Abstract
Fluorogenic RNA aptamers provide a powerful tool for study of RNA analogous to green fluorescent protein for the study of proteins. Spinach and Broccoli are RNAs selected in vitro or in vivo respectively to bind to an exogenous chromophore. They can be genetically inserted into an RNA of interest for live-cell imaging. Spinach aptamer has been altered to increase thermal stability and stabilize the desired folding. How well these fluorogenic RNA aptamers behave when inserted into structured cellular RNAs and how aptamer properties might be affected remains poorly characterized. Here, we report a study of the performance of distinct RNA Spinach and Broccoli aptamer sequences in isolation or inserted into the small subunit of the bacterial ribosome. We found that the ribosomal context helped maintaining the yield of the folded Baby Spinach aptamer; other versions of Spinach did not perform well in the context of ribosomes. In vivo, two aptamers clearly stood out. Baby Spinach and Broccoli aptamers yielded fluorescence levels markedly superior to all previous Spinach sequences including the super-folder tRNA scaffolded tSpinach2. Overall, the results suggest the use of Broccoli and Baby Spinach aptamers for live cell imaging of structured RNAs.
INTRODUCTION
Live-cell imaging of RNA can be performed using aptamer–fluorogen complexes that elicit fluorescence at different wavelengths (1–8). Spinach, a 98-nt RNA aptamer that binds 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), was developed as a small-molecule mimic of the GFP fluorophore (1). The structure of the original RNA aptamer sequence suffered from thermal instability and a propensity for misfolding that resulted in reduced brightness (9). To address these shortcomings a super-folding Spinach2 sequence was engineered. Spinach2 has fluorescence that is less context-dependent and is brighter in living cells than the original Spinach (9). iSpinach was developed for in vitro studies (10).
Crystal structures of Spinach RNA in complex with DFHBI (11,12) were used to guide miniaturization that led to the generation of ‘Baby Spinach’ (11). Baby Spinach is 51-nucleotide long, and fluorescence levels are comparable to that of the parental Spinach. Since illumination of the Spinach-DFHBI complex induces photoconversion with subsequent dissociation of the fluorogen leading to fast fluorescence decay, a new illumination scheme was developed to improve Spinach-DFHBI RNA imaging in live cells (13). A recent study demonstrated that tandem arrays of Spinach amplify the fluorescence signal and thereby facilitate imaging of low-abundance cellular mRNAs without affecting transcription, translation or degradation (14). A comparative study of the performance of Spinach aptamers in vivo has not been performed, and global strategies for tagging cellular RNAs for imaging have yet to be described.
Here, we investigated the performance of nine distinct Spinach sequences and the analogous Broccoli aptamer both as transcripts and as insertions in 16S ribosomal RNA (rRNA). Broccoli and Spinach likely have very similar overall structures (15,16). It was previously reported that Baby Spinach has fluorescence levels comparable to Spinach2 (11). As a transcript, despite use of the recommended folding protocol (11), Baby Spinach behaved poorly in our hands. In the context of the ribosomal RNA, however, it performed better than the other Spinach constructs and was markedly superior to Spinach and Spinach2 in vitro and in vivo. The short Broccoli aptamer yielded good levels of fluorescence in vitro; however, the sequence was not well tolerated inside the ribosome and drastically decreased cell growth. Unexpectedly, it provided remarkably high levels of fluorescence in vivo as Baby aptamer. Fractionation of total RNA from cells expressing Spinach-tagged 16S rRNA showed that Baby Spinach was more resistant to degradation than other Spinach sequences, which could account for its improved performance in living cells.
MATERIALS AND METHODS
Reagents and equipment
DFHBI and DFHBI-1T were purchased from Lucerna Technologies. Bacterial strains were grown in liquid Luria-Bertani (LB) broth at 37°C with shaking at 200 rpm or on LB agar plates at 37°C. In some cases, media contained 100 μg/ml ampicillin or 50 μg/ml kanamycin. Cell imaging experiments were performed using an inverted microscope Zeiss (Axio-observer Z1). Fluorescence excitation, emission spectra and fluorescence intensity measurements were performed using a Tecan Infinite 200 PRO plate reader.
Plasmid construction
Spinach sequences used in this study are summarized in Supplementary Table S1. Spinach2-mini is a truncated version of Spinach2 designed for this study that has structural characteristics similar to those of Spinach-mini. The ApaI/XbaI region of rrnB operon was subcloned between HindIII and EcoRI sites of pUC18 in order to facilitate the generation of Spinach ribosome constructs (pUC18-AX). A PstI site was created in the helix 33a region by mutating T1030G and G1032A to facilitate insertion of Spinach sequences (pUC18-AXP). The different Spinach sequences were introduced at the PstI site. Each Spinach sequence was amplified by PCR and cloned into pUC18-AXP using either an In-Fusion HD Cloning Kit (Clontech) or a Quick Ligation Kit (NEB). The ApaI/XbaI fragments harbouring Spinach sequences were then placed into pKK3535, which carries the entire rrnB rRNA operon under the control of the native, constitutively active promoter P1P2 (17). These constructs were named pKK3535-h33a-Spinach or pKK3535-h33a-Broccoli vectors. All oligonucleotides used are listed in Supplementary Table S2.
Preparation of Escherichia coli TA531 expressing Spinach-modified ribosomes
Escherichia coli strain TA531 was obtained from CGSC at Yale University. In this strain, the seven copies of the chromosomal rRNA operon have been deleted and plasmid pHC-rrnC carrying a single copy of rrnC operon serves as the sole source of rRNA genes; an encoded kanamycin resistance gene enables selection (18). Replacement of pHC-rrnC in TA531 by pKK3535-h33a-Spinach/Broccoli plasmid was achieved by transformation of the E. coli followed by growth in liquid LB medium containing ampicillin for at least two days after reaching the stationary phase. The saturated culture was diluted and inoculated onto LB plates containing ampicillin, and single colonies were isolated. Plasmid substitution was tested by evaluating resistance of colonies to ampicillin and sensitivity to kanamycin. Colonies that survived only on ampicillin LB plates were selected.
Growth of E. coli expressing Spinach/Broccoli ribosomes
E. coli TA531 harbouring pKK3535-h33a-Spinach/Broccoli plasmids were inoculated into liquid LB in Erlenmeyer flasks from glycerol stocks. The overnight cultures were diluted with 50 ml of fresh LB medium to an OD of 0.05–0.07. ODs at 600 nm were measured every 30 min until cell growth reached early stationary phase.
To confirm that DFHBI was not toxic to E. coli cells that express the Spinach-modified rRNA, cell growth was measured in presence of DFHBI with an Infinite 200 PRO microplate reader. Wells of a 96-well microtiter plate containing 190 μl of LB were inoculated with 5 μl of an overnight culture (OD600 = 2) of E. coli strain TA531. DFHBI dissolved in LB was added to the culture to result in a final concentration of 200 μM. The growth was followed by measuring the absorbance at 600 nm every 15 min at 37°C with constant shaking at 218 rpm except during measurement.
Expression and purification of Spinach/Broccoli ribosomes
Tightly coupled 70S ribosomes containing the Spinach or Broccoli aptamers were prepared from E. coli (19) by inoculation of 1 L LB with 10 ml of a saturated overnight culture of E. coli TA531 harbouring pKK3535-h33a-Spinach/Broccoli plasmid. Cells were grown to an OD600 of 0.4 and harvested by centrifugation (8000 g, 4°C for 10 min). The pellet was washed with buffer A [20 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM 2-mercaptoethanol]. Cells were frozen in liquid nitrogen and stored at −80°C until use. All subsequent steps were performed at 4°C or on ice. Cells were resuspended in buffer A and lysed using a bench-top press (Carver). Lysates were diluted with buffer A and centrifuged (15 000 g). Crude ribosomes were then prepared by sedimentation (131 000 g, 16 h) through sucrose cushions. The resulting ribosome pellet was washed twice with 1 ml of buffer A and resuspended in 250 μl of buffer A. Crude ribosomes were frozen in liquid nitrogen and stored at −80°C.
Further purification of ribosomes was performed using a cysteine-charged sulfolink resin (20). A 100-μl aliquot of crude ribosome solution was diluted with the same volume of buffer B [20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.5 mM EDTA, 2-mercaptoethanol] to adjust the concentration of NH4Cl to be 50 mM and was then loaded onto 2 ml of charged resin. The column was washed with 10 ml of buffer C [20 mM Tris-HCl (pH 7.5), 10 mM MgCl2] containing 60 mM NH4Cl and ribosomes were eluted with buffer C containing 500 mM NH4Cl. Ribosomes in the eluted fractions were collected by ultracentrifugation (213 000 g for 16 h). The ribosome pellet was resuspended in 50 μl of ribosome buffer [20 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2]. The ribosome solutions were aliquoted, quick-frozen and stored at −80°C.
In vitro transcription of Spinach aptamers
RNA fragments containing various Spinach sequences were prepared by in vitro transcription using T7 RNA polymerase (21). The template DNAs for transcription were prepared by PCR by amplifying pKK3535-h33a-Spinach/Broccoli plasmids using T7-16SrRNA-h33-F and 16SrRNA-h33-R primers (Supplementary Table S2). Transcripts were purified using Nucleospin RNA spin columns (Macherey–Nagel) with a protocol adapted for short RNAs (Macherey–Nagel) or by denaturing PAGE followed by electroelution, ethanol precipitation and dialysis against milliQ water (22).
In vitro fluorescence measurement of Spinach-DFHBI complexes
RNA transcripts were dissolved in water. Snap-cool refolding was done by heating at 90°C for 2 min and then placing on ice for 5 min after which buffer and DFHBI were added. For refolding using slow cooling, samples were heated at 90°C for 2 min and placed on ice for 2 min. Buffer and DFHBI solutions were added, and samples were kept at 65°C for 5 min. The RNA was then refolded by a slow decrease of temperature over a period of 20 min (40 cycles with a temperature decrease at a rate of 0.3°C/s for 3.3 s followed by a step of 30 s of equilibration) down to 25°C. Fluorescence measurements were performed in 384-well black microtiter plates at 25°C with the excitation at 455 nm and emission monitored at 506 nm. The background signal from DFHBI in culture medium or buffer was subtracted for each measurement.
Fluorescence measurements were performed in Tris-HCl buffer [40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 125 mM KCl] (11), ribosome-KCl buffer [20 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM 2-mercaptoethanol and 125 mM KCl] or in HEPES/KCl buffer [20 mM HEPES (pH 7.4), 100 mM KCl and 1 mM MgCl2] (9). Fluorescence of each Spinach/DFHBI complex was measured after incubating Spinach-RNA or ribosome with DFHBI for 15 min at 25°C. A 10-μl aliquot of the complex solution was introduced into a well of a 384-well microtiter plate, and fluorescence intensity was measured.
The folding efficiency of Spinach aptamers was measured based on the procedure described by Strack et al., 2013 (9) by calculating the ratio of fluorescence for 4.5 μM DFHBI with 0.09 μM RNA (or ribosomes) to 4.5 μM RNA (or ribosomes) with 0.09 μM DFHBI.
Whole-cell fluorescence measurements and live-cell imaging of E. coli strain TA531 harbouring Spinach ribosomes
To measure fluorescence of E. coli cells that express Spinach rRNA in liquid culture, overnight cultures of E. coli strain TA531 harbouring pKK3535-h33a-Spinach/Broccoli variants were inoculated into 10 ml of fresh LB containing ampicillin and grown to exponential phase. Cells were collected in 1.5-ml aliquots and washed once with the 1 ml of M9 medium (47.75 mM Na2HPO4, 22.04 mM KH2PO4, 8.56 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2). Washed cells were resuspended in M9 medium to an OD of 2.5 and incubated with 20 μM DFHBI-1T for 45 min at 37°C. Aliquots of 10 μl were transferred to a 384-well microtiter plate, and fluorescence intensity at 25°C was measured (excitation wavelength, 470 nm; emission wavelength, 525 nm).
For live-cell imaging, the cultured cells were washed once with M9 medium containing 0.4% glucose and incubated with 200 μM DFHBI-1T for 90 min at 37°C. The cells were washed with M9 medium containing 0.4% glucose and put on agarose pads. Microscope images of the E. coli cells were taken at room temperature using an Axio Observer Zeiss microscope (objective 63X, N.A. 1.46, excitation 470 ± 20 nm, emission 525 ± 25 nm) and analysed using ImageJ software.
Analysis of RNA degradation
Degradation of Spinach-tagged rRNA was investigated by fractionation on urea-PAGE. E. coli TA strains harbouring Spinach 16S rRNA plasmids were grown to an OD600 of 0.4 in LB. Total RNA was purified from a 1-ml aliquot of the culture using NucleoSpinRNA columns (Machery–Nagel). Samples of 500 ng of total RNA were loaded on 6% polyacrylamide-7 M urea gels. Gels were washed with water for 5 min three times and stained using DFHBI-1T or SYBR Gold as described (16).
Fluorescent primer extension was carried out as described previously (23). Cy5-labelled DNA primer was purchased from MWG Biotech. rRNAs (250 ng) were mixed with the Cy5-labelled primer (10 pmol) in hybridization buffer (25 mM HEPES, pH 7.0, 50 mM KCl), heated to 90°C (1–2 min), and slowly cooled to 45°C. Reverse transcription reactions were performed using SuperScript II (Invitrogen) in the supplied reaction buffer at 45°C for 40 min. The reactions were stopped by ethanol precipitation. cDNA samples were dissolved in 10 μl 7 M urea, and 1.5 to 3 μl of each sample was loaded on an 8% polyacrylamide-7 M urea gel. The gel was imaged using Typhoon Trio scanner (GE Healthcare).
RESULTS AND DISCUSSION
Design of Spinach- and Broccoli-tagged ribosomes
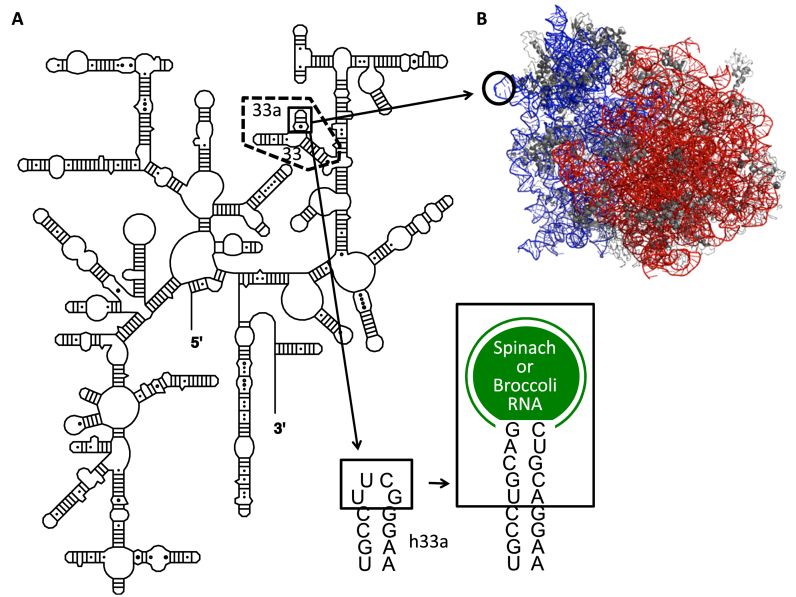
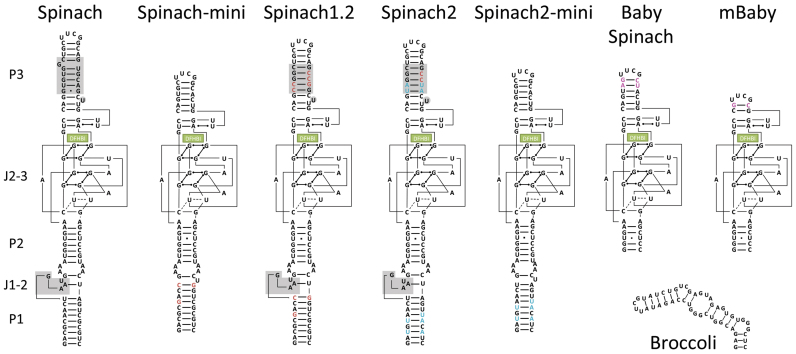
Spinach and Broccoli RNA sequences were introduced into helix 33a of 16S rRNA (Figure 1A); the UUCG tetraloop was replaced with Spinach or Broccoli RNA sequence. In crystal structures of the ribosome (24), helix 33a is not involved in any tertiary interaction and protrudes outside the ribosome structure (Figure 1B). This region of 16S rRNA is not phylogenetically conserved (25), and extension of the helix does not perturb E. coli ribosomal function (26). Spinach sequences were inserted at a PstI site, which was introduced in the helix 33a region of 16S rDNA by site-directed mutagenesis (Supplementary Figure S1A and S1B). Except for Baby Spinach, mBaby Spinach and Broccoli, Spinach sequences were inserted by conventional ligation resulting in the extension of helix 33a by five base pairs (Supplementary Figure S1A and S1B). For Baby and mBaby Spinach and Broccoli constructs, we performed cloning by recombination. We took advantage of the natural scaffold provided by helix 33a to shorten helix P2 of the Baby aptamer by two base pairs (Supplementary Figure S1C). In addition to the original Spinach, we tested nine other constructs (Figure 2): minimized Spinach (Spinach-mini) (1), a thermostable variant of Spinach (Spinach1.2), the super-folder Spinach (Spinach2) (9), the versions of Spinach1.2 and Spinach2 with tRNA scaffolds (tSpinach1.2 and tSpinach2) (1,27), the miniaturized Baby Spinach (11) and Broccoli (16). In addition to these published Spinach sequences we tested Spinach2-mini (an aptamer we designed by analogy to Spinach-mini), and a deletion mutant of Baby Spinach with the P3 stem shortened by four base pairs (mBaby Spinach).
Figure 1.
Site of insertion of Spinach/Broccoli sequences into 16S rRNA. (A) Secondary structure of 16S rRNA. The site of insertion in helix 33a (h33a) is boxed. The in vitro transcribed domain containing helices 33a and 33 is indicated by dashed lines. (B) Side view of 70S Escherichia coli ribosome (PDB code 2AW4 (50S) 2AVY (30S) (24)) with the tip of helix 33a highlighted. 16S and 23S rRNA molecules are shown in blue and red, respectively.
Figure 2.
Secondary structures of Spinach and Broccoli aptamers used in this study. DFHBI is shown as a green box. Grey boxes are the bases deleted in the ‘mini’ versions of Spinach and Spinach2. Red bases are the sites of mutations that transform Spinach into Spinach1.2 and 2. Blue and pink base pairs are those specific for Spinach2 and Baby Spinach, respectively.
In vitro fluorescence assay of the Spinach/Broccoli constructs
Prior to examining the performance of Spinach and Broccoli sequences in the context of the ribosome, we performed a comparative study of the levels of fluorescence of each aptamer construct in previously used buffers (9,11) and in the ribosome buffer (20 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2) supplemented with 125 mM KCl. The tested transcripts included helices 33 and 33a of 16S rRNA (Figure 1) and thereby closely resembled the ribosomal context. The shorter constructs rather than entire 16S rRNA transcripts were used to avoid potential complications of naked rRNA misfolding. Transcripts were generated by in vitro T7 transcription and purified over Nucleospin columns or by denaturing PAGE. The RNAs were folded by heating at 90°C for 1 min and then placing on ice. The crystal structure of Spinach revealed that K+ and Mg2+ are part of the structure of folded DFHBI-Spinach complex (11,12). For this reason, buffers used for Spinach fluorescence assays contained these cations.
Level of fluorescence in ribosome–KCl buffer in the presence of a large excess of DFHBI were almost identical to that measured in the HEPES/KCl buffer, indicating that the ribosome-KCl buffer is suitable for performing fluorescence assays using Spinach (Supplementary Figure S2). When using an equimolar amount of DFHBI to RNA, the fluorescence was higher in ribosome buffer supplemented with KCl than in HEPES/KCl buffer, suggesting that Spinach folded better in the ribosome-KCl buffer. The concentration of magnesium might contribute to this effect since an increase of fluorescence signal was observed relative to the Tris-HCl buffer (5 mM MgCl2) when compared to ribosome-KCl buffer (10 mM MgCl2; Figure 3A, inset). Our observation is also supported by a recent comparison of fluorescence levels measured in Tris-HCl/HEPES buffers at identical concentrations of MgCl2 for a Spinach aptamer; this study found Tris-HCl superior to HEPES (28). It has been reported that Tris-borate and HEPES/KCl buffers act differently as a counterion for RNA, which may result in differences in RNA folding. Buchmueller and Weeks found that Tris-borate has a weak capacity to stabilize RNA folding, which may reflect relatively unfavourable interactions between the bulky Tris-borate ion and RNA or may be due to partial coordination of RNA functional groups by borate (29). Similarly, HEPES/KCl is bulkier and more rigid than Tris-HCl. We believe therefore that this underlies the stronger capacity of Tris-HCl than HEPES/KCl to stabilize RNA folding.
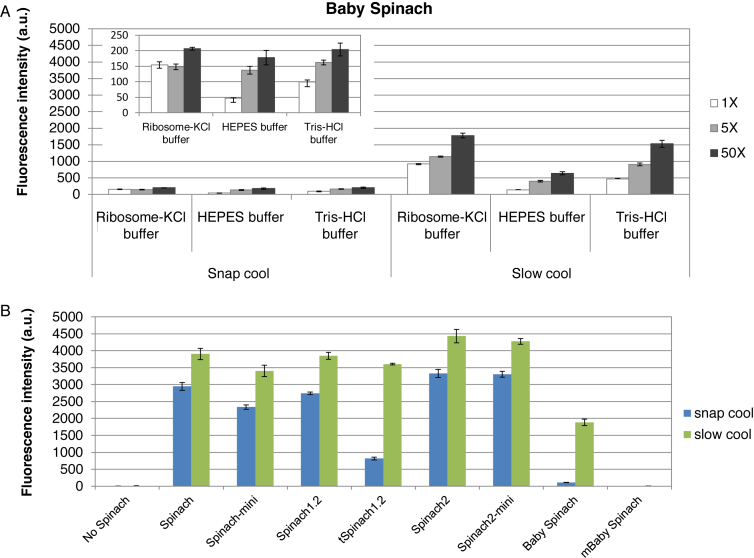
Figure 3.
Effect of folding protocols on fluorescence of Spinach transcripts. (A) Baby Spinach requires slow cooling for efficient folding. For snap cooling, the RNA transcript in water was heated to 90°C, placed on ice for 5 min and then buffer and DFHBI were added. For slow cooling, the buffer and DFHBI were added to the snap-cooled samples; samples were then incubated at 65°C for 5 min and then cooled slowly to 25°C. Fluorescence intensities were determined using 0.2 μM RNA and 0.2 μM (white bars), 1 μM (light grey bars) and 10 μM (dark grey bars) of DFHBI in indicated buffers. Tris-HCl buffer contains 40 mM Tris-HCl (pH 7.5), 125 mM KCl, 5 mM MgCl2 (11). The signals obtained with the snap cool protocol are expanded in the inset. (B) Fluorescence intensities of Spinach sequences in ribosome-KCl buffer in the presence of 50-fold molar excess of DFHBI after folding using the snap cool (blue bars) or slow cool (green bars) protocol. Error bars are s. e. m. for three independent experiments.
In the case of Baby Spinach, signals were surprisingly low using the snap cool protocol (Figure 3A, left panel). Therefore the RNA was folded using a slow cooling protocol that was reported previously to improve Baby Spinach RNA folding (11). Indeed, fluorescence levels improved markedly using this method (Figure 3A, right panel). The other constructs were subjected to the snap and the slow cooling folding protocols, and the levels of fluorescence were higher by at least 30% (4-fold higher for tSpinach1.2) using the slow cooling protocol (Figure 3B). The intensity of Baby Spinach fluorescence remained inferior to that of all other Spinach aptamers.
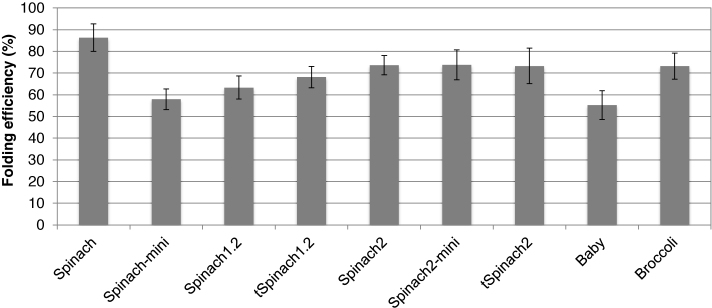
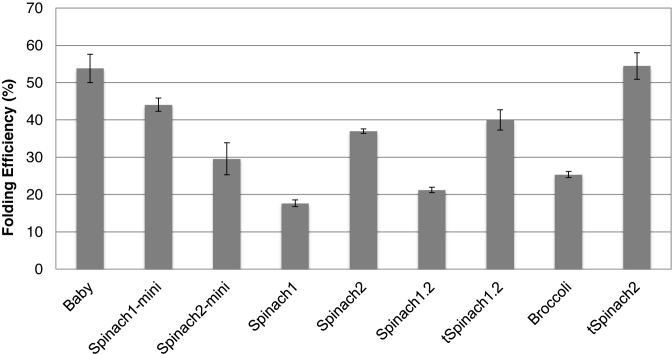
We then used the slow cooling folding protocol and estimated the folding efficiencies of the aptamers with a previously described assay (9). The estimated folding efficiencies for most of the constructs were within 60–70%, the minimum was for Baby (55 ± 7%) and the maximum was for Spinach (86 ± 6%) (Figure 4). Spinach constructs are known to suffer from misfolding during T7 transcription (9). We showed here that the in vitro refolding protocol described for Baby Spinach (11) improved folding efficiency of all tested RNA aptamers. Baby RNA remained the construct harboring the weakest folding efficiency.
Figure 4.
Folding efficiencies of the Spinach and Broccoli aptamers. Error bars are s. e. m. for three independent experiments.
Expression of 16S rRNA modified with aptamer sequences can impact bacterial cell growth
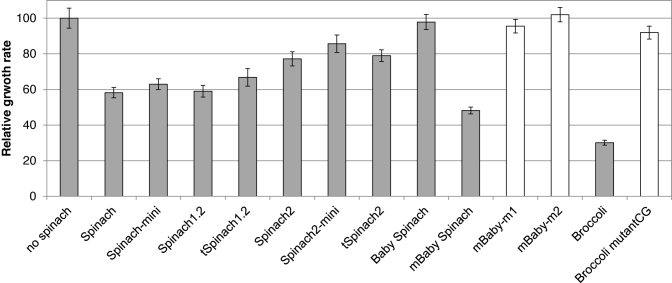
The plasmid pHC containing the rrnC operon in strain E. coli TA531, a strain from which all seven copies of the genomic rRNA operons have been deleted (18), was replaced with plasmid pKK3535 harbouring Spinach/Broccoli sequences within the 16S rDNA of rrnB operon. This strain was used because expression of a homogenous population of ribosomes facilitated evaluation of the effect of Spinach/Broccoli insertion on cell growth and allowed purification of aptamer-tagged ribosomes. As shown in Figure 5, introduction of earlier generation Spinach sequences (Spinach, Spinach 1.2) caused slight defects in E. coli growth. Expression of 16S rRNA containing Spinach1.2 with the stabilizing tRNA scaffold (tSpinach1.2) altered the physiology of E. coli TA531; aggregates of cells were observed (Supplementary Figure S3). Expression of all other constructs tested here (Spinach, Spinach-mini, Spinach2, tSpinach2, Spinach2-mini, Spinach1.2, Baby Spinach, the deletion mutant, mBaby Spinach and Broccoli) did not modify the physiology of cells grown in liquid culture (data not shown); however, insertion of most of the constructs cause a reduction in growth rate. The cells that express 16S rRNA in which super-folder Spinach2 or its variants (tSpinach2, Spinach2-mini) were inserted grew at almost the same rate as wild-type cells (Figure 5). Among the Spinach2 variants, the shortest insert, Spinach2-mini grew fastest. A wild-type level of growth was observed for cells expressing 16S rRNA with Baby Spinach, the shortest version of the constructs tested. Interestingly, insertion of Broccoli, which is also short (49 nts), drastically impaired cell growth. These results indicated that the length of the RNA inserted in helix 33a is not the only factor that influences cell growth. In agreement with this observation, insertion of mBaby Spinach, in which 8 nucleotides were removed from the P3 stem of Baby Spinach, was about as deleterious to cell growth as Broccoli.
Figure 5.
Growth of E. coli expressing Spinach-containing 16S rRNA. Rates were measured in LB liquid culture at 37°C. Values were normalized to E. coli strain TA531, which expresses wild-type 16S rRNA. Growth rates of mBaby and Broccoli mutants are indicated in white.
The very low growth rate conferred by both mBaby and Broccoli ribosomes placed a strong selection pressure on cells during the chasing step resulting in the appearance of rare fast growing bacteria. Sequencing of the plasmids from these cells identified the point mutations that restored wild-type growth (Figure 5). For mBaby Spinach, two point mutations were observed in the hairpin of the aptamer within the short sequence 5΄- UUCGCGUU -3΄ (Supplementary Figure S4A). This sequence is unique among all Spinach constructs. The fast-growing mBaby Spinach variant m1 has the sequence 5΄- U(C)CGCGUU -3΄ and the other mutant m2 has the sequence 5΄- UUCGC(A)UU -3΄ instead of the original sequence (mutations are indicated with parenthesis). We found a complementary sequence, 5΄- 968AACGCGAA975 -3΄, in helix 31 of 16S rRNA (Supplementary Figure S4B). Since a direct interaction between these two sequences in mature 30S can be excluded, we suspect that contacts between these two sequences occur during ribosomal assembly in vivo leading to altered ribosomes. For Broccoli, growth was restored in cells containing plasmids with one of two point mutations within h33 of 16S rRNA (G1018U and C1027U) or a single mutant within the Broccoli aptamer (Supplementary Figure S4C). The latter did not elicit DFHBI fluorescence (data not shown). In contrast to the sequence of mutant mBaby, no significantly homologous sequence was found in E. coli genome for Broccoli. The mutations found in the fast growing cells introduce a non-Watson–Crick base pair in stems of h33 and h33a. The reasons for the growth defects remain to be explained.
Fluorescence of the Spinach constructs in the ribosomal context
We then investigated the fluorescence intensities of the Spinach/Broccoli aptamers in the context of the ribosome. Ribosomes were purified from E. coli strain TA531 in which all ribosomes were produced from the rrnB operon carried by the plasmid pKK3535 with the Spinach or Broccoli insertion. This ensured homogeneity of aptamer-tagged ribosomes. DFHBI did not affect the growth rates of cells that expressed any of the constructs (Supplementary Figure S5), indicating that the fluorophore is non-toxic and that it does not affect ribosomal activity. Fluorescence spectra of the ribosomes were the same for all constructs (Supplementary Figure S6), and intensities were strongly stimulated by K+ (Supplementary Figure S7) as previously observed (11,12). Fluorescence intensities of the purified ribosomes were measured in ribosome–KCl buffer at two different ratios of RNA to DFHBI (9). Values provide information on the percentage of aptamer correctly folded in vivo. The trend in folding efficiencies, in the context of ribosome, was different from those of RNAs transcribed in vitro. Folding efficiencies of Spinach, Spinach1.2 and Broccoli were 17.6 ± 0.9%, 21.2 ± 0.8% and 25.4 ± 0.8%, respectively, in the context of the ribosome (Figure 6). With the help of the tRNA scaffold, the folding efficiency of tSpinach1.2 was significantly higher, at 40.0 ± 2.7%. Similarly, the recently developed ‘super-folder’ Spinach2 folded efficiently (54.5 ± 3.6%), as previously observed (9). Baby Spinach without the tRNA scaffold, which behaved poorly as an RNA transcript, had a folding efficiency of 53.8 ± 3.8% in vivo. This result suggests that rRNA serves as a scaffold that promotes Baby Spinach folding or prevents its degradation in vivo.
Figure 6.
In vivo folding efficiencies of ribosomes that contain Spinach variant 16S rRNAs. Error bars are s. e. m. for three independent experiments.
Live-cell imaging of Spinach-tagged ribosomes
Spinach-tagged ribosomes were then expressed in strain TA531, and levels of fluorescence were measured in live cells. In the course of this study, new sets of DFHBI derivatives have been reported, one of which, DFHBI-1T is more compatible with widely used microscopy filter sets for GFP than DFHBI (4), has improved brightness and is commercially available. Thus, DFHBI-1T was used for the live-cell imaging studies. In the presence of DFHBI-1T, folding efficiencies of Baby Spinach and Broccoli 16S rRNA transcripts, refolded in vitro, reached almost 100% (Supplementary Figure S8A). This increase in the folding efficiencies was also observed for the purified Spinach/Broccoli ribosomes (Supplementary Figure S8B). This suggests that complex formation in vitro is more productive with DFHBI-1T than with DFHBI.
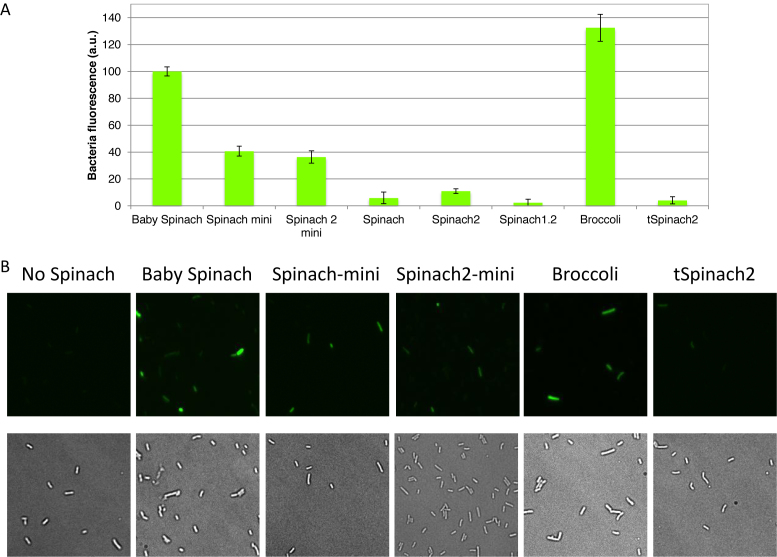
Cells were grown in LB to the exponential phase, and the culture media was exchanged with M9 media before addition of 20 μM DFHBI-1T and incubation at 37°C. M9 media contains Mg2+ and K+ cations ensuring high levels of fluorescence of Spinach derivatives during measurements. Previously, the signal from Spinach2 was found to be 3-fold higher than Spinach in eukaryotic cells and 2.1-fold higher in E. coli (9). In our experimental conditions in E. coli, Spinach2 was 1.8-fold brighter than Spinach (Figure 7A). As observed in vitro, short Spinach constructs inserted in helix 33a of 16S rRNA clearly showed higher fluorescence signals than did the longer aptamers. For example, cells expressing Spinach-mini and Spinach2-mini generated signals stronger than those expressing Spinach or Spinach2 by 6.9- and 3.3-fold, respectively. More importantly, cells that expressed Baby Spinach or Broccoli ribosomes were 9.2-fold and 12.1-fold brighter, respectively, than the super-folder Spinach2. In the case of cells expressing Spinach2 with tRNA scaffold, the intensity of cellular fluorescence was extremely low, in contrast to in vitro experiments. The RT-qPCR of purified total RNA showed that 16S rRNA expression levels containing all variants were quite homogeneous.
Figure 7.
In vivo fluorescence of cells expressing Spinach variant 16S rRNAs. (A) Fluorescence signal from E. coli TA531 cells incubated with 20 μM DFHBI-1T in M9 (2 mM Mg2+) buffer at 37°C for 45 min. Samples were excited at 470 nm in a plate reader, and fluorescence was recorded at 525 nm. Baby Spinach signal was set as 100. Error bars are s. e. m. for three independent experiments. (B) Imaging of E. coli TA531 cells expressing Spinach-variant-tagged or Broccoli-tagged ribosomes. Cells we incubated with 200 μM DFHBI-1T for 90 min at 37°C, mounted on an agar pad and imaged at room temperature under a microscope. Brightness of the images was adjusted based on Baby Spinach signal. Note heterogeneity in the levels of fluorescence at the single-cell level.
Fluorescence microscopy imaging of the E. coli cells expressing various Spinach-tagged ribosomes confirmed that those expressing Baby Spinach or Broccoli exhibited strong fluorescence (Figure 7B, Supplementary Figure S9A). For each aptamer construct, the fluorescence of cells from the same culture varied (Figure 7B, Supplementary Figure S9B). This might reflect differences, at the single-cell level, in intracellular DFHBI accumulation or differences in the quantity of intact ribosomes.
RNA degradation in vivo
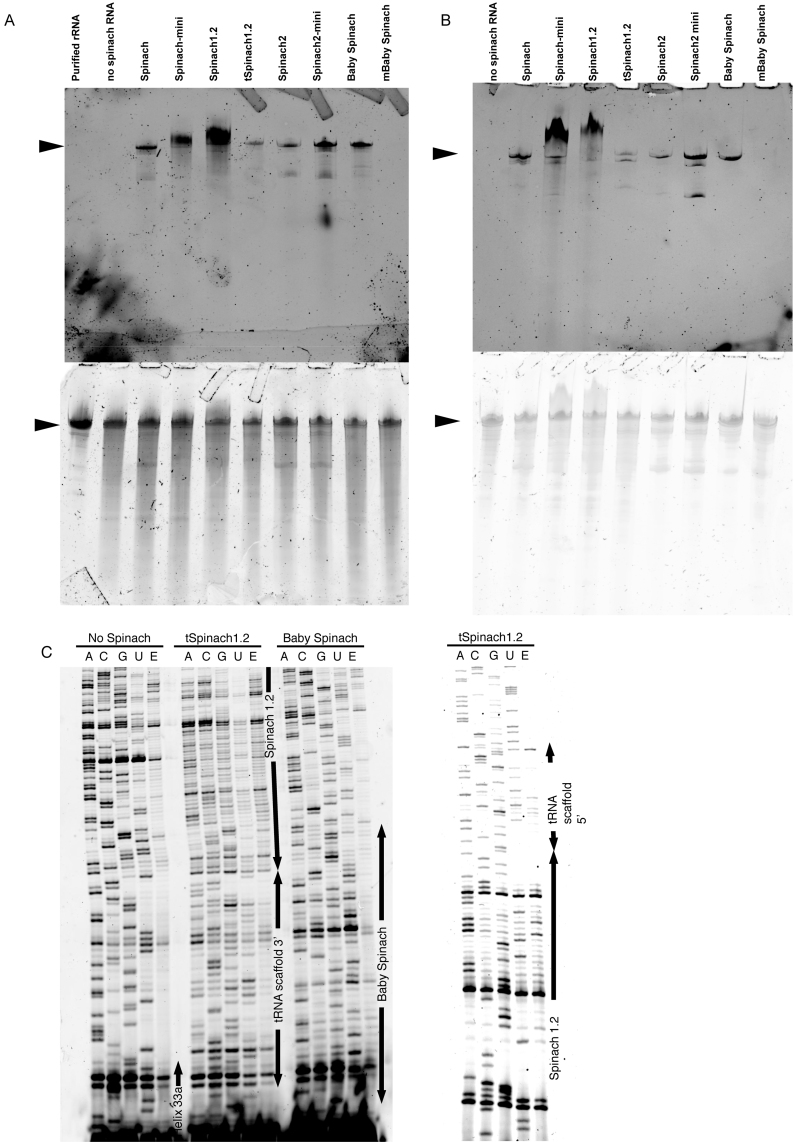
Degradation of rRNA containing the Spinach insert could account for low fluorescence signals of some of the aptamers in cells. We therefore asked whether the RNA aptamers were subjected to degradation by endo- or exonucleases in E. coli. To visualize degradation products, total cellular RNA was extracted from cells expressing Spinach-tagged 16S rRNA. Analysis on 6% polyacrylamide denaturing gel followed by DFHBI-1T or SYBR Gold staining revealed the pattern of degradation products (Figure 8A). DFHBI-1T should stain 16S rRNA containing the dye-binding aptamer. mBaby Spinach-modified RNA was undetectable on gels stained with DFHBI-1T. Other constructs showed a major band corresponding at the length expected for 16S rRNA. Lower molecular weight bands were detected for all constructs but were less intense for the sample in which rRNA was modified with Baby Spinach (Figure 8A). As expected, a control of total cellular RNA from a strain expressing non-tagged ribosome had no RNA that generated fluorescence upon staining with DFHBI-1T. The degradation bands observed with other constructs are therefore the results of cleavage of 16S rRNA molecules. Spinach-mini and Spinach 1.2 reproducibly produced a band of higher molecular weight that remains uncharacterized. A similar experiment was performed on purified Spinach-tagged ribosomes (Figure 8B). With the exception of Baby Spinach, which resulted in a single band, all other constructs gave lower molecular weight bands that stained with DFHBI-1T indicating that cleavages within 16S rRNA occurred that generated large fragments of rRNA containing the Spinach tag.
Figure 8.
Degradation of Spinach-tagged 16S rRNA. (A) Total RNA from E. coli TA531 cells expressing Spinach-tagged ribosomes was separated on urea-PAGE and stained with DFHBI-1T (top) and SYBR Gold (bottom). 16S rRNA bands are indicated by arrows. (B) Ribosomes were purified from E. coli TA531 cells expressing Spinach-tagged 16S rRNA. RNA was separated on urea-PAGE and stained with DFHBI-1T (top) and SYBR Gold (bottom). (C) Fluorescent primer extension analysis of cleavage sites in Spinach-modified 16S rRNA. Fluorescent primer extension products generated using Cy5-labelled DNA primer were analysed on urea-PAGE. Regions corresponding to helix 33a, tRNA scaffold, Spinach and Baby Spinach are indicated by arrows.
We next identified possible cleavage sites in the vicinity or within the Spinach tags on purified rRNA for three constructs. Purified ribosomes were phenol extracted to obtain rRNA samples that were subjected to fluorescently labelled primer extension as previously described (23). Cleavage by an endonuclease results in a stop of primer extension by reverse transcriptase. In the case of tSpinach1.2, multiple bands were detected in the region of the aptamer and a few bands were detected within the tRNA scaffold (Figure 8C, Supplementary Figure S10). It was recently reported that endonucleases, which normally process tRNAs in vivo, cleave the tRNA-scaffold of tBroccoli and tSpinach sequences followed by a removal of the remaining flanking regions by trimming at the 3΄ terminus by exonucleases RNase T and RNase PH (15). In the context of the ribosome, the situation seems quite different, as we did not observe a strong primer extension stop at the 3΄ processing site of tRNA within the tSpinach1.2 construct. Instead, multiple stops were observed within the aptamer sequence suggesting that endonucleolytic cleavages occur within this region with only few additional cuts in the tRNA scaffold. The difference may be due to the context of tRNA scaffold. In our study the tRNA scaffold was inserted into helix 33a of the 16S rRNA; thus, 3΄ and 5΄ regions of the tRNA are embedded in an RNA stem. In contrast the tRNA was flanked by single-stranded regions in the construct used by Filonov et al. (15). In addition, in the context of the ribosome, steric hindrance may protect the tRNA sequence from RNases. It has been shown that the presence of DFHBI induces a conformational change within the Spinach aptamer with G-quartet stabilization (11). Thus, the cleavages that we observed within the aptamer sequence may be a consequence of accessibility and partial unfolding in the absence of DFHBI in vivo. The small size of Baby Spinach aptamer and its enhanced folding propensity likely prevents this non-specific degradation (Figure 8C).
CONCLUSION
Our comparative studies of the performance of different Spinach aptamers when inserted at the same position within a large structured cellular RNA, the 16S rRNA, revealed a strategy that can be used to identify robust aptamers for bacterial live-cell imaging. The site of insertion, helix 33a, was chosen because previous studies showed that this stem-loop protrudes outside the ribosome and is tolerant of extra RNA sequences (26). Folding properties of the different aptamers were first evaluated in vitro in the context of T7 RNA transcripts that included a full domain comprising helices 33 and 33a. Surprisingly, Baby Spinach, which requires a specific folding protocol that includes slow cooling, did not bind dye as well as the other Spinach aptamers. In contrast, when the aptamers were expressed in vivo and purified in native form as Spinach-tagged ribosomes, the results were different. Clearly, the yield of folded Baby Spinach aptamer in this context was superior to yields of the other constructs. Baby Spinach was also resistant to nuclease cleavage and degradation. These results suggest that the structured environment of 16S rRNA helix 33a provides a natural scaffold that promotes folding of and/or confers stability to the Baby Spinach aptamer. The folding efficiency of the Broccoli aptamer remained low in the ribosomal context but surprisingly generated high fluorescence levels in vivo. We did not find any correlation between folding efficiencies measured on purified ribosomes and the levels of fluorescence detected in live cell imaging suggesting that in vitro conditions do not fully recapitulate the situation experienced by aptamers in vivo. Indeed, the cytoplasm of living cells is a complex mixture of biopolymers, small inorganic and organic compounds that may affect RNA folding and stability. In addition, molecular crowding was shown to drastically increase the RNA folding rate of a GAAA tetraloop-receptor system (30).
The Baby Spinach aptamer was well tolerated by the ribosome at helix 33a; however, insertion of a shorter version of the aptamer, mBaby Spinach, into this helix of the 16S rRNA was deleterious to cell growth. The Broccoli aptamer insertion drastically reduced cell growth relative to the wild-type cells but did provide very high fluorescence levels. This demonstrates that the sequence of an exogenous RNA requires fine-tuning for proper insertion and RNA imaging. In vivo, Baby Spinach and Broccoli aptamers clearly stood out from other Spinach variants tested for high levels of fluorescence. These aptamers appear to be the variants that most effectively label large, structured cellular RNAs. Their performance might be further enhanced with the use of a recently reengineered three-way junction scaffold that resists nucleases cleavage and degradation (15).
Visualizing ribosomes by labelling ribosomal RNAs with Spinach aptamers would provide unique opportunities to study prokaryotic translation. Using ribosomes tagged with fluorogenic RNA aptamers in combination with orthogonal systems (31,32) or with tethered subunits (33), for example, would allow investigating translation of a specific mRNA in vivo. This approach would be extremely powerful especially with RNA aptamers suitable for super-resolution imaging that are currently being developed (6).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S.C. Blanchard (Weill Medical College, Cornel University) for support and S. Jaffrey (Weill Medical College, Cornel University) for providing the Spinach1 sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique [CNRS to S.Y.]; JASSO, a fellowship from French embassy in Japan ‘Bourse du gouvernement français’ and a fellowship from FRM [Fondation pour la recherché Médicale-FDT20140931068) to M.O.]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R.. Fluorescence imaging of cellular metabolites with RNA. Science. 2012; 335:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pothoulakis G., Ceroni F., Reeve B., Ellis T.. The spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synthetic Biol. 2014; 3:182–187. [DOI] [PubMed] [Google Scholar]
- 4.Song W., Strack R.L., Svensen N., Jaffrey S.R.. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 2014; 136:1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolgosheina E.V., Jeng S.C., Panchapakesan S.S., Cojocaru R., Chen P.S., Wilson P.D., Hawkins N., Wiggins P.A., Unrau P.J.. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014; 9:2412–2420. [DOI] [PubMed] [Google Scholar]
- 6.You M., Jaffrey S.R.. Structure and mechanism of RNA mimics of green fluorescent protein. Annu. Rev. Biophys. 2015; 44:187–206. [DOI] [PubMed] [Google Scholar]
- 7.Strack R.L., Jaffrey S.R.. Live-cell imaging of mammalian RNAs with Spinach2. Methods Enzymol. 2015; 550:129–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You M., Litke J.L., Jaffrey S.R.. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strack R.L., Disney M.D., Jaffrey S.R.. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013; 10:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Autour A., Westhof E., Ryckelynck M.. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016; 44:2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferre-D'Amare A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han K.Y., Leslie B.J., Fei J., Zhang J., Ha T.. Understanding the photophysics of the spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 2013; 135:19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Fei J., Leslie B.J., Han K.Y., Kuhlman T.E., Ha T.. Tandem Spinach array for mRNA imaging in living bacterial cells. Sci. Rep. 2015; 5:17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filonov G.S., Kam C.W., Song W., Jaffrey S.R.. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 2015; 22:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014; 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosius J., Ullrich A., Raker M.A., Gray A., Dull T.J., Gutell R.R., Noller H.F.. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981; 6:112–118. [DOI] [PubMed] [Google Scholar]
- 18.Asai T., Zaporojets D., Squires C., Squires C.L.. An Escherichia coli strain with all chromosomal rRNA operons inactivated: Complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:1971–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazauric M.H., Seol Y., Yoshizawa S., Visscher K., Fourmy D.. Interaction of the HIV-1 frameshift signal with the ribosome. Nucleic Acids Res. 2009; 37:7654–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leshin J.A., Rakauskaite R., Dinman J.D., Meskauskas A.. Enhanced purity, activity and structural integrity of yeast ribosomes purified using a general chromatographic method. RNA Biol. 2010; 7:354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazauric M.H., Leroy J.L., Visscher K., Yoshizawa S., Fourmy D.. Footprinting analysis of BWYV pseudoknot-ribosome complexes. RNA. 2009; 15:1775–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soler N., Fourmy D., Yoshizawa S.. Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB. J. Mol. Biol. 2007; 370:728–741. [DOI] [PubMed] [Google Scholar]
- 23.Ying B.W., Fourmy D., Yoshizawa S.. Substitution of the use of radioactivity by fluorescence for biochemical studies of RNA. RNA. 2007; 13:2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H.. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005; 310:827–834. [DOI] [PubMed] [Google Scholar]
- 25.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorywalska M., Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., Puglisi J.D.. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005; 33:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponchon L., Dardel F.. Recombinant RNA technology: the tRNA scaffold. Nat. Methods. 2007; 4:571–576. [DOI] [PubMed] [Google Scholar]
- 28.Auslander S., Fuchs D., Hurlemann S., Auslander D., Fussenegger M.. Engineering a ribozyme cleavage-induced split fluorescent aptamer complementation assay. Nucleic Acids Res. 2016; 44:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchmueller K.L., Weeks K.M.. Tris-borate is a poor counterion for RNA: a cautionary tale for RNA folding studies. Nucleic Acids Res. 2004; 32:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupuis N.F., Holmstrom E.D., Nesbitt D.J.. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui A., de Boer H.A.. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rackham O., Chin J.W.. A network of orthogonal ribosome x mRNA pairs. Nat. Chem. Biol. 2005; 1:159–166. [DOI] [PubMed] [Google Scholar]
- 33.Orelle C., Carlson E.D., Szal T., Florin T., Jewett M.C., Mankin A.S.. Protein synthesis by ribosomes with tethered subunits. Nature. 2015; 524:119–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.