Abstract
The RNA interference (RNAi)-based therapeutic ARC-520 for chronic hepatitis B virus (HBV) infection consists of a melittin-derived peptide conjugated to N-acetylgalactosamine for hepatocyte targeting and endosomal escape, and cholesterol-conjugated RNAi triggers, which together result in HBV gene silencing. To characterize the kinetics of RNAi trigger delivery and 5΄-phosphorylation of guide strands correlating with gene knockdown, we employed a peptide-nucleic acid (PNA) hybridization assay. A fluorescent sense strand PNA probe binding to RNAi duplex guide strands was coupled with anion exchange high performance liquid chromatography to quantitate guide strands and metabolites. Compared to PCR- or ELISA-based methods, this assay enables separate quantitation of non-phosphorylated full-length guide strands from 5΄-phosphorylated forms that may associate with RNA-induced silencing complexes (RISC). Biodistribution studies in mice indicated that ARC-520 guide strands predominantly accumulated in liver. 5΄-phosphorylation of guide strands was observed within 5 min after ARC-520 injection, and was detected for at least 4 weeks corresponding to the duration of HBV mRNA silencing. Guide strands detected in RISC by AGO2 immuno-isolation represented 16% of total 5΄-phosphorylated guide strands in liver, correlating with a 2.7 log10 reduction of HBsAg. The PNA method enables pharmacokinetic analysis of RNAi triggers, elucidates potential metabolic processing events and defines pharmacokinetic-pharmacodynamic relationships.
INTRODUCTION
RNA interference (RNAi) is a gene silencing system that involves non-coding short RNAs and an RNA-induced silencing complex (RISC) that functions within the cell cytoplasm (1–5). RNAi gene regulation mechanisms are diverse, but one process degrades mRNA to silence gene expression (6,7). This function has been harnessed to study the role of gene expression in cellular processes and disease, and to develop novel therapies for a variety of clinical applications (8–16). Non-coding RNAi triggers (siRNA) are 21–30 base pair double-stranded oligonucleotides comprising a sense (or passenger) strand that matches sequences of mRNAs in the cell, and an antisense (guide) strand that is complementary to the mRNA. Once the RNAi trigger is loaded into RISC, the sense strand is cleaved and dissociates from the guide strand. Sequence-specific base-pairing between the remaining guide strand and its cognate mRNA results in cleavage of the mRNA by the RISC component Argonaute 2 (AGO2), and is then further degraded by cytosolic exonucleases. A single functional guide strand loaded into RISC mediates the cleavage of multiple mRNA molecules (17–25).
We previously developed a Dynamic Polyconjugate™ (DPC) platform for delivery of RNAi trigger molecules in vivo for therapeutic applications (9,26,27). The method involves intravenous co-injection of cholesterol-conjugated RNAi triggers together with a membrane-active biodegradable amphipathic reversibly-masked melittin-like peptide (MLP) (9,26,27). The MLP contains the targeting ligand N-acetylgalactosamine that binds to asialoglycoprotein receptors (ASGPR) on hepatocytes, thereby enabling hepatocyte targeting and internalization (9,26). The cholesterol-conjugated RNAi triggers are internalized by endocytosis possibly through interaction with lipoprotein receptors (10,28–30), and co-segregate into endosomes with the MLP (27). The acidification of endosomes during maturation triggers MLP unmasking, enabling the DPC to interact with and destabilize the endosome membrane. This promotes release of the RNAi trigger from endosomes into the cytoplasm where incorporation into RISC induces RNAi (26).
The DPC platform was used in ARC-520, an RNAi-based therapeutic targeting hepatitis B virus (HBV) (8,9,31). ARC-520 contains two RNAi triggers, AD0009 and AD0010, which target different sequences in the HBV genome enabling reduction of the pre-genomic RNA and silencing of all HBV transcripts. This includes the transcript encoding the viral surface antigen (HBsAg), which is a component of the virion, but also plays a role in altering host antiviral immune responses (8,9,31). In this report, the pharmacokinetics (PK) of ARC-520 RNAi triggers was evaluated in mice that transiently expressed the HBV genome in their hepatocytes to elucidate the RNAi-specific processing events involved in trigger biodistribution and metabolism, and identify potential correlations to trigger pharmacodynamic (PD) activity. To quantitate RNAi triggers in tissues after in vivo delivery, we hybridized a fluorescently-tagged peptide-nucleic acid (PNA) probe to tissue lysates or immunoisolated AGO2 followed by anion exchange high performance liquid chromatography. The PNA assay enabled separate quantitation of 5΄-phosphorylated and non-phosphorylated guide strands in mouse tissues after treatment with ARC-520. This element is key since 5΄-phosphorylation of RNAi triggers is indicative of endosomal release and cytoplasmic exposure, the subcellular compartment where CLP1 kinase resides (32). The 5΄-phosphorylated guide strand species were observed within 5 min after ARC-520 injection, and were detected for up to 4 weeks corresponding to the duration of HBV gene knockdown. The 5΄-phosphorylated guide strands in liver represented 0.1–0.3% of the total level of guide strands in hepatocytes. Approximately 16% of the total 5΄-phosphorylated guide strands were incorporated into RISC. In PK/PD analyses, this level of HBV-specific 5΄-phosphorylated guide strands in RISC correlated with >99% knockdown of serum HBsAg levels in HBV transgenic mice, highlighting the silencing potency of ARC-520.
MATERIALS AND METHODS
Dynamic Polyconjugate ARC-EX1
A 26 amino acid MLP was synthesized from FMOC-protected L-amino acids using standard solid-phase peptide synthesis methods (Bachem Americas, Inc., Torrance, CA, USA). Deprotection was performed using trifluoroacetic acid and the MLP was purified by reverse phase HPLC. Purity was >98% as determined by analytical HPLC and identity was confirmed by mass spectrometry (LC-MS). The N-acetylgalactosamine (NAG) ligand was conjugated to carboxy dimethylmaleamide (CDM) to form CDM-NAG (Sigma-Aldrich, Madison, WI, USA) according to published procedures (26). For preparation of MLP-(CDM-NAG), CDM-NAG was added to MLP in a 250 mM HEPES-buffered pH 8.5 aqueous solution at a 5:1 (w/w) ratio at room temperature and the modification reaction was allowed to proceed for 30 min. The pH of the reaction mixture was then adjusted to 9.0 with 4M NaOH. The CDM-based reagents react with the five primary amines on MLP (4 lysines and the N-terminus). The extent of the reaction was assayed by titrating residual primary amines using 2, 4, 6-trinitrobenzenesulfonic acid and was confirmed as >95% complete. MLP-(CDM-NAG) was purified by tangential flow in a 10 mM bicarbonate pH 9.0 buffer. Dextran solid (MW 1000) was added to masked, purified peptide (10%, w/w) and the material was lyophilized. The final material is referred to as ARC-EX1.
Cholesterol-RNAi triggers and standards
RNAi triggers were synthesized on MerMade 12 synthesizers (BioAutomation, Irving, TX, USA) using standard phosphoramidite chemistry. The integrity and purity of the single strands were confirmed by standard ion pairing HPLC and Q-TOF mass spectrometry. All single strands were >85% pure as determined by anion exchange high performance liquid chromatography (AEX-HPLC) and were within 2 Daltons of their theoretical mass as determined by LC-MS. At specific nucleotide positions, 2΄-OH groups on the triggers were replaced by 2΄-F or 2΄-OMe groups to increase nuclease resistance and minimize innate immune system stimulation. The cholesterol moiety was introduced into the sense strand 5΄ terminus for in vivo analysis. The HBV therapeutic ARC-520 consists of ARC-EX1 and API-520, where API-520 contains a mixture of two RNAi duplexes (AD0009 and AD0010) in 1 mM phosphate buffer, pH 7.2.
Animals and tissues
Female ICR mice (6–8 weeks old) were obtained from Harlan Laboratories (Indianapolis, IN, USA). Transiently transgenic NOD-SCID mice expressing the entire human HBV genome in the liver were generated as described previously (9). On first day of studies, animals weighed 20.0 ± 0.4 g. Throughout the studies animals had access ad libitum to Teklad Irradiated Rodent Diet 8904 (Teklad, Madison, WI, USA) and water via bottles. ARC-520 for injection was prepared as follows: lyophilized ARC-EX1 (10 mg) was dissolved with 2 ml sterile water. The ARC-EX1 solution was added to tubes containing isotonic glucose plus API-520 such that the final volume injected intravenously through the tail vein into each mouse was 200 μl at the desired ratio of API-520 to ARC-EX1. For serum and organ collection, mice were anesthetized with 2–3% isoflurane. Blood samples were collected from the submandibular area into serum separation tubes, and plasma was isolated using 500 μl Capiject K2-EDTA micro-collection tubes centrifuged at 2000 × g for 5 min. Organs were harvested, frozen in liquid nitrogen and stored at −80°C. All animal procedures were approved by the Arrowhead Pharmaceuticals IACUC.
Quantitation of RNAi triggers in plasma and tissues
Tissue was placed in liquid nitrogen and ground with a chilled mortar and pestle to a fine powder. The ground tissue was weighed and mixed with Affymetrix Lysis Buffer (Affymetrix Lysis Solution and nuclease-free water at 1:2 ratio) (Affymetrix, Santa Clara, CA, USA). The sample was then diluted with lysis buffer containing 50 μg/ml proteinase K and sonicated for 15–30 s with a probe sonicator to yield a tissue lysate. Thawed plasma samples were solubilized in lysis buffer containing 50 μg/ml proteinase K. Serial dilutions of RNAi trigger standards were also prepared in lysis buffer. These RNAi trigger standards were either 23-mer/21-mer (sense strand/antisense strand) duplexes or 20-mer/21-mer duplexes with an inverted dT on the sense strand 3΄ terminus. SDS was precipitated from the standards and samples by adding 10 μl of 3M KCl to 100 μl of the tissue sample solution. After incubating 10 min on ice, the samples were centrifuged for 15 min at 2700 × g. Quantitation of RNAi triggers was performed using the supernatant (33). Sequence-specific PNA probes were generated for RNAi triggers AD0009, AD0010 and miR122 (Panagene, Daejeon, Korea) that bind to the antisense strand of each RNAi trigger. These contain the fluorescent Atto-425 label at the N-terminus, separated from the PNA chain by a diethylene glycol linker. One PNA probe was added to each supernatant sample. Hybridization of the probe to the antisense strand was performed in 96-well conical-bottom plates. Plates were sealed and incubated at 95°C for 15 min in a thermal cycler. The temperature of the thermal cycler was gradually reduced over 15 min to a final temperature of 54°C. Samples were kept at 4°C until they were loaded onto an HPLC autosampler for analysis. HPLC analysis was carried out using a Shimadzu HPLC system equipped with an LC-20AT pump, an SIL-20AC autosampler, an RF-10Axl fluorescence detector and a CTO-20Ac column oven (Shimadzu Scientific Instruments, Columbia, MD, USA). The 96-well plate from the hybridization step was loaded onto the autosampler. One hundred microliter samples were injected onto a DNAPac PA-100 4 × 250 mm analytical column (Fisher Scientific, Pittsburgh, PA, USA) with an attached 4 × 50 mm guard column (Fisher Scientific, Pittsburgh, PA, USA). Analysis was carried out at a flow rate of 1 ml/min with a column oven temperature of 50°C. A gradient elution using mobile phase A (10 mM Tris-HCl pH 7, 100 mM NaCl, 30% acetonitrile) and mobile phase B (10 mM Tris-HCl pH 7, 900 mM NaCl, 30% acetonitrile) was used. Fluorescence detection was set to 436 nm excitation and 484 nm emission with a medium gain setting of 4. Concentrations of analytes eluted in the relevant range were calculated using a multi-point external standard calibration curve. Calibration curves were generated with PNA-hybridized full-length non-phosphorylated RNAi triggers (AD0009 and AD0010), and 5΄-phosphorylated RNAi triggers (RD10074 and RD10077, corresponding to AD0009 and AD0010, respectively).
Immunoisolation of RISC
AGO2-containing RISC was immunoisolated from liver lysates to evaluate RNAi triggers specifically loaded into RISC. Pre-weighed mouse liver sections (∼25 mg) were homogenized using a Precellys Evolution Homogenizer (Bertin Technologies, Saint-Quentin en Yveline, France) in 1 ml Cell Lysis Solution (Wako Chemicals USA, Richmond, VA, USA) supplemented with Complete Ultra Tablets containing protease inhibitors (Roche, Indianapolis, IN) at 4°C and centrifuged at 12 000 × g for 10 min. Separately, anti-mouse AGO2 antibody beads (Wako Chemicals USA, Richmond, VA, USA) were washed 2 times in the Cell Lysis Solution at 4°C and combined with the supernatant of the liver tissue lysate. The beads/liver lysate mixture was incubated overnight at 4°C with gentle mixing. The beads were washed 3 times with 1 ml Cell Lysis Solution and elution was performed by incubation of beads in 100 μl of Affymetrix Lysis Buffer (Affymetrix, Santa Clara, CA, USA).
HBsAg ELISA
HBsAg levels in serum were determined with an HBsAg EIA 3.0 kit (Bio-Rad Laboratories, Inc., Redmond, WA, USA) as described by the manufacturer. Sera were diluted 10 to 2000-fold in 5% nonfat dry milk prepared in phosphate buffered saline (PBS). Secondary HBsAg standards were prepared from serum of ICR mice (Harlan Laboratories, Indianapolis, IN, USA) that had been given a hydrodynamic tail vein injection (9) with 10 μg HBsAg-expressing plasmid pRc/CMV-HBs (Aldevron, Fargo, ND, USA). The serum with secondary standards was also diluted in the non-fat milk solution, as undiluted serum causes nonspecific binding to the ELISA plate. Serum was isolated from the mice 24 h after plasmid injection and frozen in aliquots. The level of HBsAg expression in the serum was determined relative to a standard curve using recombinant ayw subtype HBsAg protein (Aldevron, Fargo, ND, USA) diluted in nonfat milk in PBS. The lower limit of quantitation (LLOQ) for the ELISA was 0.08 ng/ml.
RESULTS
PNA method development and characterization
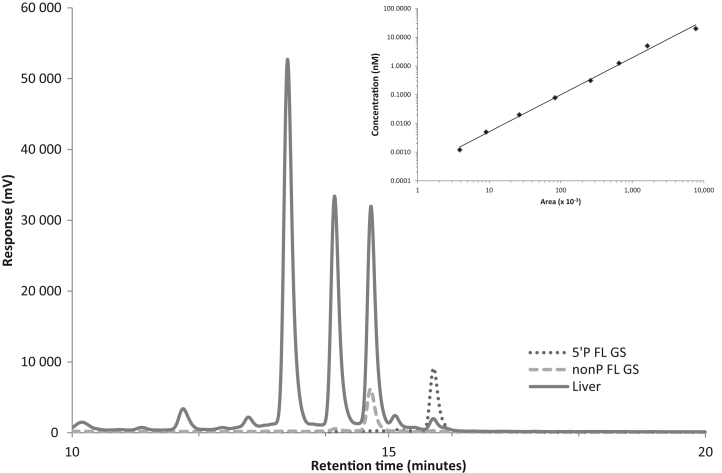
The PNA assay is based on sequence-specific hybridization of a fluorescently-tagged single stranded neutrally charged PNA probe to the anionic guide strand of the target RNAi trigger, followed by AEX-HPLC that separates free uncharged PNA probe from those duplexed to guide strands of the RNAi trigger. We first determined the LLOQ of the assay by hybridizing an Atto-425 fluorescent HBV-specific AD0009-derived guide strand PNA probe to the RNAi duplex AD0009, followed by AEX-HPLC to measure the amount of guide strands in the assay. Under the conditions used, we found the PNA assay to exhibit a dynamic range of >3 orders of magnitude with an LLOQ of 0.001 nM (Figure 1A, inset). Similar results were obtained with a PNA probe for the HBV-specific RNAi duplex AD0010 (Supplementary Figure S1).
Figure 1.
Qualitative analysis of ARC-520 RNAi trigger guide strands (AD0009) in mouse liver. Mice were treated with 8 mg/kg ARC-520, then sacrificed after 1 h and livers harvested. Anion exchange high performance liquid chromatography (AEX-HPLC) separation traces of fluorescent PNA probe hybridization mixtures were generated for non-phosphorylated full length AD0009 standard (---), 5΄-phosphorylated full length AD0009 standard RD10074 (…) and mouse liver lysate (__), and then overlaid. mV, millivolts. Inset: standard PNA assay curve for AD0009. Atto-425 fluorescently tagged PNA probe specific for HBV sequence AD0009 was hybridized to purified AD0009 duplex at increasing concentrations and evaluated by AEX-HPLC. Area under the curve for each sample was calculated by LabSolutions software (Shimadzu).
Studies suggest that RNAi triggers require a 5΄-phosphate on the guide strand for RISC loading and silencing activity (34–36). An endogenous RNA kinase present in the cell cytoplasm, known as CLP1, catalyzes the phosphorylation of 5΄-hydroxyl ends of endogenous or synthetic RNAi triggers (32). As AD0009 and AD0010 are synthesized with a 5΄-hydroxyl, the presence of a 5΄-phosphate would act as a molecular marker of cytoplasmic localization. To determine if the PNA assay distinguished between non-phosphorylated and 5΄-phosphorylated forms of the RNAi trigger guide strands, standard duplexes were generated that were either non-phosphorylated (AD0009 and AD0010) or 5΄-phosphorlyated (RD10074 and RD10077) and then subjected to the PNA assay. The results indicated that non-phosphorylated and 5΄-phosphorylated guide strands (AD0009 and RD10074) were distinguishable with baseline separation in the assay (Figure 1).
To evaluate if the assay could be used to detect RNAi trigger strands present in the livers of mice, we administered 8 mg/kg ARC-520 into transiently transgenic NOD-SCID mice expressing the entire HBV genome as previously described (9). Mice were sacrificed after 1 h, their livers harvested and liver lysates were generated for evaluation in the PNA assay. The HPLC profile indicated the presence of the full-length (FL) non-phosphorylated RNAi guide strands, FL 5΄-phosphorylated guide strands and unidentified trigger metabolites eluting with shorter retention times (Figure 1). Further, we employed an immunoisolation assay with antibodies to the AGO2 endonuclease of RISC (17,21–23) to evaluate the amount of FL guide strands associated with RISC. The results indicated that only 5΄-phosphorylated guide strands associated with RISC as demonstrated by the similar retention time to the 5΄-phosphorylated FL guide strand standard (Supplementary Figure S2). Addition of a 3΄-phosphate to the AD0009 guide strand standard (AD03550) resulted in a shorter retention time, indicating that the guide strand species in RISC was phosphorylated at its 5΄ terminus (Supplementary Figure S2). By 1 h after treatment a fraction of the guide strands had escaped from endosomes into the cytoplasm and subsequently were phosphorylated at their 5΄ ends. These data indicate that the PNA assay detects RNAi guide strands in biological matrices and distinguishes various metabolites.
Biodistribution of ARC-520 RNAi trigger guide strands in mouse tissues
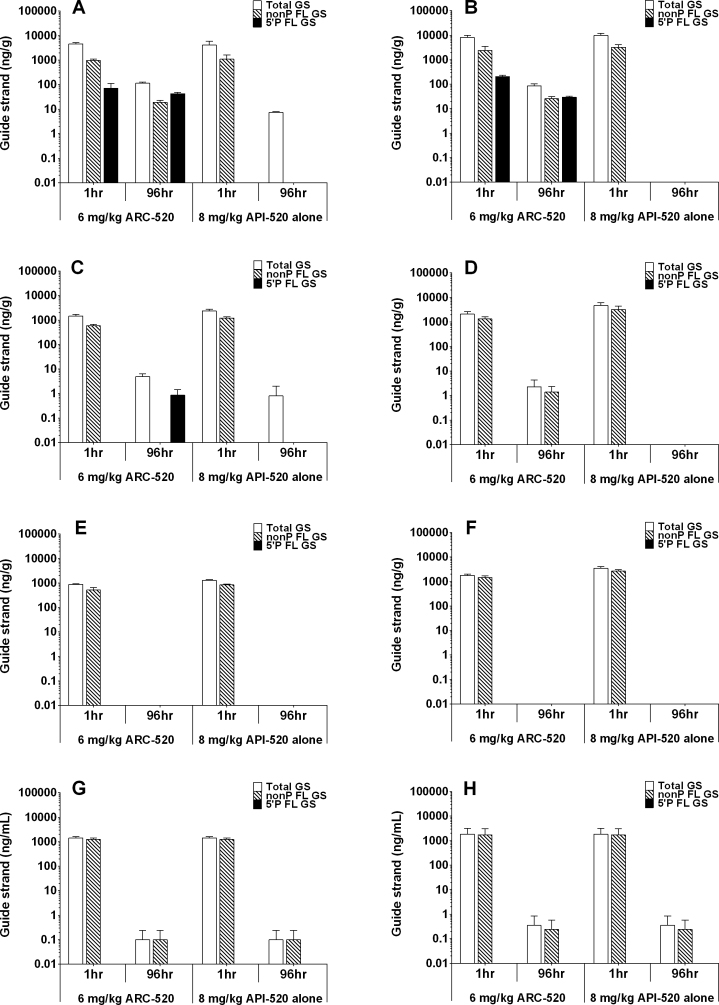
Employing the PNA assay, we evaluated the biodistribution of ARC-520 RNAi triggers in mice. Combining the AEX-HPLC peak area quantitation via the standard curve and known amounts of tissue evaluated, we aimed to quantitate levels of guide strands and a subset of their metabolites per defined amount of animal tissues or plasma. Livers, spleens, kidneys and plasma were collected from mice either 1 h or 96 h after treatment with either 6 mg/kg ARC-520 (AD0009 + AD0010 with ARC-EX1) or 8 mg/kg of API-520 alone (AD0009 + AD0010 without ARC-EX1). For both AD0009 and AD0010, the amount of total GS including metabolites was several-fold greater in liver than in the other tissues or plasma at 1 h post-injection (Figure 2), whether or not the RNAi triggers were injected alone (API-520) or with ARC-EX1 (ARC-520). The increased level of total guide strands observed in liver may be due to significant uptake and metabolism via lipoprotein association and internalization (29), while the amount detected in other tissues may have a greater proportion of non-internalized RNAi triggers. ARC-EX1 does not appear to affect these processes as the RNAi trigger biodistribution is similar with and without ARC-EX1. In addition, although appreciable amounts of full-length guide strands could be detected in the liver, spleen and kidney, most of the total guide strands detected was composed of metabolites, especially in the liver. These had shorter retention times suggesting that they were n-1, n-2 or smaller species likely generated by nucleases present in these organs (data not shown). This is in contrast to what was detected in the plasma, where most guide strands circulated as intact molecules. Guide strands possessing a 5΄-phosphate were detected in the liver only when delivered with ARC-EX1, consistent with our mechanism of active cytoplasmic delivery in the presence of ARC-EX1 with attendant 5΄-phosphorylation.
Figure 2.
Biodistribution of ARC-520 RNAi trigger guide strands in mouse tissues. Mice were administered 6 mg/kg ARC-520 or 8 mg/kg API-520, then at 1 h or 96 h post-administration mice were sacrificed and tissues collected. PNA assay was employed to evaluate non-phosphorylated full-length guide strands (non-P FL GS), 5΄-phosphorylated full-length guide strands (5΄ P FL GS) and the combination of non-P FL GS + 5΄ P FL GS + metabolites (Total GS). Quantitation of guide strands was based on a standard curve, and indicated as ng of guide strands per gram of tissue or plasma. (A) AD0009 in liver; (B) AD0010 in liver; (C) AD0009 in spleen; (D) AD0010 in spleen; (E) AD0009 in kidney; (F) AD0010 in kidney; (G) AD0009 in plasma; and (H) AD0010 in plasma.
By 96 h post-injection, the amounts of the AD0009 and AD0010 guide strands were significantly reduced in all organs examined and plasma compared to the 1 h time point. This may be due to guide strand uptake and subsequent degradation by nucleases present in the tissues. The reduction in guide strand content was greatest in mice receiving API-520 alone without ARC-EX1, suggesting that ARC-EX1 prevented or inhibited degradation of a small percentage of the guide strands. The total amount of guide strands (including all FL forms and metabolites) observed in liver exceeded that observed in the other tissues.
Additional evidence that ARC-EX1 may impact the metabolism of RNAi triggers is evidenced by the observation that only mice receiving ARC-520 had detectable levels of 5΄-phosphorylated FL guide strands of both AD0009 and AD0010 in their liver. These may represent guide strands that have been released from the endosome by the action of ARC-EX1. A minor amount of 5΄-phosphorylated AD0009 guide strands was observed in spleen after 96 h (Figure 2C), while no 5΄-phosphorylated AD0010 guide strands were detected (Figure 2D). The presence of 5΄-phosphorylated AD0009 guide strands in spleen tissue may result from lipoprotein binding and uptake through lipoprotein receptors, accompanied by limited endosomal release (29). The observation that AD0009 (but not AD0010) was detected in spleen may be due to inherent resistance of AD0009 to nuclease digestion. This is evidenced by the survival of AD0009 and not AD0010 at 96 h in mouse liver after dosing with API-520 alone (Figure 2A and B). No 5΄-phosphorylated guide strands accumulated in plasma or kidneys, despite the presence of full-length non-phosphorylated guide strands, suggesting that the RNAi triggers present in the kidney were unable to localize cytoplasmically. In addition, the absence of 5΄-phosphorylated species in plasma suggests that RNAi trigger-specific kinases are absent in this compartment.
Kinetics of RNAi trigger guide strand 5΄-phosphorylation
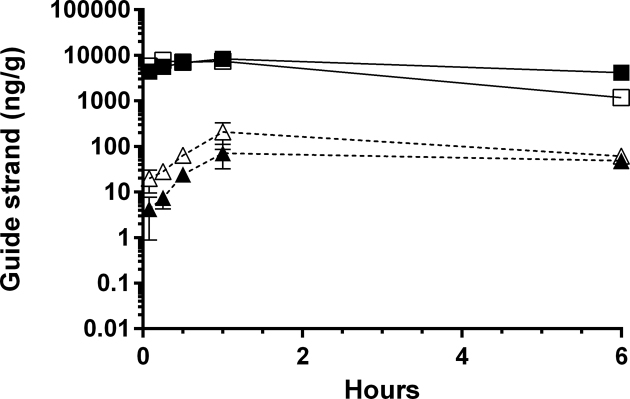
To evaluate early kinetics and quantitation of HBV-specific RNAi triggers in ARC-520 treated mice, multiple liver samples were collected over a 6 h time course. Guide strands containing a 5΄-phosphate were detected within the first 5 min after ARC-520 treatment, reaching maximum accumulation after 1 h. Quantitation of the PNA assay peaks indicated that the amount of non-phosphorylated RNAi triggers in liver was more than two orders of magnitude greater than that of the 5΄-phosphorylated RNAi triggers (Figure 3). This relative amount of 5΄-phosphorylation is consistent with that observed following RNAi trigger delivery by lipid nanoparticles and using RISC immunoisolation and PCR-based technologies (37). Within 1 h post-injection, ∼1% of the total guide strands present in the liver was 5΄-phosphorylated, indicative of rapid RNAi trigger escape from endosomes using ARC-520.
Figure 3.
Kinetics of ARC-520 RNAi trigger guide strand accumulation in mouse liver. Mice were injected with 8 mg/kg ARC-520 and sacrificed at multiple time-points for evaluation of HBV guide strands in liver by PNA assay. Quantitation of guide strands was based on a standard curve, and indicated as ng of guide strands per gram of liver tissue. Guide strands quantitated were AD0009 non-phosphorylated FL (▪), AD0010 non-phosphorylated FL (■), AD0009 5΄-phosphorylated FL (▴) and AD0010 5΄-phosphorylated FL (Δ).
HBsAg knockdown kinetics and RNAi trigger guide strand decay
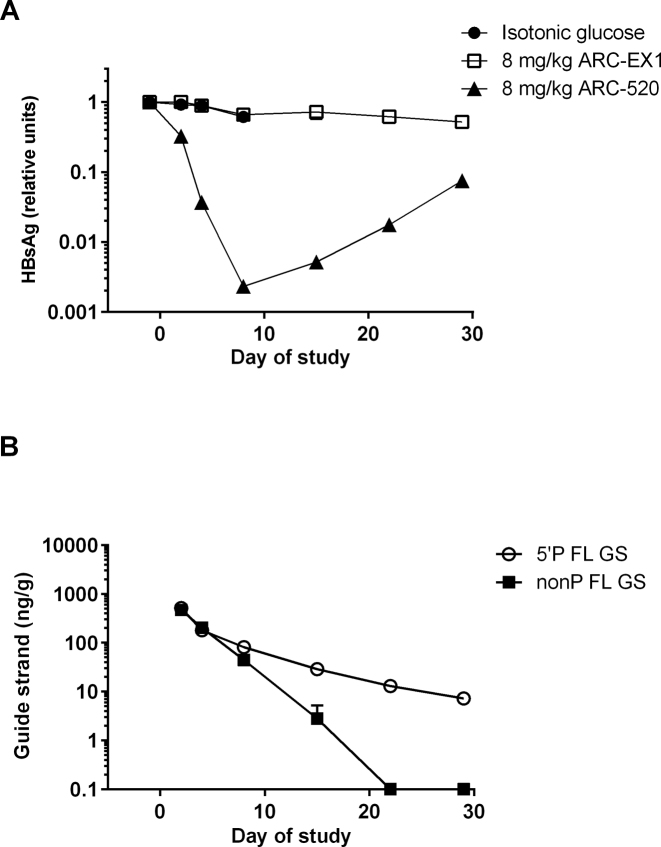
To evaluate long-term kinetics of HBV gene knockdown as a function of guide strand status, transiently transgenic NOD-SCID mice expressing the entire human HBV genome in the liver (9) were treated with ARC-520. A control group treated with ARC-EX1 alone was included to determine whether the MLP influenced the stability of the liver-expressed HBV genome over time. Serum HBsAg levels were reduced by >2 log10 by ARC-520, but were not reduced by ARC-EX1 alone (Figure 4A). After 29 days, serum HBsAg levels remained diminished by >90%. In liver, the amount of 5΄-phosphorylated FL guide strands exhibited a slow rate of decay, consistent with the long duration of HBsAg knockdown. In contrast, non-phosphorylated FL guide strands decayed rapidly to below the level of quantitation (0.1 ng/g) by 21 days after treatment (Figure 4B). The level of total guide strands was diminished by 1 log10 on Day 2 (Figure 4B) relative to the amount observed at 6 h (Figure 3). This reduction was likely due to degradation of the guide strands remaining in endosomes by cellular nucleases. However, the relative amount of 5΄-phosphorylated guide strands was similar between the two experiments, suggesting that the guide strands present in the cytoplasm were exposed to less nuclease activity. Despite the rapid induction of 5΄-phosphorylated guide strands, the reduction in serum HBsAg exhibited a lag period with its nadir at Day 8 after ARC-520 treatment. This is likely due to the time it takes pre-existing HBsAg to be cleared from serum, which the data indicated is significantly longer than the time required for 5΄-phosphorylated guide strands to induce RNAi and degrade mRNA.
Figure 4.
HBV surface antigen (HBsAg) knockdown kinetics and RNAi trigger guide strand decay in mouse liver. Isotonic glucose, ARC-520 or ARC-EX1 alone was administered as a single dose at 8 mg/kg to mice. (A) Serum levels of HBsAg were evaluated by ELISA. (B) Mice were sacrificed at multiple time points and livers evaluated by PNA assay for non-phosphorylated full-length guide strands (non-P FL GS) or 5΄-phosphorylated full-length guide strands (5΄P FL GS). Quantitation of guide strands was based on a standard curve, and indicated as ng of guide strands per gram of liver tissue. LLOQ, lower limit of quantitation (0.1 ng/g).
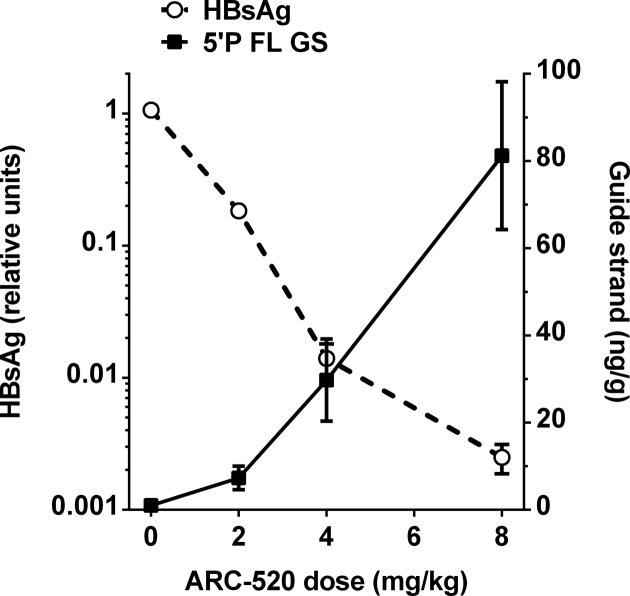
To evaluate a potential PK/PD relationship between the degree of serum HBsAg suppression and the amount of accumulating 5΄-phosphorylated guide strands in liver, a dose-response study was established in NOD-SCID mice expressing the human HBV genome in the liver. Mice were treated with isotonic glucose or different doses of ARC-520 (2–8 mg/kg), then serum and livers were harvested on day 8 post-treatment. An inverse correlation between HBsAg levels in serum and 5΄-phosphorylated FL guide strands (AD0009 + AD0010) in liver was observed, as expected for a specific molecule responsible for mRNA targeting (Figure 5). Maximal inhibition of HBsAg (>99%) was detected when the level of 5΄-phosphorylated guide strands reached its apex. The relationship between dose and 5΄-phosphorylated guide strands is nearly linear in the treatment phase of the study for doses ranging from 2–8 mg/kg. In contrast, non-linearity of HBsAg knockdown relative to the dose was observed, but is consistent with observations by others for different RNAi targets using alternate delivery systems (37,38).
Figure 5.
Dose-dependent correlation between HBsAg levels and 5΄-phosphorylated RNAi trigger guide strands. Mice were administered isotonic glucose or a single dose of 2–8 mg/kg ARC-520. On day 8, HBsAg levels were evaluated in serum and AD0009 + AD0010 5΄-phosphorylated FL guide strands were assessed in liver. Quantitation of guide strands was based on a standard curve, and indicated as ng of guide strands per gram of liver tissue.
RISC harbors 5΄-phosphorylated HBV-specific guide strands
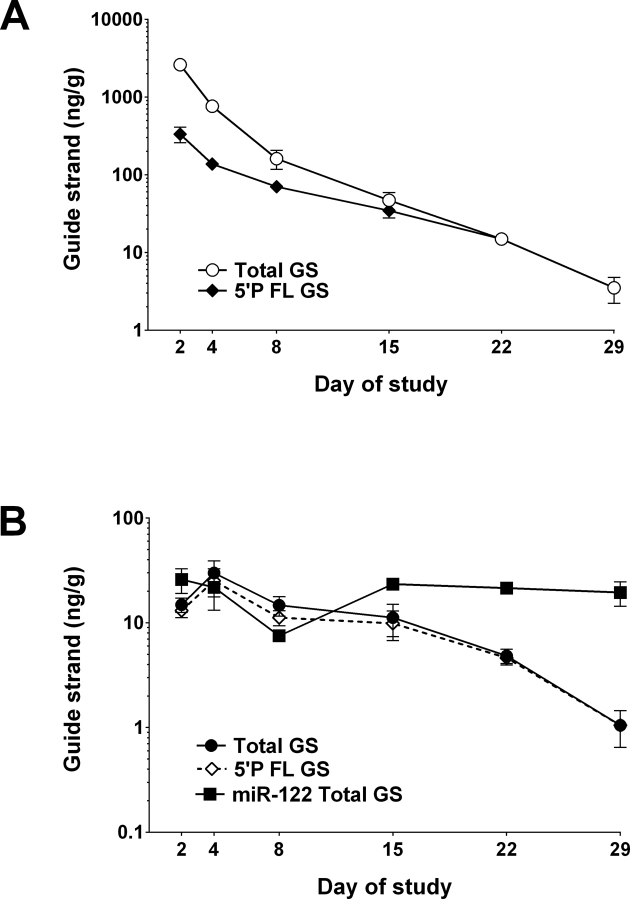
RISC is a cytoplasmic multi-component ribonucleoprotein complex that interacts with RNAi triggers leading to endonucleolytic cleavage of mRNA, thereby promoting gene silencing (17–24). We employed an immunoisolation assay with antibodies to the AGO2 endonuclease of RISC (17,21–23) to evaluate the amount of FL guide strands loaded into RISC. In a study similar to that described in Figure 4, liver samples were harvested for analysis in the PNA assay before and after immunoisolation of AGO2. The amount of 5΄-phosphorylated guide strands (AD0009) detected in RISC on Day 8 (maximal HBsAg knockdown) was 16% of the total level of 5΄-phosphorylated guide strands detected in liver lysates (Figure 6). A similar amount of 5΄-phosphorylated AD0010 guide strands was observed in RISC (data not shown). To correct for immunoisolation variance between samples, PNA evaluation of miR122 (a stably expressed miRNA abundant in mouse liver) (37,39) was undertaken for each immunoisolation sample and was relatively invariant (Figure 6B). The data indicate that the rate of decay for AD0009 in RISC was similar to that observed for total 5΄-phosphorylated guide strands (Figure 6B).
Figure 6.
RISC association and decay kinetics of HBV RNAi guide strands. ARC-520 (8 mg/kg) was administered as a single dose to mice, and AD0009 total full-length guide strands and 5΄-phosphorylated full-length guide strands were evaluated by PNA assay in liver at multiple time points over 29 days. Argonaute 2 (AGO2) was immunoisolated from liver lysates, and both AD0009 and miR122 were evaluated by PNA assay (indicated as ng of guide strands per gram of liver tissue). (A) Liver lysate comparing AD0009 total guide strands (combined non-phosphorylated, 5΄-phosphorylated and metabolites) and AD0009 5΄-phosphorylated FL guide strands. (B) AGO2 immunoisolation comparing AD0009 total guide strands (combined non-phosphorylated, 5΄-phosphorylated and metabolites), AD0009 5΄-phosphorylated FL guide strands alone and total miR122 (control).
DISCUSSION
The PNA-based system (40) described in this study is a quantitative method to evaluate the level of RNAi trigger guide strands in tissues and plasma, and the first described assay that differentiates between full-length non-phosphorylated and 5΄-phosphorylated guide strand forms. The method also detected metabolites of the guide strands that are generated by ribonuclease activity in plasma or following cell uptake of RNAi triggers. Overall, the assay enables: (i) tracking the biodistribution of RNAi triggers in vivo, (ii) assessment of 5΄-phosphorylation induction kinetics, (iii) evaluation of the decay of the guide strands relative to the non-phosphorylated forms and metabolites, (iv) identification of PK/PD relationships between gene knockdown and 5΄-phosphorylation of RNAi triggers and (v) quantitation of guide strands loaded into RISC.
In mouse studies reported here, the uptake, biodistribution and processing of HBV-specific RNAi triggers conjugated with cholesterol were evaluated. The generation and in vivo delivery of RNAi triggers, including cholesterol-conjugated versions were previously reported (9,10,27–29,41,42), and the triggers were shown to distribute predominantly to liver in mice (8,9,29,30). Additionally, minor levels of cholesterol-RNAi triggers were reported in spleen and kidney (29). However, the use of radiolabels, PCR-based methods or ELISA-based assays to track the presence of RNAi trigger guide strands in these tissues does not differentiate between non-phosphorylated and 5΄-phosphorylated forms (29,43–48). Based upon immunoisolation of the AGO2 endonuclease in RISC and subsequent RT-PCR evaluation of the associated guide strands, one could invoke that the immunoisolated guide strands were 5΄-phosphorylated (37). The mechanism of cell uptake for cholesterol-trigger conjugates involves lipoprotein particles, lipoprotein receptors and possibly other transmembrane proteins (29). This mechanism complements the uptake of the MLP into liver through binding of the N-acetylgalactosamine ligand to the C-type lectin ASGPR on hepatocytes (8,9,26,27). The liver uptake of RNAi triggers and ARC-EX1 via different receptors reduces the likelihood of receptor occupancy competition, and may explain the enhanced uptake observed in liver.
The rapid accumulation of 5΄-phosphorylated guide strands observed in this study supports the premise that the ARC-EX1 excipient promotes endosomal escape of the RNAi triggers. The onset of guide strand 5΄-phosphorylation by CLP1 (32) within 5 min after intravenous administration of ARC-520 preceded the rapid gene knockdown observed. Studies employing lipid nanoparticles (LNP) have demonstrated that guide strands were loaded into RISC at least 30 min after LNP/RNAi trigger injection in mice (37). A clear inverse correlation between hepatitis B viral antigen levels in mouse serum and 5΄-phosphorylated guide strands was observed in this study, establishing a PK/PD relationship between drug exposure and gene silencing. The dose proportional effect of 5΄-phosphorylation of guide strands relative to the degree of serum HBsAg knockdown suggests that cytoplasmic RISC remains unsaturated in the dose range evaluated. Further, the slow decay of full-length 5΄-phosphorylated HBV-specific guide strands matched the sustained knockdown of serum HBsAg after a single dose of ARC-520. The decay rate of RISC-associated HBV-specific 5΄-phosphorylated guide strands was similar to that of the total pool of guide strands regardless of their phosphorylation status. In contrast, the amount of 5΄-phosphorylated guide strands of an endogenous continually-expressed miRNA (miR122) in RISC was stable.
The PNA method enables exquisite quantitation of unlabeled non-phosphorylated RNAi triggers delivered in vivo. Biodistribution of ARC-520 guide strands predominantly to liver tissue correlated with both ASGPR targeting mechanisms of ARC-EX1 and cholesterol-conjugated RNAi triggers. In the absence of ARC-EX1, significant delivery or tissue association of non-phosphorylated guide strands was observed only at an early time point, and was unproductive in that no 5΄-phosphorylation occurred. The addition of ARC-EX1 confirmed our earlier observations that knockdown of serum HBsAg required the presence of ARC-EX1 co-injection with the cholesterol-conjugated RNAi triggers (9,27). Evaluation of guide strand accumulation kinetics and their decay provided a platform to understand the rate at which these molecules are trafficked through endosomes/lysosomes to the cytoplasm where RISC resides. Our observations suggest that uptake and endosomal release occurs within minutes after drug administration, but incorporation and residence of the HBV-specific RNAi triggers in RISC is sustained for weeks. Additionally, the robustness of this technique allows for potential characterization of RNAi trigger metabolites, an effort that may facilitate drug development regulatory requirements.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Julia Hegge and the LAR team for animal studies, Qili Chu for HBV assays, Holly Hamilton for data analysis, Lauren Almeida and the RNAi synthesis team for triggers, Eric Lund for mass spectrometry and Ingo Roehl and team at Axolabs GmbH for initial assay development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Arrowhead Pharmaceuticals, Inc. Funding for open access charge: Arrowhead Pharmaceuticals, Inc.
Conflict of interest statement. All authors are employees and shareholders of Arrowhead Pharmaceuticals, Inc.
REFERENCES
- 1.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 2.Hannon G.J. RNA interference. Nature. 2002; 418:244–251. [DOI] [PubMed] [Google Scholar]
- 3.McManus M.T., Sharp P.A.. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002; 3:737–747. [DOI] [PubMed] [Google Scholar]
- 4.Carthew R.W., Sontheimer E.J.. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbashir S.M., Lendeckel W., Tuschl T.. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001; 15:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meister G., Tuschl T.. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004; 431:343–349. [DOI] [PubMed] [Google Scholar]
- 7.Wilson R.C., Doudna J.A.. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013; 42:217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gish R.G., Yuen M., Chan H.L.Y., Given B.D., Lai C., Locarnini S.A., Lau J.Y.N., Wooddell C.I., Schluep T., Lewis D.L.. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015; 121:97–108. [DOI] [PubMed] [Google Scholar]
- 9.Wooddell C.I., Rozema D.B., Hossbach M., John M., Hamilton H.L., Chu Q., Hegge J.O., Klein J.J., Wakefield D.H., Oropeza C.E. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 2013; 21:973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama T., Butler J.S., Sehgal A., Severgnini M., Racie T., Sharman J., Ding F., Morskaya S.S., Brodsky J., Tchangov L. et al. Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Mol. Ther. 2012; 20:1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S.M., Geick A., Hadwiger P., Harborth J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004; 432:173–178. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005; 23:1002–1007. [DOI] [PubMed] [Google Scholar]
- 13.Kim D.H., Rossi J.J.. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007; 8:173–184. [DOI] [PubMed] [Google Scholar]
- 14.Dorsett Y., Tuschl T.. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004; 3:318–329. [DOI] [PubMed] [Google Scholar]
- 15.De Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J.. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007; 6:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel'in A.V., Milstein S. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 17.Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D.. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005; 123:607–620. [DOI] [PubMed] [Google Scholar]
- 18.Leuschner P.J., Ameres S.L., Kueng S., Martinez J.. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006; 7:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Tan H., Tian H., Liang C., Chen S., Liu Q.. Autoantigen La promotes efficient RNAi, antiviral response, and transposon silencing by facilitating multiple-turnover RISC catalysis. Mol. Cell. 2011; 44:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T.. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004; 15:185–197. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C.. Slicer function of Drosophila argonautes and its involvement in RISC formation. Genes Dev. 2005; 19:2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rand T.A., Petersen S., Du F., Wang X.. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005; 123:621–629. [DOI] [PubMed] [Google Scholar]
- 24.Ye X., Huang N., Liu Y., Paroo Z., Huerta C., Li P., Chen S., Liu Q., Zhang H.. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 2011; 18:650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haley B., Zamore P.D.. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004; 11:599–606. [DOI] [PubMed] [Google Scholar]
- 26.Rozema D.B., Lewis D.L., Wakefield D.H., Wong S.C., Klein J.J., Roesch P.L., Bertin S.L., Reppen T.W., Chu Q., Blokhin A.V. et al. Dynamic Polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong S.C., Klein J.J., Hamilton H.L., Chu Q., Frey C.L., Trubetskoy V.S., Hegge J., Wakefield D., Rozema D.B., Lewis D.L.. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic Acid Ther. 2012; 22:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raouane M., Desmaele D., Urbinati G., Massaad-Massade L., Couvreur P.. Lipid conjugated oligonucleotides: A useful strategy for delivery. Bioconjugate Chem. 2012; 23:1091–1104. [DOI] [PubMed] [Google Scholar]
- 29.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007; 25:1149–1157. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz C., Hadwiger P., John M., Vornlocher H.P., Unverzagt C.. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004; 14:4975–4977. [DOI] [PubMed] [Google Scholar]
- 31.Gish R.G., Given B.G., Lai C., Locarnini S.A., Lau J.Y.N., Lewis D.L., Schluep T.. Chronic hepatitis B: Virology, natural history, current management and a glimpse at future opportunities. Antiviral Res. 2015; 121:47–58. [DOI] [PubMed] [Google Scholar]
- 32.Weitzer S., Martinez J.. The human RNA kinase hClp1 is active on 3΄ transfer RNA exons and short interfering RNAs. Nature. 2007; 447:222–226. [DOI] [PubMed] [Google Scholar]
- 33.Martínez T., González M.V., Roehl I., Wright N., Pañeda C., Jiménez A.I.. In vitro and in vivo efficacy of SYL040012, a novel siRNA compound for treatment of glaucoma. Mol. Ther. 2014; 22:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker J.S., Roe S.M., Barford D.. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005; 434:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J.. Structural basis for 5΄-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005; 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lima W.F., Wu H., Nichols J.G., Sun H., Murray H.M., Crooke S.T.. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009; 284:26017–26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pei Y., Hancock P.J., Zhang H., Bartz R., Cherrin C., Innocent N., Pomerantz C.J., Seitzer J., Koser M.L., Abrams M.T. et al. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010; 16:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann T.S., Lee A.C.H., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M. et al. RNAi-mediated gene silencing in non-human primates. Nature. 441:111–114. [DOI] [PubMed] [Google Scholar]
- 39.Chang J., Nicolas E., Marks D., Sander A., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. et al. miR-122, a mammalian liver-specific microRNA, is processed for hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen P.E. Peptide nucleic acids (PNA) in chemical biology and drug discovery. Chem. Biodivers. 2010; 7:786–804. [DOI] [PubMed] [Google Scholar]
- 41.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016; 44:6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg M.S., Morris K.V.. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016; 44:6505–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landesman Y., Svzikapa N., Cognetta A., Zhang X., Bettencourt B.R., Kuchimanchi S., Dufault K., Shaikh S., Gioia M., Akinc A. et al. In vivo quantification of formulated and chemically modified small interfering RNA by heating-in-Triton quantitative reverse transcription polymerase chain reaction (HIT qRT-PCR). Silence. 2010; 1:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng A., Li M., Liang Y., Wang Y., Wong L., Chen C., Vlassov A.V., Magdaleno S.. Stem-loop RT-PCR quantification of siRNAs in vitro and in vivo. Oligonucleotides. 2009; 2:203–208. [DOI] [PubMed] [Google Scholar]
- 45.Kim E., Park T.G., Oh Y., Shim C.. Assessment of siRNA pharmacokinetics using ELISA-based quantification. J. Controlled Release. 2010; 143:80–87. [DOI] [PubMed] [Google Scholar]
- 46.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi B., Keough E., Matter A., Leander K., Young S., Carlini E., Sachs A.B., Tao W., Abrams M., Howell B. et al. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. 2011; 59:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y., Ou M., Keough E., Roberts J., Koeplinger K., Lyman M., Fauty S., Carlini E., Stern M., Zhang R. et al. Quantitation of physiological and biochemical barriers to siRNA liver delivery via lipid nanoparticle platform. Mol. Pharm. 2014; 11:1424–1434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.