Abstract
Functional characterization of the transcriptome requires tools for the systematic investigation of RNA post-transcriptional modifications. 2΄-O-methylation (2΄-OMe) of the ribose moiety is one of the most abundant post-transcriptional modifications of RNA, although its systematic analysis is difficult due to the lack of reliable high-throughput mapping methods. We describe here a novel high-throughput approach, named 2OMe-seq, that enables fast and accurate mapping at single-base resolution, and relative quantitation, of 2΄-OMe modified residues. We compare our method to other state-of-art approaches, and show that it achieves higher sensitivity and specificity. By applying 2OMe-seq to HeLa cells, we show that it is able to recover the majority of the annotated 2΄-OMe sites on ribosomal RNA. By performing knockdown of the Fibrillarin methyltransferase in mouse embryonic stem cells (ESCs) we show the ability of 2OMe-seq to capture 2΄-O-Methylation level variations. Moreover, using 2OMe-seq data we here report the discovery of 12 previously unannotated 2΄-OMe sites across 18S and 28S rRNAs, 11 of which are conserved in both human and mouse cells, and assigned the respective snoRNAs for all sites. Our approach expands the repertoire of methods for transcriptome-wide mapping of RNA post-transcriptional modifications, and promises to provide novel insights into the role of this modification.
INTRODUCTION
RNA molecules play a major role in the regulation of any cellular process. Recent reports of pervasive transcription occurring across the genome of higher metazoans (1,2) make structural and functional characterization of these transcripts a key need for deeper comprehension of their regulation and mechanisms of action. Beside the primary sequence information, cellular RNAs are modified post-transcriptionally. More than a hundred possible RNA post-transcriptional modifications have been identified so far (3), and roughly two third of them involve the addition of methyl groups, mainly on nucleobase nitrogens (4). In the last years, many efforts have been made to develop novel high-throughput methods for transcriptome-wide mapping of RNA post-transcriptional modifications, such as 5-methylcytosine (5,6) (m5C), N6-methyladenosine (7–9) (m6A) and pseudouridine (10,11) (Ψ).
2΄-O-Methylation (2΄-OMe) of the ribose moiety is one of the most abundant RNA modifications (12). Oppositely to other post-transcriptional modifications, 2΄-O-methylation occurs on the oxygen of the free 2΄-OH of the ribose moiety. Its deposition is mediated by a multi-protein complex, whose catalytic core resides in the Fibrillarin methyltransferase enzyme (Fbl). The specificity of RNA-substrate recognition by the complex is mediated by non-coding C/D box small nucleolar RNA (snoRNA) guides, that contain short complementary segments (11–20 nt) to the target RNA (4,13,14). These snoRNAs contain two conserved sequence motifs, namely box C (RUGAUGA) and box D (CUGA), and the methylation occurs on the target RNA precisely five nucleotides upstream of the box D. 2΄-OMe function is still largely unknown. Although the chemical properties of this modification do not provide any clue to specific functional roles, it is clear that it can affect sterical properties, hydrogen-bonding potential, and structural rigidity of the target RNA. Several 2΄-OMe residues have been mapped on rRNA, and they do not appear to be randomly distributed, indeed they are preferentially located within conserved regions, thus suggesting that 2΄-OMe can influence both ribosome structure and function (15). It is clear that it might influence several features of the RNA like folding, assembly or stability (16). One of the possible theories is that ribose methylation assists and guides the assembly of ribosomes by maintaining the rigidity of the RNA backbone at key regions of the rRNA (17). Interestingly, thermophilic organisms undergo higher amounts of ribose methylations than mesophilic organisms (18), in agreement with the increased structural rigidity provided by ribose methylations (19). Therefore, ribose methylation is likely to increase thermal stability of essential regions of the ribosome. Moreover, the provided rigidity might also contribute to RNA folding; indeed, NMR analysis of Escherichia coli provided strong evidence that the U2552 2΄-OMe site is critical for a proper folding of the A loop (20), maybe contributing to the precise positioning of the tRNA in the A site (15).
Thus, the development of accurate methods for mapping this modification is a key need to enable its systematic analysis. To date, three different approaches have been reported for site-specific mapping of 2΄-OMe residues (13,21,22). The first two methods are based on the increased resistance of 2΄-OMe residues toward alkaline hydrolysis and RNase H digestion, while the third is based on the specific pausing of the reverse transcriptase on these residues under limiting dNTP concentrations. Although an high-throughput approach based on alkaline hydrolysis has been recently proposed (23), no state-of-the-art method exists for the high-throughput mapping of 2΄-O-Methylated residues within RNA. Therefore, we here investigated the suitability of all the three aforementioned methodologies for genome-scale studies, using HeLa ribosomal RNAs as a benchmark, and we found that the approach based on the use of limiting dNTP concentrations is the most sensitive and specific among the three. Moreover, analysis of 2OMe-seq data for mouse and human cells revealed the existence of 12 previously unannotated rRNA 2΄-OMe sites, 11 of which are conserved between the two species.
MATERIALS AND METHODS
Cells culture
HeLa S3 cells were obtained from ATCC (cat. CCL2.2), and cultured in DMEM (4.5g/l d-glucose), supplemented with 10% FBS, 0.1 mM NEAA, 25 U/ml penicillin and 25 μg/ml streptomycin. Mouse embryonic stem cells E14 were grown on 0.1% gelatin-coated plates and maintained in DMEM (4.5g/l d-glucose), supplemented with 15% heat-inactivated FBS, 0.1 mM NEAA, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, 25 U/ml penicillin, 25 μg/ml streptomycin and 1500 U/ml LIF, as previously described (24). Cells were cultured at 37°C, in the presence of 5% CO2. Cells confluency was always kept below 80%.
RNA extraction and quality assessment
RNA was extracted using TRIzol reagent (Invitrogen), following manufacturer instructions. RNA integrity measurements were performed using Fragment Analyzer™ (Advanced Analytical). All samples had RNA Quality Number (RQN) >9.8.
Western blot
Approximately 1 × 106 cells were scraped in 1 ml of cold phosphate buffered saline (PBS) and centrifuged for 5΄ at 1000g. Cell pellets were resuspended in 200 ml of cold F-Buffer [10 mM Tris–HCl (pH 7.0), 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride and 5 mM ZnCl2]. Cells were subjected to three cycles of sonication (30 s ON, 30 s OFF, high power) using a Bioruptor Twin sonicator (Diagenode), and then stored on ice for 10’. Cell extract was then centrifuged for 10 min at 14 000g, and the pellet was discarded. Cell lysates were quantified using BCA Protein Assay Kit (Pierce), and 15 μg of total proteins were loaded on each lane of a 4–20% polyacrylamide gel.
Rabbit monoclonal antibody against Fibrillarin was obtained from Cell Signaling (cat. #2639), and used at a final dilution of 1:1000. Beta-actin was used as the loading control.
shRNAs design, cloning and transfection
Custom shRNAs were constructed using the TRC hairpin design tool (http://www.broadinstitute.org/rnai/public/seq/search), and designed to target the 3΄-UTR of the Fbl transcript (NM_007991). shRNAs with more than 14 consecutive matches to non-target transcripts were avoided. Hairpins were cloned into pLKO.1 vector (Addgene: 10878) and each construct was verified by sequencing. For mouse ESCs transfection, ∼106 cells were seeded in a 6-well plate the day prior to transfection. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen), following manufacturer protocol. Cells were maintained for 4 days in canonical medium supplemented with Puromycin (2 μg/ml for 1 day, 1 μg/ml for 3 days).
2OMe-seq library preparation #1 (without fragmentation)
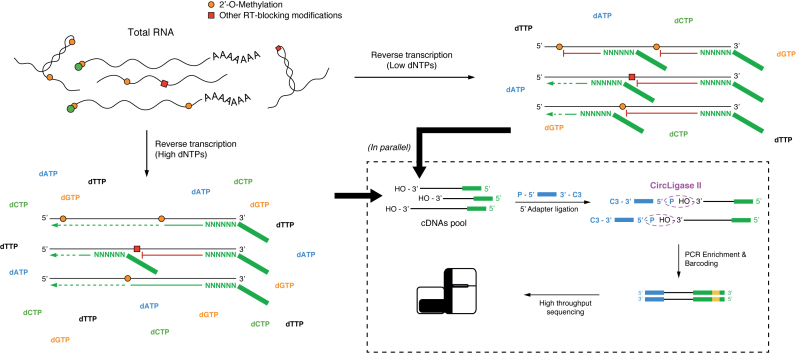
For each experiment, 2 μg of total RNA were spiked-in with 100 ng of in vitro transcribed RNAs (where specified, pooled in the amounts specified in the Generation of synthetic 2΄-O-Methylated spike-ins paragraph in Supplementary Data). 2-OMe-seq libraries were prepared with minor changes to the protocol used for CIRS-seq (25). Briefly, reverse transcription was performed in a 20 μl reaction, using either 1 mM final dNTP (high dNTP sample), or 0.004 mM final dNTP (low dNTP sample), 0.5 U/μl AMV Reverse Transcriptase (NEB) and random hexamers fused with the reverse complement of the Illumina TruSeq RNA 3΄ Adapter (CCTTGGCACCCGAGAATTCCANNNNNN). The reverse transcription reaction was conducted in 30 min at 42°C, followed by 10 min at 95°C to inactivate the AMV enzyme. Template RNA was degraded by addition of 1 μl of Ribonuclease H (Ambion) and 1 μl of RNase Cocktail Enzyme Mix (Ambion), followed by 30 min incubation at 37°C. cDNAs recovery, and excess adapter removal was performed using Agencourt Ampure XP beads (Beckman Coulter). Following cleanup, cDNAs were separated on a 10% TBE–urea PAGE gel, and a gel slice corresponding to 70–300nt fragments was cut. DNA was recovered by passive diffusion in TE buffer [10 mM Tris–HCl, 1 mM EDTA] for 16 h at 37°C with moderate shaking. Following ethanol precipitation, an adapter corresponding to the reverse complement of the standard Illumina TruSeq Small RNA 5΄ Adapter (GATCGTCGGACTGTAGAACTCTGAAC), modified with a 5΄-P group and a 3΄-C3 spacer, was ligated to cDNAs 3΄-OH termini using 200 U of CircLigase II for 6 h at 65°C. This approach allowed us to keep the strand-specificity of the library, so that each read started 1 nt downstream of the RT stopping point. The adapter-ligated cDNAs were then subjected to 15 cycles of PCR using standard Illumina TruSeq primers, and excess primers were removed using Agencourt Ampure XP beads. Libraries were pooled in equimolar amounts, and subjected to sequencing on the Illumina™ NextSeq 500 Sequencer.
Sequencing data pre-processing
Reads pre-processing (adapter clipping and trimming), mapping, and counting were performed using the rt-count utility from RNA Structure Framework v1.1.0 (26) (http://rsf.hugef-research.org), with parameters: -ca TGGAATTCTCGGGTGCCAAGG -cl 25 -cm 1 -bm 1 -b5 5 -bc 3200000 (-ca AGATCGGAAGAGCACACGTCT was used for Alkaline hydrolysis and 2OMe-seq Method #2 libraries, due to the different 3΄ adapter used). Reference was composed of 28S and 18S rRNA sequences, plus sequences of the three in vitro generated spike-ins (where included). Post-processing was performed using custom scripts.
Prediction of guide snoRNAs
To identify putative snoRNAs targeting the discovered sites, a sequence of 20 nucleotides centered on the discovered site was extracted, and passed to the Snoscan software (14) (http://lowelab.ucsc.edu/snoscan/), with default parameters. SnoRNA sequences were obtained from the latest ENSEMBL release, and from deepBase predictions (http://deepbase.sysu.edu.cn/download.php).
Compilation of known 2΄-OMe sites in rRNA
For 18S rRNA, the list of human 2΄-OMe residues was taken from Maden and coworkers data (Table III from Maden et al. (27)), by excluding all those sites for which methylation data was available only for Xaenopus laevis. For 28S rRNA, a conservative list of 2΄-OMe residues was obtained from snoRNA-LBME-db (28), by excluding all those sites for which methylation was only predicted by Laszlo et al. (13), but not experimentally validated. This yielded a final list of 31 and 51 2΄-OMe sites, respectively for 18S and 28S rRNA.
For mouse rRNA 2΄-OMe sites analysis, 18S and 28S rRNA sequences from human and mouse were aligned using the EMBOSS Needle tool (http://www.ebi.ac.uk/Tools/psa/emboss_needle/nucleotide.html), and known human sites were lifted over from human to mouse.
Data access
Sequencing data have been deposited on the NCBI Gene Expression Omnibus under accession GSE73065.
RESULTS
2OMe-seq allows high-throughput mapping of 2΄-O-Methylated residues
Given the lack of highly-specific single-base resolution methods for the mapping of 2΄-O-methylated (2΄-OMe) residues within RNA molecules, we have developed a new technique, named 2OMe-seq, that enables the identification and quantitation of the relative abundance of 2΄-OMe residues in a high-throughput fashion.
To date, three approaches have been proposed for site-specific identification of 2΄-OMe residues.
They are based respectively on the increased resistance of 2΄-OMe residues toward alkaline hydrolysis treatment (13) and RNase H digestion (22), or on the differential processivity of the reverse transcriptase (RT) through 2΄-OMe residues under high and low dNTP concentrations (27).
To test their ability to capture 2΄-OMe sites, we first prepared libraries of HeLa total RNA starting with either RNase H treatment, or alkaline hydrolysis in sodium carbonate buffer, coupled to adapter ligation and RT, and sequenced them on Illumina platform (Supplementary Data, and Supplementary Data). Since 2΄-OMe confers resistance to either treatments to the phosphodiester bond on its 3΄ side, mapped sequencing reads will accumulate one nucleotide downstream of non-methylated bases, while they will be relatively depleted one nucleotide downstream of 2΄-OMe sites. We defined a simple score metric, ranging from 0 (no protection) to 1 (total protection), which captures the local depletion of RT stops on 2΄-OMe sites with respect to their immediate context (Supplementary Data). With both methods, we measured a score increase that was slightly more pronounced with the alkaline hydrolysis than with the RNase H treatment (Supplementary Data). We then produced ROC curves to evaluate the discriminative power of the two methods. ROC curves measure the discriminative ability of a binary classifier; an area under the curve (AUC) of 0.5 corresponds to a prediction that does not perform better than random, while AUC values >0.5 represent increasingly better predictions, with 1 corresponding to a perfect prediction. From our analysis, alkaline hydrolysis treatment showed a better discriminative power compared to RNase H treatment (AUC: 0.76 and 0.87 respectively for RNase H and alkaline hydrolysis treatments, Supplementary Data). Despite this, we noticed a moderate yet significant resistance of pseudouridine (Ψ) residues to alkaline hydrolysis (but not to RNase H) treatment (Supplementary Data), in agreement with a recent report (23). Due to recent reports that Ψs are highly abundant within higher eukaryote transcriptomes (10,11), we reasoned that the ‘cross-reactivity’ exhibited by this method would not enable an accurate mapping of 2΄-OMe sites.
We next investigated the peculiar ability of 2΄-OMe residues to induce, under limiting dNTP concentrations, a specific reverse transcription (RT) stop one nucleotide downstream of the methylated site. To take advantage of this chemistry, we prepared libraries of HeLa total RNA by random-primed RT under both high (1 mM) and low (0.004 mM) dNTP concentrations, followed by adapter ligation and massive parallel sequencing on Illumina platform (Figure 1). We named this method 2OMe-seq.
Figure 1.
Schematics of 2OMe-seq library generation. Total RNA from cells is subjected to reverse transcription (RT) under either high or low dNTP concentrations, using random hexamers coupled to the 3΄ sequencing adapter. RT at low dNTP concentrations causes the RT to stall 1 nucleotide downstream of 2΄-OMe sites. cDNAs are then recovered, and ligated to the 5΄ sequencing adapter. Sequencing indexes are then introduced by PCR (Method #1, see Supplemental Information for details).
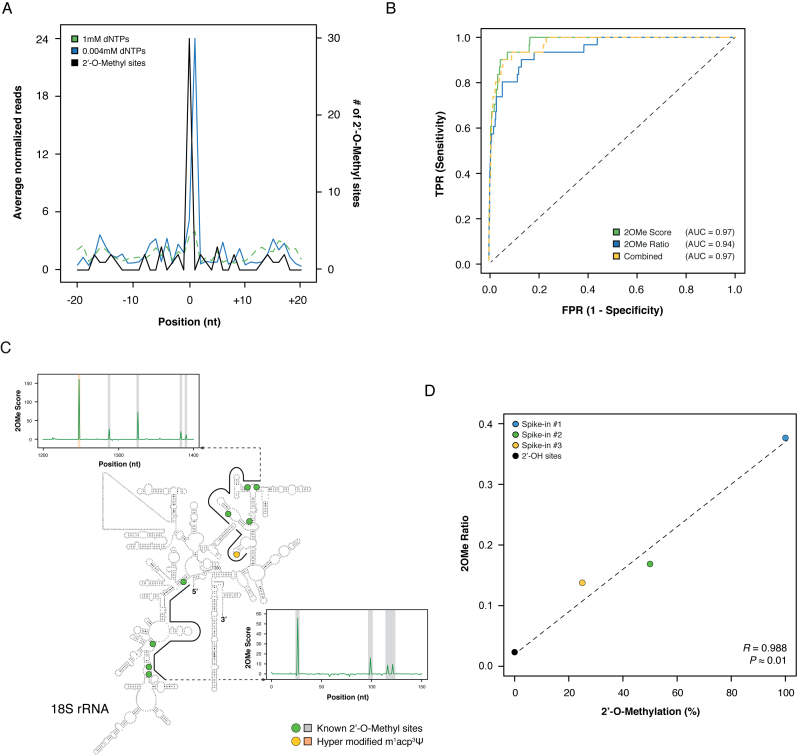
Analysis of HeLa annotated 2΄-OMe sites in human 18S rRNA (27) revealed a strong accumulation of RT-stops 1 nucleotide downstream of the annotated sites in the low dNTP sample, that was not observed at high dNTP concentrations (Figure 2A). To perform sites calling and relative quantitation, we further defined two metrics: (i) 2OMe Score, which captures the enrichment of a 2΄-OMe site in the low dNTP library versus the high dNTP library and (ii) 2OMe Ratio, which represents the fraction of reads stopping at a given position, out of the total reads covering that position in the low dNTP sample, and can thus capture the relative stoichiometry of the modification.
Figure 2.
Overview of the 2OMe-seq approach. (A) Meta-2OMe plot of the average normalized number of reads (left axis) for the 1 mM dNTPs (dashed green) and 0.004 mM dNTPs (solid blue) libraries on HeLa 18S rRNA. The number of 2΄-OMe residues at each position in the window is indicated (black, right axis). (B) ROC curves on HeLa 18S rRNA for 2OMe Score (solid green), 2OMe Ratio (solid blue), and their logistic combination (dashed yellow) trained on 28S rRNA. (C) Representative windows along HeLa 18S rRNA. Known 2΄-OMe sites are outlined by green dots on the secondary structure, and grey outlines on the 2OMe Score charts. The hyper modified m1acp3Ψ site previously reported to completely block RT (27) is indicated with a yellow dot on the secondary structure, and an orange outline on the 2OM Score chart. The secondary structure of the human 18S rRNA has been obtained from CRW website (34) (http://www.rna.icmb.utexas.edu/). (D) Scatterplot of the known methylation level versus the calculated 2OMe Ratio for the three synthetic spike-ins. Point 0 2OMe Ratio was calculated as the average of the 2OMe Ratio of all 2΄-OH bases of the three spike-ins.
Both metrics gave high discriminatory power, measured as the area under the curve (AUC, Figure 2B), between the positive set of known 2΄-OMe sites (Figure 2C), and the negative set of all other residues in 18S rRNA (AUC: 0.97, 0.94 and 0.97 respectively for 2OMe Score, 2OMe Ratio, and their logistic combination). Similar results were obtained when including also the 28S rRNA, although the measured AUC was slightly lower, suggesting the existence of unannotated sites (AUC: 0.93 and 0.90 respectively for 2OMe Score and 2OMe Ratio, Supplementary Data), thus the true accuracy of 2OMe-seq is probably underestimated.
We conducted the same analysis on a pool of three in vitro synthesized 200 nt RNAs, each one bearing a single 2΄-OMe site with a well-defined methylation level (25–50–100% methylation, Supplementary Data). This time, a near perfect discriminatory power was found for both metrics (AUC: 1.00 and 0.98, respectively for 2OMe Score and 2OMe Ratio, Supplementary Data). Notably, oppositely to the alkaline hydrolysis approach (Supplementary Data), no aspecific enrichment of RT-stops at pseudouridines was detected (Supplementary Data), further confirming the high specificity of our method.
We furthermore assessed the ability of 2OMe-seq to quantify the relative stoichiometry of the identified 2΄-OMe sites. Although it doesn't capture the absolute stoichiometry, probably due to the partial efficiency of the low dNTP-mediated RT termination, we found an exceptional correlation between the known modification levels for the 2΄-OMe sites in the 3 synthetic RNAs, and the calculated 2OMe ratio (R2 = 0.976, Figure 2D).
Additionally, to test the robustness of our method, we also performed 2OMe-seq on fragmented RNA, by direct RNA 3΄-adapter ligation, and RT using an oligonucleotide complementary to the adapter (Supplementary Data and Supplementary Data). Comparable results were obtained.
2΄-O-Methylated residues are extremely conserved across species
Since most studies on 2΄-O-methylation have been conducted on HeLa cells, we decided to investigate the conservation of this modification by performing 2OMe-seq in E14 mouse embryonic stem cells (ESCs).
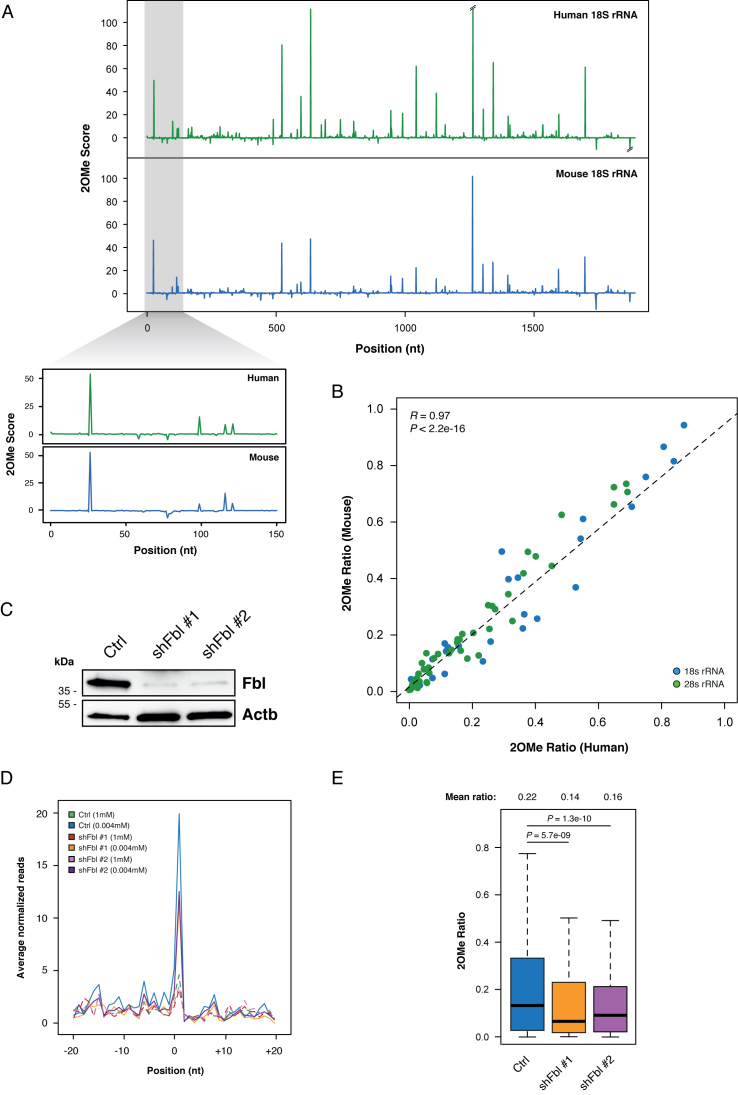
Analysis of 2OMe scores for HeLa and mouse ESCs rRNA showed nearly identical profiles (Figure 3A), thus supporting a functional role for 2΄-OMe residues at specific rRNA sites, and confirming the extremely conserved nature of this modification, as previously suggested by conservation analysis of snoRNAs (29,30).
Figure 3.
2OMe-seq captures the relative modification stoichiometry and its variation. (A) 2OMe Score profiles for HeLa and ESCs 18S rRNA. (B) Scatterplot of the 2OMe Ratio measured in HeLa versus ESCs for each 2΄-OMe residue in the 18S (blue) and 28S (green) rRNAs. (C) Western blot of Fibrillarin in control and Fbl knockdown ESCs. Beta actin was used as the loading control. (D) Meta-2OMe plot of the average normalized number of reads (left axis) for the 1 mM dNTPs (dashed lines) and 0.004 mM dNTPs (solid lines) libraries on control and Fbl knockdown ESCs rRNA. (E) Boxplot of 2΄-O-methylation abundance measured by 2OMe Ratio in control and Fbl knockdown ESCs. Significance is given by Wilcoxon Rank Sum test statistics.
Moreover, comparison of the relative methylation level of orthologous rRNA 2΄-OMe sites in HeLa and mouse ESCs measured by 2OMe ratio, revealed an extraordinarily high correlation between human and mouse (R2 = 0.94, Figure 3B and Supplementary Data), suggesting that both the presence and relative abundance of rRNA 2΄-OMe sites are under strong purifying selection.
2OMe-seq can capture methylation level variations
To assess whether 2OMe-seq was able to capture variations in the relative 2΄-O-methylation levels, we performed knockdown of the Fibrillarin (Fbl) methyltransferase in ESCs with two independent shRNAs (Figure 3C, and Supplementary Data), and performed 2OMe-seq compared to control ESCs. Notably, Fbl knockdown resulted in a strong reduction of the RT pausing 1 nucleotide downstream of annotated rRNA 2΄-OMe sites (Figure 3D), but not on the synthetic spike-in 2΄-OMe sites (Supplementary Data). The 2OMe Ratio also showed a significant reduction of the relative methylation levels in the presence of either shRNA (P-value: 5.7e-09 and 1.3e-10 respectively for Fbl sh#1 and sh#2, Figure 3E). As an independent evaluation of this result, we treated denatured total RNA from control and knockdown ESCs with the NAI reagent (31). NAI specifically forms adducts with free ribose 2΄-OH groups on flexible residues of the RNA chain, thus we expected 2΄-OMe residues to be protected. Since the adduct formed by NAI induces the RT-stop 1 nucleotide downstream of the modified site, the specific loss of 2΄-O-methylation can be monitored by the increased reactivity toward NAI. In agreement with this idea, the analysis of 2΄-OMe sites reactivity with NAI markedly increased in Fbl knockdown ESCs compared to control cells (Supplementary Data), while almost no difference was observed on 2΄-OH sites. Moreover, the observed reactivity increase was proportional to the difference captured by 2OMe-seq. This observation confirmed the reliability of the 2OMe Ratio metric in capturing methylation level variations.
2OMe-seq can identify novel bona fide 2΄-O-Methylated residues
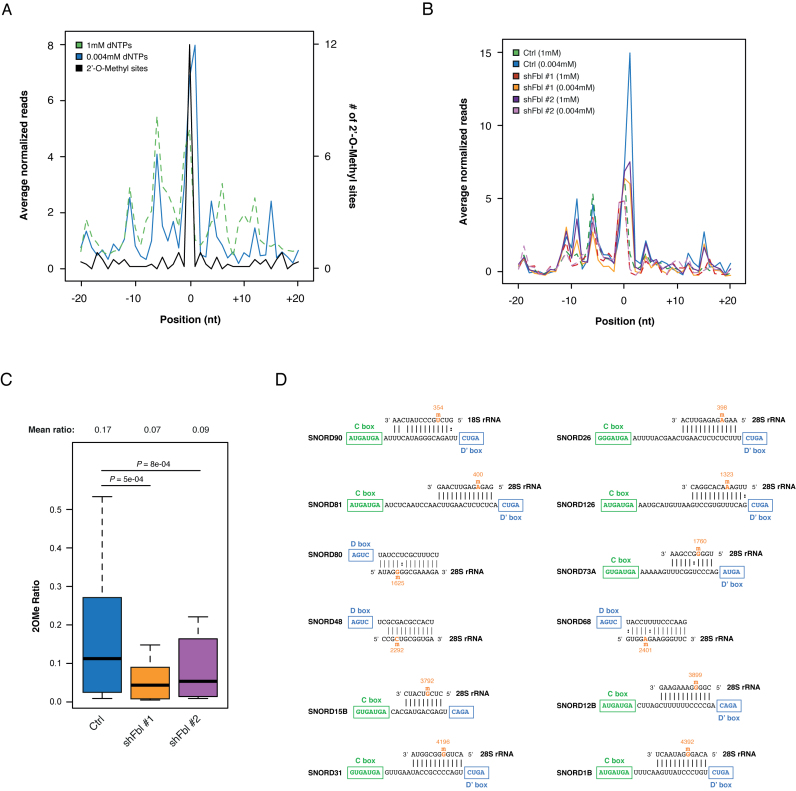
Deeper inspection of our 2OMe-seq data on HeLa cells revealed the presence of at least 12 novel putative 2΄-OMe sites across 18S and 28S rRNA. In agreement with the 2΄-OMe chemistry, these sites not only induced specific RT-stop under limiting dNTP concentrations (Figure 4A), but also showed increased resistance toward both alkaline hydrolysis and RNase H treatments (Supplementary Data). Furthermore, 11 out of 12 sites were also found in ESCs (exception made for 28S G4196), and Fbl knockdown led to an extreme reduction of the RT pausing on these sites (Figure 4B). Concordantly, both shRNAs showed significant reduction of the relative methylation levels on these sites as measured by 2OMe Ratio (P-value: 5e-04 and 8e-4 respectively for Fbl sh#1 and sh#2, Figure 4C).
Figure 4.
2OMe-seq identifies novel bona fide 2΄-OMe sites. (A) Meta-2OMe plot of the average normalized number of reads (left axis) for the 1mM dNTPs (dashed green) and 0.004mM dNTPs (solid blue) libraries on 12 novel HeLa sites. The number of 2΄-OMe residues at each position in the window is indicated (black, right axis). (B) Meta-2OMe plot of the average normalized number of reads (left axis) for the 1mM dNTPs (dashed lines) and 0.004mM dNTPs (solid lines) libraries for control and Fbl knockdown ESCs on 12 novel 2΄-OMe sites. (C) Boxplot of 2΄-O-Methylation abundance on the novel sites measured by 2OMe ratio in control and Fbl knockdown ESCs. Significance is given by Wilcoxon Rank Sum test statistics. (D) SnoRNAs predicted to guide methylation on the discovered sites.
Lack of 28S G4196 2΄-O-Methylation conservation in mouse is easily explained by analyzing SNORD31 sequence conservation, that is strong in primates, but almost absent in rodents (Supplementary Data).
All together these data demonstrate that 2OMe-seq can be used to discover novel bona fide 2΄-OMe sites. In addition, for all sites, we were able to assign the putative snoRNA guide (Figure 4D), further confirming the genuineness of these sites. It is worth to note that among these sites, 9 had been previously bioinformatically predicted (13), but not detected by conventional gel-based approaches (32), thus revealing the extreme sensitivity of our method.
DISCUSSION
In this work, we provided a new method, named 2OMe-seq, for the identification at single-base resolution and relative quantitation of 2΄-O-methylated residues. The main advantages of this method are a higher straightforwardness and sensitivity with respect to gel-based analyses, and a stronger specificity compared to alternative approaches such as RiboMeth-seq (23) due to the increased resistance of other modified residues toward partial alkaline hydrolysis treatment, such as pseudouridines. Since recent works have shown pervasive presence of pseudouridinilated residues across mammalian transcriptomes (10,11), a strong specificity constitutes a key requirement for an unbiased analysis of 2΄-OMe residues.
Using validated HeLa rRNA sites as a benchmark, 2OMe-seq was evaluated on both whole and fragmented RNA, yielding comparable results, thus suggesting the suitability of our approach also for the analysis of partially fragmented or degraded samples, such as FFPE samples. Although 2΄-OMe sites banding is not uniform across different sites, in agreement with previous gel-based analysis (21), RT-stop under limiting dNTP concentrations appears to be highly specific and sensitive. Moreover, closely spaced sites, that are quite abundant in rRNA, are well resolved too, probably thanks to the random-priming approach used in our method.
The major limitation of 2OMe-seq is represented by the impossibility to obtain an absolute quantitation RNA methylation levels. Nonetheless, analysis of synthetic RNA spike-ins bearing individual 2΄-OMe sites at well-defined stoichiometry, showed the ability of 2OMe-seq to capture the relative modification abundance. Moreover, application of 2OMe-seq to Fbl knockdown cells demonstrated its suitability for differential methylation studies.
By performing comparative analysis of 2΄-OMe levels at rRNA sites in HeLa and mouse ESCs we revealed that not only the positioning, but also the relative abundance of individual 2΄-OMe sites is strongly conserved between human and mouse. This suggests that the positioning of these residues might have a key role in regulating the structure and function of the ribosome.
Furthermore, a deeper analysis of HeLa 2OMe-seq data revealed the presence of 12 novel putative 2΄-OMe sites across 18S and 28S rRNAs. 11 out of 12 sites are strongly conserved in both human and mouse, similarly to previously annotated sites. The only exception is represented by 28S G4196, that is missing in mouse cells, and can be explained by the lack of SNORD31 conservation in rodents. As an independent validation, analysis of alkaline hydrolysis and RNase H digestion data showed an increased resistance of these sites to both treatments, in agreement with known 2΄-OMe features. Moreover, Fbl knockdown led to a significant reduction of the methylation levels for these sites in ESCs, and computational analysis allowed us to assign the putative snoRNA guides for all these sites. Furthermore, 9 of these can be experimentally validated by cross-checking with the residual 11–13 mol of 2΄-OMe that could not be unambiguously mapped by Maden and colleagues (33). Taken together these data confirms that these sites represent novel bona fide 2΄-OMe residues, and demonstrates that 2OMe-seq is suitable for the high-throughput discovery of 2΄-OMe sites.
In summary, we describe here a robust and accurate method for high-throughput mapping at single-base resolution of 2΄-O-methylated residues within RNA, and we use it to identify 12 novel conserved bona fide 2΄-OMe sites. Our approach can be easily applied to map and quantify 2΄-OMe residues in a wide range of organisms, and will greatly facilitate the study of this abundant modification on a transcriptome-wide scale.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Douglas H. Turner (University of Rochester) for helpful discussion and critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC) [IG 2014 Id.15217]; AIRC TRansforming IDEas in Oncological research (TRIDEO) [17182]. Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. . Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M., Project Consortium ENCODE . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Behm-Ansmant I., Helm M., Motorin Y.. Use of specific chemical reagents for detection of modified nucleotides in RNA. J. Nucleic Acids. 2011; 2011:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Motorin Y., Helm M.. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011; 2:611–631. [DOI] [PubMed] [Google Scholar]
- 5. Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T.. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012; 40:5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edelheit S., Schwartz S., Mumbach M.R., Wurtzel O., Sorek R.. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013; 9:e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. . Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 8. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3΄ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K. et al. . m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V.. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014; 515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. et al. . Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014; 159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall R.H. Method for isolation of 2΄-O-methylribonucleosides and N1-methyladenosine from ribonucleic acid. Biochim. Biophys. Acta (BBA). 1963; 68:278–283. [Google Scholar]
- 13. Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 14. Lowe T.M., Eddy S.R.. A computational screen for methylation guide snoRNAs in yeast. Science. 1999; 283:1168–1171. [DOI] [PubMed] [Google Scholar]
- 15. Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 16. Hengesbach M., Schwalbe H.. Structural basis for regulation of ribosomal RNA 2΄-o-methylation. Angew. Chem. Int. Ed. Engl. 2014; 53:1742–1744. [DOI] [PubMed] [Google Scholar]
- 17. Baldridge K.C., Contreras L.M.. Functional implications of ribosomal RNA methylation in response to environmental stress. Crit. Rev. Biochem. Mol. Biol. 2014; 49:69–89. [DOI] [PubMed] [Google Scholar]
- 18. Noon K.R., Bruenger E., McCloskey J.A.. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 1998; 180:2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S.. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2΄-hydroxyl group. Biochemistry. 1992; 31:1040–1046. [DOI] [PubMed] [Google Scholar]
- 20. Blanchard S.C., Puglisi J.D.. Solution structure of the A loop of 23S ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maden B.E. Mapping 2΄-O-methyl groups in ribosomal RNA. Methods. 2001; 25:374–382. [DOI] [PubMed] [Google Scholar]
- 22. Yu Y.T., Shu M.D., Steitz J.A.. A new method for detecting sites of 2΄-O-methylation in RNA molecules. RNA. 1997; 3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 23. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2015; 54:451–455. [DOI] [PubMed] [Google Scholar]
- 24. Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H.G., Oliviero S.. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013; 155:121–134. [DOI] [PubMed] [Google Scholar]
- 25. Incarnato D., Neri F., Anselmi F., Oliviero S.. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014; 15:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Incarnato D., Neri F., Anselmi F., Oliviero S.. RNA structure framework: automated transcriptome-wide reconstruction of RNA secondary structures from high-throughput structure probing data. Bioinformatics. 2016; 32:459–461. [DOI] [PubMed] [Google Scholar]
- 27. Maden B.E., Corbett M.E., Heeney P.A., Pugh K., Ajuh P.M.. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995; 77:22–29. [DOI] [PubMed] [Google Scholar]
- 28. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoeppner M.P., Poole A.M.. Comparative genomics of eukaryotic small nucleolar RNAs reveals deep evolutionary ancestry amidst ongoing intragenomic mobility. BMC Evol. Biol. 2012; 12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kehr S., Bartschat S., Tafer H., Stadler P.F., Hertel J.. Matching of Soulmates: coevolution of snoRNAs and their targets. Mol. Biol. Evol. 2014; 31:455–467. [DOI] [PubMed] [Google Scholar]
- 31. Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y.. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013; 9:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maden B.E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 1990; 39:241–303. [DOI] [PubMed] [Google Scholar]
- 33. Maden B.E. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 1988; 201:289–314. [DOI] [PubMed] [Google Scholar]
- 34. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Müller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.