpxR), ME2824 (cpxA), HW1273 (Wt), and ME2824/pJM0329.
FIG. 4.
(A) Western blotting for InvE and IpaB proteins in S. sonnei strains. Lanes: cpxR, MS2830; cpxA, MS2824; Wt, MS390; −, MS2824 carrying pACYC177; +, MS2824 carrying pJM0329. The lowest band indicates a cross-reacting irrelevant protein detected by the InvE polyclonal antibody as a loading control (L/C). (B) Northern blotting for virF and invE mRNAs. Total RNA was purified from cpxA (MS2824) and wild-type (MS390) strains in the same cultures as those used in panel A. Either 18 or 4 μg of total RNA was loaded for the blotting of virF (left panel) and invE (middle panel), respectively. The right panel indicates the ethidium bromide-stained RNA pattern after electrophoresis in a 1% agarose gel to detect virF mRNA.
FIG. 3.
(A) β-Galactosidase activities from the invE-lacZ transcriptional fusion plasmid pJM4320. (B) β-Galactosidase activities from invE-lacZ translational fusion plasmid pJM4321. The bars on the graphs indicate strains ME2830 (cpxR), ME2824 (cpxA), HW1273 (Wt), and ME2824/pJM0329. (C) Western blotting with anti-β-galactosidase antibody for the above strains carrying pJM4320 and pJM4321.
cpxA deletion affects translation of InvE.
Since the cpxA mutant maintained substantial levels of virF expression, we next examined transcription of the invE gene, the second regulatory factor of TTSS, by using invE-lacZ transcriptional fusion plasmid pJM4320. Transcription of the invE gene initiates at a single start site located at bp −54 upstream of the translation initiation site, and no additional promoters have been detected (42). The transcriptional fusion plasmid pJM4320 (containing the sequence from bp −260 to −33 upstream from the first ATG codon of invE) encodes the invE promoter that was ligated to the SD sequence from the promoter cloning vector pCB182 (36) (see Materials and Methods). The β-galactosidase activity in cpxA strain ME2824 carrying pJM4320 was similar to that of HW1273 (Fig. 3A). The difference between ME2824 and HW1273 was not significant, as determined by analysis of variance (P < 0.05).
In order to examine posttranscriptional processing of InvE expression, reporter plasmid pJM4321 carrying the invE-lacZ translational fusion gene was constructed with the same lacZYA vector backbone as the transcriptional fusion pJM4320. Plasmid pJM4321 encodes the invE sequence from bp −260 to +118 (corresponding to 40 amino acids of the InvE protein), which is fused in frame with the lacZYA gene. The cpxA strain ME2824 carrying pJM4321 showed significantly decreased (21% of that of HW1273) β-galactosidase activity from the invE-lacZ translational fusion plasmid. ME2824 carrying both pJM4321 and the cpxA-encoding plasmid pJM0329 completely recovered β-galactosidase activity to the level of the wild-type strain (Fig. 3B).
In addition to the assays for β-galactosidase activities, protein expression of β-galactosidase from the invE-lacZ fusion gene was directly detected by Western blotting with anti-β-galactosidase antiserum. As expected, the InvE-LacZ fusion protein was slightly larger size than native β-galactosidase protein in a single band, which means that no artificial initiation of translation occurred in the invE-lacZ translational fusion gene (Fig. 3C). The intensity of the bands of β-galactosidase protein from the invE-lacZ transcriptional fusion plasmid was not significantly different between the cpxA and wild-type strains; however, that of the InvE-LacZ translational fusion protein was clearly at a low level in the cpxA strain ME2824, which was comparable to expression of the native InvE protein in the cpxA strain (Fig. 3C and Fig. 1B).
Characterization of cpxA deletion in S. sonnei.
Since deletion of the cpxA gene was characterized in E. coli strain HW1273, we next constructed the same cpxA and cpxR deletions in S. sonnei strain MS390, a donor of the virulence plasmid carried by HW1273. The cpxRA genes of MS390 were sequenced before construction of the deletions. The sequences showed a striking similarity with those of E. coli except for two nucleotides that did not alter the amino acid sequence. The PCR-based gene disruption technique (9) was also applicable to S. sonnei.
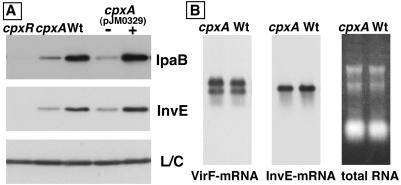
The cpxA deletion strain (MS2824), cpxR deletion strain (MS2830), and wild-type strain (MS390) were examined for expression of the InvE and IpaB proteins by Western blotting. Consistent with our previous result that CpxR is essential for virF expression (28), cpxR strain MS2830 completely lost the ability to express both InvE and IpaB (Fig. 4A). cpxA strain MS2824 expressed decreased levels of the InvE and IpaB proteins, which were similar to the levels expressed by the E. coli cpxA mutant. On the basis of the identical sequences of E. coli and S. sonnei, cpxA-encoding plasmid pJM0329 was able to restore the decreased InvE and IpaB expression in MS2824 (Fig. 4A).
Transcription of the virF and invE genes in the S. sonnei cpxA mutant MS2824 was examined by Northern blotting of the same culture that was used for Western blotting. The results obtained were similar to those obtained from the lacZ fusions in E. coli. The levels of virF and invE mRNAs were actually the same in the cpxA and wild-type strains of S. sonnei (Fig. 4B). The intact expression of invE mRNA and the apparently decreased protein expression of InvE in the S. sonnei cpxA strain indicate that the phenotype caused by deletion of cpxA was the same in S. sonnei and E. coli.
In an attempt to determine the processes that affect InvE expression in the cpxA strain, the chemical stability of invE mRNA was examined after addition of rifampin to the MS2824 and MS390 cultures. invE mRNA was potentially unstable (chemical half-life was 2.5 min on an average), and the decay rates were actually comparable between the cpxA and wild-type strains (data not shown). In addition, no significant change in the protein stability of InvE was detected by Western blotting after addition of chloramphenicol or rifampin to cultures of the cpxA and wild-type strains of S. sonnei (data not shown). Therefore, InvE expression seemed to be inhibited at a later step of posttranscriptional processing in the cpxA background.
DISCUSSION
In the previous study (27), we focused only on repression of the virF gene under low-pH conditions and found that the cpxA145::Tn10 allele affected low-pH-dependent expression of the virF gene but had little effect on expression of virF under neutral-pH conditions. Further examination of TTSS expression in the mutant was not performed. However, when we selected mutations causing decreased expression of TTSS in this study, we again isolated cpxA::mini-Tn10 mutants of E. coli HW1273, which carry the virulence plasmid of S. sonnei. Further characterization of cpxA deletion strains of E. coli and S. sonnei revealed that the loss of CpxA had little effect on transcription of the virF and invE genes under neutral-pH conditions; however, a large effect on the expression of InvE through posttranscriptional effects on protein expression was observed. This indicates that the Cpx two-component system is involved in another activation pathway for TTSS expression in E. coli and S. sonnei in addition to previously characterized transcriptional regulation of virF through CpxR (28).
In this study, we showed that CpxA and CpxR of the Cpx two-component system have phenotypically separate effects on virF expression. CpxA has little effect on virF expression; however, CpxR is essential for the transcription of virF (28). In cpxA mutants, several genes under Cpx regulation have been shown to be transcribed at levels similar to those in the wild-type strain (3, 8). These phenomena are explained by the phosphatase activity of CpxA. The phosphatase activity is thought to improve the specificity of the signal input by preventing the cognate regulator from irrelevant phosphorylation with internal low-molecular-weight phosphodonors such as acetylphosphate (3, 6, 8). Therefore, complete deletion of cpxA would not prevent CpxR from becoming autophosphorylated, in which state the genes regulated by the Cpx system should be activated to some extent (6, 8, 11). Transcription of the virF gene would be activated by the autophosphorylation of CpxR. However, we could not exclude the possibility of the existence of another sensor component interacting with the CpxR regulator. This will be resolved in future studies.
We demonstrated that mutation of the cpxA gene affected the posttranscriptional expression of InvE. However, we do not know whether the effect works through CpxR, since CpxR is essential for the transcription of virF and invE. We could not examine the translational effect on invE in the cpxR mutant. However, in this study, because virF transcription was negligibly affected by the cpxA mutation, it would be reasonable to assume that the the mutant retains CpxR activity. Considering this, the posttranscriptional regulation of InvE would be chiefly influenced by another unidentified factor(s), noncognate response regulator, or other two-component pairs through the Cpx pathway. Recently, Gubbins et al. demonstrated that TraJ, a regulator of F conjugation, is subject to posttranscriptional regulation upon high-level activation of the Cpx response (15). One possibility is that the unidentified posttranscriptional regulatory factor is activated to lower InvE expression at a high concentration of CpxR-P through loss of the phosphatase activity of CpxA. Several factors associated with posttranscriptional regulation such as RimM protein, which is known to bind to an initial ribosome-mRNA complex and maintain efficient translation (5), have been reported. These factors may be involved in the posttranscriptional regulation of InvE. Further examinations remain to be conducted.
The two HW1273 strains carrying an invE transcriptional fusion (pJM4320) or invE translational fusion (pJM4321) in the single-copy plasmid pTH18rrnBT showed a large difference in β-galactosidase activity. The HW1273 strain carrying pJM4321 expressed 11-fold higher activity than that carrying pJM4320 (Fig. 3A and B). Translational efficiency can be increased more than 10-fold by the presence of a well-matched SD sequence, the correct distance of the SD sequence from the first AUG, and the presence of consecutive G's at the second codon position (21). The translation efficiency of InvE might be potentially high due to the combination of these factors in the translation initiation sequence. Abundant production of the InvE protein supports the idea of InvE action on TTSS expression. Beloin and Dorman demonstrated that InvE alters the binding ability of DNA-binding proteins such as H-NS to the promoter regions, which in turn activates the transcription of both the mxi and ipa genes (4). Since multiple InvE binding sites have been predicted on the promoter regions of both genes (40), a considerable amount of InvE would be required for the substitution of DNA-binding proteins.
Comparison of the cpxRA sequence of S. sonnei with that of E. coli indicated 100% identity at the level of amino acid sequences. Based on the similarity in the genetic backgrounds of the two organisms (20), E. coli strain HW1273 and S. sonnei may have the same activation cascades of virulence. The Cpx two-component system is involved in the expression of various virulence genes among different species of gram-negative bacteria. Our results indicated that the Cpx two-component system is one of the key sensor regulators for virulence gene expression in S. sonnei. Further studies to resolve the details of the regulation of the Cpx system will provide us with the means to develop efficient ways to prevent shigellosis.
Acknowledgments
We are grateful to K. Datsenko and B. Wanner for the kind gift of a set of plasmids for PCR-based gene disruption techniques to our laboratory. We are grateful to Hiromi Sato for sequencing the cpxRA genes of S. sonnei. We thank S. Nakayama for generously providing the plasmid for the cpxA clone and for fruitful discussions. We thank S. Shaikh for anti-IpaB antibody. The cloning vectors pTH18cs5, pTH18kr, and pTH18cr were kind gifts from the cloning vector collection at the National Institute of Genetics (Japan).
This research was supported by a grant-in-aid from the Ministry of Health, Labor and Welfare and Ministry of Education, Science and Technology of the Japanese Government.
REFERENCES
- 1.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 3.Alves, R., and M. A. Savageau. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25-51. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., and C. J. Dorman. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825-838. [DOI] [PubMed] [Google Scholar]
- 5.Bylund, G. O., B. C. Persson, L. A. Lundberg, and P. M. Wikstrom. 1997. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J. Bacteriol. 179:4567-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 11.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 12.Dong, J., S. Iuchi, H. S. Kwan, Z. Lu, and E. C. Lin. 1993. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene 136:227-230. [DOI] [PubMed] [Google Scholar]
- 13.Dorman, C. J., N. N. Bhriain, and C. F. Higgins. 1990. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 344:789-792. [DOI] [PubMed] [Google Scholar]
- 14.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubbins, M. J., I. Lau, W. R. Will, J. M. Manchak, T. L. Raivio, and L. S. Frost. 2002. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J. Bacteriol. 184:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto-Gotoh, T., M. Yamaguchi, K. Yasojima, A. Tsujimura, Y. Wakabayashi, and Y. Watanabe. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185-191. [DOI] [PubMed] [Google Scholar]
- 17.Hromockyj, A. E., and A. T. Maurelli. 1989. Identification of an Escherichia coli gene homologous to virR, a regulator of Shigella virulence. J. Bacteriol. 171:2879-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, K., T. Nakajima, T. Sasaki, and H. Watanabe. 1991. Localization of IpaB protein in Escherichia coli K-12 MC1061 strain carrying Shigella sonnei form I plasmid pSS120. Microbiol. Immunol. 35:335-341. [DOI] [PubMed] [Google Scholar]
- 19.Kato, J., K. Ito, A. Nakamura, and H. Watanabe. 1989. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect. Immun. 57:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4:1125-1132. [DOI] [PubMed] [Google Scholar]
- 21.Lewin, B. 1994. Genes V, p. 260-271. Oxford University Press Inc., New York, N.Y.
- 22.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 30.Porter, M. E., and C. J. Dorman. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176:4187-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 32.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russel. 2002. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sasakawa, C., K. Kamata, T. Sakai, S. Makino, M. Yamada, N. Okada, and M. Yoshikawa. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasakawa, C., S. Makino, K. Kamata, and M. Yoshikawa. 1988. Isolation, characterization, and mapping of Tn5 insertions into the 140-megadalton invasion plasmid defective in the mouse Serény test in Shigella flexneri 2a. Infect. Immun. 54:32-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, K., and C. F. Beck. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene 42:37-48. [DOI] [PubMed] [Google Scholar]
- 37.Shapira, S. K., J. Chou, F. V. Richaud, and M, J. Casadaban. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene 25:71-82. [DOI] [PubMed] [Google Scholar]
- 38.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamano, K., S.-I. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supermolecular structure of Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniya, T., J. Mitobe, S. Nakayama, Q. Mingshan, K. Okuda, and H. Watanabe. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB boxA-like sequence. J. Bacteriol. 185:5158-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 42.Tobe, T., M. Yoshikawa, T. Mizuno, and C. Sasakawa. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobe, T., M. Yoshikawa, and C. Sasakawa. 1995. Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J. Bacteriol. 177:1094-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran Van Nhieu, G., and P. J. Sansonetti. 1990. Mechanism of Shigella entry into epithelial cells. Curr. Opin. Microbiol. 2:51-55. [DOI] [PubMed] [Google Scholar]
- 45.Tran Van Nhieu, G., E. Caron, A. Hall, and P. J. Sansonetti. 1999. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 18:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, H., and A. Nakamura. 1985. Large plasmids associated with virulence in Shigella species have a common function necessary for epithelial cell penetration. Infect. Immun. 48:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, H., E. Arakawa, K. Ito, J. Kato, and A. Nakamura. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of InvE with ParB of plasmid P1. J. Bacteriol. 172:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber, R. F., and P. M. Silverman. 1988. The cpx proteins of Escherichia coli K12. Structure of the cpxA polypeptide as an inner membrane component. J. Mol. Biol. 203:467-478. [DOI] [PubMed] [Google Scholar]
- 49.Zychlinsky, A., B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti. 1994. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol. 11:619-627. [DOI] [PubMed] [Google Scholar]