Abstract
All restriction enzymes examined are phosphodiesterases generating 3΄-OH and 5΄-P ends, but one restriction enzyme (restriction glycosylase) excises unmethylated bases from its recognition sequence. Whether its restriction activity involves endonucleolytic cleavage remains unclear. One report on this enzyme, R.PabI from a hyperthermophile, ascribed the breakage to high temperature while another showed its weak AP lyase activity generates atypical ends. Here, we addressed this issue in mesophiles. We purified R.PabI homologs from Campylobacter coli (R.CcoLI) and Helicobacter pylori (R.HpyAXII) and demonstrated their DNA cleavage, DNA glycosylase and AP lyase activities in vitro at 37°C. The AP lyase activity is more coupled with glycosylase activity in R.CcoLI than in R.PabI. R.CcoLI/R.PabI expression caused restriction of incoming bacteriophage/plasmid DNA and endogenous chromosomal DNA within Escherichia coli at 37°C. The R.PabI-mediated restriction was promoted by AP endonuclease action in vivo or in vitro. These results reveal the role of endonucleolytic DNA cleavage in restriction and yet point to diversity among the endonucleases. The cleaved ends are difficult to repair in vivo, which may indicate their biological significance. These results support generalization of the concept of restriction–modification system to the concept of self-recognizing epigenetic system, which combines any epigenetic labeling and any DNA damaging.
INTRODUCTION
Restriction enzymes cleave DNA that lacks proper epigenetic modification (base methylation in most of the cases studied) to abolish its biological activity (1). All known restriction enzymes hydrolyze a phosphodiester bond linking the monomer nucleotide units and generate 3΄-OH and 5΄-P ends (Figure 1, ii). They fall into several superfamilies, each characterized by a unique fold and motifs. We earlier looked for restriction enzymes of novel structure and function by taking advantage of the nature of restriction–modification (RM) systems as mobile genetic elements (2). In comparing genomes, we looked for genes that lack any known motif of restriction enzymes but move together with another gene with the motifs of DNA methyltransferases (3,4).Those restriction enzyme candidate genes were expressed in vitro and tested for DNA cleavage activity (4). The product of one such gene, from the hyperthermophilic archaeon Pyrococcus abyssi, was an endonuclease recognizing 5΄GTAC (4) and designated as R.PabI. It likely forms an RM system together with a linked DNA methyltransferase (M.PabI) generating 5΄GTm6AC (5,37). This restriction enzyme turned out to have a novel fold, designated as ‘half pipe’ (6). In addition to its preference for high temperature (85°C), this enzyme displayed several exceptional properties. One is independence from divalent metal ions (6). Structural analysis of the co-crystal with cognate DNA and biochemical analysis demonstrated that this enzyme is a DNA glycosylase that excises adenine bases from its target sequence (Figure 1, iii) (7). Its superfamily members are called restriction glycosylases (8).
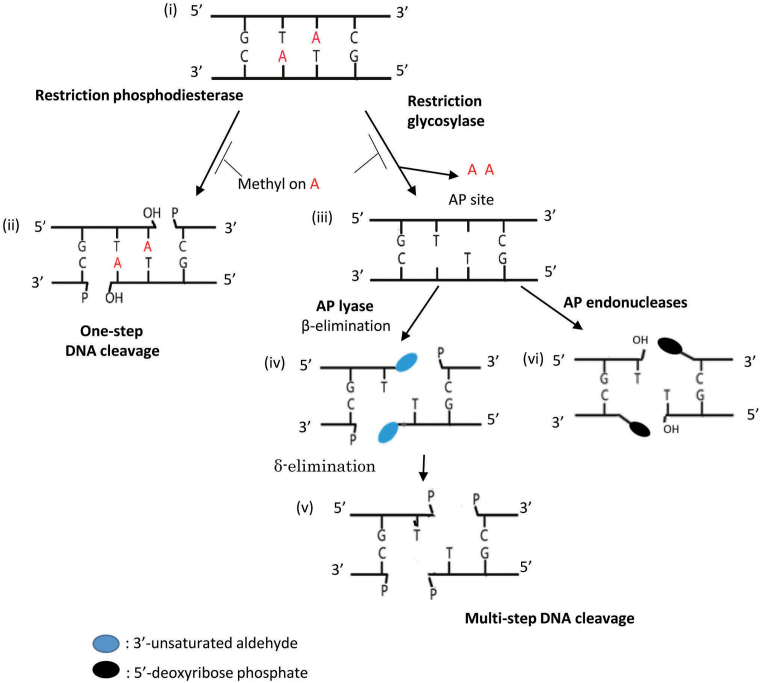
Figure 1.
DNA cleavage pathways for restriction enzymes. (i) A double-strand DNA with the recognition sequence for PabI. A in red, adenine base to be excised unless methylated by a paired modification enzyme. (ii) Hydrolysis of phosphodiester bonds to generate two 3΄-OH and 5΄-P end pairs. (iii) Generation of AP sites by base excision. (iv) Cleavage with AP lyase generates two breaks with 5΄-P and 3΄-modified sugar (blue oval) ends by β-elimination. (v) Further δ-elimination. (vi) AP site cleavage by AP endonucleases, generating 3΄-OH and 5΄- deoxyribose phosphate ends but not a sticky end.
This finding started a new phase in the study of restriction enzymes and led us to propose generalization of the concept of RM systems to general RM systems or self-recognizing epigenetic systems (9). Any agent that damages DNA with a particular epigenetic status can be called a general restriction enzyme. This generalization relates RM systems to toxin–antitoxin systems (10). Indeed, there are systems (BREX and others) that limit DNA bacteriophage propagation by means other than DNA breakage (11,12). Such generalized systems may also include uracil N-glycosylases in prokaryotes and eukaryotes.
What could be the relationship between base excision and DNA strand cleavage in the restriction process of these restriction glycosylases? In one report (7), the enzyme R.PabI was concluded not to be an endonuclease: strand cleavage both in vitro and in vivo was ascribed to heat-promoted processing of the AP sites (abasic sites). Another study, however, demonstrated that R.PabI itself has a type of endonuclease activity called AP lyase (Figure 1, iv) that generates unique ends (8). This activity is, however, weak and uncoupled with the glycosylase activity, and its role in restriction remains unclear. Indeed, DNA treated with R.PabI at a low temperature (37°C) remained free of any strand breaks but lost transformation activity with Escherichia coli cells at 37°C (8).
In the present work, we aimed to clarify the role, if any, of DNA strand cleavage in the restriction action of the restriction glycosylases. We analyzed PabI homologs from mesophilic bacteria (4,7,9,13) (see alignments there) and used an E. coli DNA transfer system at 37°C. Our results suggest the critical role of endonucleolytic breakage in the restriction process but unexpected diversity in its origin.
MATERIALS AND METHODS
For details, see Materials and Methods in detail in the supplementary materials with the same subsection names.
Bacterial strains, bacteriophages, plasmids and oligonucleotides
All E. coli strains and plasmids used here are listed in Supplementary Table S1. The synthetic oligonucleotides used are listed in Supplementary Tables S2 and S3. The synthetic genes for R.PabI homologs, R.HpyAXII and R.CcoLI, were designed considering the protein sequences in NCBI resources and codon optimization for expression in E. coli, by Funakoshi.
Restriction enzyme expression in vivo
Expression plasmid, pET28a::pabIR, pET28a::ccoLIR or pET28a::hpyAXIIR was introduced into E. coli T7 Express lysY/Iq harboring pBAD30_cviQIM.
Purification of restriction enzymes
Escherichia coli T7 Express lysY/Iq harboring pBAD30_cviQIM and one of the restriction enzyme expression plasmids, pET28a::pabIR, pET28a::ccoLIR, or pET28a::hpyAXIIR, was used. IPTG was added to a final concentration of 0.5 mM. The cells were collected by centrifugation and sonicated and centrifuged at 7 krpm for 20 min. The supernatant containing R.PabI or R.PabI (D214A) was heated at 75°C while the supernatant containing R.HpyAXII or R.CcoLI was left on ice. The supernatants were centrifuged at 7 krpm for 20 min again and filtered using a 0.45-um PDVF filter. The filtrates were bound to Ni-NTA Agarose resin and eluted. The fused His-tag was removed by thrombin digestion. The flowthrough was loaded onto a Heparin HP column, and then the proteins were eluted. Concentrated protein solution was mixed with appropriate volumes of glycerol, 100 mM EDTA and 100 mM DTT to generate a final protein stock solution in 10 mM MES pH 6.0, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol. In gel filtration, its native form gave a peak consistent with its dimer form.
DNA cleavage assay
An 861-bp linear double-stranded DNA fragment was used as a substrate for the cleavage assay (Supplementary Figure S2B). The cleavage reaction was performed in 10 ul with 0.1 M sodium phosphate buffer (pH 6.5), 2.66 pmol (266 nM) of purified enzyme and 0.38 pmol (0.38 nM) of substrate at 85°C (R.PabI/R.PabI(D214A)) or 37°C (R.HpyAXII/R.CcoLI) for 1 h.
DNA glycosylase assay
Each of the top and bottom strands of the 40-mer oligonucleotide with a single ‘GTAC’ (Supplementary Table S2) was labeled with γ32P-ATP, and annealed with its complementary oligonucleotide. 0.2 pmol of the 40-bp substrate was incubated with 0–1.4 pmol of purified enzyme (that is, with 4-fold dilution) in 20 ul of 0.1 M phosphate buffer (pH 6.5) at 60°C (R.PabI) or 37°C (R.HpyAXII, R.CcoLI) for 1 h. For R.CcoLI, we also incubated the 0.2 pmol 40-bp substrate with 0–0.2 pmol enzyme in 20 ul of 20 mM MOPS–KOH (pH 7.0) for 1 h. Half the reaction mixture was treated with 0.1 M NaOH at 70°C for 10 min to cleave DNA at the generated AP sites, and then neutralized with HCl.
AP lyase assay
To construct the AP site-containing 40-bp substrate, uracil-containing (5΄ -GTUC/3΄ -CUTG) double-stranded oligonucleotides (Supplementary Table S2) were labeled and generated, and then incubated with uracil N-glycosylase (UNG). The reaction was performed in 20 ul of 0.1 M phosphate buffer (pH 6.5) or 20 mM MOPS buffer (20 mM MOPS–KOH, pH 7.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT), containing 0.2 pmol of AP site-containing substrate (32P-labeled top or bottom strand) and 0–2 pmol of purified R.CcoLI at 37°C for 1 h.
NaBH4 trapping
R.CcoLI (0–6 pmol, 0–300 nM) and a 40-bp substrate DNA (0.2 pmol, 10 nM), containing 5΄-GT#C (# = AP site)/3΄-C#TG and a 5΄ -32P label on the top strand, were incubated in 20 ul of 20 mM MOPS buffer (20 mM MOPS–KOH, pH 7.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT) at 37°C for 20 min and then with 100 mM NaBH4 at 25°C for 30 min. DNA–R.CcoLI complexes were denatured in gel loading buffer containing 3% SDS at 90°C for 10 min and separated through 10% SDS-PAGE.
Restriction of phage propagation
Escherichia coli strains harboring (or not harboring) an RM expression plasmid was grown at 37°C overnight in LB medium with 50 ug/ml Ap, 30 ug/ml Km and 0.5% arabinose. Then the culture was diluted 100-fold and grown in tryptone broth supplemented with 0.2% maltose and 10 mM MgSO4 at 37°C for 1 h. IPTG was added to 0.1 mM, and incubated at 37°C for 3 h. 2 ml of culture was mixed with 2 ml top agar and then poured onto a bottom agar plate. Bacteriophage lambda vir was spotted on the plates. The plates were incubated at 37°C.
Restriction of the chromosome in vivo
The wild-type or the mutant strain harboring (or not harboring) an RM expression plasmid was grown overnight at 37°C in LB medium with 50 ug/ml Ap, 30 ug/ml Km and 0.5% arabinose, and the culture was diluted 100-fold in LB medium with 50 ug/ml Ap, 30 ug/ml Km and 0.5% arabinose and grown at 37°C for 2–3 h. IPTG was added to a final concentration of 0.1 mM prior to incubation at 37°C for 0.5 h. 1 ml of the culture was centrifuged at 5 krpm for 10 min at 4°C, and the cells were resuspended. The suspension was diluted and spotted on the plates. The plates were incubated overnight at 37°C before colony counting.
Restriction in transformation
Plasmid pBAD30_cviQIM (0.84 pmol) was treated with purified R.PabI (8.4 pmol) in 50 ul of 0.1 M sodium phosphate buffer (pH 6.5) at 37°C for 1 h to generate AP sites. A 200-ng equivalent of the AP site containing pBAD30_cviQIM was transferred into the original and the mutant T7 Express lysY/Iq E. coli strains by electroporation to count the colony-forming units/ml.
Motif frequency analysis
Expected motif frequency (EGTAC) was defined: EGTAC = (NTAC x NGTA)/NTA, where NGTA, NTAC and NTA are the numbers of motifs GTA, TAC and TA, respectively.
Restriction enzyme expression in vitro
Two protein-coding regions of the pabIR homologs were inserted into pEU3-NIIb, a plasmid for cell-free protein synthesis. The R.PabI homolog coding region was amplified by PCR with KOD-plus (Toyobo) from plasmids with a BamHI site attached at their 3΄ ends by using primers: CF-CcoLI-F/CF-CcoLI-R and CF-HpyAXII-F/CF-HpyAXII-R (Supplementary Table S2). The putative restriction enzymes were expressed in a wheat-germ-based cell-free protein synthesis system.
PCR analysis of deletion mutant strains
Constructed mutant E. coli strains nfo, xth, recA, recB, dinB and polB, were confirmed by colony PCR with the corresponding primers (Supplementary Table S2).
Helicobacter pylori strains for transcriptome analysis
Primers used for this experiment are listed in Supplementary Table S3. Helicobacter pylori strain P12 derivatives carrying a deletion in the pabIR homolog (= HPP12_0511) alone (strains PIK65, PIK70) or both the pabIR homolog and the pabIM homolog (= HPP12_0510) (strain PIK69) were constructed using homologous recombination to avoid post-segregational killing. The candidate transformant colonies were streaked on a selective plate, and then one of the clones that appeared was designated as PIK65 (ΔHPP12_0511).
Transcriptome analysis
Total RNA was extracted from two replicate exponential-phase cultures of PIK69 and PIK70. The rRNA-depleted samples were used for cDNA library construction. The cDNA library was sequenced on the HiSeq2500 platform.
RESULTS
Mesophilic R.PabI homologs have endonuclease (AP lyase) activity
R.PabI is from a hyperthermophilic archaeon and shows no detectable AP lyase activity at 37°C (8), a temperature at which in vivo restriction can be studied with E. coli. We therefore turned to its homologs in mesophilic bacteria (9): R.HpyAXII from Helicobacter pylori strain HPAG1, which cleaves at 5΄GTAC (13), and R.PabI homolog (locus_tag, WP_000052868.1) from Campylobacter coli. First, to test whether they possess DNA cleavage activity, we synthesized their genes and expressed them in vitro with a wheat-germ-based cell-free protein synthesis system. R.CcoLI was shown to cleave DNA (Supplementary Figure S1). Thus, we named the R.PabI homolog (locus_tag, WP_002830209.1) from Campylobacter coli as an active member of the R.PabI family of restriction glycosylases, R.CcoLI.
Next, the His-tagged versions of the two homologs were overexpressed in E. coli in the presence of a chlorella virus DNA methyltransferase recognizing the same sequence, then tag-free versions of the homologs were purified by a two-step affinity purification protocol (see Materials and methods for details). The purified enzymes were visualized as a single band in SDS-PAGE (Supplementary Figure S2 A).
The two homologs displayed cleavage activity for DNA carrying a single recognition site (5΄GTAC) (Figure 2A). An earlier study demonstrated that R.HpyAXII recognizes and cleaves this sequence (13). In reactions of R.CcoLI with a supercoiled plasmid with a single 5΄GTAC site, both the linear product and the open circle product appeared (Supplementary Figure S7). We saw single-strand breakage activity on a hemimethylated DNA ((8) and Supplementary Figure S6). From these, we inferred the presence of the major route of double-strand breakage with the minor route of single-strand breakage.
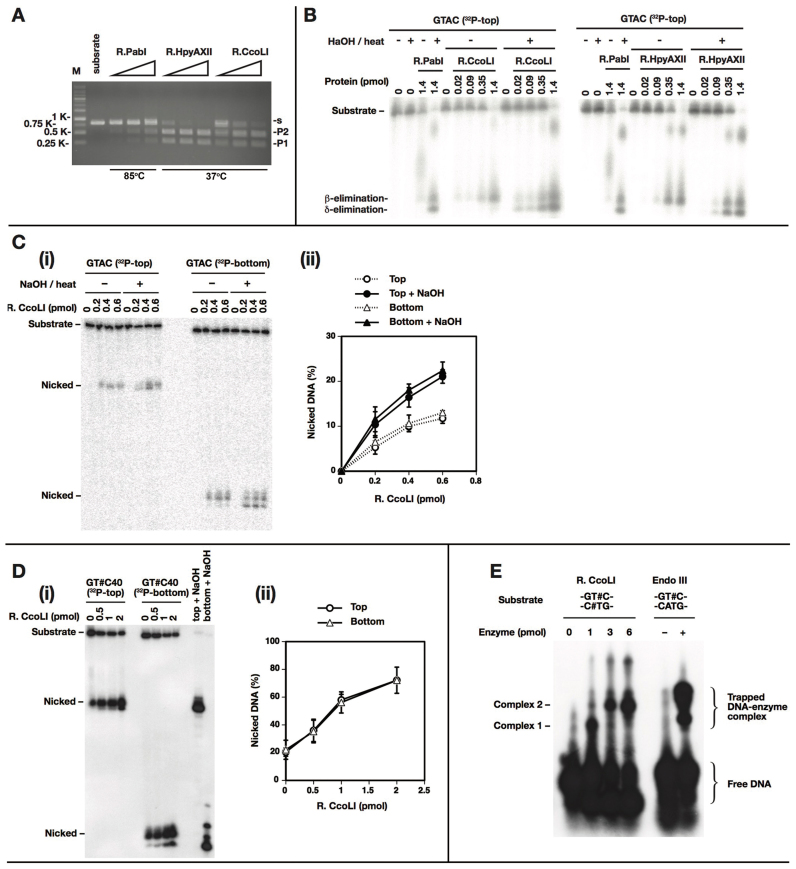
Figure 2.
Biochemical activities. (A) DNA cleavage. PCR product (861 bp) with single ‘GTAC’ (0.38 pmol, 200 ng) (Supplementary Figure S2 B) was treated with R.PabI, R.HpyAXII, or R.CcoLI (2.66 pmol and 3-fold serial dilution from that) in 100 mM phosphate buffer, pH 6.5, at 85°C or 37°C for 1 h. The substrate (s) cleavage generated P1 (552 bp) and P2 (309 bp). (B) Glycosylase and AP lyase. A 40-mer oligonucleotide with a single ‘GTAC’ (0.2 pmol) (with top strand 32P-labeled at 5΄) was treated with R.PabI (1.4 pmol), R.HpyAXII and R.CcoLI (0–1.4 pmol, 4-fold serial dilution) in 100 mM phosphate buffer, pH 6.5, at 60°C (R.PabI) or 37°C (R.HppyAXII, R.CcoLI) for 1 h. The mixture was further treated with 0.1 M NaOH, 70°C for 10 min and neutralized with HCl. The mixture was electrophoresed through 18% denaturing PAGE. (C) Glycosylase of R.CcoLI. A 40-mer oligonucleotide with single ‘GTAC’ (0.2 pmol) (top or bottom strand 32P-labeled at 5΄) was treated with 0–0.6 pmol R.CcoLI in 100 mM phosphate buffer, pH 6.5, 37°C for 1 h. It was further treated with 0.1 M NaOH at 70°C for 10 min, neutralized with HCl and electrophoresed through 18% denaturing PAGE in (i). The amounts of nicked top and bottom strands are plotted in (ii). n = 3. An error bar indicates standard deviation. (D) AP lyase of R.CcoLI. An oligonucleotide with 5΄-GTUC/3΄-CUTG was treated with uracil N-glycosylase to generate the double AP sites. A 20-ul reaction mixture with 0.1 pmol of the substrate (32P-labeled top or bottom strand at 5΄) and 0–2 pmol purified R.CcoLI in 100 mM phosphate buffer, pH 6.5 was incubated at 37°C for 1 h. The samples were electrophoresed through 18% denaturing PAGE in (i). The amounts of nicked top and bottom strands are plotted in (ii). n = 3. An error bar indicates standard deviation. (E) Trapping DNA–R.CcoLI intermediate. The double or single AP-site substrate was generated by ‘GTUC’ treatment with uracil N-glycosylase and the top strand was 32P-labeled at 5΄. A mixture of 0.2 pmol substrate with 0–6 pmol R.CcoLI or with 20 units of Endo III was incubated in buffer (20 mM MOPS–KOH, pH 7.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT) or buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT) at 37°C for 20 min and then with 100 mM NaBH4 at 25°C for 30 min. DNA–R.CcoLI complexes were denatured in a gel loading buffer containing 3% SDS at 90°C for 10 min and separated through 10% SDS-PAGE.
Treatment with R.PabI and its homologs, R.HpyAXII and R.CcoLI, of DNA carrying a single recognition site (5΄GTAC) gave the same product (by β-elimination) in phosphate buffer. When NaOH was added, another product was formed (by δ-elimination). The order of β-elimination to δ-elimination is explained by chemistry (see Supplementary Figure S4 of (8)). These results show that the homologs have glycosylase activity at 37°C (Figure 2B). An increase in the products with addition of NaOH suggests that the glycosylase and AP lyase are not coupled. The cleavage occurred with the same efficiency at the top strand and the bottom strand (Figure 2C). The specific glycosylase activity was estimated to be 0.11 pmol/pmol R.CcoLI/h.
The AP lyase activity of CcoLI was confirmed by an oligonucleotide substrate with AP sites, which was generated by excision of uracil from 5΄GTUC. The results show that R.CcoLI cleaves the AP site substrate and gives the same product at 37°C as R.PabI did at a higher temperature (Figure 2D). The efficiency was the same with the top and bottom strands. It was estimated to be 0.072 pmol/pmol R.CcoLI/h in 0.1 M phosphate buffer. The ratio of the specific activities of glycosylase to AP lyase was 1.5:1, which was much lower than the ratio (3.7:1) in R.PabI at 70°C (8). In other words, R.CcoLI displays more coupling of AP lyase with glycosylase.
We found that the optimal NaCl concentration is 50 mM and the optimal pH is 7.0 for R.CcoLI glycosylase in Tris–HCl buffer. Under these conditions, the glycosylase and AP lyase activities of R.CcoLI were approximately 8 times higher than with 0.1 M phosphate buffer (Supplementary Figure S3). However, the glycosylase/AP lyase ratio remained the same (1.4:1).
Competition of the AP lyase activity by a GTAC-containing duplex was carried out. The GTAC inhibited the cleavage of GT#C significantly, while the CTAC with a low efficiency (Supplementary Figure S8). This specific outcompetition further demonstrates the presence of lyase activity in R.CcoLI.
The presence of AP lyase in R.CcoLI was confirmed by trapping its Schiff-base intermediate by reduction with NaBH4 (Figure 2E), as with R.PabI (8). We do not know the nature of Complex 1 or Complex 2 or why their formation depended on the amount of enzyme.
Restriction of phage propagation
Whether or not R.PabI family restriction glycosylases can block phage propagation has not yet been tested. We expressed the restriction enzymes together with the chlorella methyltransferase, which methylates 5΄GTAC, in E. coli cells and measured their ability to restrict infection with bacteriophage lambda vir. The wild-type lambda carries 113 ‘GTAC’ sites. R.PabI decreased plaque formation by two orders of magnitude (Figure 3A). The restriction was not seen with R.PabI mutant D214A. As shown in previous studies (7,8), the mutant could not cleave DNA (Supplementary Figure S2B). R.CcoLI and R.HpyAXII displayed the same level of restriction as wild-type R.PabI.
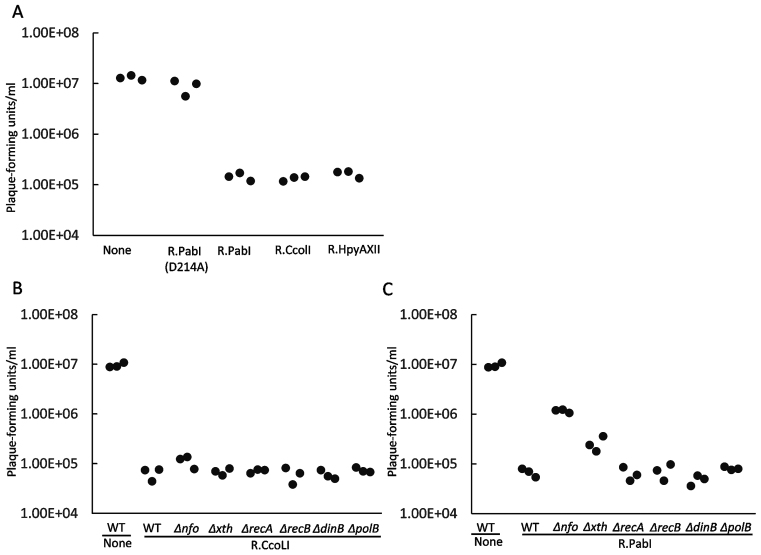
Figure 3.
Restriction in vivo of bacteriophage λ propagation. Infection with bacteriophage λ of E. coli and mutant strains expressing a PabI family restriction enzyme. Expression of M.CviQI, which methylates 5΄GTAC, and of restriction enzyme was induced by arabinose and IPTG, respectively. Plaques in a spot of phage suspension on a cell lawn were counted. (A) Restriction by PabI family. R−, with methyltransferase CviQI but without a restriction enzyme; R.PabI (D214A), with M.CviQI and R.PabI (D214A) with a mutation inactivating the glycosylase. (B) Restriction by R.CcoLI in E. coli with a deletion mutation in various genes. (C) Restriction by R.PabI in E. coli mutants. The control strains labeled ‘None’ contained pET28a (empty vector) in addition to pBAD30_cviQIM.
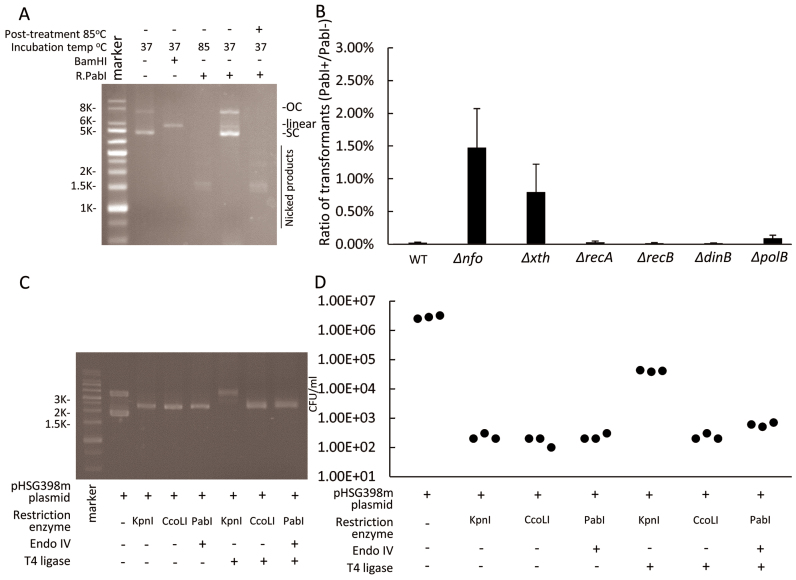
Because DNA glycosylases initiate base excision repair, we examined whether cellular DNA repair systems could enhance or alleviate the restriction mediated by R.PabI family enzymes. E. coli carry two major AP endonucleases, endonuclease IV encoded by nfo (15,16) and exonunuclease III encoded by xth (17). For R.CcoLI (Figure 3B), the restriction level was not affected by deletion of these genes for AP endonucleases or by deletion of dinB or polB implicated in repair DNA synthesis (18,19). The recA and recB deletions had no detectable effect. Lack of homologous recombination repair is expected because of the condition of single infection.
R.PabI, however, showed a different response to the mutations. Deletion of either of the two AP endonuclease genes weakened the restriction (Figure 3C). This indicates that the AP endonuclease promotes, as opposed to diminishes, the restriction, possibly by introducing a DNA strand break. The rec mutations or DNA polymerase mutations did not have a detectable effect.
Restriction of endogenous bacterial chromosomes
Restriction enzymes attack endogenous bacterial chromosomes under specific conditions (20). For Type II RM, such an attack takes place in cells that have lost the RM genes and is called post-segregational killing (2). The chromosome breakage is repaired by homologous recombination involving RecA and RecBCD proteins (21). We tried to replicate chromosome attack by suppressing expression of the chlorella methyltransferase gene under arabinose inducible promoter by the addition of glucose. As shown in Figure 4A, the suppression led to a decrease in viable cell counts by three orders of magnitude. In recA mutant and recB mutant strains, the decrease was larger. We did not detect such a decrease in the control with expressed methyltransferase (Figure 4B). This indicates that the chromosomal damage was repaired by homologous recombination, presumably between sister chromosomes as with Type II restriction phosphodiesterases (21).
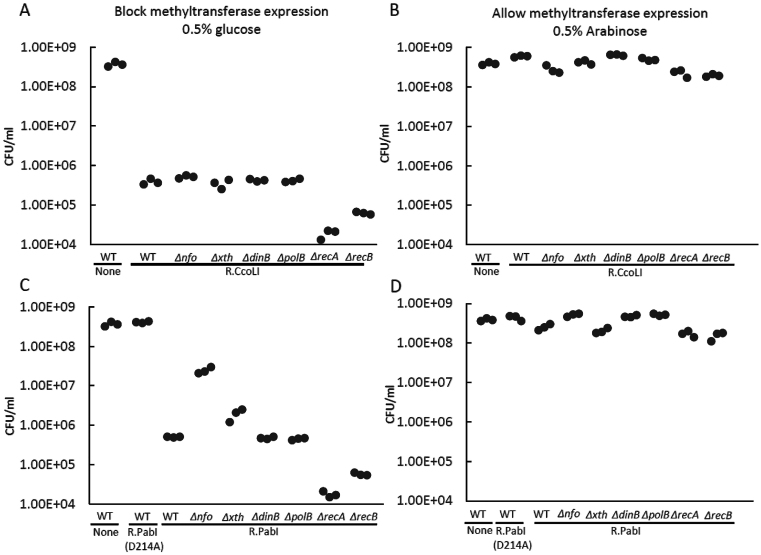
Figure 4.
Restriction of endogenous bacterial chromosome in vivo. M.CviQI and restriction enzyme expression were induced by arabinose and IPTG, respectively. After removal of the inducers, the remaining restriction enzyme molecules attacked the host chromosome at newly replicated, unmethylated sites to kill the cells. (A, C) Replacing arabinose and IPTG by glucose. (B, D) Allowing methyltransferase expression with arabinose. (A, B) E. coli wild-type and mutant strains carrying CcoLI restriction enzyme gene. (C, D) E. coli wild-type and mutant strains carrying PabI restriction enzyme gene. R−: Strain carrying M.CviQI plasmid only. The control strains labelled ‘None’ contained pET28a (empty vector) in addition to pBAD30_cviQIM.
With R.PabI, we observed a comparable decrease in viable cell counts, which was eliminated by the D214A mutation (Figure 4C). The decrease was again more severe in the recA and recB mutants, which suggests recombination repair of the damage. The mutations in two AP endonuclease genes diminished the cell killing. The effect was stronger with nfo mutant (Figure 4C, D). This indicates that AP endonucleases promote cell killing likely through chromosome cleavage at AP sites.
Restriction of transforming plasmid
An earlier study demonstrated the HpyAXII RM system restricts incoming plasmid and chromosomal DNA in H. pylori (13). When a plasmid preparation was treated with R.PabI at 37°C, it remained supercoiled, which means that it contained no strand breaks at all, but it had lost the ability to transform into E. coli (8). This suggests that base excision is responsible for the restriction.
In order to see what gene function is involved in this restriction, we repeated this experiment with E. coli mutated in the above genes (Figure 5A and B). We found that the mutations in the two AP endonuclease genes increased transformation efficiency (Figure 5B). This suggests again that the AP endonucleases promote restriction likely by introducing strand cleavage at the AP sites on the incoming plasmid.
Figure 5.
Restriction of transforming plasmid. (A) An unmethylated plasmid with three 5΄GTAC/3΄CATG sites, pBAD30_cviQIM (methylated or unmethylated), was treated with R.PabI or BamHI at 37°C for 1 h. (B) The supercoiled plasmid treated with R.PabI at 37°C in A was purified and electroporated into E. coli wild type and its mutant derivatives expressing R.PabI or not (empty vector) in the absence of arabinose. The ratio [transformant number with host with R.PabI plasmid]/[transformant number with host with empty vector] was plotted. n = 3. An error bar indicates standard deviation. (C) A plasmid with a single site for PabI family (5΄GGTACC) ( = a site for KpnI) (pHSG398m) (Supplementary Figure S5) was treated with R.PabI, R.CcoLI or KpnI at 37°C for 1 h. The product with R.PabI was further treated with endonuclease IV (an AP endonuclease) for strand cleavage. The three linearized plasmids were purified and treated with T4 DNA ligase. (D) The products in C were used for transformation into E. coli HST08. The colony-forming units were counted. SC, supercoil; OC, open circle.
Strong inactivation is observed in (8) (Figure 4) and here (Figure 5A). The inactivation is explained by change of opposite abasic sites at 5΄GTAC/3΄CATG to a double-strand break by various endonucleases as described (Figure 1, iv, v, vi.). A linearized plasmid loses transformation efficiency (36).
Difficulty in repair by rejoining the cleaved ends
As expected from the atypical end structure, the AP lyase-generated ends are not rejoined by DNA ligase (22). Indeed, DNA treated with R.PabI was not easily rejoined by DNA ligase in vitro (6). We examined whether this is also the case in vivo. We prepared a plasmid carrying a single site for PabI homologs and KpnI (5΄GGTACC) (Supplementary Figure S5). When we treated the plasmid with KpnI or CcoLI (Figure 5C), the transformation efficiency decreased by four orders of magnitude (Figure 5D). Treatment with DNA ligase moderated this decrease for KpnI but not for CcoLI (Figure 5 D).
When the plasmid was treated with R.PabI and then with AP endonuclease (endonuclease IV), we expected the end structure illustrated in Figure 1, vi. We found the same decrease in transformation efficiency. This decrease was not moderated by treatment with DNA ligase, as expected (Figure 5C, D).
This and the above results suggest that the biological significance of the PabI family of restriction glycosylases may lie in the difficulty of repair by end joining, especially for incoming, unmethylated DNA. We carried out relevant analyses as discussed in the two following sections.
Avoidance of the recognition sequence in the genomes
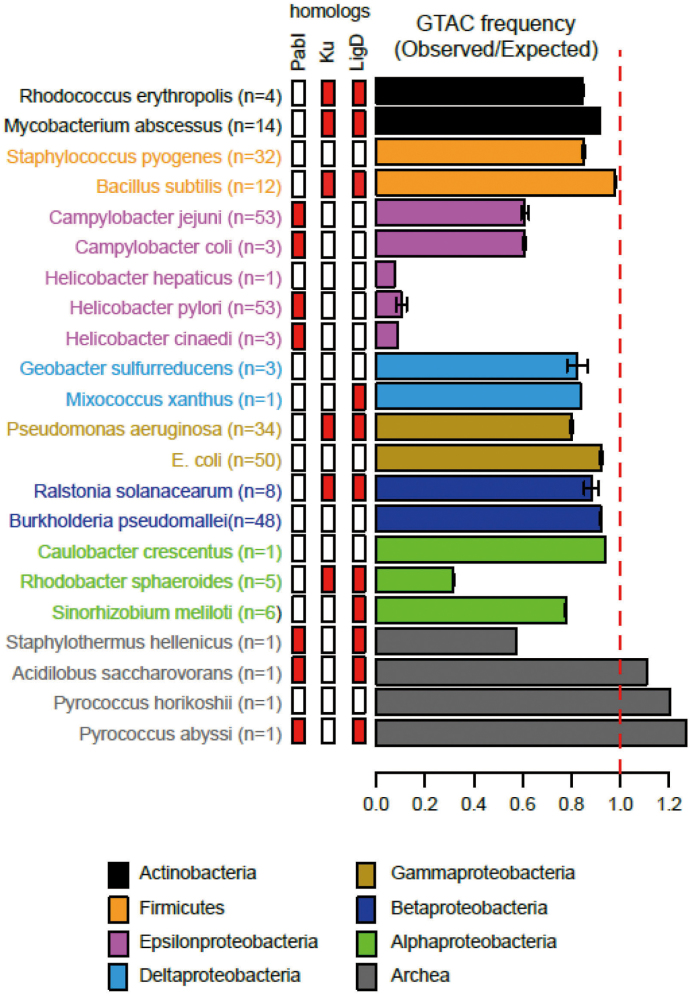
An earlier report revealed strong avoidance of the target sites (5΄GTAC) in H. pylori (13). We calculated the ratio of the observed number of sites to the expected number of sites for related bacteria by a Markov method (14). We found intermediate avoidance in Campylobacter and very severe avoidance in Helicobacter (Figure 6). This is compatible with long-term inheritance of the PabI family in Helicobacter and in Campylobacter (9). The restriction avoidance roughly correlates with the presence/absence of PabI family members. Ancestors of H. hepaticus may have been exposed to the PabI family as PabI family members are widely distributed in the genus Helicobacter (9). The strong avoidance of their recognition sites may be caused by selection by the toxicity against incoming non-self (unmethylated) DNA. In contrast, we had noticed lack of GTAC avoidance in Pyrococcus abyssi (37). This could be due to recent acquisition of PabI family by Pyrococcus (23), weaker AP lyase activity of R.PabI, or a special chromatin structure of Pyrococcus (37).
Figure 6.
Avoidance of 5΄GTAC on the chromosomes of Campylobacter and Helicobacter. Species belonging to different taxa are shown in distinct colors. Bars indicate the mean and SD. Side bars indicate presence (red) or absence (white) of a PabI family RM system, Ku homologs and LigD homologs. n indicates the number of complete genomes analyzed.
The 5΄GTAC site is relatively frequent on rRNA genes compared to other parts of the genome (13). Presumably, the damage there may be easily repaired by homologous recombination between two ectopic copies as well as between sister chromosomes. Some plasmids found in H. pylori strains appear not to have experienced severe restriction avoidance (Supplementary Figure S9). This may be explained by (i) the short period in which the plasmid was exposed to the restriction, (ii) recombination repair between multiple copies, (iii) maintenance of the endogenous multi-copy plasmid not being affected very much by loss of a copy or (iv) some advantage provided by the strong restriction activity to the plasmid or its operon.
Effect on the transcriptome
To address the impact of GTAC methylation on the transcriptome, H. pylori strain P12 derivatives carrying a deletion of the pabIR homolog (= HPP12_0511) alone (strains PIK65, PIK70), or a deletion of both the pabIR homolog and the pabIM homolog (= HPP12_0510) (strain PIK69), were constructed (Materials and methods) and their transcriptomes were compared by the RNA-seq method. The R.PabI homolog (HPP12_0511 not HPP12_0512) was co-deleted to bypass the effects of the restriction enzyme gene on transcriptome (24). Only few genes were judged to be differentially expressed (Supplementary Table S4). These include one specificity gene of a type I RM system. This suggests communication of PabI RM and this type I RM system. It also affected a gene for a membrane protein.
A gene in the microcin (a bacteriocin) operon on a plasmid was suppressed in the knockout (Supplementary Figure S10). This gene is in a region with many 5΄ GTAC copies. We do not know the underlying mechanisms or its biological significance.
DISCUSSION
These results indicate that endonucleases play a crucial role in the restriction action by the PabI family. Tighter coupling of AP lyase to glycosylase activity in a mesophilic enzyme demonstrates their close collaboration in vitro and suggests that in vivo. It is of interest which amino acid is involved in the trapped Schiff intermediate in the lyase reaction (Figure 2E). We did not detect a decrease in AP lyase activity in R.PabI mutants at either of the two conserved Lys residues (K73A, K 202A) (7).
The E. coli AP endonucleases assisted restriction in E. coli by R.PabI, which shows undetectable AP lyase activity at 37°C (8). AP endonucleases are involved in DNA damage repair (25) but also in DNA destruction in mammalian apoptosis (26). Xth (Exo III) and Nfo (Endo IV) account for the major and minor AP endonuclease activities, respectively, in uninduced E. coli. Both the xth and nfo single mutants exhibit moderate sensitivity to methylmethane sulfonate that generates abasic sites, and the xth nfo double mutant exhibits extreme hypersensitivity to methylmethane sulfonate (38). This result indicates that together with Xth (Exo III), Nfo (Endo IV) contribute significantly to the repair of AP sites derived from methylated purines in cells. Thus, the effects of the nfo mutation on the survival of phage, plasmid, and cells observed in this study is consistent with the known role of Nfo (Endo IV). The mild effect of the xth mutation relative to nfo remains elusive.
Our results suggest that the AP lyase and AP endonucleases promote restriction by introducing a DNA strand break. However, we cannot exclude the contrasting, but not necessarily exclusive, possibility that the endonucleases release the restriction glycosylase from DNA after base excision and allow its reaction on another substrate.
In AP endonuclease mutants, R.PabI-mediated restriction of the incoming DNA or chromosomal DNA is modulated. This suggests that AP sites are repaired or tolerated by other mechanisms such as translesion DNA synthesis. In relation to this, we do not know whether the PabI family acts as a mutator, an anti-mutator, or neither.
What could be the biological significance of these restriction glycosylases? Type II RM systems attack the host bacterial chromosome in post-segregational killing. When their genes are lost from a cell, its descendant will carry fewer methylase molecules to protect hemimethylated sites generated after chromosome replication (2,21,27). The remaining restriction enzyme will cleave exposed sites. Bacterial homologous recombination systems (RecA, RecBC) assisted repair of restriction cleavage of chromosomal DNA likely through recombination between sister chromosomes but did not help repair of a single invading DNA (21,28). As in type IIP systems, this represents the interaction of three selfish elements: the bacterium, the RM system, and the invading DNA. The bacterial genome protects its chromosomes but not the invading DNA.
These glycosylases may discriminate between incoming DNA and endogenous chromosomal DNA in another way. R.PabI and R.CcoLI can cleave hemimethylated DNA, present on newly-replicated chromosomal DNA, to generate a single-strand break (8) (Supplementary Figure S6) as several Type IIP restriction phosphodiesterases. Otherwise, R.CcoLI also have a minor route to generate single strand break on unmethylated DNA (Supplementary Figure S7). A single-strand break of hemimethylated DNA and unmethylated by a minor activity or will be readily repaired by the base excision repair machinery (25,29) as well as by homologous recombination (30–32). On the other hand, incoming DNAs are likely to be either fully methylated or unmethylated. The latter will suffer a double-strand break, which cannot be repaired by base excision repair or by recombination repair in single infection (28). The unique feature of these restriction glycosylases is the generation of ends with atypical structures lacking bases via AP lyase or AP endonucleases (Figure 1, iv, v, vi), which are not readily repaired by end joining (Figure 5). RM systems consisting of a restriction glycosylase and methylase are, therefore, very effective means of combating invading non-self DNAs but are helpful in repairing endogenous bacterial chromosomes, once restriction avoidance has prevailed in the chromosomes (Figure 6). Our results (Figures 3–5) are consistent with such a concept of distinguishing endogenous and invading DNAs. The small effect of the H. pylori homolog on gene expression (Supplementary Table S4, Figure S10), possibly in part due to the low number of GTACs in the genome, highlights their role as ‘a strong poison to invaders’ as opposed to a gene regulator. There may be many other factors that can affect efficiency of restriction. Clearly, we need more experiments to test this hypothesis.
Non-homologous end joining (NHEJ) is an important mechanism for DNA double-strand break repair. The key factors of NHEJ are Ku, terminus-bridging protein with 5΄-dRP/AP lyase activity (33), and LigD, terminus-ligating protein. Ku homolog often accompanies LigD homologs, but there is apparent incompatibility between PabI homolog and Ku homolog in a given genome (Figure 6). The PabI homologs and Ku homologs might have some functional overlap, which results in loss of the Ku homologs.
The difficulty in repair by end joining may be useful in applications. For example, in treating with a mixture of an R.PabI family enzyme and a type II restriction phosphodiesterase in vitro, only ends created by the latter will be easily rejoined by a ligase. In vivo, a specific target DNA may be effectively destroyed by these restriction glycosylases. These effects may be useful in genome manipulation and in gene therapy.
In these experiments, the bacteria are not in their natural habitat where many other phenomena can occur. These bacteria were grown outside of the body and were not directly taken from their natural habitat such as human gastroenteric tracts. In order to assess biological significance of these restriction glycosylases, we also need to address their natural history and their evolution inscribed in the genomes and the methylomes (18,34,35).
These overall results substantiate the concept of restriction glycosylase and, furthermore, support generalization of the concept of restriction–modification system to the concept of self-recognizing epigenetic system, or epigenetic immune systems, which combines any mode of epigenetic labeling and any mode of DNA damaging. This big picture in the prokaryotic immune systems and in epigenetics will be detailed elsewhere.
ACCESSION NUMBER
NGS data was submitted under DRA accession no. DRA004356.
Supplementary Material
ACKNOWLEDGEMENTS
pEU3-NIIb plasmid was kindly provided by Yaeta Endo. We are grateful to Hisaji Maki and Pablo Radicella for discussion.
Author contributions: I.K. and H.I. designed research; Y.Z., T.M., H.Y., T.N., M.F., N.T., K. I., Y.F. and H.I. performed research; Y.Z. and I.K. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MEXT KAKENHI [221S0002 to I.K.]; JSPS KAKENHI [26650123 and 15K14572 to I.K., 15K18665 to H.Y.]; Ichiro Kanehara foundation of promotion of medical sciences and medical care. Funding for open access charge: Jean d'Alembert scholarship from Université Paris-Saclay.
Conflict of interest statement. None declared.
REFERENCES
- 1.Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G., Murray N.E.. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014; 42:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naito T., Kusano K., Kobayashi I.. Selfish behavior of restriction–modification systems. Science. 1995; 267:897–899. [DOI] [PubMed] [Google Scholar]
- 3.Chinen A., Uchiyama I., Kobayashi I.. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction–modification genes with large genome polymorphisms. Gene. 2000; 259:109–121. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa K., Watanabe M., Kuroita T., Uchiyama I., Bujnicki J.M., Kawakami B., Tanokura M., Kobayashi I.. Discovery of a novel restriction endonuclease by genome comparison and application of a wheat-germ-based cell-free translation assay: PabI (5΄-GTA/C) from the hyperthermophilic archaeon Pyrococcus abyssi. Nucleic Acids Res. 2005; 33:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe M., Yuzawa H., Handa N., Kobayashi I.. Hyperthermophilic DNA methyltransferase M.PabI from the archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 2006; 72:5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazono K., Watanabe M., Kosinski J., Ishikawa K., Kamo M., Sawasaki T., Nagata K., Bujnicki J.M., Endo Y., Tanokura M. et al. Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 2007; 35:1908–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazono K., Furuta Y., Watanabe-Matsui M., Miyakawa T., Ito T., Kobayashi I., Tanokura M.. A sequence-specific DNA glycosylase mediates restriction–modification in Pyrococcus abyssi. Nat. Commun. 2014; 5:3178. [DOI] [PubMed] [Google Scholar]
- 8.Fukuyo M., Nakano T., Zhang Y., Furuta Y., Ishikawa K., Watanabe-Matsui M., Yano H., Hamakawa T., Ide H., Kobayashi I.. restriction–modification system with methyl-inhibited base excision and abasic-site cleavage activities. Nucleic Acids Res. 2015; 43:2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima K.K., Kobayashi I.. Transmission of the PabI family of restriction DNA glycosylase genes: mobility and long-term inheritance. BMC Genomics. 2015; 16:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mruk I., Kobayashi I.. To be or not to be: regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 2014; 42:70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb T., Sberro H., Weinstock E., Cohen O., Doron S., Charpak-Amikam Y., Afik S., Ofir G., Sorek R.. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015; 34:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoskisson P.A., Sumby P., Smith M.C.. The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology. 2015; 477:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humbert O., Salama N.R.. The Helicobacter pylori HpyAXII restriction–modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008; 36:6893–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha E.P., Viari A., Danchin A.. Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res. 1998; 26:2971–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demple B., Johnson A., Fung D.. Exonuclease III and endonuclease IV remove 3΄ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:7731–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puri R.V., Singh N., Gupta R.K., Tyagi A.K.. Endonuclease IV Is the major apurinic/apyrimidinic endonuclease in Mycobacterium tuberculosis and is important for protection against oxidative damage. PLoS One. 2013; 8:e71535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souza L.L., Eduardo I.R., Padula M., Leitao A.C.. Endonuclease IV and exonuclease III are involved in the repair and mutagenesis of DNA lesions induced by UVB in Escherichia coli. Mutagenesis. 2006; 21:125–130. [DOI] [PubMed] [Google Scholar]
- 18.Hastings P.J., Hersh M.N., Thornton P.C., Fonville N.C., Slack A., Frisch R.L., Ray M.P., Harris R.S., Leal S.M., Rosenberg S.M.. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One. 2010; 5:e10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becherel O.J., Fuchs R.P.. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8566–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusano K., Naito T., Handa N., Kobayashi I.. restriction–modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:11095–11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handa N., Ichige A., Kusano K., Kobayashi I.. Cellular responses to postsegregational killing by restriction–modification genes. J. Bacteriol. 2000; 182:2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman I.R. DNA ligase: structure, mechanism, and function. Science. 1974; 186:790–797. [DOI] [PubMed] [Google Scholar]
- 23.Rusinov I., Ershova A., Karyagina A., Spirin S., Alexeevski A.. Lifespan of restriction–modification systems critically affects avoidance of their recognition sites in host genomes. BMC Genomics. 2015; 16:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asakura Y., Kobayashi I.. From damaged genome to cell surface: transcriptome changes during bacterial cell death triggered by loss of a restriction–modification gene complex. Nucleic Acids Res. 2009; 37:3021–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiederhold L., Leppard J.B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A.E., Izumi T., Prasad R., Wilson S.H. et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004; 15:209–220. [DOI] [PubMed] [Google Scholar]
- 26.Madlener S., Strobel T., Vose S., Saydam O., Price B.D., Demple B., Saydam N.. Essential role for mammalian apurinic/apyrimidinic (AP) endonuclease Ape1/Ref-1 in telomere maintenance. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17844–17849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichige A., Kobayashi I.. Stability of EcoRI restriction–modification enzymes in vivo differentiates the EcoRI restriction–modification system from other postsegregational cell killing systems. J. Bacteriol. 2005; 187:6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handa N., Kobayashi I.. Type III restriction is alleviated by bacteriophage (RecE) homologous recombination function but enhanced by bacterial (RecBCD) function. J. Bacteriol. 2005; 187:7362–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mol C.D., Hosfield D.J., Tainer J.A.. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3΄ ends justify the means. Mutat. Res. 2000; 460:211–229. [DOI] [PubMed] [Google Scholar]
- 30.Meselson M.S., Radding C.M.. A general model for genetic recombination. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi I. Mechanisms for gene conversion and homologous recombination: The double-strand break repair model and the successive half crossing-over model. Adv. Biophys. 1992; 28:81–133. [DOI] [PubMed] [Google Scholar]
- 32.Pagès V. Single-strand gap repair involves both RecF and RecBCD pathways. Curr. Genet. 2016; 62:519–521. [DOI] [PubMed] [Google Scholar]
- 33.Roberts S.A., Strande N., Burkhalter M.D., Strom C., Havener J.M., Hasty P., Ramsden D.A.. Ku is a 5΄-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010; 464:1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima K.K., Furuta Y., Yahara K., Fukuyo M., Shiwa Y., Nishiumi Y., Yoshida M., Azuma T., Yoshikawa H., Kobayashi I.. Population evolution of Helicobacter pylori through diversification in DNA methylation and interstrain sequence homogenization. Mol. Biol. Evol. 2016; 33:2848–2859. [DOI] [PubMed] [Google Scholar]
- 35.Furuta Y., Namba-Fukuyo H., Shibata T.F., Nishiyama T., Shigenobu S., Suzuki Y., Sugano S., Hasebe M., Kobayashi I.. Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity. PLoS Genet. 2014; 10:e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi N., Kobayashi I.. Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:2790–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi I., Ishikawa K., Watanabe M.. Chromatin as anti-restriction adaptation: a hypothesis based on restriction enzymes of a novel structure. Proc. Int. Symp. Extremophiles Appl. 2007; 205:167–174. [Google Scholar]
- 38.Cunningham R.P., Saporito S.M., SPITZER S.G., Weiss B.. Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol. 1986; 168:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.