Abstract
The DEAH box helicase Prp43 is a bifunctional enzyme from the DEAH/RHA helicase family required both for the maturation of ribosomes and for lariat intron release during splicing. It interacts with G-patch domain containing proteins which activate the enzymatic activity of Prp43 in vitro by an unknown mechanism. In this work, we show that the activation by G-patch domains is linked to the unique nucleotide binding mode of this helicase family. The base of the ATP molecule is stacked between two residues, R159 of the RecA1 domain (R-motif) and F357 of the RecA2 domain (F-motif). Using Prp43 F357A mutants or pyrimidine nucleotides, we show that the lack of stacking of the nucleotide base to the F-motif decouples the NTPase and helicase activities of Prp43. In contrast the R159A mutant (R-motif) showed reduced ATPase and helicase activities. We show that the Prp43 R-motif mutant induces the same phenotype as the absence of the G-patch protein Gno1, strongly suggesting that the processing defects observed in the absence of Gno1 result from a failure to activate the Prp43 helicase. Overall we propose that the stacking between the R- and F-motifs and the nucleotide base is important for the activity and regulation of this helicase family.
INTRODUCTION
Helicases are NTP-dependent molecular motors characterized by conserved sequence motifs and harboring a wide range of biochemical activities such as NTP-dependent unwinding of double stranded nucleic acids, chaperoning of RNA folding or remodeling of ribonucleoprotein (RNP) complexes (1). These proteins are involved in almost all the biological processes involving nucleic acids such as replication, translation, editing and splicing (2,3). Helicases have been classified in six superfamilies (SF) on the basis of sequence and structural conservation (1). Despite the conservation of the structural core, there are fundamental differences in the function of these helicases and in how they act on nucleic acids.
The DEAH/RHA helicases, named after the sequence of motif I (Asp-Glu-Ala-His) and RNA helicase A (RHA), a representative member of the family, are part of the SF2 superfamily which also contains the DEAD-box and Ski2-like helicases. The 15 members of this family in humans (7 in yeast) are involved in transcription, translation, ribosome biogenesis, pre-mRNA splicing or RNA sensing. Yeast DEAH helicase Prp43 (DHX15 in humans) is the only member for which a complete structure is available (4,5). It is also remarkable because it is involved in several distinct biological processes and because it is activated by at least six different protein partners (6,7). In splicing, Prp43 ensures lariat-spliceosome dissociation (8,9). In ribosome biogenesis, Prp43 is one of the few factors implicated in several steps of the process and involved in the maturation of the two subunits through ribosomal RNA and snoRNA remodeling (10–13). The human homologue DHX15 is also involved in viral RNA sensing and triggers the NF-κB signaling pathway (14).
All superfamilies 1 and 2 (SF1 and SF2) helicases share a conserved helicase core, composed of two RecA-like domains, but differ in their C-terminal and N-terminal domains (15). The helicase conserved core harbors the two main activities common to all SF1 and SF2 helicases: ATP hydrolysis and nucleic acid binding. The NTP binding pocket is located at the interface between the two RecA ‘lobes’ and the single stranded nucleic acid binding site lies on top of these two domains. Helicases perform their function because NTP binding/hydrolysis and nucleic acid binding affinity are coupled through the global reorientation of the RecA domains. NTP binding induces a ‘closed’ conformation because coordination of the gamma phosphate requires residues from the two RecA domains (16). In contrast, the NDP bound or apo state promotes an open conformation. These two conformations modulate the structure of the nucleic acid binding site by modifying the distance between the residues from the two RecA domains involved in nucleic acid binding (17). This in turn modulates the specificity and/or affinity of the helicase for the nucleic acid. This property is used by helicases to achieve very different biochemical functions, but the most common and studied activity in vitro is double strand unwinding (18).
SF2 helicases separate double-stranded nucleic acids (NA) through a wide variety of mechanisms. Since ATP hydrolysis leads to the transition between closed and open conformations, the repeated hydrolysis of ATP would lead to a reciprocating motor-like motion. This has led to models in which the helicase translocates along a single nucleic acid strand and displaces the complementary strand or bound protein. However, the detailed mechanisms proposed vary in different helicase families. DEAD-box helicases are now thought to act as ATP dependent single stranded RNA binding proteins which unwind NA by a double-strand invasion mechanism rather than to translocate on the RNA (19). The UvrD (SF1) displays a processive and directional ‘wrench-and-inchworm’ mechanism (20,21), where one ATP is consumed by NA base pair unwound. The SF2 Hel308 DNA helicase threads a single strand through a ring-like structure formed by the RecA, the winged-helix (WH) and the Ratchet domains and separates the strands using a β-hairpin wedge protruding from RecA2 domain (22).
DEAH/RHA helicases contain an architecture analogous to Ski2-like helicases Hel308, Ski2 and Mtr4: they form a ring-like structure formed by RecA, WH and Ratchet domains and a β-hairpin wedge and all contain additional auxiliary domains (23,24). A striking property of DEAH/RHA helicases is that at least two of their members, Prp43 and Prp2, are regulated by proteins containing a glycine-rich domain of 50 amino acids called the G-patch domain (6). The ability to stimulate the in vitro ATPase and helicase activities of Prp43 has been demonstrated for Pfa1 (25), Ntr1 (26,27), Gno1/PinX1 (28), RBM5 (29), GPATCH2 (30) and Cmg1 (7) while Spp2 is the only known activator of Prp2 (31–33). In pre-mRNA splicing, Ntr1p recruits Prp43 to the spliceosome (27,34). In contrast, Gno1/PinX1 and Pfa1/Sqs1, which both act in the ribosome biogenesis pathway, are not responsible of the recruitment of Prp43 to the pre-ribosomal particles (25). Gno1/PinX1 stimulates Prp43 function in early 90S and early pre-60S particles (28), while Pfa1 participates in the maturation of the late pre-40S particles (25,35).
Amongst the SF2 helicases, DEAH/RHA helicase family displays a unique nucleotide binding mode. The majority of helicases from the SF2 family contain a Q-motif. This motif, located in the first RecA domain, contains a glutamine that specifically binds to the adenine moiety of the ATP substrate and confers specificity for ATP over all the other NTPs (36). The structure of the Prp43 (5) and MLE (37) DEAH/RHA helicases, and also of the Hepatitis C virus NS3 helicase (16), revealed the absence of the Q-motif and a completely different conformation of the nucleotide: the conformation of the phosphate backbone and the helicase catalytic machinery is conserved, but the ribose/base moiety is rotated 180 degrees in respect to other helicases. In Prp43, the adenine base is stacked between residues R159 and F357 of the first and second RecA domains respectively and since there are no specific H-bonds with the base, DEAH/RHA helicases display in vitro activity with all four NTPs.
The detailed mechanism by which Prp43 is activated by G-patch proteins is still unknown. In this work, we explore the contribution of the unique nucleotide binding mode of DEAH/RHA helicases to the regulation of their activity. We have used a combination of X-ray structures, enzymatic assays, Isothermal Titration Calorimetry (ITC) measurements and in vivo analyses to highlight a novel role of base stacking in the modulation of Prp43 activity. Based on these results, we propose a model for DEAH/RHA activity modulation by G-patch activators.
MATERIALS AND METHODS
Protein expression and purification
Escherichia coli Rosetta 2 DE3 strain were transformed by electroporation with the plasmid pSL18 coding Prp43-his (25), pSL18 coding Prp43 F357A or pSL18 coding Prp43 R159A. Transformed bacteria were grown at 37°C under agitation in 2× TY medium complemented by ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). When O.D. at 600 nm reached 0.8, protein expression was induced with 0.5 mM isopropyl-β-d-galactopyranoside and the culture was incubated at 15°C during 16 h under agitation. Bacteria transformed with pSL18 and variants were harvested by centrifugation and resuspended in lysis buffer containing 20 mM Tris–HCl pH 8, 200 mM NaCl (buffer A), 1 mM MgCl2 and 5 mM β-mercaptoethanol and stored at −20°C.
The lysis was completed by sonication and the lysate was centrifugated at 20 000g during 30 min. Prp43-his was purified from the soluble fraction on Ni-NTA resin and eluted with increasing imidazole concentrations. The nickel affinity chromatography was followed by a gel filtration step on a Superdex 200 16/60 column connected to an ÄKTA pure (GE Healthcare) using buffer A + 10 mM β-mercaptoethanol. Proteins were purified for ITC measurement in buffer A + 2.5 mM MgCl2. The Prp43 R159A and F357A mutants were purified as the wild-type and the absence of major conformational change in these mutants was checked by CD spectroscopy (data not shown).
Pfa1 was expressed from pSL20 Pfa1 (25), and Prp22 was expressed from pET-16 Prp22 (38) with the same protocol described above for Prp43 and its variants. Lysis was performed in buffer B (500 mM NaCl, 20 mM Tris–HCl pH 8), 1 mM MgCl2, 5 mM β-mercaptoethanol. The nickel affinity chromatography was performed on a 5 ml HisTrap (GE Healthcare) in buffer B + 5 mM β-mercaptoethanol containing a linear gradient of imidazole ranging from 20 to 500 mM. Gel filtration was performed in buffer B + 10 mM β-mercaptoethanol. All the proteins were concentrated with Amicon (Millipore) with a molecular weight cut-off of 30 000 Da.
Crystallization, data collection and processing
Prp43 (6 mg/ml) crystallized at 4°C by the hanging drop vapor diffusion method from a 1:1 mixture of protein, 1 mM CDP, 1 mM MgCl2 and precipitant containing 6–7.25% PEG 4000, 100 mM ammonium acetate and 50 mM sodium cacodylate pH 6.5. Crystals were transferred in the mother liquor containing 7.5%, 15% and 30% glycerol before flash freezing. X-ray diffraction data were all collected on the Proxima 1 beamline at SOLEIL synchrotron (Gif-sur-Yvette, France). Diffraction data were processed using MOSFLM (39) and SCALA (40) and the XDS package (41). The crystal belongs to the P3221 space group with two molecules per asymmetric unit. The cell parameters and data collection statistics are reported in Table 1.
Table 1. Data collection and refinement statistics.
| Ligand | CDP (PDB 5JPT) |
|---|---|
| Space group | P3221 |
| Unit cell parameters | a = 118.07 Å. |
| b = 118.07 Å. | |
| c = 252.15 Å | |
| α = β = 90° | |
| γ = 120° | |
| Beamline | SOLEIL (PX1) |
| Wavelength (Å) | 1.0882 |
| Resolution (Å) (outer shell) | 48.30–2.93 (3.04–2.93) |
| Reflections measured | 287 332 |
| Unique reflections | 44 183 |
| Redundancy | 6.5 |
| Completeness (%) (outer shell) | 99.6 (96.5) |
| Rmerge (%) | 11.7 (72.1) |
| (I)/<σ(I)> | 14.8 (2.6) |
| Refinement statistics | |
| Phasing | Molecular replacement |
| Molecules/UA | 2 |
| Rwork/Rfree (%) | 20.3/25.7 |
| Number of atoms non-H | 12 040 |
| <B> (Å2) | 55.5 |
| r.m.s.d. bonds (Å) | 0.005 |
| r.m.s.d. angles (°) | 0.97 |
| Ramachandran statistics | |
| Most favored (%) | 96.55 |
| Allowed (%) | 3.31 |
Structure resolution and refinement
Molecular replacement for the data was performed with Prp43 without water and ligand (PDB: 2XAU) with Phaser (42) from the CCP4i program suite. The molecular replacement solution structure was automatically rebuilt using Phenix (43). Manual reconstruction and ligand fitting were achieved using Coot (44). Refinement was performed with Phenix.
NTPase assays
Reaction mixtures (50 μl) contained 10 nM Prp43 (wild-type or mutant), 0.2 mM polyA, 100 mM KCl, 25 mM HEPES–KOH pH 8, 2.5 mM Mg(CH3COO)2 and 0.2 mM DTT. Enzymatic assays were carried in 96-well microplate and free phosphate assays were performed with BIOMOL GreenTM reagent (Enzo Life Science). At each time, reaction was quenched by the addition of 0.5 M EDTA pH 8.8. At the end of the experiment, 200 μl BIOMOL GreenTM reagent were added in each well, the absorbance at 620 nm was monitored after an incubation of 25 min at 25°C. The quantity of free phosphate was calculated by interpolations of the values on a phosphate standard curve. All measurements were performed in triplicates. Kinetic parameters were determined by non-linear fit.
Isothermal titration calorimetry
ITC experiments were performed on a MicroCal iTC200 (Malvern) with a 40 μM protein solution in the cell and a 0.8–2 mM NDP solution in the syringe. We typically performed 30 injections of 0.8–1.5 μl and monitored heat release after injection. Data processing was performed on Origin 7.0 (OriginLab). Thermodynamic parameters ΔH, ΔG and ΔS were determined with the binding isotherm, from these parameters, Kd was calculated. All measurements were performed in triplicate at least. The measurement of CDP isotherms, due to its low binding affinity, was performed by increasing the concentration of the protein to the 40 μM solubility limit of Prp43. A precise affinity from the CDP isotherms could not be measured in these conditions but a minimum Kd value could be estimated.
Helicase activity assays
In a final volume of 10 μl, the helicase substrate (1 nM) was pre-incubated with 100 nM Prp43 and/or 500 nM Pfa1, 10 min on ice, in the reaction buffer (20 mM HEPES pH8, 5 mM Mg(CH3COO)2, 0.2 mM DTT, 200 μg/ml BSA, 20 nM trap oligonucleotide). The reaction mixtures were complemented or not with 1 mM nucleotide, and incubated at 30°C for 30 min. The reactions were stopped by addition of 3 μl of a solution containing 1 mg/ml proteinase K, 1.25% SDS, 10 mM Tris–HCl pH8, 0.06% bromophenol blue, 0.06% xylene cyanol and 40% glycerol in the presence of 600 nM of the trap oligonucleotide. The samples were analyzed by native electrophoresis on 12% polyacrylamide, 5% glycerol gels using 1× TBE, 5% glycerol as running buffer, at 4°C. Gels were scanned with a Licor Odyssey, and analysed with Image Studio Lite software. The 3΄-flanking DNA/RNA duplex used as a substrate was the same as that used in (25), with a fluorescent (Alexa680) instead of radioactive labelled DNA.
Yeast strains and media
The yeast strains were derived from the BMA64 background (ura3-1, trp1-Δ2, ade2-1, leu2-3,112, his3-11,15, can1-100). Wild-type or mutant PRP43 ORFs were cloned into plasmid pHA113 (centromeric vector with TRP1 auxotrophy marker) under the control of the GAR1 promoter and upstream of a sequence encoding the ZZ tag (ZZ domains from protein A of S. aureus). These plasmids were transformed into a GAL::PRP43 strain, expressing endogenous PRP43 under the control of the conditional galactose promoter. The resulting strains were propagated on a synthetic medium lacking tryptophan and containing galactose as a carbon source. Upon shift to glucose-containing medium and growth for 27 h to repress expression of chromosomal PRP43 allele, the phenotypes induced by the plasmid-born copies of PRP43 were analysed. The wild-type and Δgno1 strains were previously described (28).
RNA extractions and Northern blot experiments
RNAs were extracted from yeast cell pellets or sucrose gradient fractions as previously described (45). RNA electrophoresis was performed according to ‘Molecular Cloning’, Sambrook and Russell, CSHL Press (‘Separation of RNA According to Size: Electrophoresis of Glyoxylated RNA through Agarose Gels’). Following electrophoresis, RNAs were transferred to Amersham Hybond N+ membranes and hybridized with 32P-labeled oligonucleotide probes using Rapid-hyb buffer (GE Healthcare). The sequence of the probes are available upon request.
Sedimentations on sucrose gradients
Yeast cell pellets were broken with glass beads in buffer K (20 mM Tris–HCl pH 7.4, 50 mM KCl, 10 mM MgCl2, 50 μg/ml cycloheximide, 1 mM dithiothreitol, 1× Roche Complete EDTA-free protease inhibitor cocktail, 0.1 U./μl Promega RNasin). Extracts were clarified by centrifugation, calibrated, loaded on 10–50% sucrose gradients and centrifuged at 39 000 rpm for 150 min on a Beckman-Coulter Optima L-100 XP ultracentrifuge using a SW41 rotor. Fractions were collected using a Foxy Jr. gradient collector (Teledyne Isco).
RESULTS
ATP base stacking modulates Prp43 ATPase activity
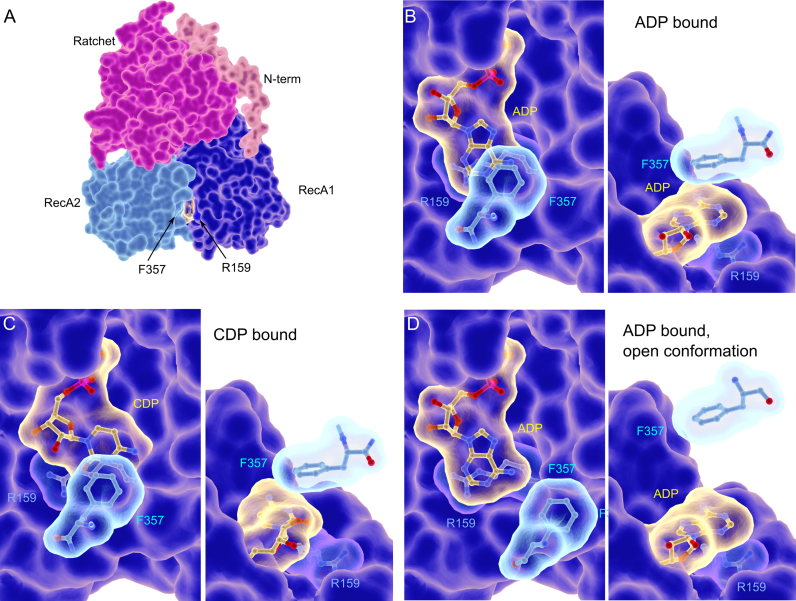
It is now well established that the relative orientation of the two RecA domains is linked to the presence of ADP or ATP in the nucleotide binding pocket. Interactions of the gamma phosphate with residues of motifs in the opposing RecA domains contribute to this structural reorganization. Binding of the ADP nucleotide in the Prp43 ‘closed’ conformation showed a novel nucleotide mediated interaction between residues of the two RecA domains (Figure 1A). We based our analysis on the ADP with an anti conformation of the base found in two Prp43 structure (5,46) rather than the syn conformation found only in one report (4).The R159 arginine (RecA1)-ATP interaction involves parallel stacking of the guanidinium on the adenine base and is compatible with cation-pi interaction (Figure 1A and B). On the other side of the adenine plane, the aromatic cycle of F357 (RecA2) is involved in a stacking interaction with the adenine base (Figure 1B). The adenine and phenyl aromatic rings do not totally overlap, but are in a parallel offset position that has been found to be an energetically favorable position in model π-π stacking studies (47).
Figure 1.
Prp43 structure. (A) Overall structure of Prp43 in complex with ADP. (B–D) Close up representation of the stacking of the the R- and F- motifs on the ADP nucleotide (B), the CDP nucleotide (C), the ADP nucleotide with F357 in the open conformation modelled from the MLE structure (D). Two orthogonal orientations are shown and the RecA2 domain has been removed for clarity. The figures of the structures are performed with UCSF Chimera (51), ePMV (52) and Blender (http://www.blender.org).
The adenine therefore seems sandwiched between the residues of the two RecA domains, but the relative strength of these two interactions is not known (Figure 1A and B). The overall binding mode is similar to the one observed in the Hepatitis C Virus (HCV) NS3 helicase. In HCV NS3 RecA1, tyrosine 241, located on the ‘spring’ helix and referred to as the Y-motif, is in the same position as R159 and stacks on the adenine base. In RecA2 a threonine 419 from motif V in the same position as F357 interacts on the other side of the adenine plane (16).
To determine if stacking to F357 and R159 is conserved in DEAH/RHA helicases we performed a sequence alignment of Prp43/DHX15 from different organisms. This shows that F357 and R159 are conserved (Supplementary Figure S1A). In addition sequence alignment of the four spliceosomal DEAH/RHA helicases from Saccharomyces cerevisiae and from Homo sapiens, and of RHA/DHX9 shows that these residues are conserved in other members of the DEAH/RHA family (Supplementary Figure S1B). The conservation of these residues suggests that all DEAH/RHA helicases have the same base binding mode as observed in Prp43. Since both the arginine on the ‘spring’ helix and the phenylalanine are conserved in DEAH/RHA helicases, we will refer to these residues as the R-motif and the F-motif, respectively.
The MLE DEAH/RHA helicase, crystallized in the presence of RNA and ATP transition state analogues has a different orientation of the RecA domains compared to the ADP-bound Prp43 (37). These two conformations have been used to propose a translocation mechanism for DEAH helicases. In this model, RecA1 acts as a ‘rigid’ ATP binding module, and rotation of the RecA2 domain induces a cascade of conformational changes that lead to RNA displacement. In the MLE conformation, the F-motif is swung out and the stacking interaction to the adenine base is lost. Modeling of Prp43 in the MLE conformation by superposition of the RecA domains also shows that F357 would not be stacked on the nucleotide base in this conformation (Figure 1D). We focused our analysis on the interaction between the nucleotide base and the RecA2 domain via F357 and evaluated its importance for the catalytic activity.
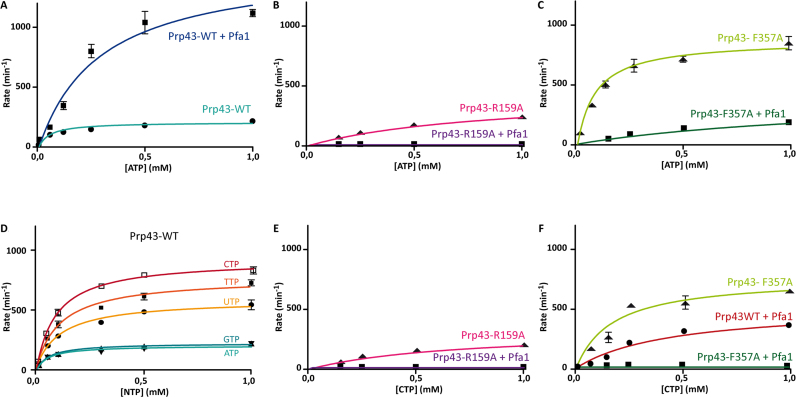
In order to evaluate the effect of adenine base stacking on the biochemical activity of Prp43, we performed ATPase assays with R- and F-motif mutants in order to evaluate the contribution of each residue in NTPase activity regulation (KM and kcat are reported in Table 2). We were unable to measure NTPase activity of either Prp43 or its mutants without an RNA cofactor. All subsequent assays were performed in the presence of PolyA. In agreement with previous results, wild type (wt) Prp43 had a weak ATPase activity (KM = 55 ± 12 μM, kcat = 3.4 ± 0.2 s−1) which is stimulated 7-fold by the addition of the Pfa1 activator (KM = 295 ± 45 μM, kcat = 25.4 ± 1.5 s−1) (Figure 2A).
Table 2. NTPase activities of Prp43 constructs.
| Proteins | NTP | kcat (s−1) | KM (μM) | kcat/KM (104.M−1.s−1) | R2 |
|---|---|---|---|---|---|
| Prp43-WT | ATP | 3.4 ± 0.2 | 55 ± 12 | 6.14 ± 0.44 | 0.88 |
| CTP | 15.4 ± 0.4 | 90 ± 8 | 16.74 ± 2.65 | 0.98 | |
| TTP | 12.8 ± 0.5 | 115 ± 15 | 11.25 ± 0.11 | 0.97 | |
| UTP | 9.7 ± 0.4 | 120 ± 15 | 8.15 ± 0.42 | 0.97 | |
| GTP | 3.7 ± 0.2 | 60 ± 10 | 6.33 ± 0.85 | 0.92 | |
| Prp43-WT + Pfa1 | ATP | 25.4 ±1.5 | 295 ± 45 | 8.67 ± 0.72 | 0.98 |
| CTP | 8.6 ± 1.1 | 445 ± 130 | 1.93 ± 0.14 | 0.94 | |
| Prp43–R159A | ATP | 7.3 ± 0.6 | 845 ± 175 | 0.85 ± 0.23 | 0.94 |
| CTP | 6.1 ± 0.3 | 935 ± 150 | 0.65 ± 0.13 | 0.95 | |
| Prp43–R159A + Pfa1 | ATP | No activity | |||
| CTP | No activity | ||||
| Prp43–F357A | ATP | 14.5 ± 0.6 | 90 ± 15 | 16.72 ± 0.31 | 0.95 |
| CTP | 12.7 ± 0.5 | 175 ± 20 | 7.32 ± 0.72 | 0.98 | |
| Prp43–F357A + Pfa1 | ATP | 6.9 ± 1.0 | 1385 ± 390 | 0.50 ± 0.26 | 0.88 |
| CTP | no activity |
Figure 2.
NTPase activities: role of Pfa1 and base stacking. Enzymatic assays were performed with BiomolGreen reagent. All conditions were tested three times and the kinetic parameters were determined and represented by non-linear fit to the Michaelis–Menten equation. Comparison of Prp43 WT (A), Prp43–R159A (B) and Prp43–F357A (C) ATPase activity in absence and presence of the G-patch co-factor Pfa1. (D) Comparison of puric and pyrimidic NTPase activities of Prp43. (E, F) CTPase activity of Prp43–R159A and Prp43–F357A in absence and presence of Pfa1.
The R159A mutant, which abolishes the packing of the adenine base with RecA1, showed a decrease in ATPase activity, marked by a 15-fold increase in the KM (KM = 845 ± 175 μM, kcat = 7.3 ± 0.6 s−1), indicating that this interaction is important for nucleotide binding and/or catalytic activity (Figure 2B).
Surprisingly, the F357A mutant, that abolishes the stacking interaction with the RecA2 domain, displayed an increased ATPase activity compared to wt Prp43 (KM = 90 ± 15 μM, kcat = 14.5 ± 0.6 s−1) (Figure 2C). This activity is comparable to the ATPase activity of the wt Prp43–Pfa1 complex. It therefore appears that disruption of the base stacking from the adenine to the RecA2 domain ‘lifts’ the auto-inhibited state of the Prp43 helicase. This is consistent with the hypothesis that the stacking of the adenine to F357 could influence the relative orientation of the RecA domains and could favor a nonproductive conformation of the RecA domains.
Prp43 has higher NTPase activity in the presence of pyrimidic compared to puric NTPs
In order to confirm that the effect of the F357A mutant is due to the loss of stacking interactions with the nucleotide base, we took advantage of the fact that Prp43 is able to hydrolyze all the NTPs due to the lack of specific contacts to the nucleotide base (8). No evidence that the enzyme uses other NTPs in vivo exists. Since the staking to F357 involves the adenine phenolic aromatic ring, a nucleotide triphosphate with a pyrimidic base would exhibit less stacking to F357 while preserving stacking interactions with R159. We therefore used NTP hydrolysis with pyrimidic bases as a convenient in vitro tool to probe the effect of F357 stacking and measured all the kinetic parameters of the NTPase activity of Prp43 in the presence of all the NTPs (Figure 2D).
The measured NTPase activity of the wt Prp43 for puric nucleotide triphosphates ATP and GTP are comparable, with a turnover of 3.4 and 3.7 s−1 (mol phosphate/mol active site/s) and a KM of 55 and 60 μM, respectively (Figure 2D). In contrast, the turnover of the NTPase activity of pyrimidic nucleotides is twice for UTP (9.7 s−1) to five times higher for CTP (15.4 s−1). The KM constant is increased two fold, suggesting that the absence of stacking to F357 decreased affinity for pyrimidic nucleotides.
Interestingly, compared to wt Prp43, the ATPase activity of the F357A mutant displays a four times increase of kcat (14.5 versus 3.4 s−1) and only a 2-fold increase of KM (90 versus 55 μM) (Figure 2A and C). The ATPase activity of the F357A mutant is therefore similar to the CTPase activity of wild-type Prp43 (kcat = 15.4 s−1). Although the cation-pi interaction of R159 to puric or pyrimidic bases could also contribute to the observed differences, these results seem to indicate that the main contribution comes from the reduced stacking to the nucleotide base by either the removal of F357 or by the use of a pyrimidine base, which have similar effects on catalytic activity.
To determine if other members of the DEAH/RHA helicases also have the same enzymatic properties as Prp43, we measured ATPase and CTPase activities of Prp22 in the presence of 0.2 mM polyA and 1 mM ATP or CTP (data not shown). The ATP hydrolysis rate is 3.7 s−1 and the CTP hydrolysis rate is 9.3 s−1. The NTPase activity with a pyrimidic base is therefore twice that of a puric base. The activity of a DHX9 construct containing only the two RecA-like domains of the helicase in the presence of pyrimidic NTP was previously found to be twice to five times that of DHX9 in the presence of puric NTP (48). These data confirm that the nature of the nucleotide base influences catalysis in other DEAH/RHA helicases. We propose that modulation of the NTPase activity through the stacking of the F-motif to the base could be a common feature of the DEAH/RHA family.
Structural analysis of CDP bound Prp43
NTPase activity assays seem to indicate that when nucleotide base stacking to the RecA2 domain is weakened, the NTPase activity is more important. In order to understand the structural features of this increase of activity in the presence of pyrimidic NTPs, we solved the structure of Prp43 in complex with CDP at 2.93 Å resolution. The complex crystallized in the same conditions as the two structures of Prp43 in complex with ADP previously solved (4,5). The structure has been solved by molecular replacement with Prp43 structure without ligand and water as a search model. Data processing and refinement statistics are presented in Table 1. In the two molecules of the asymmetric unit, a residual density corresponding to CDP is present in the active site after molecular replacement.
The overall structures of Prp43 in complex with ADP or CDP are the same with a calculated r.m.s.d. of 0.58 Å and therefore display the same relative orientation of the RecA domains. The active site of the CDP bound structure (Figure 1C) can be compared with the ADP bound (Figure 1B). The positions of magnesium and phosphates of ADP and CDP are superimposable, indicating no differences in binding of the phosphates in puric and pyrimidic nucleotides. This conservation in phosphate conformation is not surprising since the enzyme is able to hydrolyze all the NTPs, and suggests that the difference in NTPase activity is not due to a difference in phosphate binding and/or hydrolysis.
The most interesting difference between all our structures is the difference in stacking of the nucleotide base between R159 and F357. The adenine is stacked between the two residues, and establishes π-π and cation-π interactions with F357 or R159. In contrast, cytosine is translated 2 Å in the opposite direction to F357 compared to puric rings. In this position and due to the reduced size of the pyrimidic base, the cytosine loses the stacking interaction to F357 (Figure 1C). We therefore used cytosine-containing nucleotides as probes of the effect of stacking interactions to the RecA2 domain in our biochemical assays, since it mimics the effect of the F357 mutant.
Affinity of Prp43 for the nucleotide is impaired without base stacking
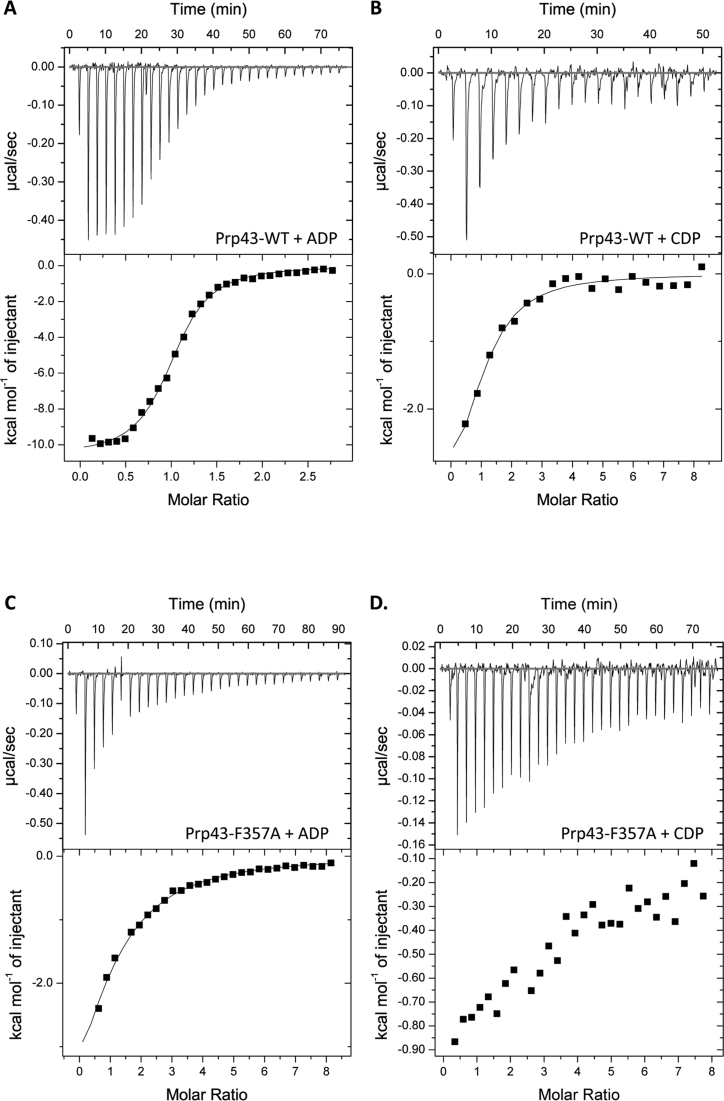
In order to further characterize Prp43 enzymatic activity and to determine what is the influence of base stacking on the affinity of the enzyme for its ligand, we measured binding affinities using ITC. The wild-type protein displays a Kd of 1.45 μM for ADP (Figure 3A). The Kd of Prp43 for CDP is superior to 25 μM (Figure 3B). We next performed the same experiment with Prp43 F357A. The Prp43–F357A mutant is strongly impaired in ADP binding, with a Kd superior to 45 μM (Figure 3C). No detectable binding of Prp43–F357A to CDP is measurable in our conditions (Figure 3D). No detectable binding of either ADP or CDP is observed with the Prp43–R159A mutant (data not shown).
Figure 3.
Affinity of Prp43 for ADP and CDP is impaired without base stacking. The affinity of Prp43-WT and Prp43–F357A for NDPs was assessed by ITC and each measure was performed three times. Affinities of Prp43-WT for ADP (A) and CDP (B) and of Prp43–F357A for ADP (C) and CDP (D).
These results show the R-motif is essential for NDP binding. Because the stacking of R159 on the NDP/NTP base is not influenced by the orientation of the RecA domains (Figure 1D), the R-motif is important for binding the nucleotide in the closed or open conformation. In contrast, the ITC experiments showed that stacking of the base of the nucleotide by F357 increases the affinity of Prp43 for NDP. Our experiments have been performed on nucleotide di-phosphates, which are the product of the reaction. The KM measured in the enzymatic assays for ATP and CTP are of the same order for Prp43 wild-type and Prp43 F357A, which suggests that base stacking has no influence on the affinity of Prp43 for its substrate. We propose that F357 has an essential role in binding the product but not the substrate. This could explain why the turnover of the enzyme increases when the stacking of F357 is lost: according to this model, the product is less retained in the active site without F357 stacking.
Affinity of Prp43 for RNA is modulated by nucleotide base stacking interaction
The affinity of helicases for their RNA substrates is often strongly dependent on the nucleotide occupancy state. For example, DEAD-box proteins alternate from a low to a high affinity RNA binding conformation in their ATP unbound/bound states. We therefore investigated the effect of base stacking interactions on the affinity of Prp43 (wt, F357A and R159A mutants) for a 21-mer single stranded RNA by filter binding experiments (Table 3 and Supplementary Figure S2).
Table 3. Affinity constants (Kd) of Prp43 (WT, R159A and F357A) for ssRNA in the presence of different NTPs.
| Nucleotide | Prp43 | Kd (nM) |
|---|---|---|
| No NTP | WT | 42 ± 17 |
| R159A | 20 ± 4 | |
| F357A | 14 ± 1 | |
| ATP | WT | 308 ± 55 |
| R159A | 18 ± 2 | |
| F357A | 87 ± 18 | |
| CTP | WT | 209 ± 42 |
| R159A | 8 ± 1 | |
| F357A | 43 ± 3 | |
| AMPPNP | WT | 8 ± 3 |
| R159A | 3 ± 0.8 | |
| F357A | 6 ± 1 | |
| ATP (10mM) | WT | 191 |
| ADP | WT | 144 |
| CDP | WT | 57 |
Binding affinities of Prp43-WT and the R159A and F357A mutants for a 21 nt ssRNA determined by filter-binding assays without or with different NTP at 1mM concentration unless otherwise stated (data in Supplementary Figure S2).
We find that all Prp43 constructs bind the RNA with ∼20 nM affinity in the absence of nucleotides (Supplementary Figure S2A). The affinity is even higher (<8 nM) for all constructs in the presence of a non-hydrolyzable ATP analog (Supplementary Figure S2B). In contrast, for wt Prp43 in the presence of ATP or CTP, the binding affinities decreased 7.5- and 5-fold respectively (Kd > 200 nM, Supplementary Figure S2C and D). It should be noted that the assays were performed in the absence of Pfa1, so that Prp43 has low helicase and ATPase activity, but a high CTPase activity. The observed low affinity of Prp43 in the presence of nucleotides could be due to a low helicase activity sufficient to dissociates the RNA during the assay. Saturating amounts of ATP (10 mM) do not affect the observed RNA affinity (191 versus 308 nM). However, in the presence ADP the binding is of the same order of magnitude as with ATP (144 versus 308 nM), whereas the RNA binding of wt Prp43 with CDP is 4-fold higher than in the presence of CTP (57 versus 209 nM).
Wt Prp43 shows higher binding affinity in the presence of CDP (57 nM) than ADP (144 nM), demonstrating the importance of binding of the F-motif to the nucleotide on RNA affinity (Supplementary Figure S2E). The Prp43–F357 mutant shows an intermediate binding affinity for RNA with ATP and CTP (87 and 43 nm respectively). In contrast, loss of stacking to R159 has a pronounced effect: Prp43–R159A mutant binds with nanomolar affinity to the RNA in the absence or presence of any nucleotide, suggesting that it is trapped in a high affinity binding state.
As could be expected, these results show an important contribution of the nucleotide occupancy on the RNA binding affinity of the enzyme. This is however the first time that stacking of two amino-acids on the nucleotide base is shown to significantly contribute to RNA binding affinity. The contribution of the F-motif to RNA affinity in the various nucleotide binding states therefore undoubtedly influences the helicase activity during the catalytic cycle.
The activation of Prp43 ATPase activity by Pfa1 is influenced by nucleotide base stacking interaction
We were able to stimulate the NTPase activity of Prp43 either by mutating F357 or by using pyrimidic nucleotides. Our working model is that this effect arises from the disruption of the stacking of the nucleotide base to F357. We therefore wanted to evaluate the effects of the G-patch protein partners in both systems. We used the G-patch containing Pfa1 protein as a model of G-patch protein partner because it is able to stimulate both ATPase and helicase activities.
In agreement with previous observations, ATPase activity of Prp43 is stimulated seven fold when Pfa1 is added (Figure 2A) (25). Surprisingly, although the Prp43 CTPase activity is of the same order of magnitude as the Prp43/Pfa1 ATPase activity (kcat of 25.4 s−1), the CTPase activity of Prp43/Pfa1 is reduced 2-fold (kcat of 8.6 s−1) (Figure 2F). In agreement with these results, the ATPase activity of Prp43–F357A/Pfa1 is also reduced by a factor of 2 (6.9 s−1) and is accompanied by a drastic increase of the KM (1385 μM with Pfa1 versus 90 μM without Pfa1). These results show that disruption of stacking interaction of the nucleotide base with F357, either through mutation of F357 or use of pyrimidic nucleotides, has the same unexpected effect. Pfa1 is unable to further stimulate the NTPase activity which, on the contrary, is decreased (Table 2).
The CTPase activity of Prp43–F357A/Pfa1 is not detectable in our conditions, presumably because it accumulates the effect of both the absence of F357 and a decreased stacking to the pyrimidic base. Likewise, although Prp43–R159A had detectable NTPase activity, we did not detect any ATPase or CTPase activity of the Prp43-R159/Pfa1 complex (Figure 2B and E). Taken together, our results show that the stacking of the nucleotide base to both RecA domains is essential for the activation of the enzyme by the G-patch protein Pfa1.
Influence of stacking on the helicase activity of Prp43
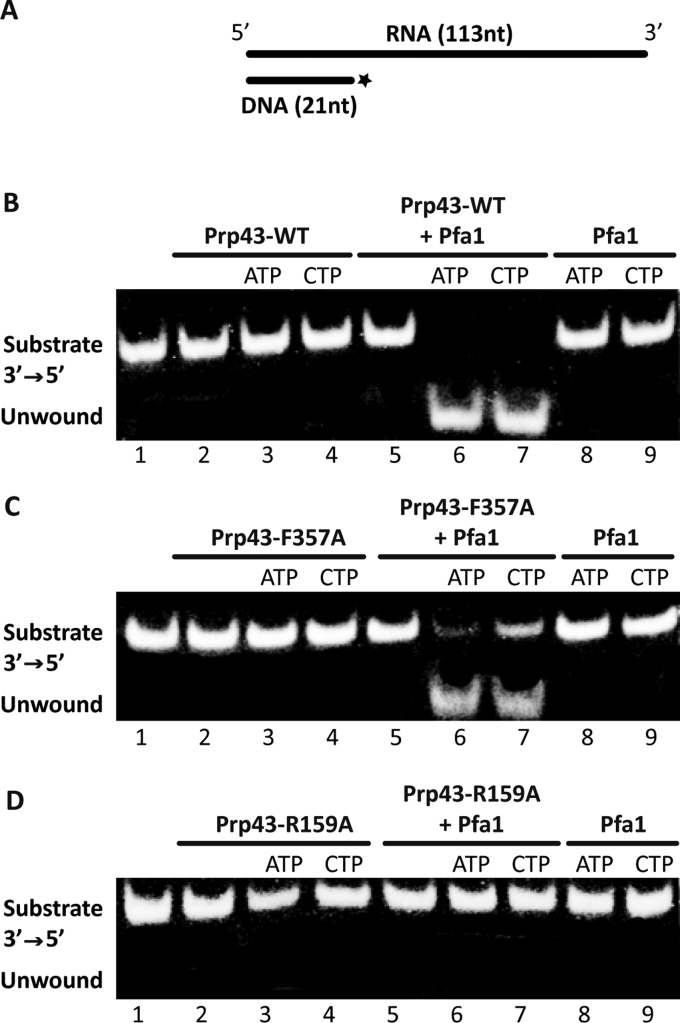
Helicases have the capacity to couple ATP hydrolysis or binding to nucleic acid unwinding. We wanted to investigate how the stacking of the nucleotide base by the residues of the RecA1 and RecA2 domains is correlated to nucleic acid unwinding. Prp43 is able to unwind short duplexes of RNA/DNA with a flanking 5΄ or 3΄ extremity (25,27). We have performed bulk helicase activities on DNA-RNA substrates as previously described (25) (Figure 4A), with a Alexa680 fluorescent labeled, instead of radioactive labeled, DNA.
Figure 4.
Influence of stacking on Prp43 helicase activity. Helicase assays were performed with a 3΄-5΄ substrate (A) which is a RNA–DNA hybrid substrate. The 113 nucleotide-long RNA probe corresponds to the 5΄ stem loop and H box of snR5 snoRNA. It was annealed with a 21 nucleotide-long fluorescently labelled DNA oligonucleotide. Unwinding of a DNA–RNA substrate by Prp43WT (B), Prp43–F357A (C) and Prp43–R159A (D) was assessed following incubation with ATP or CTP and with or without Pfa1 as indicated. Positions of the RNA/labelled DNA substrate and the unwound oligonucleotide are indicated.
In agreement with previous work, no helicase activity could be measured in the absence of NTP for any of the samples tested: Prp43 (Figure 4B lane 2), Prp43–F357A (Figure 4C lane 2) Prp43–R159A (Figure 4D, lane 2), and the same conditions in the presence of Pfa1 alone (Figure 4B, C and D, lanes 8 and 9). In our conditions, the helicase activity of wild-type Prp43, Prp43–F357A and Prp43–R159A is not detectable in the presence of either ATP or CTP (Figure 4B and C and D, lanes 3 and 4). Therefore, although the CTPase activity of Prp43 or the ATPase activity of Prp43–F357A is upregulated, this increased NTPase activity does not lead to an increased helicase activity.
In agreement with previous work, we find that Prp43/Pfa1 is able to unwind the duplex in the presence of ATP (Figure 4B, lane 6 and Supplementary Figure S3B). However, despite the observation that Prp43/Pfa1 CTPase and Prp43–F357A/Pfa1 ATPase activity is weaker than Prp43/Pfa1 ATPase activity in the enzymatic assays (Figure 2A, C and F), only a minor difference is observed in the helicase activity in our conditions (Figure 4B and C and Supplementary Figure S3B and D).
Surprisingly, we could detect helicase activity of Prp43–F357A/Pfa1 in the presence of CTP (Figure 4C, lane 7), although no CTPase activity could be detected (Figure 2F). The amount of unwound RNA is however reduced by a factor of two. Therefore, in our assay, we only see a difference in helicase activity with the combination of F357A mutation and the use of CTP as a co-factor.
In contrast, Prp43–R159A, that also has no detectable ATPase or CTPase activity in the presence of Pfa1 (Figure 2B and E), has barely detectable unwinding activity without or with Pfa1 and independently of the nature of the NTP base (Figure 4D, lanes 3, 4, 6 and 7 and Supplementary Figure S3C). Our results demonstrate that in contrast to the stacking to the RecA2 domain, stacking to the RecA1 domain mediated by the R159 residue is essential to maintain NTPase and helicase activities.
Effect of loss of nucleotide stacking in vivo
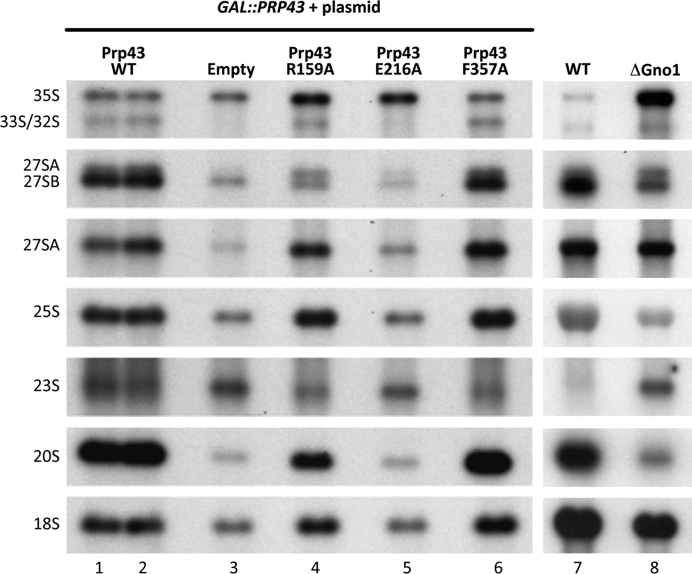
In order to observe the effect of nucleotide stacking on the in vivo activity of Prp43, we performed yeast complementation assays with Prp43 mutants. Since Prp43 is essential in yeast (10), we constructed a strain in which the chromosomal PRP43 ORF is placed under the control of a galactose promoter (GAL::PRP43), and we transformed this strain with plasmids expressing wild-type or mutant versions of PRP43, or with the empty vector as a control. The resulting cells were shifted to a glucose-containing medium to repress the expression of the chromosomal PRP43 allele and the phenotypes resulting from the expression of the plasmid-borne copies were analyzed. Complementation with the Prp43-E216A mutant, which is catalytically dead in vitro, or the empty vector, could not support growth on glucose. Pre-rRNA processing analyses indicate that in these two conditions, there is accumulation of 35S and 23S, and strong depletions of all other pre-rRNA intermediates and mature rRNA compared to wild-type (Figure 5, lanes 1, 2, 3 and 5).
Figure 5.
Prp43 R159 is important for rRNA processing. Strain GAL::PRP43 expressing the chromosomal PRP43 open reading frame under the control of the glucose-repressible promoter was transformed with pHA113 plasmids carrying PRP43-WT (lanes 1 and 2), Prp43–R159A (lane 4), PRP43-E216A (lane 5) or Prp43–F357A (lane 6), or with the empty pHA113 vector as a control (lane 3). The resulting strains were grown on a galactose-containing medium (chromosomal PRP43 expressed) and shifted to glucose for 27 h to deplete endogenous Prp43 proteins. Cells were harvested, total RNAs were extracted and the accumulation levels of the indicated pre-rRNAs were analysed by northern blot using specific oligonucleotide probes. The processing defects observed with PRP43–R159A can be compared to those observed in absence of Gno1 (lane 8, see (28)). The 27SA2 intermediate is detected using two different probes hybridizing either within ITS2 (upper signal, detection of 27SA2 and 27SB intermediates) or within ITS1 between sites A2 and A3 (detection of 27SA2 alone).
Since stacking of the nucleotide on the R- and F-motifs influences both the ATPase and the helicase activities, we analyzed the complementation with the Prp43–R159A and Prp43–F357A mutants. Both mutants are viable and Western blotting analysis indicated that the two mutant proteins are expressed and stable in yeast (Supplementary Figure S4). Prp43–F357A mutant displayed no observable pre-rRNA processing defects (Figure 5, lane 6). In contrast, the strain expressing Prp43–R159A displayed a processing defect characterized by an accumulation of the 35S pre-rRNA, a strong depletion of the 27SB pre-rRNAs but not of the 27SA2 intermediate, and a depletion of the 20S (Figure 5, lane 4). The main difference between this processing defect (R159A) and that observed under conditions of full depletion of Prp43 (EV), or expression of a catalytically dead mutant (E216A), is that the accumulation levels of the 27SA2 are not affected. This result indicates that Prp43 R159A retains some activity that allows proper production and maturation of the 27SA2 intermediate. Surprisingly, this processing defect is highly reminiscent of the defect observed in the absence of the G-patch protein Gno1 (Figure 5, lane 8, and (28)). This result shows that the Prp43–R159A mutation phenocopies the absence of Gno1, strongly suggesting that the processing defects observed in the absence of Gno1 result from a failure to activate Prp43.
Since the F357A and R159A mutants are viable in our complementation assays, we constructed yeast strains expressing these mutants as sole source of Prp43 protein to more precisely characterize their growth rate. We observed that Prp43–F357A mutant displayed wild type growth phenotype (Supplementary Figure S5C) consistent with the absence of processing defect observed in the complementation assay. In contrast, Prp43–R159A displayed a small growth defect in rich medium (Supplementary Figure S5B).
In agreement with the in vitro results, the stacking of the nucleotide to the RecA1 or to the RecA2 domains has different consequences in vivo. Loss of stacking to the RecA2 domain (F357) does not seem to affect the in vivo activity in agreement with the mild helicase activity defect in vitro. In contrast, loss of stacking to the RecA1 domain (R159), which strongly reduces the helicase activity in vitro, induces a growth defect (Supplementary Figure S5B). Interestingly, the rRNA processing phenotype of this mutant is less severe than that of an active site mutant (Figure 5, compare lanes 4 and 5) which may suggest that some functions of Prp43 in pre-rRNA processing are differently affected in this mutant.
G-patch activation of nucleotide stacking mutants in vivo
Because the Prp43–F357A mutant displayed an ATPase activity similar to the Prp43-wt in the presence of the activator G-patch protein Pfa1 in vitro, we evaluated whether this mutant could rescue in vivo the growth and pre-rRNA processing defects caused by the absence of a G-patch protein. The absence of Gno1 induces a processing defect previously described in Figure 5, lane 8. Since Pfa1 is not essential, we performed these experiments in a ΔPfa1, GAL::LTV1 strain which displays a strong accumulation of the 20S precursor to the mature 18S rRNA. Because we could not know which activity of Prp43 would be most affected, we tried to rescue the growth defect of cells lacking either Pfa1 or Gno1 by overexpression of Prp43–F357A. In both cases, overexpression of the Prp43–F357A mutant is unable to rescue the growth and pre-rRNA processing defects due to the absence of Pfa1 or Gno1 (data not shown). This indicates that although the ATPase activity of Prp43–F357A is similar to that of wt Prp43+Pfa1, Prp43–F357A cannot function in vivo without Pfa1. This is in agreement with the observation that Prp43–F357A does not have measurable helicase activity in the absence of Pfa1 in vitro. The F357A mutation therefore seems to uncouple the ATPase and helicase activities of Prp43, and cannot up regulate its activity in vivo.
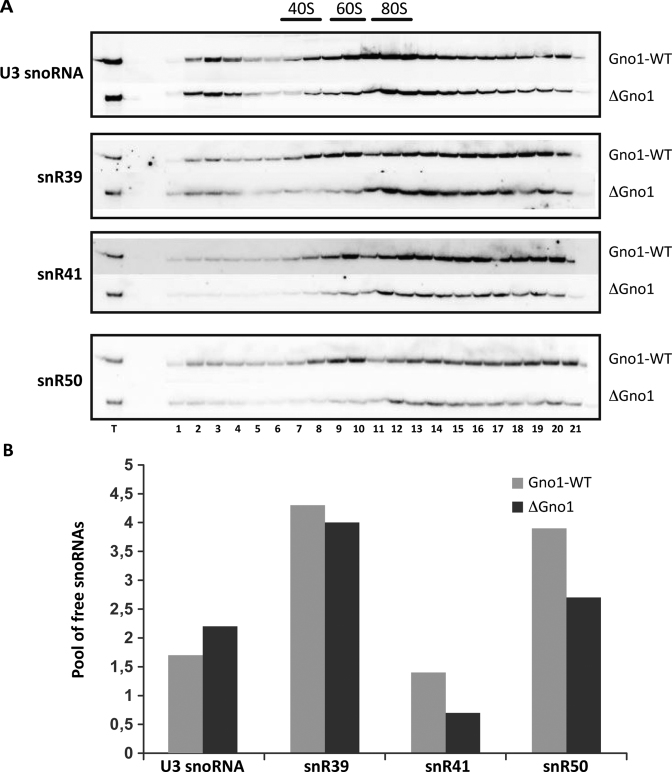
Since the R159A mutant displays rRNA processing defects similar to the Δgno1 strain, we investigated further the functional link between Prp43–R159A and Gno1. Prp43 activity is necessary for releasing certain snoRNAs such as snR39, snR41 and snR50 from pre-ribosomes, targeting the methylation of 25S rRNA nucleotides within 90S or early pre-60S pre-ribosomes (13). This was inferred from the analysis of the sedimentation profiles of snoRNPs on sucrose gradients, Prp43 depletion inducing a retention of some snoRNPs into dense fractions containing large pre-ribosomal particles and a concomitant reduction of the pool of free snoRNPs. We have repeated sedimentations on sucrose gradients with a Δgno1 strain and with Prp43 mutants (Figure 6 and Supplementary Figure S6) and analyzed the retention of snR39, snR41 and snR50 snoRNAs, and U3 as a negative control. We found that the absence of Gno1 depletion affected the sedimentation profiles of snR39, snR41 and snR50 (Figure 6) and also that of U3 but to a lesser extent. The most striking difference is the depletion of snR39, snR41 and snR50 from the fractions corresponding to pre-60S particles (lanes 8–9–10) compared to the wild type profiles. This observation indicates that in wild type cells, these snoRNAs remain associated with early or intermediate pre-60S particles, a result fully consistent with the fact that several snoRNAs guiding 25S rRNA nucleotide modifications are found associated with Npa1, a component of early pre-60S particles interacting predominantly with the 27SA2 pre-rRNA (49). On this basis, the release of snR39, snR41 and snR50 from pre-ribosomes by Prp43 is expected to occur within early or intermediate pre-60S particles. If Gno1 is required for this release, an accumulation of these snoRNAs within pre-60S particles is expected to occur in absence of Gno1. We observed on the contrary a decrease in the accumulation of these snoRNAs in fractions 8–9–10. We reported previously that Gno1 depletion affects the production of early pre-60S particles and induces partial degradation of intermediate pre-60S particles containing the 27SA3 and 27B precursors (28). This most likely explains the depletion of snR39, snR41 and snR50 from pre-60S particles containing fractions. Interestingly however, quantifications showed that, as reported upon Prp43 depletion, the pool of free snR39, snR41 and snR50 snoRNAs decreases in absence of Gno1 suggesting retention into pre-ribosomes. This behavior is not common to all snoRNAs since, on the contrary, the pool of free U3 increases. This result is in favor of a role of Gno1 in the activation of Prp43 to release these snoRNAs, but the fact that Gno1 is required for efficient maturation of 90S particles and for the production and stability of intermediate pre-60S particles complicates the interpretations.
Figure 6.
Sedimentation profiles of snR39, snR41, snR50 and U3 in absence of Gno1. (A) Sedimentation profiles of different snoRNAs: snR39, snR41, snR50 and U3 snoRNA as a reference, using extracts from cells expressing or not the G-patch co-factor Gno1. (B) PhosphorImager quantifications of the pool of free snoRNAs (snoRNAs present in fractions 1–5) relatively to the signal corresponding to the snoRNAs in the input sample (T).
We next tested the effect of Prp43 R159A and F357A mutations using sucrose gradient sedimentations (Supplementary Figure S6). Although the effects are less pronounced than in the absence of Gno1, the pool of free snR39, snR41 and snR50 snoRNAs are also reduced in the R159A mutant compared to the wild type profile (Supplementary Figure S6). As observed upon full depletion of Prp43 (13) the sedimentation profile of U3 is also modified to some extent, possibly because of indirect effects due to perturbations of other stages of the processing pathway. Similarly to the phenotype observed in the absence of Gno1, the Prp43–R159A mutant also displayed a reduced 60S peak suggesting that stacking of the nucleotide base to the RecA1 domain is required for efficient production of the 60S subunit (Supplementary Figure S6B).
CONCLUSION
The nucleotide base couples RecA domains
In Prp43, the base of the nucleotide is sandwiched between R159 (R-motif, RecA1) and F357 (F-motif, RecA2) of the two RecA-like domains, a binding mode of the base which is specific to the DEAH/RHA family (5). We show that the NTPase activity of Prp43 is higher when the base stacking to the RecA2 domain is impaired, and that this interaction is important in the regulation by G-patch proteins. In the translocation model of DEAH/RHA helicase proposed from the structures of Prp43 and MLE, the RecA-like domains alternate between an open and a closed conformation (5,37). We reasoned that this interaction of Prp43 with the ATP base is implicated in a sort of structural communication between the two RecA-like domains and could be involved in auto-inhibition of the enzyme.
In order to evaluate the contribution of base stacking to the function of Prp43, we used Prp43 mutants impaired in stacking with the NTP base either on the RecA1 domain (R159A) or on the RecA2 domain (F357A). We also used CTP as the helicase substrate in our in vitro assays, since we showed that the smaller cytosine base is less stacked to F357 compared to adenine. We show that base stacking of the NTP on the R-motif of the RecA1 domain is necessary for the basal NTPase activity, while stacking to the F-motif on the RecA2 domain seems to inhibit the activity of the enzyme. More importantly, stacking of the nucleotide base to the R- or F-motif also affects Prp43 activation by Pfa1: when stacking to one of these two motifs is removed, Pfa1 fails to activate Prp43 NTPase activity.
Surprisingly, the effect of these mutations on NTPase activity does not translate in similar effect on the helicase activity. For example, although the ATPase activity of the Prp43–F357A mutant is similar to the ATPase activity of the Prp43/Pfa1 complex, the helicase activity of this mutant in absence of Pfa1 is undetectable in vitro. This seems to indicate that the F357A mutation enables the protein to hydrolyze the nucleotide, but has lost the ability to couple ATP hydrolysis to unwind the nucleic acid duplex, much in the way a motor can consume its fuel, but runs with a broken driving belt. This shows that caution should be used when assessing the activity of helicases on the basis of their ATPase activity alone. Interestingly, there is a strict correlation between the NTPase activity of the Prp43/Pfa1 complex and its helicase activity (ie Prp43–F357A + ATP > Prp43–F357A + CTP > Prp43–R159A + NTP), suggesting that the G-patch effectively couples the ATPase and helicase activities.
Although the R159A mutation strongly affects Prp43 helicase activity in vitro, this mutation only induces a mild growth defect in vivo suggesting that the residual ATPase and helicase activities of Prp43–R159A (Figure 2 and Supplementary Figure S3) are sufficient to fulfill some of the functions of Prp43 in vivo. Interestingly, the extent of the pre-RNA processing defects induced by the different mutations in vivo correlates with the extent of the defects observed in vitro (E216A > R159A > F357A).
In the work of Warkocki et al. (33), the authors used in vivo purified spliceosome complexes as substrates of the DEAH helicase Prp2. In order to activate Prp2 and no other helicase, they very cleverly used the fact that in contrast to other helicases, Prp2 is able to hydrolyze UTP, and performed activity tests using UTP as the sole NTP source. Although in the conditions used the ATP and UTP activities seem to be similar, our work suggests that using a pyrimidic nucleotide for these assays could lead to potentially unforeseen effects in the NTPase activity, the helicase activity or the activation by G-patch proteins.
Model of Prp43 activation
The mechanism by which G-patch proteins activate DEAH/RHA helicases remains poorly understood. In addition, the precise mechanism for RNA unwinding also remains controversial, as the structural elements favor a processive helicase mechanism, while it was suggested that the Dhr1 DEAH/RHA helicase functions with a DEAD-box like mechanism (50). In this work, we highlight a novel mechanism that contributes to the regulation of the activity of this family of helicases. We show that the stacking of the nucleotide base between two opposing residues of the two RecA domains influences Prp43 ATPase and helicase activities, its activation by G-patch proteins in vitro, and its activity in vivo.
We propose that the nucleotide base couples the ATPase and helicase activities by locking the conformation of the two RecA domains through the interaction with R- and F-motifs. This interaction would act as an autoinhibition mechanism that would keep the helicase in a non productive state, by preventing the reorientation of the two RecA domains and keeping the helicase in a single conformational state. In our model, interaction of the G-patch protein would release the inhibition mechanism and allow the protein to separate nucleic acid strands. This model is compatible with both a processive or DEAD-box helicase mechanism. In the processive mechanism, the RecA domains have to cycle through an open and closed conformation, while in the DEAD-box like mechanism, the enzyme alternates between a high and low RNA binding state. The precise mechanism for DEAH/RHA helicase function and regulation will therefore need the structures of RNA bound states and of a complex with G-patch proteins.
Unwinding of snoRNA/pre-rRNA base-pairings by Prp43-Gno1 in vivo
Prp43 has been detected by CRAC at the vicinity of several nucleotides in the 25S rRNA known to be methylated by box C/D snoRNAs such as snR39, snR41 or snR50 (13). Depletion of Prp43 induces retention in the pre-ribosomal particles of these specific snoRNAs, leading to the proposal that Prp43 unwinds the base-pairing between these snoRNAs and the pre-rRNA. Our data show that these snoRNAs, and very likely several others modifying 25S rRNA nucleotides also, co-sediment with 90S and pre-60S particles on sucrose gradients in wild type cells, indicating that they are incorporated into 90S pre-ribosomes and that their release by Prp43 occurs at the stage of intermediate pre-60S particles. In absence of Gno1, we observed a reduction of the pool of free snR39, snR41 and snR50 snoRNAs but not of U3, suggesting that Gno1 is required for their dissociation from pre-ribosomes. However, these snoRNAs do not accumulate in the fractions of the gradient corresponding to pre-60S particles, most likely as a consequence of partial degradation of intermediate pre-60S particles containing all 27S species except 27SA2. Since Gno1 activates Prp43 in vitro (28), these data suggest that unwinding of the snoRNAs by Prp43 requires activation by Gno1 in yeast cells. This is further supported by the fact that the R159A mutation phenocopies the absence of Gno1 both in terms of snoRNA retention and global processing. The R159A mutation prevents activation by a G-patch cofactor in vitro and strongly affects the helicase activity of Prp43. Our data therefore suggest that the helicase activity of Prp43 is required for snoRNA unwinding. The absence of Gno1 or expression of Prp43 R159A affects the maturation of the pre-60S particles, suggesting that a failure to unwind snoRNAs from the pre-rRNA perturbs the maturation of the pre-ribosomal particles. It is very likely however that both Prp43 and Gno1 fulfill additional functions, in the maturation of 90S and pre-60S particles.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline scientists on the PROXIMA I and II beamlines.
Author contributions: J.R.P. and S.R. expressed and purified, crystallized the Prp43/CDP complex and solved the crystal structure. J.R.P. and F.C. performed the NTPase assays. J.R.P. and L.D. performed the ITC experiments. F.C. and M.B. performed the helicase assays. S.L. performed RNA protein binding assays. M.H. constructed yeast expression vectors, generated yeast strains, performed yeast complementation assays (growth curves, serial dilutions, westerns) and pre-rRNA processing analyses by northern blot. M.H. and A.H. performed the sedimentations on sucrose gradients and downstream analyses. R.C. expressed and purified Prp43 proteins and O.H. performed helicase assays for Supplementary Figure S3. N.L., J.R.P. and S.R. designed the studies, interpreted data and wrote the paper with contributions from S.L., M.B., A.H., Y.H.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Work at UPD was supported by CNRS, University Paris Descartes; Nanogears grant from USPC; RNPGenesis grant from the Agence Nationale de la Recherche (ANR-11-JSV8-0004), the Institut Universitaire de France (to N.L.). Work in the Henry/Henras group is supported by CNRS, University of Toulouse III (Paul Sabatier) and Agence Nationale de la Recherche grant RIBOPRE40S (ANR-10-BLAN-1224). J.R.P. and M.H. were supported by Ph.D. fellowships from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche. Funding for open access charge: University Paris Descartes.
Conflict of interest statement. None declared.
REFERENCES
- 1.Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 2.Tanner N.K., Linder P.. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001; 8:251–262. [DOI] [PubMed] [Google Scholar]
- 3.Jankowsky E., Fairman M.E.. RNA helicases — one fold for many functions. Curr. Opin. Struct. Biol. 2007; 17:316–324. [DOI] [PubMed] [Google Scholar]
- 4.He Y., Andersen G.R., Nielsen K.H.. Structural basis for the function of DEAH helicases. EMBO Rep. 2010; 11:180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walbott H., Mouffok S., Capeyrou R., Lebaron S., Humbert O., van Tilbeurgh H., Henry Y., Leulliot N.. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010; 29:2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert-Paganin J., Rety S., Leulliot N.. Regulation of DEAH/RHA Helicases by G-Patch Proteins. BioMed Res. Int. 2015; e931857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heininger A.U., Hackert P., Andreou A.Z., Boon K.-L., Memet I., Prior M., Clancy A., Schmidt B., Urlaub H., Schleiff E. et al. Protein cofactor competition regulates the action of a multifunctional RNA helicase in different pathways. RNA Biol. 2016; 13:320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka N., Schwer B.. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry (Mosc.). 2006; 45:6510–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourmann J.-B., Schmitzová J., Christian H., Urlaub H., Ficner R., Boon K.-L., Fabrizio P., Lührmann R.. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013; 27:413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebaron S., Froment C., Fromont-Racine M., Rain J.-C., Monsarrat B., Caizergues-Ferrer M., Henry Y.. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005; 25:9269–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Combs D.J., Nagel R.J., Ares M., Stevens S.W.. Prp43p is a DEAH-Box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 2006; 26:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeds N.B., Small E.C., Hiley S.L., Hughes T.R., Staley J.P.. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 2006; 26:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohnsack M.T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D.. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009; 36:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosallanejad K., Sekine Y., Ishikura-Kinoshita S., Kumagai K., Nagano T., Matsuzawa A., Takeda K., Naguro I., Ichijo H.. The DEAH-Box RNA helicase DHX15 activates NF-κB and MAPK signaling downstream of MAVS during antiviral responses. Sci. Signal. 2014; 7:ra40–ra40. [DOI] [PubMed] [Google Scholar]
- 15.Fairman-Williams M.E., Guenther U.-P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu M., Rice C.M.. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikrishnan K., Powell B., Cook N.J., Webb M.R., Wigley D.B.. Mechanistic Basis of 5΄-3΄ Translocation in SF1B Helicases. Cell. 2009; 137:849–859. [DOI] [PubMed] [Google Scholar]
- 18.Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008; 37:317–336. [DOI] [PubMed] [Google Scholar]
- 19.Pan C., Potratz J.P., Cannon B., Simpson Z.B., Ziehr J.L., Tijerina P., Russell R.. DEAD-Box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol. 2014; 12:e1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohman T.M., Tomko E.J., Wu C.G.. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008; 9:391–401. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.Y., Yang W.. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006; 127:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Büttner K., Nehring S., Hopfner K.-P.. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007; 14:647–652. [DOI] [PubMed] [Google Scholar]
- 23.Jarmoskaite I., Russell R.. RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 2014; 83:697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson S.J., Jackson R.N.. Ski2-like RNA helicase structures. RNA Biol. 2013; 10:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebaron S., Papin C., Capeyrou R., Chen Y.-L., Froment C., Monsarrat B., Caizergues-Ferrer M., Grigoriev M., Henry Y.. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 28:3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christian H., Hofele R.V., Urlaub H., Ficner R.. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2013; 42:1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka N., Aronova A., Schwer B.. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007; 21:2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y.-L., Capeyrou R., Humbert O., Mouffok S., Kadri Y.A., Lebaron S., Henras A.K., Henry Y.. The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res. 2014; 42:7330–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu Z., Jin W., Zhang L., Li X.. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012; 586:977–983. [DOI] [PubMed] [Google Scholar]
- 30.Lin M.L., Fukukawa C., Park J.H., Naito K., Kijima K., Shimo A., Ajiro M., Nishidate T., Nakamura Y., Katagiri T.. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 2009; 100:1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy J., Kim K., Maddock J.R., Anthony J.G., Woolford J.L.. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995; 1:375–390. [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman E.J., Maeda A., Wei J., Smith P., Beggs J.D., Lin R.J.. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004; 24:10101–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warkocki Z., Schneider C., Mozaffari-Jovin S., Schmitzová J., Höbartner C., Fabrizio P., Lührmann R.. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015; 29:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai R.T., Fu R.H., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C.. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005; 19:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pertschy B., Schneider C., Gnädig M., Schäfer T., Tollervey D., Hurt E.. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009; 284:35079–35091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P.. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003; 11:127–138. [DOI] [PubMed] [Google Scholar]
- 37.Prabu J.R., Müller M., Thomae A.W., Schüssler S., Bonneau F., Becker P.B., Conti E.. Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol. Cell. 2015; 60:487–499. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka N., Schwer B.. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-Box splicing dactor Prp22†. Biochemistry (Mosc.). 2005; 44:9795–9803. [DOI] [PubMed] [Google Scholar]
- 39.Leslie A.G., Powell H.R.. Processing diffraction data with Mosflm. Evolving Methods for Macromolecular Crystallography. 2007; Springer; 41–51. [Google Scholar]
- 40.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2005; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 45.Tollervey D., Mattaj I.W.. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987; 6:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauchert M.J., Fourmann J.-B., Christian H., Lührmann R., Ficner R.. Structural and functional analysis of the RNA helicase Prp43 from the thermophilic eukaryote Chaetomium thermophilum. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016; 72:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobza P., Selzle H.L., Schlag E.W.. Potential energy surface for the benzene dimer. Results of ab Initio CCSD(T) calculations show two nearly isoenergetic structures: T-shaped and parallel-displaced. J. Phys. Chem. 1996; 100:18790–18794. [Google Scholar]
- 48.Schütz P., Wahlberg E., Karlberg T., Hammarström M., Collins R., Flores A., Schüler H.. Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-Box variant protein. J. Mol. Biol. 2010; 400:768–782. [DOI] [PubMed] [Google Scholar]
- 49.Dez C., Froment C., Noaillac-Depeyre J., Monsarrat B., Caizergues-Ferrer M., Henry Y.. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 2004; 24:6324–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sardana R., Liu X., Granneman S., Zhu J., Gill M., Papoulas O., Marcotte E.M., Tollervey D., Correll C.C., Johnson A.W.. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015; 13:e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 52.Johnson G.T., Autin L., Goodsell D.S., Sanner M.F., Olson A.J.. ePMV embeds molecular modeling into professional animation software environments. Struct. Lond. Engl. 2011; 19:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.