Abstract
Structural and biochemical features suggest that the almost ubiquitous bacterial YbeY protein may serve catalytic and/or Hfq-like protective functions central to small RNA (sRNA)-mediated regulation and RNA metabolism. We have biochemically and genetically characterized the YbeY ortholog of the legume symbiont Sinorhizobium meliloti (SmYbeY). Co-immunoprecipitation (CoIP) with a FLAG-tagged SmYbeY yielded a poor enrichment in RNA species, compared to Hfq CoIP-RNA uncovered previously by a similar experimental setup. Purified SmYbeY behaved as a monomer that indistinctly cleaved single- and double-stranded RNA substrates, a unique ability among bacterial endoribonucleases. SmYbeY-mediated catalysis was supported by the divalent metal ions Mg2+, Mn2+ and Ca2+, which influenced in a different manner cleavage efficiency and reactivity patterns, with Ca2+ specifically blocking activity on double-stranded and some structured RNA molecules. SmYbeY loss-of-function compromised expression of core energy and RNA metabolism genes, whilst promoting accumulation of motility, late symbiotic and transport mRNAs. Some of the latter transcripts are known Hfq-binding sRNA targets and might be SmYbeY substrates. Genetic reporter and in vitro assays confirmed that SmYbeY is required for sRNA-mediated down-regulation of the amino acid ABC transporter prbA mRNA. We have thus discovered a bacterial endoribonuclease with unprecedented catalytic features, acting also as gene silencing enzyme.
INTRODUCTION
The ybeY gene is almost ubiquitous in bacteria and has been included in the minimal prokaryotic genome set (1–3). These evidences support a rather fundamental physiological role of its protein product. Accordingly, YbeY loss-of-function is lethal in some bacterial species and in others can result in pleiotropic changes (3–8). Structural studies of YbeY orthologs uncovered a conserved three histidine H(X)3H(X)4DH motif shared by metallo-hydrolases and global similarity to the MID domain of eukaryotic AGO proteins involved in small RNA-directed gene silencing (9–12). These features suggest that YbeY could serve catalytic and/or RNA-binding/chaperone functions in bacteria. However, the biochemical activity of this protein has remained elusive until recently.
Escherichia coli YbeY (from here on EcoYbeY) has been shown to be required for efficient translation, optimal activity of the 30S ribosomal subunit and accurate ribosome assembly, particularly upon a temperature upshift (5,13–15). The severe defects of EcoYbeY deletion mutants in ribosome biogenesis have been mainly attributed to a failure in rrn transcription anti-termination and abnormal maturation of the rRNA precursors, which together result in accumulation of misprocessed 16S, 23S and 5S rRNA species (5,13,16). Similar effects on rRNA maturation have been also described for YbeY orthologs of the human pathogens Yersinia enterocolitica and Vibrio cholerae (7,8). Supporting these observations it has been also reported that the purified EcoYbeY acts as a single strand-specific metallo-endoribonuclease (13). The H114 residue within the histidine triad involved in metal binding and the arginine R59 integrating another conserved cluster of amino acids are required for EcoYbeY activity in vivo and in vitro (5,12,13). Processing of the rRNA precursors to their functional mature forms in vivo involves the concerted activity of additional RNases, including RNase III, RNase R, RNase E, RNase G and PNPase (5,13,17). Indeed, YbeY and RNase R are major components of a remarkable late-ribosome quality control system, particularly important for stress survival, which mediates removal of assembled 70S ribosomes specifically containing defective 30S subunits (13).
Structural modeling of the YbeY protein encoded by the nitrogen-fixing symbiotic bacterium Sinorhizobium meliloti evidenced a positively charged cavity resembling the AGO MID domain that anchors si/miRNAs to the RNA-induced silencing complex (10,18,19). In bacteria, a large fraction of the known small non-coding RNAs (sRNAs) regulates translation and/or stability of trans-encoded target mRNAs (20). To date, the chaperone activity assisting trans-sRNA function has been almost exclusively attributed to the widespread bacterial RNA-binding protein Hfq (21,22). However, nearly half of the sequenced bacterial genomes do not encode a recognizable Hfq homolog and several well-characterized trans-sRNAs have been shown to be Hfq-independent (23–25). For example, in S. meliloti only 14% of the annotated trans-acting sRNAs bind to Hfq (26). These observations suggest that other proteins may assist sRNA-mediated post-transcriptional control of gene expression. YbeY has been proposed to fulfil this important role since: (i) lack or depletion of YbeY results in differential accumulation of subsets of sRNAs and their predicted mRNA targets in E. coli, S. meliloti and V. cholerae (8,10,27) and (ii) YbeY and Hfq similarly influence S. meliloti sensitivity to a number of stress agents (e.g. paraquat, SDS, ethanol, NaCl or heat) and the symbiotic interaction with its legume host, alfalfa (3,10,28). However, these studies did not provide data supporting an active role of YbeY in the sRNA–mRNA interplay.
In this work, we have purified and characterized S. meliloti YbeY (from here on SmYbeY) regarding its RNA-binding ability and biochemical activity. Furthermore, we have also investigated the influence of SmYbeY loss-of-function in the S. meliloti transcriptome. Our results show that this protein acts as a metal-dependent single and double-strand endoribonuclease rather than as Hfq-like RNA chaperone. SmYbeY activity profoundly impacts on conserved fundamental chromosomally-encoded functions as well as on certain acquired plasmid-encoded S. meliloti pathways. We also provide evidences of the involvement of SmYbeY in the Hfq-dependent riboregulation of amino acid uptake.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions and oligonucleotides
Bacterial strains and plasmids used in this work along with their relevant characteristics are listed in Supplementary Table S1. Oligonucleotide sequences are provided in Supplementary Table S2. E. coli strains were routinely grown in Luria–Bertani medium (LB) at 37°C and rhizobia in either complex tryptone-yeast (TY) or defined MM media at 30°C (29,30). When required, growth media were supplemented with the appropriate antibiotic(s) at the following final concentrations (μg/ml) unless otherwise stated: streptomycin (Sm) 250, ampicillin (Ap) 200, tetracycline (Tc) 10, erythromycin (Er) 100, gentamicin (Gm) 45 and kanamycin (Km) 50 for E. coli and 180 for rhizobia.
For co-immunoprecipitation (CoIP) experiments wild-type and SmybeYFLAG strains were subjected to five different growth/stress conditions in 50 ml broth. Exponential and stationary cultures were obtained in TY medium upon bacterial growth to OD600 0.6 and 2.8, respectively. Cold and heat shocks were applied to exponentially growing bacteria in TY during 1 h by shifting the temperature to 20°C and 42°C, respectively. The salt shock was imposed during 1 h by addition of 0.4 M NaCl to exponential cultures in MM medium.
Construction of S. meliloti mutants and derivative strains
SmYbeY is encoded by the gene annotated as SMc01113 in the genome of the reference Rm1021 strain (31). S. meliloti SmΔybeY and SmybeYFLAG derivatives were both generated in the ExpR+ Sm2011 derivative strain Sm2B3001 (32). To create an in-frame deletion of ybeY, 938-bp and 921-bp DNA fragments flanking the SMc01113 ORF were PCR amplified from genomic DNA with primer pairs YbeY_F1/YbeY_iR and YbeY_iF/YbeY_R1. Primers YbeY_iR and YbeY_iF carry HindIII sites at their 5΄-ends. The resulting PCR products were thus restricted with HindIII and ligated to each other. The ligation reaction was used as a template for a second PCR with primers YbeY_FS and YbeY_RE that yielded a 1871-bp DNA fragment flanked by EcoRI sites and containing the ybeY deletion with the junction sequence ATGACGGCGAAGCTTTAA. This fragment was digested with EcoRI and inserted into the suicide vector pK18mobsacB (33) to yield pK18ΔybeY, which was mobilized to the Sm2B3001 strain by a biparental mating involving E. coli S17-1 (34). Recombinants that underwent single and double crossover events were subsequently isolated by Km resistance and counterselection in 10% sucrose as described in (35). Deletion of ybeY in the selected double recombinant was confirmed by PCR with primers YbeY_MutF and YbeY_MutR followed by HindIII restriction of the PCR product as well as by full re-sequencing of parent and mutant strain genomes on an Illumina MiSeq System applying a TruSeq, V2 Chip (2 × 250 bp) (Supplementary Figure S1).
Mutant SmΔybeY was complemented with plasmid pJBYbeY expressing SmYbeY from its own promoter. For that, an 810-bp genomic DNA fragment containing the SmYbeY coding sequence along with 266 nt of its upstream region was PCR-amplified with the primers pair YbeY_PrF/YbeY_PrR. The PCR product was first cloned into the pGEM-T® Easy vector (Promega Corporation), then retrieved by HindIII-EcoRI restriction and finally inserted into the low copy vector plasmid pJB3Tc19 (36) to generate pJBYbeY, which was conjugated into the SmΔybeY mutant by biparental mating.
The loci encoding the trans-sRNAs AbcR1 and AbcR2 were deleted in both the parent Sm2B2001 and the SmΔybeY strains by allelic replacement mediated by plasmid pK18EryΔR1/2 (37) to generate the double Sm2B2001ΔR1/2 and triple SmΔR1/2ΔybeY mutants.
Tagging of ybeY with the FLAG epitope (Sigma-Aldrich) to generate the SmybeYFLAG strain was done as follows. The full-length SmYbeY coding sequence (devoid of its TAA stop codon) along with 935 nt of its upstream genomic region was amplified by PCR with the pair of primers YbeY_F1/YbeY_XbaI, the latter adding a XbaI restriction site to the 3΄-end of the fragment. The PCR product was first cloned into pGEM-T® Easy, retrieved as a SacII (genomic site internal to the PCR product)-XbaI fragment and finally inserted upstream of the DNA sequence coding for three tandem FLAG epitopes in the previously constructed pKS3xFLAG plasmid (35) to generate pSK5΄YbeYFlag. A second 921-bp genomic DNA fragment, starting with the SmYbeY stop codon, was generated by PCR with primers YbeY_iF and YbeY_RK that add HindIII and KpnI restriction sites to its 5΄ and 3΄ ends, respectively. This PCR product was cloned into pGEM-T® Easy, then excised as a HindIII-KpnI DNA fragment and inserted immediately downstream of the 3xFLAG DNA sequence in pSK5΄YbeYFlag to yield pKSYbeY3xFlag. Finally, a 2372-bp DNA fragment encoding the C-terminal FLAG-tagged SmYbeY was amplified from pKSYbeY3xFlag with the primers pair YbeY_FS/YbeY_RE, then digested with EcoRI and inserted into pK18mobsacB to generate the suicide plasmid pK18YbeY3xFlag. This plasmid was mobilized to the Sm2B3001 strain by biparental mating for replacement of wild-type ybeY by the modified allele. Presence of ybeYFLAG in several double recombinants selected as previously described was confirmed by PCR on genomic DNA with primers YbeY_MutF and YbeY_MutR, XbaI restriction of the PCR products and Western blot analysis with commercial ANTI-FLAG monoclonal antibodies (Sigma-Aldrich) following a published protocol (35).
All PCR reactions required for cloning were performed with the proofreading Phusion™ High-Fidelity DNA polymerase (Thermo Scientific). Plasmid inserts were always checked by sequencing to confirm the absence of PCR-introduced mutations.
CoIP-RNA preparation, RNAseq and data analysis
Exponential, stationary, cold, heat and salt stressed cultures of both Sm2B3001 (control) and SmybeYFLAG strains were obtained as described, and 50 ml of each culture were pooled before bacterial lysis by sonication. CoIP–RNA was obtained from both control and SmybeYFLAG lysates using the ANTI-FLAG M2 affinity gel (Sigma) followed by organic extraction as described (26). This procedure was performed twice and equivalent quantities of CoIP–RNA from each replicate and strain were finally pooled.
Control and SmybeYFLAG CoIP–RNA pools were further processed by Vertis Biotechnologie AG to generate two strand-specific cDNA libraries as previously described (26). Libraries were sequenced on an Illumina MiHiSeq System applying a TruSeq, V3 Chip (2 × 300 bp).
After sequencing, reads were demultiplexed based on their sequence indices and mapped with Bowtie2 version 2.1.0 (38), using standard parameters after quality trimming, to the S. meliloti Rm1021 reference sequence (31). Data visualization and analysis based on an updated version of the S. meliloti public GenDB project including annotations of identified sRNAs (39,40) were done with the ReadXplorer software (41). Within ReadXplorer, the Express test and DESeq2 (42) tools were used to identify transcripts differentially represented in Control and SmybeYFLAG CoIP–RNA.
Overexpression, purification and cross-linking of recombinant SmYbeY and SmYbeY-R69A
The S. meliloti ybeY gene was amplified by PCR from genomic DNA of strain Sm2B3001 with Phusion High-Fidelity DNA polymerase using the primers YbeY_F2 and YbeY_R2. The purified 529-bp PCR product was double digested with NdeI and BamHI and cloned into the histidine tag-containing pET15b vector (Novagen) to yield pET15b-SmYbeY. The point mutation R69A was introduced into pET15b-SmYbeY by site-directed mutagenesis using primers R69A_F and R69A_R, originating plasmid pET15b-SmR69A. Both constructs were checked by sequencing (STAB Vida, Portugal).
Both plasmids were transformed into E. coli BL21(DE3) (Novagen) for the expression of the recombinant proteins. Cells were grown at 37°C to an OD600 of 0.5 in 100 ml LB medium supplemented with 100 μg/ml ampicillin. At this point, protein expression was induced by addition of 0.5 mM IPTG and bacteria were grown for a further 4 h. Cells were pelleted by centrifugation and stored at −80°C. The culture pellet was resuspended in 3 ml of Buffer A (10 mM Tris-HCl, 200 mM potassium acetate, 5 mM β-mercaptoethanol, pH 7.5). Cell suspension was lysed using a French Press at 1000 psi in the presence of 0.1 mM PMSF. The crude extract was treated with Benzonase (Sigma) to degrade the nucleic acids and clarified by a 30 min centrifugation at 10 000 g. Purification was performed in an ÄKTA FPLC™ system (GE Healthcare). The cleared lysate was subjected to a histidine affinity chromatography in a HisTrap HP column (GE Healthcare) equilibrated in Buffer A. Proteins were eluted by a continuous imidazole gradient up to 500 mM in Buffer A. The fractions containing the purified protein were pooled together, concentrated and buffer exchanged to Buffer B (20 mM Tris-HCl pH7.5) using a HiTrap Desalting column (GE Healthcare). The proteins were then added to a HiTrap Q HP column (GE Healthcare) equilibrated in Buffer B to perform anion exchange. Protein elution was achieved by a continuous NaCl gradient up to 1 M in buffer B. Eluted proteins were concentrated by centrifugation at 4°C with Amicon Ultra Centrifugal Filter Devices of 10 000 MWCO (Millipore), and buffer exchanged to Buffer A. The purity of the proteins was verified in a 15% SDS-PAGE gel followed by BlueSafe staining (Nzytech, Portugal). Proteins were quantified using the Bradford Method and 50% (v/v) glycerol was added to the final fractions prior storage at −20°C. RNase E from Salmonella enterica serovar Typhimurium, used as a control in the activity assays, was purified as described (43).
To verify if SmYbeY is able to form oligomers, 0.5 μg of the purified recombinant protein were incubated in a 10 μl cross-linking reaction with 10 mM Hepes pH 7.4, 250 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT and increasing concentrations (1–20 μg) of disuccinimidyl suberate. The reaction was performed at room temperature for 30 min and quenched by adding 1 μl of Tris-HCl 1M pH 7.5 and SDS loading buffer. The samples were boiled during 5 min and then analyzed in a 15% SDS-PAGE gel. RNase III, which is active as a dimer, was used as a positive control.
In vitro production of RNA substrates and SmYbeY activity assays
The single-stranded RNA substrates used in the in vitro activity assays were the synthetic 16-mer and 30-mer oligoribonucleotides (Supplementary Table S2), which were labeled at their 5΄ end with [γ-32ATP] and T4 Polynucleotide Kinase (Ambion) in a standard reaction. Both RNA molecules were also hybridized to a complementary non‐labeled 16-mer oligoribonucleotide added in excess (molar ratio 1:5) in order to obtain a perfect 16-16ds duplex, and a 30–16ds double stranded substrate with a 3΄poly(A) tail. The hybridization was performed during 10 min at 80°C followed by 45 min at 37°C.
The R1.1 RNA, the unprocessed 3΄ terminus of the 16S rRNA (39-mer), and the complementary 3΄ terminus 16S rRNA were generated using a synthetic DNA template (R1.1 REV, 3΄ 16S-rRNA Sm (39-mer) REV and 3΄ 16S-rRNA Sm complementary, respectively) and a commercial promoter oligonucleotide (T7 FW) (StabVida, Portugal) for in vitro transcription, following a previously described method (44). Briefly, the synthetic DNA template (0.5 μM) and the promoter oligonucleotide (0.6 μM) were annealed in 10 mM Tris-HCl pH 8.0 by heating for 5 min at 70°C, followed by 30 min incubation at 37°C. In vitro transcription was carried out using the ‘Riboprobe in vitro Transcription System’ (Promega) and T7 RNA polymerase with a molar excess of [32P]-α-UTP over non-radioactive UTP. In order to remove the DNA template, 1 U of DNase (Promega) was added and incubated 30 min at 37°C. RNA transcripts generated from in vitro transcription were purified by electrophoresis on an 8.3 M urea/polyacrylamide gel. The gel slice was crushed and the RNA was eluted overnight at room temperature with elution buffer [3 M ammonium acetate pH 5.2, 1 mM EDTA, 2.5% (v/v) phenol pH 4.3]. The RNA was ethanol precipitated, resuspended in RNase free water and quantified using the Biophotometer Plus (Eppendorf). tRNA-Ser, AbcR2, 5΄-prbA, ugpA and SMa_asRNA_345 RNAs were similarly generated, however, the corresponding DNA templates for in vitro transcription were produced by PCR using genomic DNA from S. meliloti Rm2011 strain (primer pairs on Supplementary Table S2). The phage T7 RNA polymerase promoter sequence was included in the forward primers. ybeY mRNA was generated using pET15b-SmYbeY (Supplementary Table S1) as template for in vitro transcription. The phage T7 RNA polymerase promoter sequence is included in the plasmid. In all cases the yield of the labeled substrates (cpm/μl) was determined by scintillation counting.
The activity assays were performed in a final volume of 50 μl containing the activity buffer 25 mM Tris-HCl pH 7.5, 150 mM NaCl or KCl and 10 mM MgCl2, MnCl2 or CaCl2 for SmYbeY or 25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 60 mM KCl, 100 mM NH4Cl, 0.1 mM DTT and 5% (v/v) glycerol for RNase E and the RNA substrate (concentrations indicated in the figure legends). As a control, an aliquot of each reaction (without the enzyme) was incubated until the end of the assay. The reactions were started by the addition of the enzyme, and further incubated at 37°C in case of SmYbeY. Aliquots of 5 μl were withdrawn at different time-points, and the reactions were stopped by the addition of formamide containing dye supplemented with 10 mM EDTA. Reaction products were resolved in a 7M urea/polyacrylamide gel at different concentrations and visualized by PhosphorImaging (FLA-2000, Fuji, Stamford, CT, USA). Each activity assay was performed at least in triplicate.
Radioactive RNA molecules used as substrates in gel shift assays are described above. Non-radioactive complementary 3΄ 16S rRNA was transcribed in the same conditions but using equimolar concentrations of all four ribonucleotides. In vitro transcription products were run on a polyacrylamide gel, identified by ethidium bromide staining and the full-length RNA molecules were cut out from the gel. RNA was eluted from the gel slice as described above. The hybridization between labeled and non-labeled substrates was always performed in the Tris component of the activity buffer by incubation for 10 min at 80°C, followed by 45 min at 37°C. A molar excess of the non-labeled substrates were added to the labeled RNA. The molar ratio of unlabeled to labeled substrate molecules was 5:1. The binding reactions were then mixed with loading buffer (48% glycerol, 0.01% bromophenol blue) and electrophoresed on native polyacrylamide gels in 0.5X TBE buffer at 200V in a cold room. Gels were analyzed using PhosphorImaging (FLA-2000, Fuji, Stamford, CT, USA).
Microarray-based transcriptomics
Total RNA was obtained from four independent exponential (OD600 0.5) and stationary (OD600 2.5) cultures of each strain, i.e. Sm2B3001 and the SmΔybeY mutant (eight preparations per strain), with the RNeasy Mini Kit (Qiagen). cDNA synthesis, Cy3- and Cy5-labeling, hybridization to Sm14kOLI microarrays, image acquisition and data analysis were performed as previously described (45). The Sm14kOLI microarray (ArrayExpress Accession No. A-MEXP-1760) carries 50mer to 70mer oligonucleotide probes directed against coding regions and both strands of the intergenic regions (Sinorhizobium meliloti 1021 Sm14kOLI) (46). Probes in intergenic regions were separated by ∼50 to 100 nt. Normalization and t-statistics were carried out using the EMMA 2.8.2 microarray data analysis software (47). Genes and 5΄-/3΄-UTRs with P-value ≤0.05 and M ≥1.0 or ≤−1.0 were included in the analysis. The M value represents the log2 ratio between both channels. Transcriptome data are available at ArrayExpress under accession number E-MTAB-5233. Functional categories of the differentially expressed genes were established according to the S. meliloti Rm1021 genome sequence annotation (31) and the KEGG database (http://www.genome.jp/kegg/).
Fluorescence reporter assays
Influence of SmYbeY and AbcR2 sRNA on the post-transcriptional regulation of the prbA mRNA was further investigated by a double-plasmid reporter assay as described (26,48). Briefly, the reporter plasmid pRprbA::egfp (26), which constitutively expresses a translational fusion of the 5΄ region of prbA mRNA (i.e. 211 nt spanning from its native transcription start site to the 18th codon) to eGFP, or the control pBBsyn-eGFP (48) were first transferred to the Sm2B3001 strain and its mutant derivative SmΔybeY. A second series of reporter strains was generated by conjugation of pRprbA::egfp into Sm2B2001ΔR1/2 and SmΔR1/2ΔybeY harboring pSRK-R2, expressing AbcR2 constitutively, or the empty control plasmid pSRK_C (37). pSRK-R2b, which expresses an AbcR2 variant mutated for base-pairing with prbA, was generated by overlapping PCR with primers PCR3_F and PCR1_R of DNA fragments amplified from pSRK-R2 with the primer pairs PCR3_F/AbcR2_R and AbcR2b_F/PCR1_R, followed by BamHI/SacI restriction of the full-length product and insertion into pSRK-C. eGFP-mediated fluorescence in normalized exponential cultures of three double transconjugants for each plasmid combination was measured in the Infinite M200 Pro microplate reader (Tecan) as described (26).
RESULTS
Genome-wide profiling of RNAs bound to SmYbeY
As global approach to explore the proficiency of SmYbeY to bind RNA we profiled the RNA species co-immunoprecipitated (CoIP-RNA) with a C-terminal FLAG-tagged variant of the protein (SmYbeYFLAG) expressed from the chromosome of the Sm2B3001 strain. Unlike the ybeY deletion mutant (SmΔybeY), the SmybeYFLAG derivative strain exhibited wild-type growth in complete TY and minimal MM media, indicating that tagging did not compromise SmYbeY function (Supplementary Figure S2). The untagged wild-type strain was used as control to assess unspecific RNA recovery. Control and SmYbeYFLAG CoIP–RNA, both obtained from bacterial pools representing five different growth conditions (i.e. exponential and stationary cultures and salt, heat and cold shocks), served as the templates to generate strand-specific cDNA libraries that were subjected to paired-end sequencing on an Illumina platform. Before organic extraction of the CoIP–RNA, the presence of SmYbeYFLAG in the RNA–protein complexes was verified by Western-blot (Supplementary Figure S3). RNaseq delivered an average of 4 200 000 reads per library of which 1 150 974 (C-wt library) and 1 406 485 (SmYbeYFLAG-derived library) mapped to unique locations within the reference S. meliloti Rm1021 genome. We then applied the DESeq2 and Express tests, both implemented in the ReadXplorer software (41), to the sets of uniquely mapped reads in order to identify transcripts differentially represented in both libraries. Transcripts covered by a minimum of 30 reads and enriched at least 2-fold in the SmYbeYFLAG library with respect to the control were scored as SmYbeY-bound (i.e. SmYbeY RNAs).
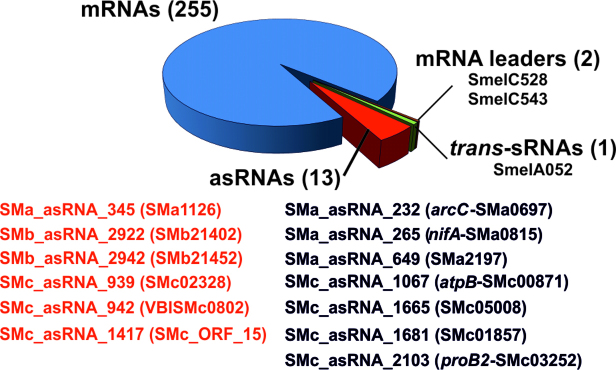
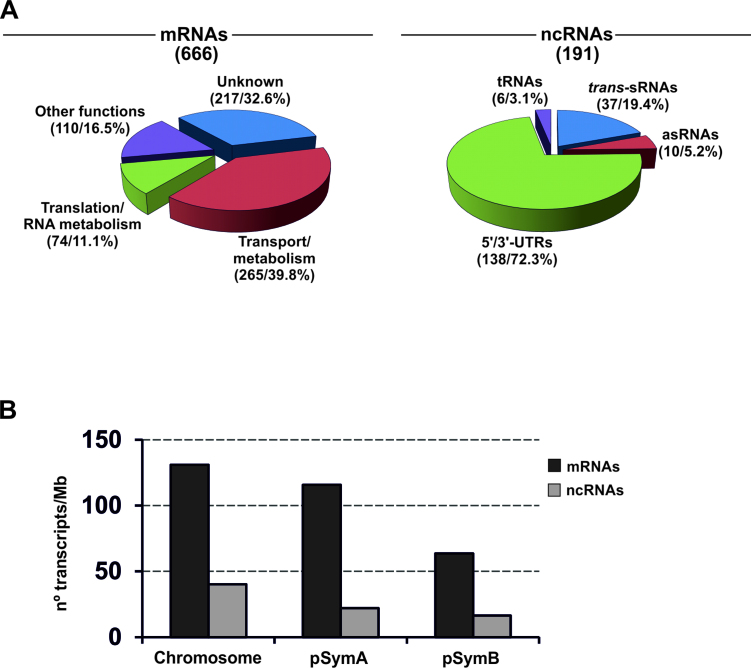
The combination of both tests rendered a catalog of 271 SmYbeY RNAs (Supplementary Table S3). Of those, 255 (94%) derived from mRNAs, 13 (4.8%) were annotated antisense sRNAs (asRNAs) and 3 (1.2%) represent other sRNAs (2 mRNA leaders and 1 trans-sRNA) (Figure 1). It is worth noting that the mRNA partners of 6 out of the 13 SmYbeY asRNAs were catalogued as SmYbeY-bound. This observation hints at certain affinity of SmYbeY for asRNA–mRNA duplexes.
Figure 1.
Identification of SmYbeY-binding transcripts. The diagram shows the number of different RNA species, i.e. mRNAs, asRNAs, mRNA leaders and trans-sRNAs, enriched ≥2-fold in SmYbeYFLAG CoIP–RNA with respect to the control. The identified asRNA/mRNA pairs are in red color.
If we compare the SmYbeY and Hfq CoIP–RNAs, the latter obtained using a similar experimental setup in the same culture conditions (26), we conclude that SmYbeY does not have Hfq-like RNA chaperone features.
SmYbeY is a metal-dependent single- and double-strand endoribonuclease
We have purified SmYbeY and performed the first characterization of its biochemical activity. Cross-linking of the recombinant protein with increasing amounts of disuccinimidyl suberate did not promote oligomerization, indicating that SmYbeY acts as a monomer of ∼20 kDa (Supplementary Figure S4).
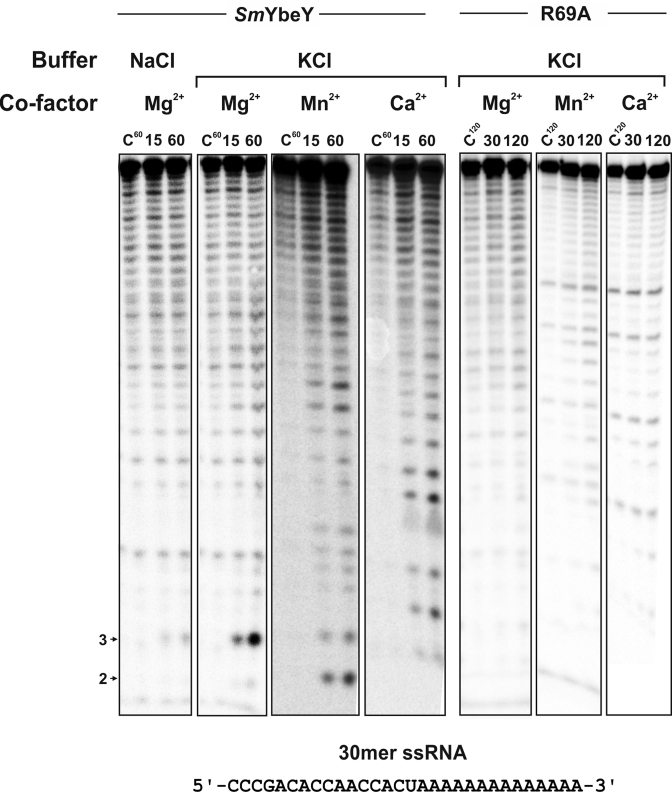
It has been reported that EcoYbeY acts as a metallo endoribonuclease that specifically cleaves single-stranded RNA (ssRNA) substrates (13). We therefore assayed purified SmYbeY for RNase activity. The monomeric SmYbeY (5 μM) was first incubated with a 5΄ end-labeled 30mer ssRNA oligonucleotide in Tris-buffered solutions containing NaCl or KCl and magnesium (Mg2+) as co-factor (Figure 2, left two panels). SmYbeY barely cleaved this substrate in NaCl-containing buffer. However, the presence of KCl enhanced cleavage efficiency, as revealed by a marked formation of a major 3-nt degradation product. The incubation of 30mer ssRNA with a higher enzyme concentration (10 μM) resulted into an increased accumulation of several additional reaction products (Supplementary Figure S5A). We have also tested the activity of SmYbeY with a smaller ssRNA, the 16mer oligoribonucleotide. This RNA was also cleaved by SmYbeY, although with less efficiency when compared to the 30mer (Supplementary Figure S5A), suggesting that SmYbeY has preference for longer substrates. We next tested SmYbeY activity on the 30mer ssRNA substrate in KCl solution with either manganese (Mn2+) or calcium (Ca2+) as alternative divalent metal co-factors (Figure 2). These new incubation conditions promoted 30mer ssRNA cleavage at different positions, most likely in a non-specific manner. Of note, the substitution of the ultraconserved arginine R69 (Supplementary Figure S6) by an alanine residue (SmYbeY-R69A) rendered SmYbeY almost inactive on this generic ssRNA substrate, which further demonstrates that the observed cleavage was protein-dependent.
Figure 2.
Activity of SmYbeY on single-stranded RNA (ssRNA). In vitro reactivity of purified wild-type SmYbeY and the mutant variant SmYbeY-R69A on a 5΄-labeled 30mer ssRNA oligonucleotide (0.02 pmol/μl) whose sequence is indicated on bottom. Enzymes were used at a concentration of 5 μM in all assays. Buffering conditions and incubation times are indicated on top of the panels. Numbers to the left indicate sizes of the major reaction products. Reactions were analyzed on 7 M urea/20% polyacrylamide gels. C, control reactions.
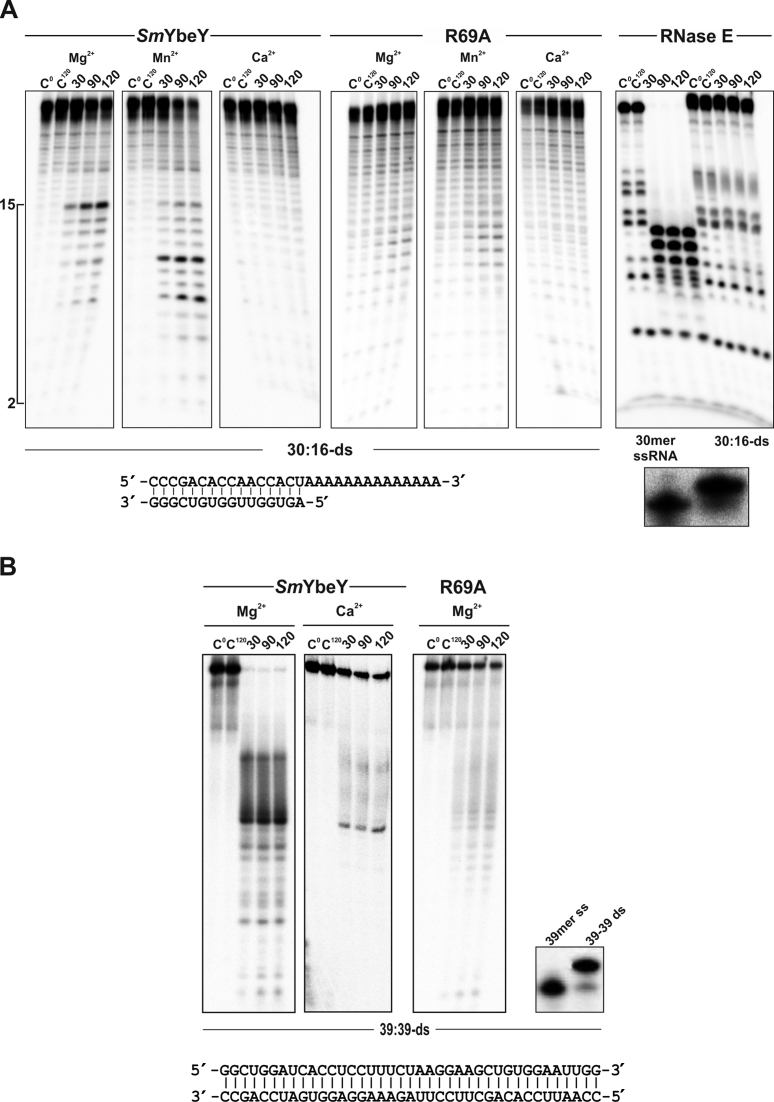
It has been also shown that EcoYbeY does not cleave double-stranded RNA (dsRNA) (13). We tested the activity of SmYbeY (5 μM) on a partially dsRNA with a single-strand 3΄-poly(A) tail (30:16-ds) as well as on fully double-stranded 16mer (16:16-ds) and 39mer (39:39-ds) RNA substrates (Figure 3 and Supplementary Figure S5B). Efficient duplex formation was checked by binding shift assays in each case. SmYbeY cleaved the 30:16-ds RNA unspecifically at multiple sites within its double-stranded portion, but more efficiently if Mn2+ was used as co-factor (Figure 3A, left two panels). In contrast, Ca2+ precluded SmYbeY activity on this RNA substrate. The SmYbeY-R69A mutant enzyme showed extremely low activity on this substrate in the presence of Mg2+ and Mn2+, with Ca2+ acting also as an inhibitor, with Ca2+ acting also as an inhibitor (Figure 3A, central panels). To further demonstrate that cleavage on the 30:16-ds RNA was SmYbeY-dependent and not the result of substrate breathing we tested the activity of the single-strand specific endoribonuclease RNase E on both the 30mer ssRNA and the 30:16-ds RNA molecules at 37°C, i.e. the SmYbeY reaction temperature (Figure 3A, right two panels). RNase E fully degraded the ssRNA whereas the double-stranded substrate remained unaltered at the end of the assay. The shorter 16:16-ds RNA was also cleaved by SmYbeY at several positions when Mg2+ was used as co-factor (Supplementary Figure S5B). Remarkably, the activity of the protein was more evident on a longer dsRNA molecule (39:39-ds), which was totally consumed after 30 min of incubation of the reaction mixtures in a Mg2+-containing buffer (Figure 3B). Once again, the activity of SmYbeY was dramatically reduced in the presence of Ca2+ (Figure 3B). Similar to what was observed with the other RNA substrates, the SmYbeY-R69A mutant enzyme showed residual activity on the 39:39-ds (Figure 3B).
Figure 3.
Activity of SmYbeY on dsRNA. Enzymes were used at a concentration of 5 μM in all assays. Reactions were incubated in KCl-containing buffer in the presence of the co-factor indicated on top of the panels. Reaction times are also indicated. Cleavage products were separated on 7 M urea/20% polyacrylamide gels. C, control reactions. (A) In vitro reactivity of purified wild-type SmYbeY and the mutant variant SmYbeY-R69A on a partially dsRNA substrate (30:16-ds) (0.02 pmol/μl) whose sequence is indicated on bottom. The bracket to the left indicates the size range of breakdown products. RNase E was assayed with the 30:16-ds and 30mer ssRNA substrates (0.02 pmol/μl) as control of cleavage specificity on dsRNA (right panel). The gel of a shift assay (15% polyacrylamide) showing effective duplex formation is shown on bottom. (B) Reactivity of SmYbeY and SmYbeY-R69A on the 39:39-ds dsRNA substrate (0.24 pmol/μl), whose sequence is shown on bottom. Gel of the shift assay (15% polyacrylamide) showing formation of the duplex is also shown.
Collectively, these results revealed that SmYbeY has ribonuclease activity on both ssRNA and dsRNA substrates, although the enzyme has preference for longer dsRNA molecules. SmYbeY-mediated catalysis is supported by Mg2+ and Mn2+ as divalent metal co-factors. Nonetheless, each co-factor influenced cleavage efficiency and breakdown patterns in a different manner. Ca2+ selectively impaired SmYbeY ability to cleave dsRNA.
SmYbeY cleaves structured and CoIP-enriched RNAs
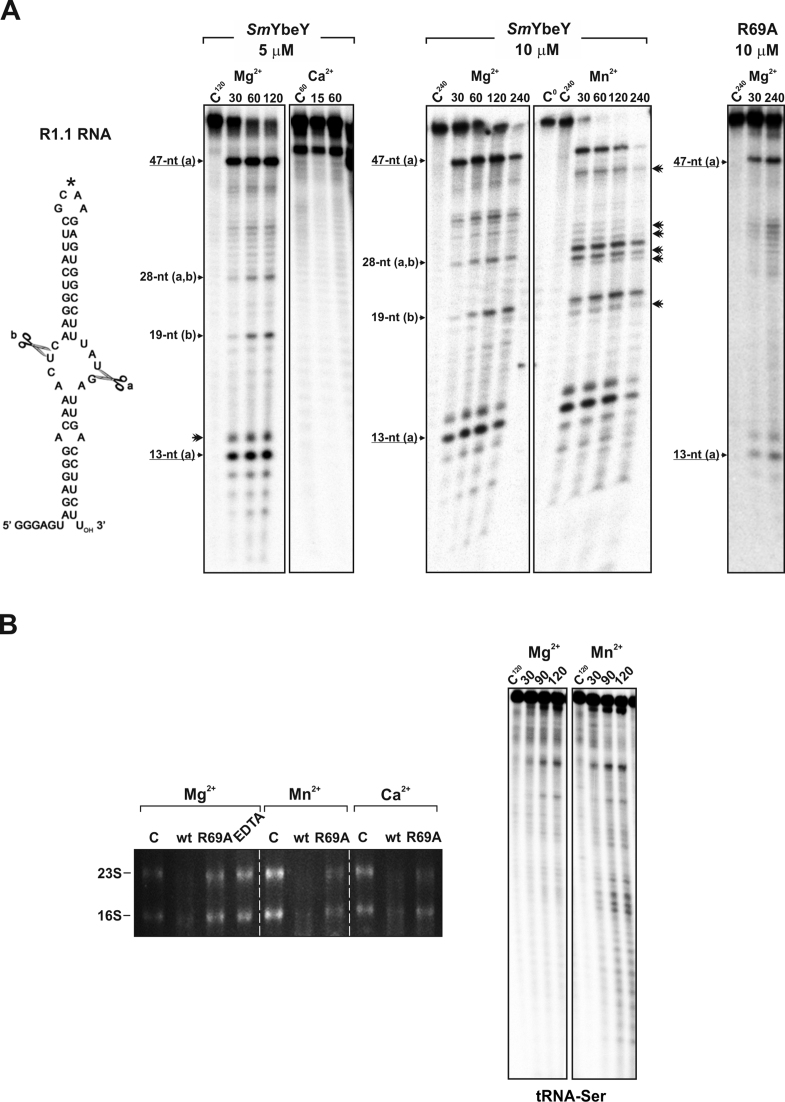
We next tested the ability of SmYbeY for cleaving a series of structured RNA molecules namely, the generic R1.1 RNA substrate, commonly used to assay the in vitro activity of the double-strand endoribonuclease RNase III (49), total rRNA and a transfer RNA (tRNA-Ser) (Figure 4). Incubation of SmYbeY (5 μM) with the internally labeled 60-nt R1.1 RNA in KCl-containing buffer and Mg2+ as co-factor rendered two major 47-nt and 13-nt long reaction products along with a series of less abundant molecules (including 19-nt and 28-nt long products) (Figure 4A, left panels). Remarkably, the 47-nt, 28-nt, 19-nt and 13-nt long RNA species derived from cleavage at positions already reported to be preferred by RNase III within the main loop of the R1.1 substrate (50,51). There were also additional minor products that may result from cleavages on the small loop (on the top) of R1.1 (indicated with an asterisk in Figure 4A). Ca2+ inhibited SmYbeY activity when assayed in the same concentration and buffering conditions. A higher SmYbeY concentration (10 μM) and longer incubation of the reaction in the presence of Mg2+ resulted in a similar R1.1 breakdown pattern (Figure 4A, central panels). However, the use of Mn2+ as co-factor in the assays promoted almost depletion of the full-length R1.1 substrate shortly upon reaction initiation (30 min) as well as minor cleavage at additional sites within R1.1. Interestingly, although less efficiently SmYbeY-R69A retained the ability of the wild-type protein for cleaving R1.1 at the major reaction sites (Figure 4A, right panel).
Figure 4.
Activity of SmYbeY on structured RNA substrates. (A) Reactivity patterns of SmYbeY and SmYbeY-R69A on the R1.1 RNA (1.12 pmol/μl), the canonical substrate for RNase III. Sequence and secondary structure of the substrate is shown to the left. Enzyme concentrations, metal co-factor and reaction times are indicated on top of the panels. Reactions were analyzed on 7 M urea/15% polyacrylamide gels. Known cleavage sites (A and B) of RNase III on the same substrate are indicated. The asterisk (*) indicates an alternative minor cleavage site within R1.1. Additional minor reactions products when Mn2+ was used as co-factor are indicated by double arrowheads. C, control reactions. (B) Left panel, activity of SmYbeY and SmYbeY-R69A on rRNA (500 ng). Enzymes (20 μM) were incubated for 2 h at 37°C in the presence of EDTA (50 mM) and/or the co-factors indicated on top. RNA was analyzed on a 1.5% agarose gel. The positions of the 23S and 16S rRNA are indicated. C, control reactions. Right panels, activity of SmYbeY (10 μM) on tRNA-Ser. Metal co-factors used in the assays and the time-course of the reactions are indicated on top of the panels. The reaction products were separated on 7 M urea/10% polyacrylamide gels. C, control reactions.
On the other hand, SmYbeY (20 μM) was also very active in degrading total rRNA in the presence of Mg2+, Mn2+ and Ca2+ (Figure 4B, left panel). We have shown that Ca2+ inhibited cleavage of SmYbeY on the structured R1.1. RNA but not on ssRNA, therefore, this result suggests that the enzyme likely recognizes the larger stems within the complex structured rRNAs as single-stranded substrates. The R69A mutation strongly attenuated reactivity of SmYbeY on rRNA whereas EDTA (50 mM) totally blocked RNase activity, further supporting that SmYbeY is a metal-dependent endoribonuclease. Given that EcoYbeY has been shown to be involved in 16S rRNA maturation (13) we also assayed the activity of SmYbeY on an in vitro synthetized 39-nt RNA substrate that mimics the sequence of the 3΄ terminus of the S. meliloti 16S rRNA precursor (Supplementary Figure S7). SmYbeY reacted poorly with this RNA molecule in the conditions tested (Mg2+ as co-factor) (Supplementary Figure S7A). Consistent with this finding, the SmΔybeY mutant showed an unaltered wild-type rRNA profile even upon a temperature upshift (Supplementary Figure S7B). Similarly, only minor products were detected after incubation of SmYbeY with the structured 89-nt long tRNA-Ser substrate (Figure 4B, right panels).
These findings confirmed the previous observations that SmYbeY is able to cleave a diversity of structured RNA molecules. Moreover, and contrary to what was observed in E. coli, SmYbeY does not seem to be involved in 16S rRNA maturation.
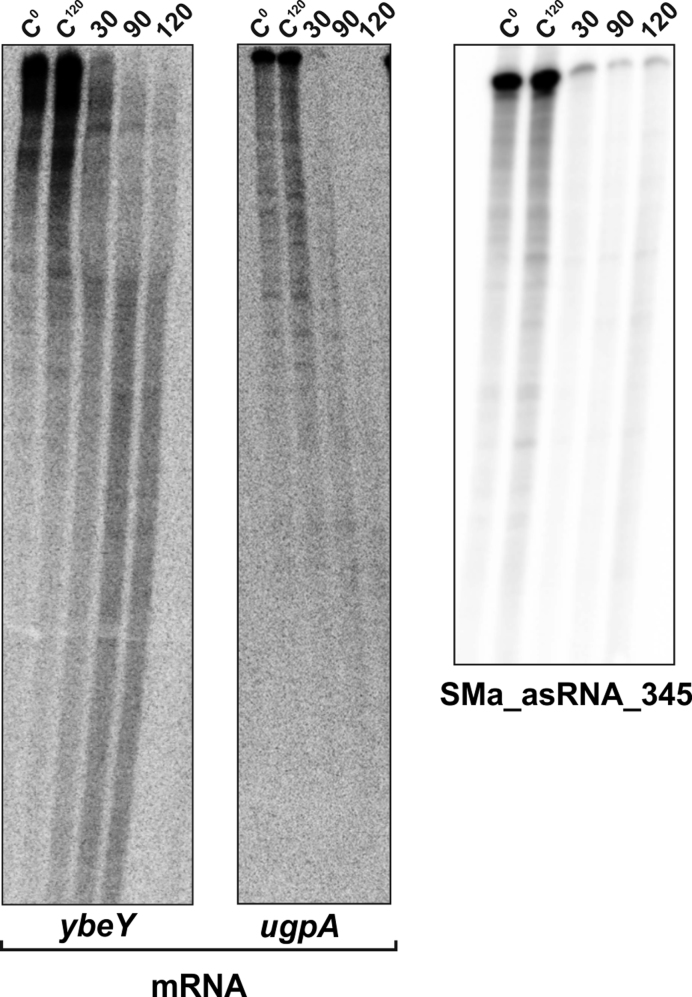
Finally, in a new series of in vitro experiments we also incubated SmYbeY with endogenous S. meliloti substrates, i.e. the ybeY and ugpA mRNAs, and the SMa_asRNA_345 asRNA (Figure 5). The two latter transcripts were selected because of their enrichment in CoIP-RNA, i.e. 3.20- and 2.07-fold with respect to the control, respectively. SmYbeY was extremely effective in cleaving all these three RNA molecules, with total consumption of the substrates early (30 min) after the start of the reactions. These results validate the CoIP–RNA described above as a resource to identify SmYbeY substrates.
Figure 5.
Activity of SmYbeY on endogenous S. meliloti substrates. SmYbeY efficiently cleaves ybeY (0.03 pmol/μl) and ugpA (0.2 pmol/μl) mRNAs (left panel) and the asRNA SMa_asRNA_345 (0.02 pmol/μl) (right panel). Reactions were analyzed on 7 M urea/8% polyacrylamide (mRNAs) or 7 M urea/10% polyacrylamide (asRNA) gels. Concentration of the enzyme in the assays was 10 μM and the reaction mixtures were incubated in the presence of Mg2+. Incubation times are indicated on top of the panels. C, control reactions.
SmYbeY influences chromosome-linked and symbiotic plasmid pathways
We also investigated the SmYbeY-dependent molecular responses of S. meliloti by profiling the transcriptomes of the Sm2B3001 strain and its deletion mutant derivative SmΔybeY on Sm14kOLI microarrays (Supplementary Tables S4 and S5). To unequivocally demonstrate that the observed changes in the transcriptome were exclusively due to the lack of SmYbeY we sequenced the genomes of the parent and mutant strains, which confirmed the in-frame deletion of ybeY and the absence of second site suppressor mutations in SmΔybeY (Supplementary Figure S1). Accordingly, the growth phenotypes of the SmΔybeY mutant in complete TY and minimal MM media were complemented with plasmid pJBYbeY, which expresses ybeY from its own promoter (Supplementary Figure S2). Total RNA was then obtained from bacteria grown to exponential (log RNA) and stationary (stat RNA) phase in complete TY medium. These experiments identified 543 and 209 SmYbeY-dependent mRNAs (i.e −1 ≥ M ≥ 1) in log and stat RNA samples, respectively, with 86 of those common to both growth states. Therefore, in our experimental conditions lack of SmYbeY altered the expression of 666 protein-coding genes (∼11% of the S. meliloti Rm1021 ORFs) (Supplementary Table S4). Functional clustering of these genes revealed that 39.8% encode metabolic functions, 11.1% are related to translation and RNA turnover and 16.5% represent widely diverse cellular processes (e.g. motility, signal transduction and transcription, transposition or nitrogen-fixation) (Figure 6A and Supplementary Figure S8). The remaining 32.6% have unpredictable function. Differential accumulation of a subset of representative mRNAs displaying M values ranging from −2.5 to 2.0 was further confirmed by qRT-PCR analysis (Supplementary Table S6).
Figure 6.
SmYbeY-dependent alteration of the S. meliloti transcriptome. (A) Number and functional categories of mRNAs and non-coding RNAs (ncRNAs) differentially accumulated in the SmΔybeY mutant. (B) Impact of SmYbeY on the accumulation of chromosomal, pSymA and pSymB mRNAs and ncRNAs. The histogram shows the number of differentially expressed transcripts per Mb in each replicon.
Sm14kOLI microarrays can also probe most of the recently identified S. meliloti non-coding RNAs (ncRNAs). Specifically, we have estimated that at least 478 known trans-sRNAs have oligonucleotide probes in these microarrays. Analysis of the hybridization signals on this set of probes revealed that lack of SmYbeY altered the expression of 131 and 77 ncRNAs during exponential and stationary bacterial growth, respectively, with 17 of those present in both data sets (Supplementary Table S5). The majority (72.3%) of these ncRNAs corresponded to putative cis-acting sense transcripts (i.e. 5΄/3΄-UTRs of mRNAs), whereas trans-sRNAs represented 19.4%, asRNAs 5.2% and tRNAs 3.1% (Figure 6A and Supplementary Figure S8). Therefore, SmYbeY does not seem to have a major role at least in trans-sRNA turnover. Our experimental approach underestimates the expression of asRNAs since only a subset of these transcripts (i.e. those antisense to 5΄/3΄-UTRs of mRNAs) are represented in the Sm14kOLI microarrays, which precludes a conclusion about the influence of SmYbeY in asRNA accumulation.
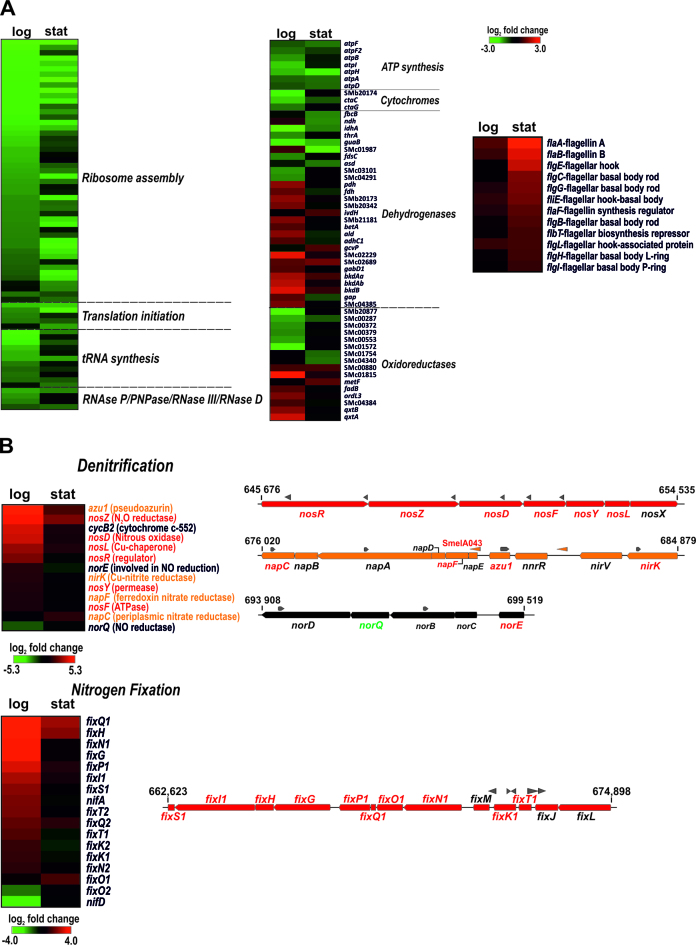
The S. meliloti genome consists of three large replicons, the chromosome (3.7 Mb) and the symbiotic megaplasmids pSymA (1.4 Mb) and pSymB (1.7 Mb). Interestingly, distribution of the differentially accumulated mRNAs in this genome revealed a similar relative impact of SmYbeY activity on the expression of chromosomal and pSymA-borne genes (Figure 6B). Chromosomal SmYbeY-dependent genes included those related to ribosome biogenesis, translation, RNA turnover, energy metabolism and flagellum assembly (Figure 7A). Specifically, SmYbeY loss-of-function resulted in pervasive down-regulation during both exponential and stationary growth of a set of genes coding for most of the ribosomal proteins, a set of elongation/translation initiation factors and tRNA synthetases as well as the ribonucleases RNase P, PNPase, RNase III and RNase D. A similar expression profile was exhibited by a number of genes involved in ATP synthesis and cytochrome C oxidase assembly. However, transcripts encoding substrate-dependent dehydrogenases and oxidoreductases showed variable accumulation in the SmΔybeY mutant. Microarray data also revealed up-regulation in this mutant of gene clusters coding for flagellar structural elements, particularly upon entry of bacteria into stationary phase. This misregulation of flagella biosynthesis most likely explains the reduced swimming motility of the SmΔybeY mutant with respect to the parent strain (Supplementary Table S7).
Figure 7.
Core and plasmid pathways influenced by SmYbeY. Changes in mRNA abundance in exponential (log) and stationary (stat) cultures are visualized in heatmaps generated with the MeV tool (http://www.tm4.org/mev.html). In the color scale (log2 fold changes) green and red stand for down- and up-regulation in the SmΔybeY mutant, respectively. (A) Chromosomal genes related to translation and RNA turnover (left), energy metabolism (middle) and flagella biosynthesis (right). (B) Genes of symbiotic plasmid pSymA involved in anaerobic denitrification (upper panel) and nitrogen-fixation (bottom panel). Gene clusters specifying each pathway are depicted to the side of each panel. Grew arrowheads stand for annotated asRNAs. The names of differentially expressed genes within each operon are colored.
S. meliloti symbiotic plasmid pSymA mostly encodes functions contributing to ecological specializations of this bacterium, e.g. microaerobic nitrate respiration and symbiotic nitrogen fixation (Figure 7B). A number of genes coding for proteins involved in the denitrification pathway including the nitrate, nitrite and nitrous oxide reductases NapC, NirK and NosZ, respectively, were strongly up-regulated in exponentially growing SmΔybeY mutant bacteria. Similar influence of SmYbeY was observed on the expression of genes coding for the master regulators of nitrogen fixation (i.e. FixK1/2 and NifA) and the elements of the electron transport chain associated with the nitrogenase activity, i.e. most of the genes integrating the fixNOQPGHIS operon. The accumulation profiles of these two sets of transcripts indicate an involvement of SmYbeY in the post-transcriptional silencing of denitrification and nitrogen fixation under free-living non-symbiotic conditions.
Discrete overlap between the Hfq- and SmYbeY-dependent genes
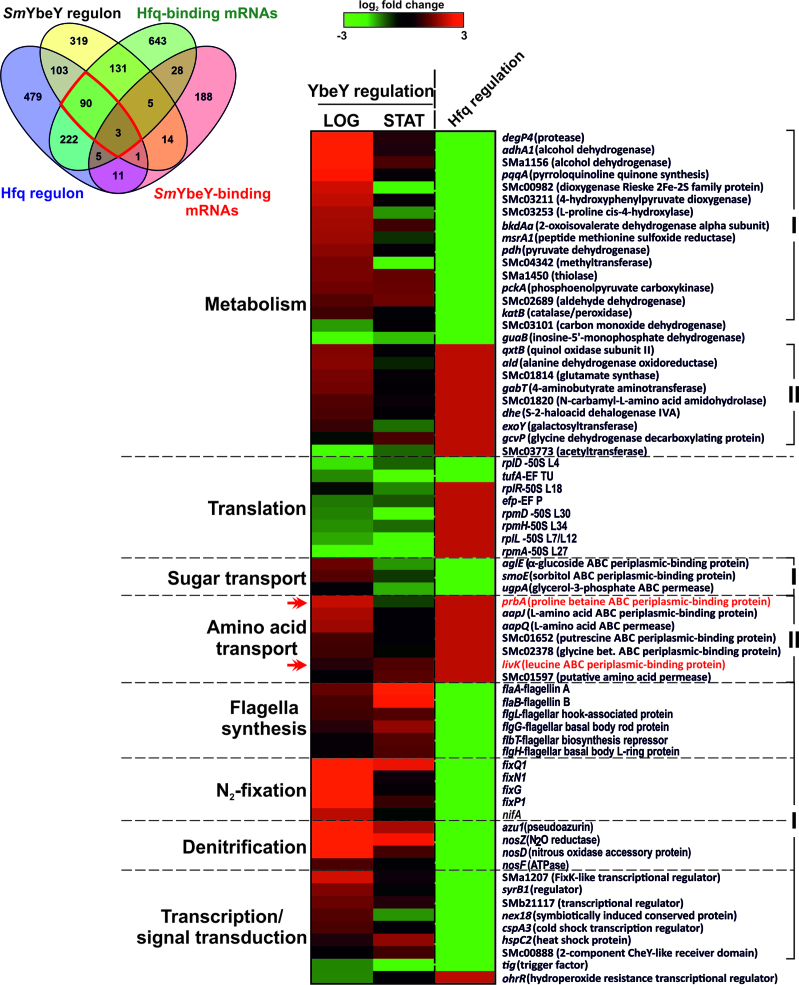
Published data have evidenced large similarities between the physiological phenotypes associated to SmYbeY and Hfq loss-of-function in S. meliloti, which has been interpreted as the consequence of a strong functional relation between the two proteins (3,10,28). To further explore these commonalities at the molecular level we have compared the Hfq- and SmYbeY-dependent gene sets (Figure 8). The latter included the 255 SmYbeY-bound mRNAs identified in the CoIP experiments (Supplementary Table S3), of which 23 (9%) were also scored as differentially expressed in the SmΔybeY mutant. Accumulation of almost half of these 23 transcripts, including the ugpA mRNA (Figure 5), was negatively influenced by SmYbeY and therefore could be preferred substrates for this endoribonuclease (Supplementary Table S4). Nonetheless, the reduced overlap between these two data sets could be mostly explained by the differences in the experimental setups, i.e. transcriptomics were only performed in two out of the five culture conditions used in the CoIP experiments.
Figure 8.
Overlap between the Hfq- and SmYbeY-dependent gene sets in S. meliloti. On the left, Venn-diagram representing Hfq- and SmYbeY-binding mRNAs and the transcripts differentially accumulated in the respective mutant strains. Hfq data sets were compiled from the literature. The red box indicates the 93 Hfq-SmYbeY co-regulated mRNAs that are known to bind the Hfq chaperone. On the right, heatmap illustrating accumulation in the SmYbeY and Hfq mutants of a subset of these 93 mRNAs (69) that encode proteins with predictable function. Functional categories are indicated to the left and the identity of each gene to the right of the panel. In the color scale (log2 fold changes) green and red stand for down- and up-regulation in the mutants, respectively. Due to the heterogeneity of Hfq data sets fixed values of −3 and 3 were applied for the Hfq-dependent genes. I, genes inversely regulated by Hfq and SmYbeY; II, genes negatively influenced by both proteins. Double arrowheads in red indicate prbA and livK mRNAs, which are experimentally confirmed targets of the homologous trans-sRNAs AbcR1 and AbcR2.
The compilation of existing transcriptomics and proteomics data uncovered a large Hfq regulon integrated by 917 protein-coding genes (35,52–54) of which 197 (∼21%) were also scored in this study as differentially expressed in the SmΔybeY mutant. Interestingly, 93 of these genes code for mRNAs that have recently been reported to bind Hfq (26). Conversely, most of the 202 SmYbeY mRNA ligands have been previously catalogued as Hfq-independent. Within the subset of 93 Hfq/SmYbeY co-regulated genes, 69 encode proteins with predictable function, namely 26 involved in metabolism, 8 in translation, 10 in nutrient uptake, 6 in flagella biosynthesis, 5 in symbiotic nitrogen-fixation, 4 in denitrification and 9 in transcription and/or signal transduction (Figure 8). These 69 genes can be grouped into two major categories according to their expression patterns in the Hfq and SmYbeY mutants; (i) genes that exhibited opposite dependence on Hfq and SmYbeY activity, being mostly up-regulated in SmΔybeY and consistently down-regulated in the different Hfq mutant strains (41 genes), and (ii) genes whose expression is negatively influenced by both proteins (17 genes) and therefore have been catalogued as up-regulated in the respective mutants. The first category includes large fractions of metabolic and regulatory genes and the full subsets of genes coding for proteins involved in denitrification, nitrogen fixation, biosynthesis of flagella and sugar transport. These mRNAs may be protected by Hfq from SmYbeY-mediated degradation. The second major group of Hfq/SmYbeY-dependent genes codes for either metabolic proteins or amino acid transporters. SmYbeY would be involved in the decay of these mRNAs upon translational inhibition by Hfq-dependent trans-acting sRNA partners.
Overall, this comparative analysis revealed a discrete overlap between Hfq- and SmYbeY-dependent genes. Nonetheless, it enabled the prediction of putative Hfq-dependent and independent SmYbeY substrates among the mRNAs exhibiting increased steady-state levels in the SmΔybeY mutant.
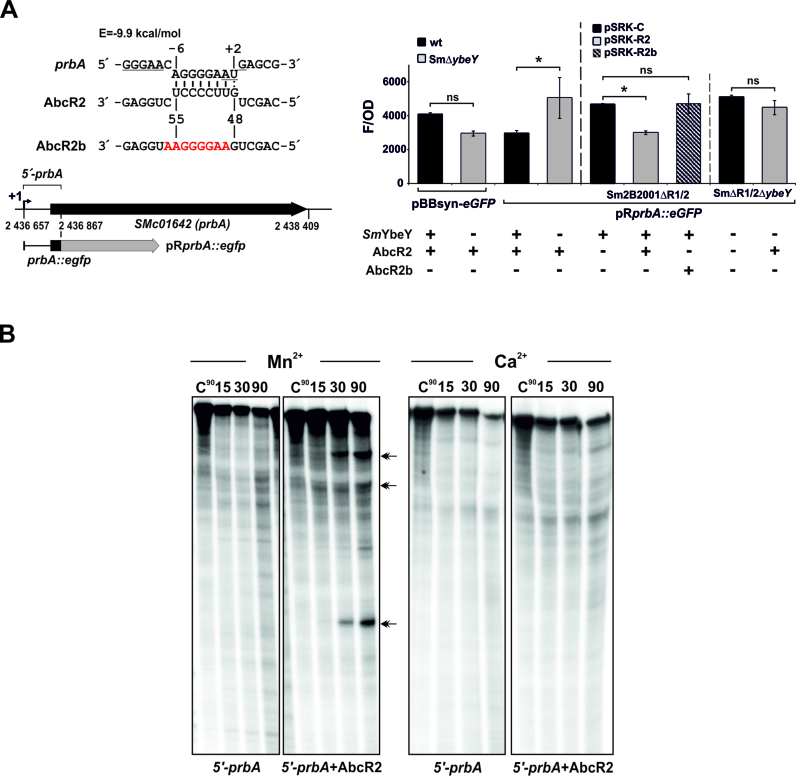
SmYbeY is required for the sRNA-mediated silencing of amino acid ABC transporters
Accumulation of some known Hfq-binding mRNAs encoding periplasmic components of amino acid ABC transporters is negatively influenced by both Hfq and SmYbeY. The Hfq-dependent trans-sRNAs AbcR1 and AbcR2 have among their experimentally confirmed targets two of these mRNAs, namely prbA and livK, which code for proline betaine and branched-chain amino acid transporters, respectively (26,37). In particular, our transcriptomics profiling revealed that levels of the prbA mRNA increased more than 5-fold in the SmΔybeY mutant with respect to the wild-type strain during exponential growth, suggesting that the post-transcriptional silencing of prbA requires SmYbeY. However, the prbA mRNA was not recovered in the CoIP–RNA. Therefore, we further investigated the putative role of SmYbeY in the regulation of prbA mRNA and if this layer of regulation is AbcR2-dependent.
Northern hybridization of total RNA from rifampicin-treated cells confirmed that lack of SmYbeY has no effect on either processing or stability of AbcR2 (Supplementary Figure S9). Thus, we hypothesized that SmYbeY may influence prbA decay upon its predicted antisense interaction with AbcR2. To test this hypothesis we measured the SmYbeY- and AbcR2-dependent fluorescence of exponential cultures of a series of reporter strains expressing constitutively from pRprbA::eGFP (26) a translational fusion of 211-nt of the prbA mRNA 5΄ region, which contains the predicted AbcR2 interaction site, to eGFP (Figure 9A). First, the Sm2B3001 strain and its SmΔybeY derivative were transformed with the control plasmid pBBsyn–eGFP, expressing constitutively eGFP with a prbA-unrelated 5΄-UTR or pRprbA::eGFP. Fluorescence of the control construct was ∼25% (statistically non-significant) lower in the mutant than in the wild-type background, probably due to eGFP mistranslation. In contrast, fluorescence of pRprbA::eGFP increased more than 50% in the SmΔybeY strain. These results confirmed the SmYbeY-dependent post-transcriptional regulation of prbA and rendered the 5΄ region of the mRNA as the specific target of this regulation. We next mobilized pRprbA::eGFP to the AbcR1/2 deletion mutant Sm2B2001ΔR1/2 and to the SmΔR1/2ΔybeY triple mutant (i.e. devoid of the AbcR1/2 and ybeY loci), both harboring the compatible control empty plasmid pSRK-C or pSRK-R2, the latter expressing AbcR2 constitutively (Figure 9A, right histogram). Fluorescence of exponentially growing Sm2B2001ΔR1/2 bacteria co-expressing AbcR2 and prbA::eGFP decreased by 35% (statistically significant) with respect to that of the same strain co-transformed with the target fusion and the empty vector pSRK-C. Further, an AbcR2 mutant variant (AbcR2b) carrying nucleotide substitutions in the predicted prbA interaction site and stably expressed from pSRK-R2b (Supplementary Figure S10) had no effect on the fluorescence of the reporter fusion. These results confirmed the AbcR2-mediated post-transcriptional down-regulation of prbA, already reported in the closely related S. meliloti strain Rm1021 (26). In contrast, lack of SmYbeY almost abrogated repression of prbA by AbcR2, i.e in the triple mutant background fluorescence of pRprbA: eGFP hardly decreased to 12% (statistically non-significant) with respect to that of the control upon AbcR2 expression.
Figure 9.
SmYbeY is required for the AbcR2-mediated silencing of the prbA mRNA. (A) Lack of SmYbeY abrogates AbcR2-dependent post-transcriptional repression of prbA in vivo. The AbcR2-prbA antisense interaction as predicted by IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) is shown on top. The Shine–Dalgarno sequence and AUG start codon of prbA are underlined. Numbers indicate positions relative to the prbA start codon and AbcR2 transcription start site. The predicted minimum hybridization energy (E) of the duplex is indicated. Nucleotide substitutions in the AbcR2b mutant variant disrupting base-pairing with prbA are colored in red. The schematics below show the prbA genomic region and the reporter fusion used in the in vivo assays. Numbers stand for coordinates in the genome of the reference strain Rm1021. The transcription start site of prbA is indicated (+1). Histogram; fluorescence of the Sm2B3001 strain and its SmΔybeY mutant derivative transformed with the control plasmid pBBsyn-eGFP or the reporter pRprbA::eGFP (left part) and fluorescence of the double Sm2B2001ΔR1/2 and triple SmΔR1/2ΔybeY mutants co-transformed with pRprbA::eGFP and pSRK-C (control), pSRK-R2 or pSRK-R2b. Values reported are means and standard deviation of 27 fluorescence measurements normalized to the OD of the culture, i.e. three determinations of three independent exponential cultures of three transconjugants for each reporter strain. The statistical significance level was set at P < 0.05 and indicated with an asterisk. ns, non-significant. SmYbeY, AbcR2 and AbcR2b genotypes of the reporter strains are indicated on the bottom of the histogram. (B) SmYbeY cleaves the AbcR2-prbA duplex in vitro. Reactivity patterns of SmYbeY (5 μM) on an internally labeled 5΄-prbA RNA (0.1 pmol/μl) either alone or in the presence of a molar excess of AbcR2. Double arrow heads indicate major cleavage products. Reactions were incubated in the presence of Mn2+ or Ca2+ as co-factor. Incubation times are indicated on top of the panels. Reactions were analyzed on 7 M urea/15% polyacrylamide gels. C, control reactions.
Finally, we tested the in vitro activity of SmYbeY (5 μM) on an internally labeled RNA substrate that mimics the 5΄ prbA region (5΄-prbA) fused to eGFP both alone and upon its incubation with a molar excess of the AbcR2 transcript in the presence of Mn2+ as co-factor (Figure 9B). In these conditions, SmYbeY hardly reacted with the 5΄-prbA molecule, further suggesting that the prbA mRNA is not a preferred ligand of this protein. In contrast, addition of AbcR2 to the reaction mixture favored cleavage of the complex at precise sites within 5΄-prbA, yielding three major cleavage products. As expected, Ca2+ blocked SmYbeY activity on the partially double-stranded prbA-AbcR2 molecule.
We therefore conclude that post-transcriptional regulation of prbA mRNA requires SmYbeY, which could be most likely involved in the decay of the message upon its antisense interaction with the Hfq-dependent trans-acting AbcR2 sRNA.
DISCUSSION
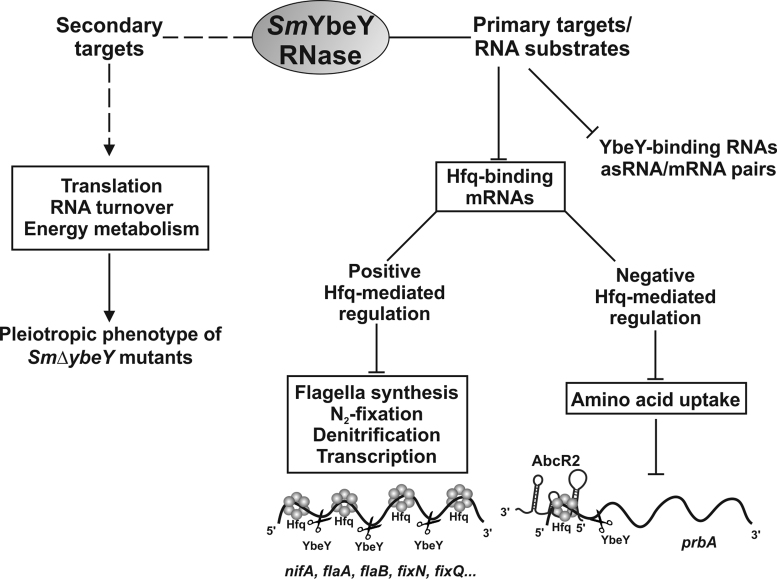
Here, we have shown that the S. meliloti YbeY protein is a divalent ion-dependent single- and double-strand endoribonuclease whose activity influences core RNA metabolism, energy producing pathways and plasmid-encoded symbiotic functions. Profiling of the SmYbeY-dependent genes and RNA ligands envisaged a number of putative substrates for this RNase. Although our data revealed that Hfq and SmYbeY participate in largely independent RNA networks, we provide evidences that the Hfq-dependent sRNA-mediated silencing of amino acid transport requires SmYbeY. Figure 10 summarizes the insights into the cellular pathways influenced by SmYbeY as revealed by our study.
Figure 10.
The SmYbeY mRNA network. The scheme summarizes the impact of SmYbeY on the S. meliloti transcriptome and its functional consequences. See discussion for details.
SmYbeY acts as versatile single- and double-strand endoribonuclease
Hfq is now viewed as a major RNA chaperone in bacteria that binds to and promotes stability of large sets of mRNA and sRNA transcripts (21). Given the apparent functional overlap between Hfq and SmYbeY in S. meliloti and the homology of SmYbeY to the MID RNA-binding domain of AGO proteins (10), we profiled the subpopulation of transcripts co-immunoprecipitated with a tagged version of SmYbeY. This approach has been proved successful for the generation of accurate and reliable genome-wide atlas of Hfq RNA ligands and sRNA–mRNA regulatory pairs in S. meliloti and other phylogenetically distant bacterial species (26,55–60). SmYbeY CoIP–RNA was barely enriched in RNA species in comparison to the number of transcripts stably bound by Hfq in the same conditions. This enrichment profile supports a major function of SmYbeY as a catalytic enzyme rather than an Hfq-like role as stabilizer and facilitator of RNA–RNA interactions in vivo. Therefore, other methods with enhanced sensitivity are required for the accurate genome-wide mapping of SmYbeY contacts of catalytic nature on RNA substrates.
To date, only the E. coli and V. cholerae YbeY orthologs have been demonstrated to have RNase activity in vitro (8,13), both exhibiting similar catalytic features. Although performed under experimental conditions different to those described for the former enzymes (e.g. reaction buffer or enzyme/substrate ratio), our assays similarly revealed catalytic activity of SmYbeY on generic and endogenous RNA molecules, but uncovered remarkable differences in its substrate specificity with respect to the well-characterized EcoYbeY. The latter behaves as a single-strand specific endoribonuclease unable to degrade dsRNA but exhibiting activity on short RNA hairpins and complex structured RNA (e.g. rRNA) (8,13). SmYbeY was proficient in cleaving ssRNA, dsRNA and a number of structured RNA substrates. To the best of our knowledge this versatility is an unprecedented feature among bacterial endoribonucleases (61). Nonetheless, the in vitro characterization of SmYbeY activity suggests a preference of the enzyme for longer double-stranded and structured RNA substrates. Its cleavage pattern on ssRNA and short dsRNA is reminiscent of non-specific endoribonucleases, such as RNase E. However, despite of being non-specific, RNAse E has a bias for cleaving AU-rich regions (62). In contrast, our experimental data did not reveal any preferred cleavage site of SmYbeY in these substrates. Of note, the substitution of the ultraconserved R69 residue (R59 in EcoYbeY), which inhibits EcoYbeY activity, similarly impaired SmYbeY cleavage on most substrates. However, this mutated version retained reliable residual activity in some structured RNA molecules tested (e.g. R1.1). Together with the finding that SmYbeY is also a double-strand endoribonuclease, this suggests that the catalytic mechanisms of both E. coli YbeY and the S. meliloti homologue may differ.
Under optimal assay conditions SmYbeY completely degraded the hairpin-structured R1.1 RNA substrate at sites recognized by the prototypical double-strand endoribonuclease RNase III (50,51). The catalytically competent form of RNase III is a homodimer in which each subunit coordinates a divalent metal ion (preferably Mg2+) and contributes to the hydrolytic cleavage of one strand of the duplex RNA substrate (63). YbeY and RNase III polypeptides are structurally unrelated (9,10,52). Indeed, our results indicated that SmYbeY does not dimerize and thus, a ∼20 kDa monomer is likely the active form of the enzyme. Co-factor requirements of EcoYbeY have not been stablished yet. However, we have shown that the in vitro reactivity patterns and performance of SmYbeY largely depend on the divalent metal ion used in the assays. Both Mg2+ and Mn2+ supported SmYbeY-mediated catalysis, but Mn2+ increased the overall cleavage efficiency. On the other hand Ca2+, which is a known inhibitor of RNase III activity (49,63), similarly blocked reactivity of SmYbeY on dsRNA and generic structured RNA but did not compromise ssRNA cleavage. It is therefore tempting to speculate on Ca2+ and Mg2+/Mn2+ availability as major environmental determinants of substrate selectivity and post-translational modulation of SmYbeY activity in vivo. In this regard, it is worthy to note that the alteration of Mn2+ homeostasis sensitizes both bacterial intracellular pathogens and plant endosymbionts to the oxidative burst induced in the host during infection (64–66). SmYbeY was able to degrade very efficiently the in vitro transcribed ybeY mRNA and therefore we cannot exclude the possibility of its autoregulation in vivo. Some RNases such as RNase E, RNase III and PNPase are also regulated at the post-transcriptional level by feedback loops involving decay of their own mRNA (61). This hypothesis of SmYbeY autoregulation should be similarly explored in the future.
SmYbeY activity influences fundamental and symbiotic functions
Absence or depletion of YbeY in bacteria decreases growth rate, alters rRNA/ribosome profiles, enhances cell sensitivity to stress and affects virulence and symbiotic traits (7,8,10,13–15,27,28). To explore the molecular basis of this pleiotropic phenotype in S. meliloti we profiled the SmYbeY-dependent transcriptome on oligonucleotide-based microarrays. Contrary to the expected effect of the removal of an endoribonuclease on the RNA steady-state levels, down-regulated transcripts far outnumbered up-regulated transcripts in the S. meliloti YbeY mutant. However, this has been a common finding of similar studies addressing the influence of the activity of diverse endo and exoribonucleases in the bacterial transcriptome, including the E. coli and Thermus thermophilus YbeY orthologs, particularly upon stress exposure (27,67–70). Down-regulated genes could be mostly regarded as secondary molecular targets of SmYbeY whose expression is positively influenced by this RNase in an indirect manner, e.g. involving alteration of regulatory intermediates or other yet unknown mechanisms. Nonetheless, a handful of SmYbeY-bound mRNAs identified in our CoIP experiments were also found among this group of down-regulated transcripts. This finding suggests a direct interaction of SmYbeY with these mRNAs in a protective mode that remains to be explored. Such a minor residual protective role in the stabilization of dsRNA and sRNAs has been already proposed for the long considered strictly catalytic ribonucleases RNase III and PNPase, respectively (61,71,72). Genes positively influenced by SmYbeY included those encoding relevant protein components of energy producing pathways, a number of RNases (e.g. PNPase, RNase III or RNase D) and key elements of the translation machinery. It is well-known that down-regulation of these fundamental physiological functions severely compromises bacterial growth and recovery upon stress exposure.
Protein mistranslation has been reported in E. coli YbeY mutants and was mainly attributed to the involvement of YbeY in rRNA maturation and ribosome quality control (5,13–16). Indeed, misprocessing of the 16S rRNA is a major molecular phenotype linked to YbeY loss-of-function in several bacterial species (5,7,8,13). In contrast, our results did not evidence an obvious involvement of SmYbeY in rRNA maturation. This was an unexpected finding since it has been shown that a number of bacterial YbeY orthologs can indistinctly rescue the pleiotropic physiological phenotypes of different ΔybeY mutants (5). The differences in substrate specificity and probably in activity mechanisms highlighted above may explain this apparent discrepancy. However, rRNA maturation involves the concerted activity of a suite of RNases some of which are not essential, can exhibit functional overlap or are interchangeable (61,67,68). Large genomes such as that of S. meliloti are typical sources of genetic redundancy that confers robustness to fundamental physiological processes (31). Therefore, we cannot rule out a role of SmYbeY in rRNA maturation that may be efficiently complemented by functionally related RNases.
Our transcriptomic analyses evidenced that SmYbeY influenced on chromosomal and pSymA-encoded functions to a similar extent. S. meliloti pSymA is a megaplasmid of mosaic origin that mostly host accessory acquired genes that specify relevant strain-specific and symbiotic traits (31). Major late symbiotic functions are coordinated via the two-component regulatory system FixLJ and the master regulators of nitrogen fixation NifA and FixK under microoxic conditions within the root nodule (73). Notably, we found that the nifA and fixK genes and a number of FixK-dependent gene clusters encoding the microaerobic denitrification pathway and elements of the electron transport chain associated with the nitrogenase complex were up-regulated in the SmYbeY mutant, particularly during exponential growth. Interestingly, YbeY has been also shown to severely disturb regulation of the Yersinia virulence plasmid pYV (7). These independent findings hint at a major universal role of YbeY in the post-transcriptional control of prokaryotic gene networks relevant to the interaction with eukaryotic hosts.
Extensive comparison of the Hfq and SmYbeY post-transcriptional regulons revealed a discrete overlap between the arrays of molecular targets of the two proteins. However, a number of known Hfq-binding mRNAs (26) exhibited accumulation patterns in the Hfq and SmYbeY defective mutants compatible with a role of this RNase in the decay of these transcripts. It has been reported that Hfq actively competes for binding to the sites of the major endoribonuclease RNase E (typically AU-rich regions) within sRNAs and mRNAs, thereby protecting these transcripts from RNase E cleavage and further exoribonucleolytic degradation (74,75). Our results revealed that sets of mRNAs with increased steady-state levels in the SmΔybeY mutant are encoded by genes positively regulated by Hfq, namely metabolic, carbohydrate transport, regulatory, flagellar, nitrogen-fixation and denitrification genes. Further supporting an Hfq-mediated stabilization, these mRNAs have been shown to be mostly recovered in their entire length by pull down with Hfq (26). Therefore, it can be hypothesized that SmYbeY has a role in the silencing of non-functional transcriptional output of flagellar and oxygen-regulated FixLJ-dependent mRNAs during stationary growth and under free-living non-symbiotic conditions, respectively (76,77).
SmYbeY mediates silencing of sRNA-regulated mRNAs
Ribonucleases are also key active players at different levels in the post-transcriptional gene silencing mediated by antisense and trans-acting sRNAs (78). RNase E and PNPase are known to affect sRNA turnover whereas RNase III can be recruited to the sRNA-target mRNA interplay for the cleavage of the double-stranded RNA duplex with the consequent degradation of the message in a mechanism that resembles eukaryotic RNA interference (43,78,79). We have shown that SmYbeY efficiently cleaves dsRNA and that asRNA–mRNA duplexes were remarkably abundant in CoIP–RNA. Thus, SmYbeY-mediated silencing of some Hfq-protected mRNAs can be triggered by antisense interaction with asRNAs. In this regard, recent RNAseq-based surveys of the S. meliloti transcriptome have uncovered functionally significant pervasive antisense transcription of pSymA-borne symbiotic genes (39,40,80). Therefore, the involvement of SmYbeY in the asRNA-mediated decay of nitrogen fixation mRNAs is a plausible scenario that merits further investigation.
RNase E has been already shown to be required for trans-sRNA regulation of cell-cycle and quorum-sensing mRNAs in S. meliloti (48,81). In contrast to what has been described for E. coli (27), transcriptomics data uncovered a scarce influence of SmYbeY on the steady-state levels of trans-sRNAs. However, among the transcripts up-regulated in the SmYbeY mutant we found a number of mRNAs coding for amino acid transporters that are putative targets of the Hfq-dependent homologous α-proteobacterial AbcR1 and AbcR2 sRNAs (37,54,82–84). Co-IP with Hfq typically recovers a specific stretch of these mRNAs, mostly derived from their 5΄ regions, rather than the full-length transcripts (26). This likely indicates that these mRNAs undergo ribonucleolytic degradation upon antisense interaction with their sRNA partners at sites within the stretch bound by Hfq. We have tested this hypothesis with the proline betaine prbA mRNA, which is an experimentally confirmed target of both AbcR1 and AbcR2 trans-sRNAs (26). Fluorescence of relevant reporter strains along with in vitro assays suggested that post-transcriptional silencing of prbA could be initiated by SmYbeY-mediated cleavage of the message at discrete positions upon AbcR2 interaction in the vicinity of the ribosome binding site. Previous works in S. meliloti, E. coli and V. cholerae had anticipated a major role of YbeY in riboregulation (8,10,27). However, this conclusion was entirely based on the misregulation of sets of sRNAs and their predicted mRNA targets upon deletion or depletion of YbeY, which does not demonstrate a direct role of this protein, either protective or catalytic, in the establishment of specific experimentally probed sRNA–mRNA interactions. Therefore, our results add SmYbeY to the repertoire of bacterial ribonucleases involved in RNA-mediated silencing.
In summary, we have shown that the highly conserved S. meliloti YbeY protein is a versatile metal-dependent double and single-strand endoribonuclease that influences turnover of bulk and sRNA-regulated mRNAs. The SmYbeY-dependent mRNA network presented here provides a solid resource for the forthcoming investigation of SmYbeY activity mechanisms underlying the post-transcriptional regulation of core RNA metabolism, energy producing pathways and late symbiotic functions in S. meliloti.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Omar Torres-Quesada for early genetic constructs to generate the YbeY mutant, Vicenta Millán and Teresa Baptista da Silva for their invaluable technical assistance, Andreas Kautz for help in the transcriptome experiments, Jochen Blom and Oliver Rupp for bioinformatics services, Bernadette Boomers for genome sequencing, and the core facilities of EEZ-CSIC for routine sequencing of plasmid inserts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitividad [ERDF-cofinanced grant BFU2013-48282-C2-2-P to J.I.J.-Z.]; LOEWE Program of the State of Hesse [SYNMIKRO to A.B]; German Research Foundation [CRC 987 to A.B.]; Fundação para a Ciência e Tecnologia (FCT) [Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular - R&D Unit, UID/CBQ/04612/2013) funded by FEDER funds through COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI) and by national funds to C.M.A.]; FCT [project PTDC/BIA-MIC/1399/2014 to C.M.A]; European Union's Horizon 2020 Research and Innovation Programme [grant agreement no. 635536 to C.M.A.]; Consejo Superior de Investigaciones Científicas (CSIC) [JAEDoc Program contract to A.P.]; FCT Post-Doctoral Fellowships [SFRH/BPD/109464/2015 to M.S., SFRH/BPD/75887/2011 to R.G.M.]; Ministerio de Economía y Competitividad [Program of Formación Post-doctoral contract to M.R.]; BMBF [FKZ 031A533 within the de.NBI network]. Funding for open access charge: Ministerio de Economía y Competitividad.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gil R., Silva F.J., Peretó J., Moya A.. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004; 68:518–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. et al. Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies B.W., Walker G.C.. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J. Bacteriol. 2008; 190:1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akerley B.J., Rubin E.J., Novick V.L., Amaya K., Judson N., Mekalanos J.J.. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies B.W., Köhrer C., Jacob A.I., Simmons L.A., Zhu J., Aleman L.M., RajBhandary U.L., Walker G.C.. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol. Microbiol. 2010; 78:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P. et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leskinen K., Varjosalo M., Skurnik M.. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology. 2015; 161:285–299. [DOI] [PubMed] [Google Scholar]
- 8.Vercruysse M., Köhrer C., Davies B.W., Arnold M.F.F., Mekalanos J.J., RajBhandary U.L., Walker G.C.. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog. 2014; 10:e1004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oganesyan V., Busso D., Brandsen J., Chen S., Jancarik J., Kim R., Kim S.-H.. Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:1219–1223. [DOI] [PubMed] [Google Scholar]
- 10.Pandey S.P., Minesinger B.K., Kumar J., Walker G.C.. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011; 39:4691–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003; 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan C., Fedorov E.V., Shi W., Ramagopal U.A., Thirumuruhan R., Manjasetty B.A., Almo S.C., Fiser A., Chance M.R., Fedorov A.A.. The YbeY protein from Escherichia coli is a metalloprotein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005; 61:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacob A.I., Köhrer C., Davies B.W., RajBhandary U.L., Walker G.C.. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol. Cell. 2013; 49:427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasouly A., Davidovich C., Ron E.Z.. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J. Bacteriol. 2010; 192:4592–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasouly A., Schonbrun M., Shenhar Y., Ron E.Z.. YbeY, a heat shock protein involved in translation in Escherichia coli. J. Bacteriol. 2009; 191:2649–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grinwald M., Ron E.Z.. The Escherichia coli translation-associated heat shock protein YbeY is involved in rRNA transcription antitermination. PLoS One. 2013; 8:e62297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulthana S., Basturea G.N., Deutscher M.P.. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA. 2016; 22:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boland A., Tritschler F., Heimstädt S., Izaurralde E., Weichenrieder O.. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010; 11:522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D.. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009; 33:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storz G., Vogel J., Wassarman K.M.. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011; 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel J., Luisi B.F.. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011; 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobrero P., Valverde C.. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012; 38:276–299. [DOI] [PubMed] [Google Scholar]
- 23.Romby P., Charpentier E.. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 2010; 67:217–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X., Zhulin I., Wartell R.M.. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002; 30:3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardill J.P., Hammer B.. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA Biol. 2012; 9:392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres-Quesada O., Reinkensmeier J., Schlueter J.-P., Robledo M., Peregrina A., Giegerich R., Toro N., Becker A., Jimenez-Zurdo J.I.. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014; 11:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey S.P., Winkler J.A., Li H., Camacho D.M., Collins J.J., Walker G.C.. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics. 2014; 15:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies B.W., Walker G.C.. Identification of novel Sinorhizobium meliloti mutants compromised for oxidative stress protection and symbiosis. J. Bacteriol. 2007; 189:2110–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beringer J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974; 84:188–198. [DOI] [PubMed] [Google Scholar]
- 30.Robertsen B.K., Aman P., Darvill A.G., McNeil M., Albersheim P.. Host-symbiont interactions: V. the structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981; 67:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galibert F., Finan T.M., Long S.R., Puhler A., Abola P., Ampe F., Barloy-Hubler F., Barnett M.J., Becker A., Boistard P. et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001; 293:668–672. [DOI] [PubMed] [Google Scholar]
- 32.Bahlawane C., McIntosh M., Krol E., Becker A.. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 2008; 21:1498–1509. [DOI] [PubMed] [Google Scholar]
- 33.Schafer A., Tauch A., Jager W., Kalinowski J., Thierbach G., Puhler A.. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994; 145:69–73. [DOI] [PubMed] [Google Scholar]
- 34.Simon R., Priefer U., Puhler A.. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1983; 1:784–791. [Google Scholar]
- 35.Torres-Quesada O., Oruezabal R.I., Peregrina A., Jofre E., Lloret J., Rivilla R., Toro N., Jimenez-Zurdo J.I.. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol. 2010; 10:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blatny J.M., Brautaset T., Winther-Larsen H.C., Haugan K., Valla S.. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 1997; 63:370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres-Quesada O., Millán V., Nisa-Martínez R., Bardou F., Crespi M., Toro N., Jiménez-Zurdo J.I.. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS One. 2013; 8:e68147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlüter J.P., Reinkensmeier J., Daschkey S., Evguenieva-Hackenberg E., Janssen S., Janicke S., Becker J.D., Giegerich R., Becker A.. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010; 11:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlüter J.P., Reinkensmeier J., Barnett M.J., Lang C., Krol E., Giegerich R., Long S.R., Becker A.. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics. 2013; 14:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilker R., Stadermann K.B., Doppmeier D., Kalinowski J., Stoye J., Straube J., Winnebald J., Goesmann A.. ReadXplorer—visualization and analysis of mapped sequences. Bioinformatics. 2014; 30:2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viegas S.C., Silva I.J., Saramago M., Domingues S., Arraiano C.M.. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2010; 39:2918–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrania J., Vorhölter F.-J., Niehaus K., Pühler A., Becker A.. Identification of Xanthomonas campestris pv. campestris galactose utilization genes from transcriptome data. J. Biotechnol. 2008; 135:309–317. [DOI] [PubMed] [Google Scholar]
- 46.Becker A., Barnett M.J., Capela D., Dondrup M., Kamp P.B., Krol E., Linke B., Ruberg S., Runte K., Schroeder B.K. et al. A portal for rhizobial genomes: RhizoGATE integrates a Sinorhizobium meliloti genome annotation update with postgenome data. J. Biotechnol. 2009; 140:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dondrup M., Albaum S.P., Griebel T., Henckel K., Jünemann S., Kahlke T., Kleindt C.K., Küster H., Linke B., Mertens D. et al. EMMA 2 – A MAGE-compliant system for the collaborative analysis and integration of microarray data. BMC Bioinformatics. 2009; 10:50–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robledo M., Frage B., Wright P.R., Becker A.. A stress-induced small RNA modulates alpha-rhizobial cell cycle progression. PLoS Genet. 2015; 11:e1005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H.L., Chelladurai B.S., Zhang K., Nicholson A.W.. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993; 21:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross G., Dunn J.J.. Structure of secondary cleavage sites of E. coli RNAaseIII in A3t RNA from bacteriophage T7. Nucleic Acids Res. 1987; 15:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barra-Bily L., Fontenelle C., Jan G., Flechard M., Trautwetter A., Pandey S.P., Walker G.C., Blanco C.. Proteomic alterations explain phenotypic changes in Sinorhizobium meliloti lacking the RNA chaperone Hfq. J. Bacteriol. 2010; 192:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao M., Barnett M.J., Long S.R., Teplitski M.. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol. Plant Microbe Interact. 2010; 23:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobrero P., Schluter J.P., Lanner U., Schlosser A., Becker A., Valverde C.. Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011. PLoS One. 2012; 7:e48494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sittka A., Lucchini S., Papenfort K., Sharma C.M., Rolle K., Binnewies T.T., Hinton J.C.D., Vogel J.. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008; 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sittka A., Sharma C.M., Rolle K., Vogel J.. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009; 6:266–275. [DOI] [PubMed] [Google Scholar]
- 57.Berghoff B.A., Glaeser J., Sharma C.M., Zobawa M., Lottspeich F., Vogel J., Klug G.. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol. Microbiol. 2011; 80:1479–1495. [DOI] [PubMed] [Google Scholar]
- 58.Dambach M., Irnov I., Winkler W.C.. Association of RNAs with Bacillus subtilis Hfq. PLoS One. 2013; 8:e55156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J.. An atlas of Hfq-bound transcripts reveals 3΄ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012; 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saadeh B., Caswell C.C., Chao Y., Berta P., Wattam A.R., Roop R.M., O'Callaghan D.. Transcriptome-wide Identification of Hfq-associated RNAs in Brucella suis by deep sequencing. J. Bacteriol. 2016; 198:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arraiano C.M., Andrade J.M., Domingues S., Guinote I.B., Malecki M., Matos R.G., Moreira R.N., Pobre V., Reis F.P., Saramago M. et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010; 34:883–923. [DOI] [PubMed] [Google Scholar]
- 62.Kaberdin V.R., Walsh A.P., Jakobsen T., McDowall K.J., von Gabain A.. Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli ribonuclease E1. J. Mol. Biol. 2000; 301:257–264. [DOI] [PubMed] [Google Scholar]
- 63.Li H., Nicholson A.W.. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996; 15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 64.Davies B.W., Walker G.C.. Disruption of sitA Compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J. Bacteriol. 2007; 189:2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papp-Wallace K.M., Maguire M.E.. Manganese transport and the role of manganese in virulence. Ann. Rev. Microbiol. 2006; 60:187–209. [DOI] [PubMed] [Google Scholar]
- 66.Anderson E.S., Paulley J.T., Gaines J.M., Valderas M.W., Martin D.W., Menscher E., Brown T.D., Burns C.S., Roop R.M.. The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect. Immun. 2009; 77:3466–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pobre V., Arraiano C.M.. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli. BMC Genomics. 2015; 16:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stead M.B., Marshburn S., Mohanty B.K., Mitra J., Castillo L.P., Ray D., van Bakel H., Hughes T.R., Kushner S.R.. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011; 39:3188–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohanty B.K., Kushner S.R.. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003; 50:645–658. [DOI] [PubMed] [Google Scholar]
- 70.Ohyama H., Sakai T., Agari Y., Fukui K., Nakagawa N., Shinkai A., Masui R., Kuramitsu S.. The role of ribonucleases in regulating global mRNA levels in the model organism Thermus thermophilus HB8. BMC Genomics. 2014; 15:386–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bandyra K.J., Sinha D., Syrjanen J., Luisi B.F., De Lay N.R.. The ribonuclease polynucleotide phosphorylase can interact with small regulatory RNAs in both protective and degradative modes. RNA. 2016; 22:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006; 124:355–366. [DOI] [PubMed] [Google Scholar]
- 73.Bobik C., Meilhoc E., Batut J.. FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 2006; 188:4890–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Folichon M., Allemand F., Regnier P., Hajnsdorf E.. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005; 272:454–463. [DOI] [PubMed] [Google Scholar]
- 75.Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U.. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003; 9:1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sourjik V., Muschler P., Scharf B., Schmitt R.. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 2000; 182:782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rotter C., Mühlbacher S., Salamon D., Schmitt R., Scharf B.. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J. Bacteriol. 2006; 188:6932–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saramago M., Bárria C., dos Santos R.F., Silva I.J., Pobre V., Domingues S., Andrade J.M., Viegas S.C., Arraiano C.M.. The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 2014; 18:105–115. [DOI] [PubMed] [Google Scholar]
- 79.Andrade J.M., Pobre V., Matos A.M., Arraiano C.M.. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA. 2012; 18:844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robledo M., Jiménez-Zurdo J.I., Becker A.. Antisense transcription of symbiotic genes in Sinorhizobium meliloti. Symbiosis. 2015; 67:55–67. [Google Scholar]
- 81.Baumgardt K., Šmídová K., Rahn H., Lochnit G., Robledo M., Evguenieva-Hackenberg E.. The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol. 2015; 13:486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilms I., Voss B., Hess W.R., Leichert L.I., Narberhaus F.. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol. Microbiol. 2011; 80:492–506. [DOI] [PubMed] [Google Scholar]
- 83.Caswell C.C., Gaines J.M., Ciborowski P., Smith D., Borchers C.H., Roux C.M., Sayood K., Dunman P.M., Roop Ii R.M.. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol. Microbiol. 2012; 85:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Overlöper A., Kraus A., Gurski R., Wright P.R., Georg J., Hess W.R., Narberhaus F.. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014; 11:624–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.