Abstract
The expression of iron regulated genes in bacteria is typically controlled by the ferric uptake regulator (Fur) protein, a global transcriptional repressor that regulates functions as diverse as iron acquisition, oxidative stress, and virulence. We have identified a fur homologue in Dichelobacter nodosus, the causative agent of ovine footrot, and shown that it complements an Escherichia coli fur mutant. Homology modeling of the D. nodosus Fur protein with the recently solved crystal structure of Fur from Pseudomonas aeruginosa indicated extensive structural conservation. As Southern hybridization analysis of different clinical isolates of D. nodosus indicated that the fur gene was present in all of these strains, the fur gene was insertionally inactivated to determine its functional role. Analysis of these mutants by various techniques did not indicate any significant differences in the expression of known virulence genes or in iron-dependent growth. However, we determined several Fur regulatory targets by two-dimensional gel electrophoresis coupled with mass spectrometry. Analysis of proteins from cytoplasmic, membrane, and extracellular fractions revealed numerous differentially expressed proteins. The transcriptional basis of these differences was analyzed by using quantitative reverse transcriptase PCR. Proteins with increased expression in the fur mutant were homologues of the periplasmic iron binding protein YfeA and a cobalt chelatase, CbiK. Down-regulated proteins included a putative manganese superoxide dismutase and ornithine decarboxylase. Based on these data, it is suggested that in D. nodosus the Fur protein functions as a regulator of iron and oxidative metabolism.
Dichelobacter nodosus is a fastidious gram-negative anaerobe that is the causative agent of footrot in sheep, goats, and other ruminants. Ovine footrot is a debilitating disease that results in the hoof separating from the underlying soft tissue, leading to lameness, loss of condition, and reduced wool growth. The three recognized forms of the disease, virulent, intermediate, and benign, differ in their clinical pathologies, with less virulent isolates causing a lower percentage of severe lesions under optimal climatic conditions (54). The virulence of an individual strain depends on factors such as its type IV fimbriae, extracellular serine proteases, and potentially on two genomic regions, the virulence-related locus (vrl) and the virulence-associated protein (vap) regions, that are preferentially associated with virulence (7, 34). Until recently, the molecular analysis of D. nodosus has been hindered by a lack of genetic tools, but we have now developed methods for the genetic manipulation of D. nodosus (33, 34).
Iron is an important micronutrient and is essential for the growth of most living cells. The levels of iron available to a cell can vary greatly depending upon the immediate environment, and since excess levels of iron can be toxic due to the formation of oxygen radicals, the uptake of iron needs to be appropriately regulated (2). The expression of iron-regulated genes in bacteria is typically controlled by the ferric uptake regulator (Fur) protein. Fur is a small, 17-kDa, global transcriptional repressor that in the presence of iron regulates functions as diverse as iron acquisition, oxidative stress, and virulence (23). The coordinated expression of virulence factors by iron availability is not surprising, since the concentration of iron at most host infection sites is low, due to stringently regulated transport systems. These low iron levels signal to bacteria that they have entered the host and subsequently lead to increased expression of iron acquisition systems as well as virulence factors. Furthermore, in some bacterial species, fur mutants have shown a reduction in virulence (29, 41, 47). Fur also acts to positively regulate genes involved in the acid tolerance response in Salmonella enterica serovar Typhimurium and regulates superoxide dismutase in Escherichia coli. The regulation of superoxide dismutase has been attributed to a small RNA molecule that is negatively regulated by Fur (2).
Fur is a zinc metalloprotein, rich in histidines, that contains one zinc and one iron binding site per monomer. It contains an N-terminal DNA recognition domain and a C-terminal domain that is involved in dimerization (16). The recent elucidation of the crystal structure of the Fur protein from Pseudomonas aeruginosa (FurPa) pinpointed the location of the metal binding sites and provided an insight into the architecture of the Fur-DNA interaction (42). The N-terminal region contains a winged-helix motif that binds to regions of DNA referred to as Fur boxes that are normally located between the −10 and −35 promoter region of Fur-repressed genes (15, 42). DNA binding occurs when Fur is complexed with Fe2+, and it is thought to inhibit the binding of RNA polymerase (16). The original E. coli Fur (FurEc) box consensus consisted of a 19-bp palindromic sequence, GATAATGATAATCATTATC, which has now been refined to three hexameric repeats of 5′-NAT(A/T)AT-3′ (16).
Fur homologues have been identified in both gram-negative and gram-positive bacteria. Many of these homologues are able to complement a furEc mutant, demonstrating that the molecular mechanism underlying transcriptional regulation by Fur is highly conserved (16). However, little has been done to describe the role of Fur in an anaerobic organism. In this study we aimed to identify and determine the regulatory role of a Fur homologue from D. nodosus. We report the successful cloning and functional analysis of the D. nodosus fur gene, the construction of a D. nodosus fur mutant, and the use of proteomics to identify proteins whose expression in D. nodosus is Fur regulated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are listed in Table 1. E. coli strains were grown at 37°C on 2× YT medium (yeast extract, tryptone, and sodium chloride) (51) supplemented with 100 μg of ampicillin per ml or 150 μg of erythromycin per ml. D. nodosus strains were grown at 37°C in an anaerobic chamber (Coy Laboratory Products, Inc.) in an atmosphere of 10% (vol/vol) H2 and 10% (vol/vol) CO2 in N2 on Eugon (Difco) yeast extract (EYE) agar with 5% defibrinated horse blood (Equicell), supplemented with 1 μg of erythromycin per ml for the selection of transformants, or in Eugon (Difco) broth with yeast extract. Iron-replete and iron-limiting conditions were achieved by the addition of 100 μM ferric chloride and 50 μM deferoxamine mesylate (Desferal; Sigma) or 200 μM 2,2′-dipyridyl (Sigma), respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−endA1 hsdR17 (rk−mk−) thi-1 λ−recA1 gyrA96 RelA1 PhoA suipE44 deoR φ80dlacZΔM15 Δ(lacZYA argF)U169 | Invitrogen |
| H1698 | AraD139Δ (argF-lac)U169 rpsL150 relA1 FlbB5301 deoC1 ptsF25 rbsR fiu::λpLacMu53 | 24 |
| H1780 | AraD139Δ (argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR fur fiu::λpLacMu53 | 24 |
| D. nodosus | ||
| VCS1703A | Serogroup G, transformable, virulent strain | J. Egerton, University of Sydney |
| JIR3764 | VCS1703A furΩerm(B) | Natural transformation |
| JIR3765 | VCS1703A furΩerm(B) | Natural transformation |
| Plasmids | ||
| pBluescript SK+ | Apr, lacZ cloning vector | Stratagene |
| pUC18 | Apr, lacZ, cloning vector | 63 |
| pJIR2236 | pUC18 EcoR1/XbaIΩ(EcoRI/XbaI VCS1703A PCR product, fur gene ∼700bp) | Recombinant |
| pJIR2281 | pBluescript SK+ containing E. coli fur gene | Recombinant |
| pJIR2300 | pBluescript SK+ with 2kb ClaI fragment containing D. nodosus fur gene | Recombinant |
| pJIR2417 | pBluescript SK+ containing erm(B) flanked by N-terminal and C-terminal region of D. nodosus fur | Recombinant |
Library screening.
The D. nodosus library was screened by using H1780, a furEc mutant containing an in-frame insertion of lacZ into the Fur-regulated fiu gene (24). In this strain expression of β-galactosidase is constitutive, and therefore red colonies are observed on iron-replete MacConkey (MAC) agar. If a fur gene is provided in trans, the translated protein should actively bind to the promoter and repress the transcription of lacZ, resulting in yellow-orange colonies.
Phenotypic analysis.
Protease phenotypes were determined on EYE agar containing 2% (wt/vol) skim milk powder (34). The elastase test (55) and twitching motility assays (34) were performed as previously described. β-galactosidase activity was determined by using cultures treated with sodium dodecyl sulfate (SDS) and chloroform as previously described (40). Each assay was repeated at least four times with separate cultures. Statistical analysis was performed by using a Student's t test.
Molecular techniques.
Unless otherwise stated, molecular techniques were performed by using standard procedures (51). Reverse transcriptase PCR (RT-PCR) of fur was carried out as described (34) by using primers 16329 and 16441 (Table 2). Oligonucleotide primers (Table 2) were synthesized by using a 393 DNA-RNA synthesizer (Applied Biosystems). Sequencing was performed with an Applied Biosystems 373A automated sequencer and analyzed by using Sequencher version 3.0 (Gene Codes Corporation). Nucleotide and amino acid comparisons were accomplished by using the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST) (1) and resources available at TIGR (http://www.tigr.org). Theoretical values for molecular mass and isoelectric points were determined by using the ProtParam tool located on the ExPASy molecular biology server (http://us.expasy.org). Signal peptides were identified by using SignalP (http://www.cbs.dtu.dk/services/SignalP).
TABLE 2.
Oligonucleotides used in this study
| Primer | Gene | Sequence (5′-3′)a | Use |
|---|---|---|---|
| 16329 | fur | CGTAATGGTGTTTTTGTGCAAT | RT-PCR, PCR |
| 16441 | fur | AAGAAATTGGTTTGGCAACG | RT-PCR, PCR |
| 16661 | fur | GCTCTAGAAAGTCTGGCAGGAAATACGC | PCR |
| 16662 | fur | CGGAATTCGCCTTGCCGTTGTAAATGTAA | PCR |
| 16875 | fur | CATGCCCGCCATCATTGTGC | PCR |
| 16944 | fur | GACAGATGAAAGCGTTGACC | PCR |
| 18156 | fur | GCAATTTGAACAAGCGGGCAT | PCR |
| 18162 | fur | GGAATTCCCAATTTCTTCATCGGCAGCC | PCR |
| 18163 | fur | CCATCGATGAAAATTGCGACCACAAAAGCG | PCR |
| 18164 | fur | TCCCCCGCGGCGCGCAGTTCAACACGAAAT | PCR |
| 25946 | 16S rRNA | CGGAATGACTGGGCGTAAA | QRT-PCR |
| 25948 | 16S rRNA | CTAGGTTGAGCCCAGGGATTT | QRT-PCR |
| 25619 | sodA | GCAGCGCTATCCGCAATAAT | QRT-PCR |
| 25620 | sodA | CGCATCGCGTTGAAATAAAGA | QRT-PCR |
| 25606 | yfeA | ACAGAAGGCGCTTTTTCCTATTTA | QRT-PCR |
| 25621 | yfeA | CGGCCACACATAGGCTTCA | QRT-PCR |
| 25608 | cbiK | AATGGGAAATGAATAAAACTAGAAAGGA | QRT-PCR |
| 25604 | cbiK | GCATGGTTGAAATACCGCAAT | QRT-PCR |
| 25616 | speF | CGCTCCGGGTGATATGGTT | QRT-PCR |
| 25617 | speF | GCACCGTGGCAAACAGATTT | QRT-PCR |
Underlining represents sequence for cloning purposes.
Quantitative RT-PCR (QRT-PCR).
RNA was isolated from D. nodosus cells grown in EYE broth with 100 μM ferric chloride, by using Trizol (Invitrogen) according to the manufacturer's instructions. RT reactions were performed as described previously (34) by using primers designed with Primer Express (Applied Biosystems) (Table 2). DNA amplification was determined by using the fluorescent dye SYBR Green. Reactions were performed in final volumes of 25 μl with the SYBR Green PCR master mix (Applied Biosystems), cDNA, and 50 nM primers on an ABI PRISM 7700 sequence detector. Samples were run in triplicate on multiple runs and calibrated against genomic DNA standards. The data were normalized to 16S rRNA levels, and reactions were the result of single products as determined by the analysis of dissociation curves.
Mutant construction and analysis.
The fur suicide vector, pJIR2417, was constructed in a pBluescript SK+ derivative that contained the erm(B) gene. Fragments generated by PCR representing the 5′ fur region and its upstream sequence (primers 18162 and 18163) and the 3′ region of fur and its downstream region (18156 and 18164) were each cloned on opposite sides of erm(B) in the genomic orientation. Natural transformation of D. nodous cells was then performed as previously described (34), and a capillary PCR (33) was used with primers 16944 and 16875 to screen transformants for an appropriately sized insertion into the D. nodosus fur gene. Two mutants were then screened further by PCR with 16S rRNA gene primers (35) to confirm that the transformants were derived from D. nodosus and with erm(B) primers (17) to verify that the erm(B) gene was present. Southern hybridizations were carried out as described (34) by using NruI (New England Biolabs)-digested chromosomal DNA isolated from wild-type and mutant strains with a fur-specific probe, constructed by using primers 16329 and 16441 (Table 2), an erm(B)-specific probe (17), and a 16S rRNA gene probe (35). HindIII (Roche)-digested DNA was hybridized with a vap-specific probe (31). To confirm that the antibiotic-resistant colonies were derived from the wild-type strain, Southern analysis with the 16S rRNA and vap probes, combined with PCR-restriction fragment length polymorphism analysis of the gene encoding an outer membrane protein of D. nodosus (omp1) (20) was performed.
Homology modeling.
The structure of the D. nodosus fur (FurDn) protein was modeled on the crystal structure of the FurPa protein (42) by using SWISS-MODEL and DeepView (Swiss-PdbViewer) (22). The final model was verified by using a Ramachandran plot and various other analytical tools contained in the WhatIf suite of programs (http://www.cmbi.kun.nl/gv/servers/WIWWWI).
Preparation of proteins for proteomic analysis.
Cell lysates were prepared from 150-ml cultures of D. nodosus grown for 24 h in iron-replete EYE broth. Cells were pelleted at 6,000 × g before being washed with 1× TE buffer (10 mM Tris-HCl[pH 8.0], 1 mM EDTA), resuspended in immobilized pH gradient (IPG) rehydration solution (8 M urea, 0.03% dithiothreitol [wt/vol], 2% [wt/vol] CHAPS [3-([3-cholamidopropyl]-dimethylammonio)-1-propanesulfonate], 0.5% IPG buffer [vol/vol; Amersham Biosciences], 0.002% [wt/vol] bromophenol blue) plus 1 mg of DNaseI (Promega) per ml and 0.25 mg of RNase (Roche) per ml, and subjected to three 30-s bursts of sonication (Sonifier B-12; Branson). Cellular debris was removed by centrifugation.
Membrane-enriched samples were obtained from 24-h, 150-ml D. nodosus iron-replete EYE broth cultures as previously described (10) before solubilization in IPG buffer, pH 4 to 7. To obtain secreted proteins, D. nodosus was grown in 150 ml of iron-replete EYE broth plus 1 mM CaCl2 for 48 h. After centrifugation, 20 ml of the supernatant was filtered through a 0.45-μm-pore-size filter (Millipore) and then precipitated overnight at 4°C with 10% (wt/vol) trichloroacetic acid. Secreted proteins were pelleted at 10,000 × g before being washed with cold acetone and resuspended in IPG buffer, pH 3 to 10. Protein concentrations were determined by using a modified Bradford method (44).
Two-dimensional gel electrophoresis.
Samples were loaded by passive rehydration onto 24-cm IPG strips (Amersham Biosciences) according to the manufacturer's instructions. Isoelectric focusing was carried out by using a gradient-based protocol for a total of 40 and 60 kVh for pH 3 to 10 and pH 4 to 7 IPG strips, respectively, by using a Multiphor II electrophoresis unit (Amersham Biosciences). Before SDS-polyacrylamide gel electrophoresis, IPG strips were equilibrated for 15 min in SDS equilibration buffer (6 M urea, 50 mM Tris-HCl [pH 8.8], 30% glycerol [vol/vol], 2% SDS, 0.002% bromophenol blue [wt/vol]) containing 65 mM dithiothreitol and then for 15 min in equilibration buffer containing 135 mM iodoacetamide. The strips were analyzed on large ExcelGel XL 12 to 14% gradient gels (Amersham Biosciences) for the cell lysate and membrane preparations or on smaller ExcelGel 12.5% homogeneous gels (Amersham Biosciences) for the extracellular proteins by using a Multiphor II apparatus according to the manufacturer's instructions (Amersham Biosciences). Two-dimensional gels loaded with analytical quantities of protein were visualized by silver staining as previously described (3). Proteins to be identified by mass spectrometry (MS) were stained with colloidal Coomassie brilliant blue G-250 according to the manufacturer's instructions (Sigma). Gel analysis was performed in triplicate, and only proteins differentially expressed across all replicates were identified by MS. Since proteomics had not previously been done in D. nodosus, proteins were initially resolved over a pH 3 to 10 gradient. This process revealed that the majority of observable proteins were located in the acidic to neutral portion of the gel; therefore, further work was carried out by using a pI range of 4 to 7.
Identification of proteins by MS.
The proteins were excised from a gel, and the gel slices were washed for at least 1 h in both double-distilled H2O and then 50% acetonitrile, before dehydration in acetonitrile for 15 min. The gel pieces were lyophilized before rehydration overnight at 37°C with 25 mM ammonium bicarbonate containing 10 mg of sequencing grade trypsin (Promega) per ml. The supernatant from this step was retained while two further 1-h peptide extractions were performed with 50% acetonitrile-0.1% (vol/vol) trifluoroacetic acid (TFA). The peptide extracts were then pooled, lyophilized, and reconstituted in 1% TFA. The peptides were then concentrated and desalted by using C18 Zip Tips (Millipore), according to the manufacturer's recommendations, and eluted with 50% acetonitrile-0.1% TFA. Matrix assisted laser desorption ionization-time of flight MS was performed on a Voyager De STR mass spectrometer (Applied Biosystems) by using the crushed crystal method with α-cyano-4-hydroxycinnamic acid. Peptide mass fingerprint spectra were automatically submitted to MS-Fit, part of the ProteinProspector (9) package, and searched against a D. nodosus set of open reading frames (ORFs) that we predicted by using Glimmer (12) from our completed but unpublished genome sequence (http://www.tigr.org). Typically, a mass tolerance of 50 ppm was used in these analyses. Glimmer, based on interpolated Markov models, was trained with ORFs larger than 600 bp from the genomic sequence, as well as with the D. nodosus genes available in the GenBank database.
Nucleotide sequence accession number.
The sequence of the D. nodosus fur gene has been deposited in the GenBank database under the accession number AY569334.
RESULTS
Cloning and sequence analysis of a D. nodosus fur homologue.
As part of a larger study aimed at identifying response regulator genes by using degenerate oligonucleotide PCR on DNA from the transformable D. nodosus strain VCS1703A, we identified a recombinant plasmid, pJIR2236, that appeared to encode a product with similarity to the C-terminal 90 amino acids of Fur proteins. Southern blotting showed that this gene region was localized to a 2-kb chromosomal ClaI fragment. A chromosomal VCS1703A ClaI library was constructed in pBluescript SK+ and then screened with the E. coli reporter strain H1780. Approximately 20 of the 40,000 colonies that were screened were pink on iron-replete MAC agar and red on iron-limiting MAC agar, indicative of Fur-mediated repression in the reporter strain (see Materials and Methods). The recombinant plasmids all carried the same 2-kb ClaI fragment, and sequence analysis of one of these plasmids, pJIR2300, confirmed that a fur homologue was present. Southern hybridization analysis performed by using the ClaI insert in pJIR2300 as a probe showed that the insert hybridized to a 2-kb chromosomal ClaI fragment as expected (data not shown).
The D. nodosus fur gene was 420 bp and encoded a putative 16.2-kDa, 140-amino-acid cytoplasmic protein with a predicted isoelectric point of 5.42. No helix-turn-helix motif (14) was present. A putative sigma70 consensus binding site was located upstream of the translational start site, and there was a potential transcriptional-termination stem-loop structure (ΔG = −7.2 kcal/mol) downstream of the translational stop codon.
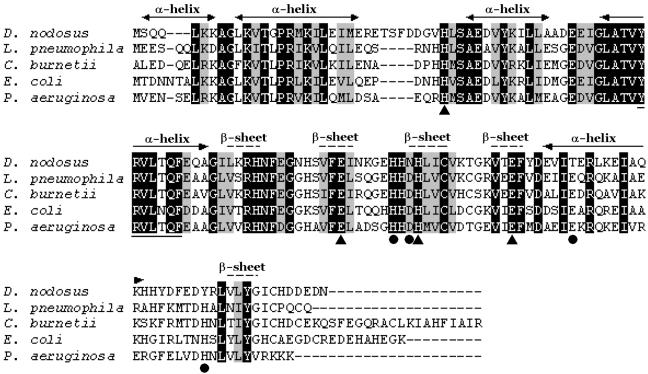
The amino acid sequence of the FurDn protein had a high degree of similarity to many Fur proteins (Fig. 1), with the most closely related proteins including those from Legionella pneumophila (60% identity) and E. coli (61% identity). These proteins have discrete areas of similarity (Fig. 1), generally corresponding to the regions that are predicted to contain secondary structures (42). One of these regions, helix 4, contains a highly conserved motif Y55-F61 (numbering from E. coli) that is believed to be involved in DNA binding (21). A putative metal binding motif in the C-terminal end of the protein, H-H-X-H-X2-C-X2-C was only partially conserved, with the FurDn protein having a threonine residue in place of the second cysteine. Located between the predicted second and third alpha helices there was also an inserted sequence (TSFD) that was unique to the FurDn protein (Fig. 1).
FIG. 1.
Alignment of D. nodosus Fur homologues. The FurDn protein was aligned by using ClustalW (59) to its two highest scoring matches as determined by BLASTP (1) as well as to the E. coli and P. aeruginosa Fur proteins. Regions predicted to contain secondary structures are shown (42). Identical residues are shaded black, with similar residues shaded gray. Metal binding site 1 residues are indicated with circles, and site 2 residues are indicated with triangles, as identified from the crystal structure of P. aeruginosa Fur. Residues underlined highlight a motif thought to be involved in DNA binding. Legionella pneumophila (27), accession no. AAA19656.1; Coxiella burnetii (R. Seshadri and J. Samuel, unpublished results), accession no. AAK00303.1; E. coli (52), accession no. P06975; and P. aeruginosa (43), accession no. Q03456.
The genetic organization of the regions flanking the fur gene did not show a high degree of similarity to regions flanking other fur genes. Upstream of the fur gene in D. nodosus there was a gene encoding a homologue of the signal recognition receptor protein FtsY and a gene (rpoH) encoding a sigma32 homologue. In Shewanella oneidensis Fur was shown to regulate rpoH (58). Downstream of fur was a gene encoding a conserved hypothetical protein and a gene (focA) encoding a protein with similarity to formate or nitrite transporters.
The D. nodosus and P. aeruginosa Fur proteins have significant structural similarity.
The recent elucidation of the crystal structure of the FurPa protein provided considerable insight into its mechanism of activation and DNA binding (42). The FurDn protein was modeled by using the structure of the FurPa protein as a reference point, and when the two structures were superimposed, they were virtually identical, which corresponded well with their predicted secondary structure similarities (data not shown). The only region that deviated significantly was the turn-loop region between helices two and three. This extended loop region contained the TSFD residues that were not present in any other Fur homologues. The structural effect of this extended loop was not profound, since it appeared to bend back toward the third helix and created only a small increase to the surface of the protein in this area. This region was not on the DNA binding site of the Fur protein, nor was it close to the dimerization domain (16).
The FurPa protein was crystallized in the presence of zinc, and subsequently two metal binding sites were identified. Binding site 1 represents the putative iron binding regulatory site and is coordinated by amino acids H86, D88, E107, and H124 (42). These residues are conserved among known Fur homologues (Fig. 1) but were not totally conserved in the FurDn protein. In the FurDn protein a change in charge was observed at position 107, where glutamate was replaced by threonine, and a significant change in charge occurred at position 124, with tyrosine in place of histidine. Site 2, coordinated by H32, E80, H89, and E100, was conserved in all of the Fur homologues, including the FurDn protein(Fig. 1) (42).
The D. nodosus fur gene complements an E. coli fur mutant.
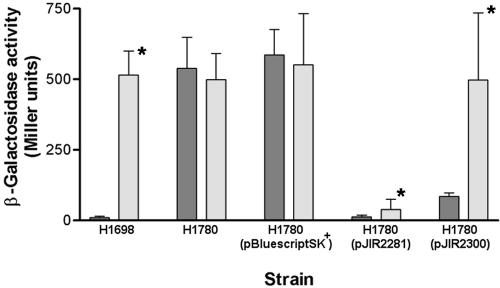
The ability of H1780(pJIR2300) to form pink colonies on iron-replete MAC agar and red colonies on iron-limiting MAC agar suggested that furDn was able to complement the furEc mutation. To quantitate the extent of this complementation, the furDn gene located on the high-copy-number vector pBluescript SK+ was introduced into the reporter strain H1780, and β-galactosidase activity was assayed in the presence and absence of Desferal. Plasmid pJIR2281, which carried furEc on pBluescript SK+, was used as a multicopy-positive control. Strain H1698 was also assayed as it carries a functional fur gene, and in the presence of iron the resultant Fur protein represses the lacZ reporter gene. When iron is chelated with Desferal, apo-Fur is no longer able to bind to the fiu promoter, resulting in the production of β-galactosidase. The results showed that in the furDn construct, β-galactosidase activity was highest when iron in the medium was limited, indicating iron-dependent regulation (Fig. 2). The control strains H1780 and H1780(pBluescript SK+) showed high levels of β-galactosidase activity irrespective of the iron concentration. When the fur gene from E. coli was added, the presence of multiple copies of this gene in the reporter system resulted in β-galactosidase repression regardless of iron content, as observed elsewhere (37, 43, 56). In comparison to iron-limited cultures, the FurDn protein was also able to significantly repress (P < 0.05) the fiu promoter when supplemented with iron (Fig. 2). The degree of repression that was observed correlated well with the observation of pink colonies on MAC media. The ability of the furDn gene to complement a furEc mutant suggested that the Fur protein was likely to be functional in D. nodosus. In subsequent studies it was shown that a fur transcript could be detected in D. nodosus by RT-PCR and that D. nodosus strains encompassing various serogroups, as well as those representing virulent and benign isolates, all had the same 2-kb ClaI fragment that hybridized to a furDn probe in Southern blot assays (data not shown).
FIG. 2.
Iron-dependent repression of a Fur-regulated promoter by the FurEc and FurDn proteins. The β-galactosidase activity of the fiu-lacZ fusion in E. coli is shown after growth under iron-replete (100 μM FeCl3; dark gray) and iron-limiting (50 μM Desferal; light gray) conditions. Shown are the fur+ (H1698) and fur mutant (H1780) reporter strains, H1780 containing the E. coli (pJIR2281), and D. nodosus (pJIR2300) fur genes on pBluescript SK+ and a vector control. Each assay was performed at least four times in four independent experiments. Mean values are displayed with standard deviations as error bars. Asterisks indicate samples that were statistically different (P < 0.05) when grown under iron-replete and iron-limiting conditions.
Construction and analysis of a D. nodosus fur mutant.
To determine the functional role of the fur gene in D. nodosus, a chromosomal fur mutant was constructed by allelic exchange. The suicide vector pJIR2417 was introduced into strain VCS1703A by natural transformation, and erythromycin-resistant colonies were selected. These resistant colonies were screened by using PCR, and two of these mutants (JIR3764 and JIR3765) were subjected to detailed analyses (see Materials and Methods) that confirmed they were derived from D. nodosus VCS1703A by double-crossover events that insertionally inactivated the fur gene (data not shown). All further studies were carried out on the fur mutant JIR3764.
To determine the potential biological effect of the fur mutation, the wild-type and mutant strains were grown under iron-replete and iron-limiting conditions. Although the growth rate of the fur mutant was the same as that of the wild-type in iron-replete medium, the use of the chelator 2,2′-dipyridyl to obtain iron-limiting conditions inhibited the already naturally poor growth of D. nodosus such that any differences were not able to be observed. In other bacteria, Fur has been shown to regulate the production of secreted proteins (16, 60). To see if the fur mutation had any effect on such proteins in D. nodosus, the wild-type and mutant strains were analyzed for their ability to secrete proteases, that is, to hydrolyze casein and elastase, as well as to carry out twitching motility, a property that is dependent on the production of type IV fimbriae. No difference was observed between the wild-type and mutant strains in these assays (data not shown).
Identification of Fur-regulated proteins by proteomic analysis.
To determine the regulatory targets of Fur, a broader proteomic approach was employed, combining two-dimensional gel electrophoresis and MS. As Fur is known to regulate genes with a wide variety of functions, proteins from cell lysate, total membrane-enriched, and extracellular fractions were analyzed. The number of proteins that were observed over the three cellular fractions constituted a high proportion of the predicted D. nodosus proteome. The cell lysate had almost 1,000 protein spots, the membrane preparations had over 100 spots, and the secretome contained a surprisingly high number of at least 60 reproducible spots.
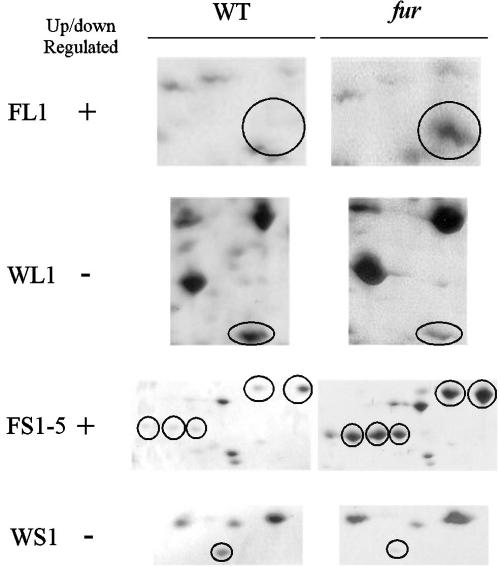
By comparing the proteome of the wild-type strain to that of the fur mutant JIR3764, we were able to observe and identify through MS several up-regulated and down-regulated proteins in the fur mutant (Fig. 3 and Table 3). We were able to visualize other differences; however, from these spots we were either unable to obtain reproducible mass spectra or unable to identify a significant match to the database. These proteins were primarily from the membrane samples. QRT-PCR was subsequently used to verify that these regulatory effects occurred at the transcriptional level. Only those proteins for which there was agreement between the proteomic and QRT-PCR data are reported here. There were four other proteins for which the proteomic and transcriptional profiles were at variance, presumably as a result of differences in posttranslational modification.
FIG. 3.
Differentially expressed proteins in the D. nodosus fur mutant JIR3764. Shown are sections of representative gels of the wild-type strain VCS1703A and its fur mutant JIR3764. Spot numbers are designated according to the following key: F, up-regulated in fur mutant; W, down-regulated in fur mutant; L, identified in the cell lysate; and S, identified in the supernatant. Relevant bands are circled. Numbering for FS1 to FS5 is right to left, working down. Gels used for analytical purposes were loaded with 100, 45, and 30 μg of protein for cell lysate, total membrane, and secreted proteins, respectively. These values increased to 300, 100, and 200 μg, respectively, for successful micropreparative loadings.
TABLE 3.
Differentially expressed proteins in the D. nodosus fur mutant JIR3764 and their expression ratios as determined by QRT-PCR
| Spot no.a | Protein identification | Identity (%) | No. of peptides matched (% coverage)c | Relative gene expressiond |
|---|---|---|---|---|
| FL1 | YfeA, iron transport protein | 64 | 10 (46) | 1.44 ± 0.35 |
| FS1 | YfeA, iron transport protein | 64 | 11 (60) | 1.44 ± 0.35 |
| FS2 | YfeA, iron transport protein | 64 | 11 (48) | 1.44 ± 0.35 |
| FS4 | CbiK domain, ABC-type cobalt transport | 29 | 15 (67) | 4.28 ± 0.97 |
| FS5 | CbiK domain, ABC-type cobalt transport | 29 | 19 (61) | 4.28 ± 0.97 |
| FS6 | CbiK domain, ABC-type cobalt transport | 29 | 11 (45) | 4.28 ± 0.97 |
| WL1 | Ornithine decarboxylaseb | 64 | 16 (23) | 0.38 ± 0.24 |
| WS1 | SodA, manganese superoxide dismutase | 58 | 9 (48) | 0.35 ± 0.02 |
F, protein up regulated in fur mutant; W, protein present in wild-type, down-regulated in fur mutant; L, present in cell lysate; and S, present in secreted sample.
Spot corresponded to a truncated form of a larger ORF.
Number of experimentally derived mass peaks that match to the in silico derived peaks for that protein (degree of coverage of proteins conferred by the residues represented by matching peaks).
Expressed as the ratio of the specific gene expression level in the fur mutant compared to wild-type normalized to the level of expression of the 16S rRNA gene.
In the cell lysate the FL1 protein was identified as being up-regulated in the fur mutant. Significant matches were obtained to the YfeA proteins of Pasteurella multocida (39) and Yersinia pestis (5) and SitA from S. enterica serovar Typhimurium (67). YfeA or SitA represents the periplasmic binding protein of an ABC-type iron and manganese transport system (4, 5, 32) and is typically encoded within an operon, yfeABCD or sitABCD. YfeB is predicted to be an ATP-binding protein, and YfeC and YfeD are predicted to be integral membrane proteins (5). Levels of yfeA expression were higher in the fur mutant than in the wild type (Table 3).
One protein, WL1, was identified in the cell lysate as being down-regulated in the fur mutant (Fig. 3 and Table 3). This protein had a high degree of identity to the inducible version of ornithine decarboxylase, particularly that from P. multocida (64% identity) (39). QRT-PCR of the gene encoding the ornithine decarboxylase, speF, showed that levels of transcription were considerably reduced (Table 3).
BprV is one of the three extracellular serine proteases of D. nodosus and has a predicted pI of 9.5 (36). Since in other bacteria Fur has been shown to regulate the production of secreted proteins (16, 60), the secreted proteins from D. nodosus were analyzed over a pH 3 to 10 (not pH 4 to 7) range so that any potential effects on BprV could be detected, in addition to the two other acidic proteases. However, no differences in protein expression were detected in the pI 9.5 region. The secretome had five proteins, FS1 to FS5, clustered around 30 kDa, with pI values of 5.5, that were significantly up-regulated at the transcriptional level in the fur mutant (Fig. 3 and Table 3). MS showed that FS1 and FS2 were isoforms of the same protein previously detected in the cell lysate, FL1, which had similarity to the iron transport protein YfeA. All three of these proteins, FL1, FS1, and FS2 are encoded by the same ORF but feature various migratory patterns. The presence of this protein in the two cellular compartments is not surprising, considering that it is predicted to have a signal sequence. Proteins FS3, FS4, and FS5 were isoforms of another protein, which also had an N-terminal signal sequence. These bands of protein spots had different isoelectric points, indicating that the protein undergoes some form of posttranslational processing or degradation. Although this protein only had moderate (29%) identity to a hypothetical protein from Neisseria meningitidis, it was identified as containing a conserved CbiK domain. CbiK is the periplasmic component of an ABC-type cobalt transport system (46). Consistent with the increased level of protein, expression of this gene, which we have designated as cbiK, showed a 4.28-fold increase in the fur mutant (Table 3).
A secreted protein, WS1, was observed to be down-regulated in the fur mutant compared to the wild type. WS1 had amino acid sequence similarity to a manganese superoxide dismutase (MnSOD), SodA, but did not have a signal sequence. The D. nodosus MnSOD homologue contained the signature pattern of iron and manganese containing SODs and had 58% identity to MnSOD from Vibrio cholerae (26). Analysis of the structural gene sodA by QRT-PCR showed that its expression was reduced nearly threefold in the fur mutant (Table 3).
An attempt was made to identify potential Fur boxes upstream of those genes that were negatively regulated by the FurDn protein. Neither alignment of these upstream regions nor processing with motif finding programs revealed any significant similarities. Attempts at identifying putative Fur targets in the D. nodosus genome by using the E. coli consensus sequence did not reveal any obvious targets such as ferrocheletases or the ferrous iron transporters FeoA and FeoB that are encoded on the D. nodosus genome. The only significant match obtained was found upstream of the cbiK gene (encoding the FS3 to FS5 proteins), which was the gene shown to be highly regulated by the FurDn protein at the transcriptional level (Table 3). It had 16 out of 19 bases conserved from the E. coli Fur box.
DISCUSSION
In the presence of iron Fur acts as a global transcriptional repressor, regulating a wide spectrum of genes that are involved in processes as diverse as iron metabolism, transport systems, oxidative stress, and virulence (16). We have identified the D. nodosus fur gene and used genetic and proteomic analysis to show that the FurDn protein is functional and is involved in the regulation of genes involved in iron and manganese homeostasis and oxidative metabolism in D. nodosus (Table 3). These findings represent the first report of a functional regulatory protein in this important animal pathogen.
The classical role of Fur is to coordinate iron metabolism with iron availability by regulating iron uptake and acquisition systems (16). In this study two D. nodosus proteins were identified that potentially have similar functions. In other bacteria YfeA is the periplasmic binding protein component of an ABC transporter system that is involved in iron uptake. YfeA (or SitA) has been shown to be regulated by Fur in all of the organisms studied to date, including Y. pestis (5), S. enterica serovar Typhimurium (67), and Shigella flexneri (50). The proteins encoded by the yfe (or sit) operon also display similarity to manganese transporters, and subsequently the system has been shown to transport both iron and manganese (4, 32). The same operon structure observed with the yfe genes (yfeABCD) was also observed in D. nodosus (data not shown). Detailed analysis of the D. nodosus genome sequence did not reveal any other iron- or manganese-specific ABC transport systems (unpublished data). Therefore, we postulate that YfeABCD may represent the primary transportation system by which iron or manganese enters the cell. The importance of this transport system is illustrated by reports that null mutants of the equivalent operon are attenuated for virulence in Y. pestis and S. enterica serovar Typhimurium (4, 30).
A protein (FS3, FS4, or FS5) containing a CbiK domain was also up-regulated in the D. nodosus fur mutant. CbiK is a cobalt chelatase that is involved in the anaerobic pathway of vitamin B12 (cobalamin) synthesis and in some species has been shown to invoke a human immune response (45, 49). Since CbiK from S. enterica serovar Typhimurium has been shown in vivo to act as a ferrochelatase by binding to Fe2+ (46), it is possible that in D. nodosus the CbiK homologue is a metal ion acquisition protein that may have a role in iron acquisition or iron and/or other metal ion homeostasis.
The cbiK gene was located only three genes downstream of the fur gene, after the putative formate or nitrate transport gene focA. No operon-like structure was evident. The only high-similarity match observed in searches based on the E. coli Fur box consensus sequence was cbiK. This Fur box conservation, in addition to the high level of Fur-dependent expression (Table 3), indicates that the FurDn protein functions in a similar manner to the FurEc protein.
The organization of the D. nodosus fur region was different from that of other characterized fur regions, although some conservation exists among members of the γ subclass of the Proteobacteria (37), to which D. nodosus belongs (13). The presence of an rpoH gene upstream of the fur gene was unique to D. nodosus, and although rpoH was not part of an operon with fur and there was no sigma32 consensus binding site in the fur promoter region, we cannot rule out the possibility that Fur may regulate rpoH, causing a discrete set of genes to be expressed, as observed in S. oneidensis (58).
In other bacteria, Fur links iron metabolism with oxidative metabolism and stress. Fur regulates SODs and is itself regulated by the oxidative stress regulators OxyR and SoxRS (23). This association with oxidative stress relates back to maintaining iron at a nontoxic level, as under aerobic conditions in the presence of iron-destructive hydroxy radicals can be produced via the Fenton reaction (2). This by-product of aerobic metabolism is not a concern for D. nodosus since it grows only under anaerobic conditions, which may explain why we did not observe a growth difference between the wild type and the fur mutant at different iron concentrations. A similar observation was made when S. oneidensis was grown anaerobically (58). Typically, a reduced growth rate is observed under aerobic iron-replete conditions, as seen in Y. pestis (53), Neisseria gonorrhoeae (57), and Staphylococcus aureus (29). This difference is less profound or is absent under microaerophilic or anaerobic conditions due to reduced radical toxicity and the increased solubility of iron, as demonstrated in P. aeruginosa (25), Helicobacter pylori (6) and S. oneidensis (58).
SODs catalyze the dismutation of oxygen radicals to hydrogen peroxide. In E. coli, Fur acts as a transcriptional repressor of the MnSOD gene, sodA, and as an indirect positive regulator of the iron-dependent superoxide dismutase (FeSOD) gene, sodB (16, 38). In D. nodosus, we observed a novel Fur regulatory effect whereby MnSOD expression in the fur mutant was reduced, resulting from a 2.8-fold reduction in sodA transcription (Table 3). This scenario is analogous to the situation with sodB in E. coli. Our analysis of the D. nodosus genome sequence did not reveal the presence of a FeSOD gene. Therefore, we postulate that in D. nodosus MnSOD may represent a functional homologue of FeSOD and may be regulated by both iron and manganese levels. In E. coli, the regulation of sodB is undertaken by a small RNA molecule, RyhB, that is regulated by Fur (38). This molecule is, in turn, regulated by the RNA chaperone Hfq (62). A RyhB homologue was not present in D. nodosus; however, a Hfq homologue with 62% identity to E. coli Hfq was present. This analysis does not preclude the presence of a functional RyhB homologue in D. nodosus, as observed in P. aeruginosa (65), since similarity between small RNA molecules of distantly related organisms is not high (64, 65). As D. nodosus is an aerotolerant anaerobe, the role of SOD may only be apparent at the site of infection, before an anaerobic environment is established, or as protection from phagocytic cells (48). We did not observe any alteration in growth of the fur mutant after 4 h of exposure to air (data not shown), although at this point in time nothing is known about the mechanisms of aerotolerance in D. nodosus. The identification of MnSOD in the culture supernatant is unusual, considering that it is a cytoplasmic protein lacking a signal peptide. Its presence is likely to be the result of cellular leakage and extracellular stability, as previously reported in other organisms (28, 61).
In other bacteria the effects of fur mutations on virulence have been mixed. A fur mutant of Campylobacter jejuni was restricted in its ability to colonize chickens (41), and in S. aureus and Listeria monocytogenes attenuation was observed in murine models (29, 47). In S. enterica serovar Typhimurium mutation of fur gave various results, depending on the route of delivery (19). By contrast, no effect on virulence was observed in P. multocida (8) or avian strains of E. coli (68). The presence of the fur gene in both virulent and benign strains of D. nodosus would suggest that it is not specifically required for virulent footrot. However, Fur may still regulate genes in virulent strains that have an effect on virulence or may regulate genes that are common to virulent and benign strains but, when differentially expressed, have an effect on virulence. Further studies are required to determine if the D. nodosus homologues of the Fur, YfeA, MnSOD, or CbiK proteins are involved in virulence.
The FurDn protein was able to significantly repress the fiu promoter in the reporter strain in an iron-dependent manner but not to the same degree as the FurEc protein (Fig. 2). The difference in levels of repression may be due to the heterologous nature of the system and fundamental differences in Fur binding preferences. This potential binding site preference may also reflect the scarcity of identifiable Fur boxes in the D. nodosus genome. Since Fur proteins are capable of binding other metal ions (11), the FurDn protein may also bind other metals instead of, or in addition to, iron. Although the sequence for metal binding site 1 is not absolutely conserved in the FurDn protein, the complementation data indicate that sufficient structural and functional similarity is present to enable the FurDn protein to complement the E. coli system and to be responsive to iron levels, indicating that the activity of the FurDn protein is associated with iron. The absence of an E. coli-like Fur binding consensus sequence has been observed elsewhere (18, 41). With other Fur proteins the degree of repression observed in this fiu reporter system is usually analogous to that of E. coli (56, 66). Only in P. aeruginosa has a degree of repression similar to the results of this study been observed (43). Both the FurDn protein (61%) and the FurPa protein (58%) have similar identity to the FurEc protein. The only major structural difference between the FurPa and FurDn proteins is in the loop region between the second and third helices, where the FurDn protein has the unique motif TSFD. The second cysteine in the C-terminal H-H-X-H-X2-C-X2-C metal binding site is changed to a threonine residue in both the P. aeruginosa and D. nodosus proteins, which may explain the altered repression observed in E. coli. These observations are in agreement with previous studies (11) which showed that Fur activity in E. coli was compromised when the same cysteine residue was changed to serine, with the derepression/repression ratio being comparable to that seen in this study and with the FurPa protein(43).
In summary, we have identified and then characterized through insertional inactivation the first transcriptional regulator to be functionally analyzed in D. nodosus. The FurDn protein had extensive structural homology to the FurPa protein and was able to complement an E. coli fur mutant. Comparison of the proteomes of the wild-type strain VCS1703A and its fur mutant JIR3764 identified several differentially expressed genes and implicated Fur as a regulator of iron and oxidative metabolism in this anaerobe.
Acknowledgments
We thank Klaus Hantke for E. coli strains H1698 and H1780, Ehmke Pohl for the Fur crystal file, and Simon Harris at the Monash University MALDI-MS core facility for carrying out the MS analysis.
D.P. is the recipient of an Australian Postgraduate Award and Monash Faculty of Medicine, Nursing and Health Sciences Postgraduate Excellence Award. This work was supported by grants from the Australian Research Council and the U.S. Department of Agriculture.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 5.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bereswill, S., S. Greiner, A. H. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billington, S. J., J. L. Johnston, and J. I. Rood. 1996. Virulence regions and virulence factors of the ovine footrot pathogen, Dichelobacter nodosus. FEMS Microbiol. Lett. 145:147-156. [DOI] [PubMed] [Google Scholar]
- 8.Bosch, M., R. Tarrago, E. M. Garrido, S. Campoy, A. R. F. de Henestrosa, A. M. P. de Rozas, I. Badiola, and J. Barbe. 2001. Expression of the Pasteurella multocida ompH gene is negatively regulated by the Fur protein. FEMS Microbiol. Lett. 203:35-40. [DOI] [PubMed] [Google Scholar]
- 9.Clauser, K. R., P. R. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (± 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 10.Comeau, D. E., and M. Inouye. 1986. A rapid procedure for fractionation of bacterial cells utilizing the TL-100 Tabletop ultracentrifuge. Beckman TL-100 News. Beckman Coulter, Fullerton, Calif.
- 11.Coy, M., and J. B. Neilands. 1991. Structural dynamics and functional domains of the Fur protein. Biochemistry 30:8201-8210. [DOI] [PubMed] [Google Scholar]
- 12.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewhirst, F. E., B. J. Paster, S. La Fontaine, and J. I. Rood. 1990. Transfer of Kingella indologenes (Snell and Lapage 1976) to the genus Suttonella gen. nov. as Suttonella indologenes comb. nov.; transfer of Bacteroides nodosus (Beveridge 1941) to the genus Dichelobacter gen. nov. as Dichelobacter nodosus comb. nov.; and assignment of the genera Cardiobacterium, Dichelobacter, and Suttonella to Cardiobacteriaceae fam. nov. in the gamma division of Proteobacteria on the basis of 16S rRNA sequence comparisons. Int. J. Syst. Bacteriol. 40:426-433. [DOI] [PubMed] [Google Scholar]
- 14.Dodd, I., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escolar, L., V. de Lorenzo, and J. Perez-Martin. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26:799-808. [DOI] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrow, K. A., D. Lyras, and J. I. Rood. 2000. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm(B) genes. Antimicrob. Agents Chemother. 44:411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman, Y. E., and M. R. O'Brian. 2003. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J. Biol. Chem. 278:38395-38401. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-del Portillo, F., J. W. Foster, and B. B. Finlay. 1993. Role of acid tolerance response genes in Salmonella typhimurium virulence. Infect. Immun. 61:4489-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghimire, S. C., and J. R. Egerton. 1999. PCR-RFLP of outer membrane proteins gene of Dichelobacter nodosus: a new tool in the epidemiology of footrot. Epidemiol. Infect. 122:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez de Peredo, A., C. Saint-Pierre, J.-M. Latour, I. Michaud-Soret, and E. Forest. 2001. Conformational changes of the ferric uptake regulation protein upon metal ctivation and DNA Binding; first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310:83-91. [DOI] [PubMed] [Google Scholar]
- 22.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 24.Hantke, K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135-139. [DOI] [PubMed] [Google Scholar]
- 25.Hassett, D. J., P. A. Sokol, M. L. Howell, J. F. Ma, H. T. Schweizer, U. Ochsner, and M. L. Vasil. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178:3996-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg, J. K., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodaon, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonal, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey, E. K., and N. P. Cianciotto. 1994. Cloning and sequencing of the Legionella pneumophila fur gene. Gene 143:117-121. [DOI] [PubMed] [Google Scholar]
- 28.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 29.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 31.Katz, M. E., P. M. Howarth, W. K. Yong, G. G. Riffkin, L. J. Depiazzi, and J. I. Rood. 1991. Identification of three gene regions associated with virulence in Dichelobacter nodosus, the causative agent of ovine footrot. J. Gen. Microbiol. 137:2117-2124. [DOI] [PubMed] [Google Scholar]
- 32.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennan, R. M., S. J. Billington, and J. I. Rood. 1998. Electroporation-mediated transformation of the ovine footrot pathogen Dichelobacter nodosus. FEMS Microbiol. Lett. 169:383-389. [DOI] [PubMed] [Google Scholar]
- 34.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosuss is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Fontaine, S., and J. I. Rood. 1996. Organization of ribosomal RNA genes from the footrot pathogen Dichelobacter nodosus. Microbiology 142:889-899. [DOI] [PubMed] [Google Scholar]
- 36.Lilley, G. G., D. J. Stewart, and A. A. Kortt. 1992. Amino acid and DNA sequences of an extracellular basic protease of Dichelobacter nodosus show that it is a member of the subtilisin family of proteases. Eur. J. Biochem. 210:13-21. [DOI] [PubMed] [Google Scholar]
- 37.Lowe, C. A., A. H. Asghar, G. Shalom, J. G. Shaw, and M. S. Thomas. 2001. The Burkholderia cepacia fur gene: co-localization with omlA and absence of regulation by iron. Microbiology 147:1303-1314. [DOI] [PubMed] [Google Scholar]
- 38.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Assay of β-galactosidase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pohl, E. P., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 43.Prince, R. W., C. D. Cox, and M. L. Vasil. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol. 175:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramagli, L. S., and L. V. Rodriguez. 1985. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 6:559-563. [Google Scholar]
- 45.Raux, E., H. L. Schubert, and M. J. Warren. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 57:1880-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raux, E., C. Thermes, P. Heathcote, A. Rambach, and M. J. Warren. 1997. A role for Salmonella typhimurium CbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J. Bacteriol. 179:3202-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rea, R. B., C. G. M. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roggenkamp, A., T. Bittner, L. Leitritz, A. Sing, and J. Hessemann. 1997. Contribution of the Mn-cofactored superoxide dismutase (SodA) to the virulence of Yersinia entercolitica serotype O8. Infect. Immun. 65:4705-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross, B. C., L. Czajkowski, D. Hocking, M. Margetts, E. Webb, L. Rothel, M. Patterson, C. Agius, S. Camuglia, E. Reynolds, T. Littlejohn, B. Gaeta, A. Ng, E. S. Kuczek, J. S. Mattick, D. Gearing, and I. G. Barr. 2001. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine 19:4135-4142. [DOI] [PubMed] [Google Scholar]
- 50.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Schäffer, S. K., K. Hantke, and V. Braun. 1985. Nucleotide sequence of the iron regulatory gene fur. Mol. Gen. Genet. 201:204-212. [DOI] [PubMed] [Google Scholar]
- 53.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart, D. J. 1989. Footrot of sheep, p. 5-45. In J. R. Egerton, W. K. Yong, and G. G. Riffkin (ed.), Footrot and foot abscess of ruminants. CRC Press, Boca Raton, Fla.
- 55.Stewart, D. J. 1979. The role of elastase in the differentiation of Bacteroides nodosus infections in sheep and cattle. Res. Vet. Sci. 27:99-105. [PubMed] [Google Scholar]
- 56.Thomas, C. E., and P. F. Sparling. 1994. Identification and cloning of a fur homologue from Neisseria meningitidis. Mol. Microbiol. 11:725-737. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, C. E., and P. F. Sparling. 1996. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 178:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates III, C. C. Brandt, J. M. Tiedje, and J. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsujibo, H., K. Miyamoto, T. Okamoto, H. Orikoshi, and Y. Inamori. 2000. A serine protease-encoding gene (aprII) of Alteromonas sp. strain O-7 is regulated by the iron uptake regulator (Fur) protein. Appl. Environ. Microbiol. 66:3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tullius, M. V., G. Harth, and M. A. Horwitz. 2001. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect. Immun. 69:6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897-909. [DOI] [PubMed] [Google Scholar]
- 63.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 64.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyllie, S., and J. E. Raulston. 2001. Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40:1027-1036. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu, C., M. Ngeleka, A. A. Potter, and B. J. Allan. 2002. Effect of fur mutation on acid-tolerance response and in vivo virulence of avian septicemic Escherichia coli. Can. J. Microbiol. 48:458-462. [DOI] [PubMed] [Google Scholar]