Abstract
MiRNAs post-transcriptionally regulate gene expression by recruiting the miRNA-induced silencing complex (miRISC) to target mRNAs. However, the mechanisms by which miRISC components are maintained at appropriate levels for proper function are largely unknown. Here, we demonstrate that Caenorhabditis elegans TEG-1 regulates the stability of two miRISC effectors, VIG-1 and ALG-1, which in turn affects the abundance of miRNAs in various families. We demonstrate that TEG-1 physically interacts with VIG-1, and complexes with mature let-7 miRNA. Also, loss of teg-1 in vivo phenocopies heterochronic defects observed in let-7 mutants, suggesting the association of TEG-1 with miRISC is necessary for let-7 to function properly during development. Loss of TEG-1 function also affects the abundance and function of other microRNAs, suggesting that TEG-1's role is not specific to let-7. We further demonstrate that the human orthologs of TEG-1, VIG-1 and ALG-1 (CD2BP2, SERBP1/PAI-RBP1 and AGO2) are found in a complex in HeLa cells, and knockdown of CD2BP2 results in reduced miRNA levels; therefore, TEG-1's role in affecting miRNA levels and function is likely conserved. Together, these data demonstrate that TEG-1 CD2BP2 stabilizes miRISC and mature miRNAs, maintaining them at levels necessary to properly regulate target gene expression.
INTRODUCTION
The identification of non-coding RNAs (ncRNAs) and the elucidation of their cellular functions have enhanced our understanding of post-transcriptional gene regulation. The microRNA (miRNA) family of ncRNAs consists of 21–24 nucleotide-long RNAs that bind to complimentary sequences in messenger RNAs (mRNAs) and regulate their expression. MiRNAs were first identified in Caenorhabditis elegans, with lin-4 and let-7 being the first miRNAs described to coordinate temporal developmental events (1,2). Since then, miRNAs have been identified in most plants and animals studied (3), and are predicted to regulate over half of human genes (4). Additionally, mis-regulation of miRNAs has been implicated in many human diseases, including neuro-degeneration, cardiovascular disease and cancer (5).
MiRNAs repress the expression of target mRNAs through the miRNA-induced silencing complex (miRISC). MiRNAs provide the sequence specificity for targeting, while proteins in miRISC provide the mRNA silencing activity, with an Argonaute protein being a core component of the miRISC. Loss of the Caenorhabditis elegans Argonaute proteins ALG-1 and ALG-2 results in embryonic lethality, emphasizing the importance of miRNA mediated gene regulation in development (6). Several effectors, such as AIN-1, AIN-2, CGH-1, NHL-2 and VIG-1, have also been identified in association with miRISC in C. elegans (7–11). VIG-1 is a C. elegans ortholog of Drosophila Vasa Intronic Gene and binds to miRISC (8,9). Reduction of VIG-1 levels suppresses let-7 expression in adult animals (9). The VIG-1 protein is detected in many tissues, including the pharynx, body wall muscles, hypodermis, reproductive system, and nervous system, throughout the larval and adult stages of C. elegans (12). We previously reported the involvement of the teg-1 (tumorous enhancer of glp-1(gf)) gene in regulating stem cell proliferation in the C. elegans germ line (13). Here, we report the identification of TEG-1 CD2BP2 as a conserved effector of miRISC in C. elegans and human cells that maintains miRISC proteins at appropriate levels, which in turn affects the abundance of diverse miRNAs families. teg-1 mutants display developmental timing phenotypes and mis-expression of uterine COG-1, an Nkx6 homeodomain transcription factor that regulates vulval differentiation. Moreover, we found that TEG-1 physically interacts with VIG-1 and is in complex with miRISC. We also found that the levels of both the VIG-1 and ALG-1 proteins, but not their mRNAs, are reduced in teg-1 mutants, suggesting that TEG-1 regulates miRISC stability, which in turn controls miRNA abundance. Furthermore, a complex containing CD2BP2 (human TEG-1 ortholog), SERBP1/PAI-RBP1 (human VIG-1 ortholog), and AGO2 (human ALG-1 ortholog) is detected in HeLa cells, and knockdown of CD2BP2 reduces the levels of let-7a, miR-24 and miR-26a, suggesting the association and function between TEG-1 CD2BP2 and the miRISC is conserved from nematodes to humans.
MATERIALS AND METHODS
C. elegans growth conditions and strains
Bristol (N2) C. elegans maintenance and manipulation was performed as previously described (14). The following alleles and markers were used in this study - Linkage Group II (LGII): vig-1(tm3383); LGIII: teg-1(oz189), teg-1(oz230); LGIV: wIS78[ajm-1::gfp; scm::gfp; unc-119(+); F58E10(+)], jcIS1[Pajm-1::gfp; unc-29(+)], PH7310 [otIs193 (cog-1p::lsy-6hp + rol-6(su1006)) syIs63]; LGX: alg-1(gk214), let-7(mg279). PS3662 [syIs63 (cog-1::gfp + unc-119(+))]. OH3646 [otIs114 (lim-6p::gfp + rol-6(su1006))I; lsy-6(ot150) V]. XB624 teg-1(oz230); jcIs1[ajm-1::gfp, unc-29(+), rol-6(su1006)]; ugEx24 was generated by co-injecting pDH122 (Pteg-1::TAP::teg-1) (13) (30 ng/μl), pCFJ90 (Pmyo-2::mCherry) (2.5 ng/μl), pGH8 (Prab-3::mCherry) (10 ng/μl), and pTG96 (sur-5::gfp) (30 ng/μl) into XB594[teg-1(oz230)/hT2g; jcIs1] animals. This array rescues the sterile phenotype of teg-1(oz230). RNAi was performed by feeding worms with Escherichia coli HT115(DE3) strain expressing the appropriate double-stranded RNAi constructs as previously described (15). Lactacystin (Cayman Chemicals) dissolved in DMSO, or DMSO alone, was added to N2 (wild-type) animals synchronized at L1 stage with M9 buffer (3 g/l KH2PO4, 6 g/l Na2HPO4, 5 g/l NaCl, 1 ml of 1 M MgSO4) containing E. coli OP50 bacteria and grown at 20°C for 2−3 days.
Microscopy, gonad dissections and indirect Immunofluorescence staining
Gonad dissection and antibody staining were performed as previously described (16). Briefly, dissected gonads were fixed with 3% paraformaldehyde for 10 min. followed by fixation/permeabilization with 100% methanol (−20°C) for >1 h. The gonads were blocked with 1% BSA for 1 h. Affinity purified rabbit anti-TEG-1 (N-terminal) antibodies were used at 1:500 dilution (13), rat anti-VIG-1 antibodies were used at 1:5000, and rabbit anti-UAF-2 antibodies were used at 1:1000 dilution (17). DNA was visualized by DAPI. DIC and fluorescence images were collected by a Zeiss Image Z1 microscope equipped with an Axiocam MRM digital camera.
Relative quantitative real-time PCR (qPCR) of mature miRNAs
Wild-type (N2) animals, homozygous teg-1(oz189) or teg-1(oz230) mutants at late L4-stage were collected using a COPAS Biosort according to manufacturer's instructions (Union Biometrica). The teg-1(oz189) and teg-1(oz230) alleles were balanced over hT2[bli-4(e937)let-?(q782) qIs48] (referred to as hT2g), allowing homozygous teg-1(oz189) and teg-1(oz230) animals to be isolated by sorting for non-GFP positive animals. Total RNA was extracted as previously described (18). 10 ng of total RNA was used to analyze the levels of mature miRNAs with TaqMan microRNA assays following the manufacturer's protocol (Applied Biosystems). The TaqMan probes used in this study are: let-7a (assay ID: 000377), lin-4 (assay ID: 000258), miR-24 (assay ID: 000402), miR-26a (assay ID: 000405), miR-48 (assay ID: 000208), miR-58 (assay ID: 000216), miR-61 (assay ID: 462167), miR-62 (assay ID: 000219), U18 (assay ID: 001764), and U48 (assay ID: 001006). Each qPCR reaction was performed in triplicate, and three biological replicates were performed using three independently isolated total RNA samples for each strain. The data were analyzed with 2−ΔΔCt analysis (19), and the samples obtained from C. elegans and human tissue culture cells were normalized with U18 and U48, respectively. Student's t-tests were performed to statistically compare the fold change of miRNA expression in mutants relative to wild-type (N2) expression.
Northern blotting
20 μg of each total RNA was separated on 15% SequaGel (National Diagnostics), and transferred onto a Genescreen Plus membrane (Perkin-Elmer). Northern blotting was performed as previously described (20,21). The following oligonucleotides were used as probes for Northern blotting:
| let-7 | 5΄-AACTATACAACCTACTACCTCA/Starfire/-3΄ |
| lin-4 | 5΄-TCACACTTGAGGTCTCAGGGA/Starfire/-3΄ |
| miR-58 | 5΄-ATTGCCGTACTGAACGATCTCA/Starfire.-3΄ |
| miR-62 | 5΄-CTGTAAGCTAGATTACATATCA/Starfire.-3΄ |
| tRNAGly | 5΄-GCTTGGAAGGCATCCATGCTGACCATT/Starfire/-3΄ |
Generation of anti-VIG-1 antibodies
A full-length cDNA encoding the VIG-1 protein was sub-cloned into pET28a vector at EcoRI and XhoI sites (pDH207) with oligonucleotides (V37: 5΄-AAAGAATTCATGAGCACTGAATACGGTTGCCAG-3΄ and V38: 5΄-AAACTCGAGTTACTTGGCACCGAGAGCTG-3΄) using PCR, and expressed in E. coli BL21(DE3). The fusion protein was purified with a Ni-NTA column (QIAGEN) according to the manufacturer's instructions, and subjected to SDS-PAGE. The gel pieces that contained the VIG-1 recombinant protein were excised and the protein was collected through gel electro-elution and used as the immunogen for two Sprague Dawley rats at the University of Calgary Animal Research Facility.
Immunoblotting
Immunoblotting was performed as previously described (13). The primary antibodies used were: (i) anti-TEG-1 (N-terminal) antibodies at 1:1000 (13); (ii) anti-VIG-1 antibodies at 1:30 000; (iii) anti-ALG-1 antibodies (Thermo Scientific) at 1:2500; (iv) anti-AGO2 antibodies (Cell Signaling) at 1:1000; (v) anti-GFP antibodies (Rockland) at 1:2000; (vi) anti-SERBP1 antibodies (anti-PAI-RBP1, Abnova) at 1:1000; (vii) anti-actin antibodies (Sigma) at 1:1000; (viii) rabbit anti-ubiquitin antibodies (Sigma) at 1:100 and (ix) anti-paramyosin antibodies (MH16, hybridoma cell line from Developmental Studies Hybridoma Bank, University of Iowa) tissue culture supernatant. Immunoblots were developed with SuperSignal WestPico substrate (Thermo Scientific) or Amersham ECL Select Western Blotting Detection Reagent (GE Healthcare). Blots were exposed to Kodak BioMax MR film.
Co-immunoprecipitation (Co-IP)
For the TEG-1 VIG-1 co-IP, ∼0.75 ml of packed N2 worms was subjected to French press. For each immunoprecipitation, 10 mg of the extract was incubated overnight at 4°C with ∼20 μg of pre-immune sera, anti-VIG-1 antibodies, or affinity purified rabbit anti-TEG-1 (C-terminal) antibodies coupled to protein G beads (Sigma). The beads were washed three times with CSK lysis buffer (100 mM PIPES, pH 6.0, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.3 M sucrose, 0.5% Triton X-100, 1 mM PMSF, and EDTA-free complete protease inhibitor cocktail (Roche)) and resuspended in 2× Laemmli loading buffer. Bound proteins were analyzed by immunoblotting as described above.
Bacterial pull-down
Bacterial pull-down experiments for TEG-1 and VIG-1 were performed as previously described (13). Recombinant full-length GST-TEG-1 was expressed using the pDH204 plasmid, while the GST-tagged TEG-1 lacking the putative GYF domain (ΔGYF) was expressed using the pDH205 plasmid. Similarly, His6X-VIG-1 was expressed using the pDH207 plasmid in E. coli BL21(DE3). In the TEG-1 IP experiment, 10 mg of RNase A was added to each reaction to determine if the interaction between TEG-1 and VIG-1 occurs in an RNA-independent manner.
let-7 pull-down
The pull-down of mature let-7 associated proteins was performed as previously described (22) with slight modifications. Synchronized XB624 (ugEx24) animals were harvested at one day past the L4 stage. Following the lysis of 0.75 ml of the worm pellet, 10 mg of total lysate was pre-cleared with 120 μl of streptavidin magnetic beads couple to biotinylated unrelated 2΄-O-methyl oligo (5΄-Bio-mCmAmUmCmAmCmGmUmAmCmGmCmGmGmAmAmUmAmCmUmUmCmGmAmAmAmUmGmUmC-3΄) at 1 μM concentration. The pre-cleared lysate was then split into two equal fractions and each incubated with 30 μl of streptavidin magnetic beads (Invitrogen), coupled with 60 pmol of either biotinylated 2΄-O-methylated unrelated oligo or mature let-7 complementary oligo (5΄-Bio-mUmCmUmUmCmAmCmUmAmUmAmCmAmAmCmCmUmAmCmUm-AmCmCmUmCmAmAmCmCmUmU-3΄) for 2 hours by end-over-end rotation at 4°C. After washing 5× with lysis buffer, the beads were suspended in 2× Laemmli loading buffer and subjected to immunoblot analysis.
HeLa cell culture conditions and transfections
The HeLa-CCL2 cell line was maintained in Dulbecco's Modified Eagle Medium containing 3.7 g/ml NaHCO3, pH 7.2, 10% (v/v) fetal bovine serum, 4 mM l-glutamine, 75 μg/ml penicillin and treptomycin at 37°C with 5% CO2. The plasmids expressing GFP or GFP-CD2BP2 (23) were prepared using the QIAGEN EndoFree maxi plasmid kit and transfected into HeLa-CCL2 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 24 hours after transfection, cells were harvested and lysed in HEPES lysis buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM pyrophosphate, 50 mM NaF, 1.5 mM Na3VO4, and protease inhibitor cocktail (Roche)) on ice for 30 min. with intermittent vortexing. The whole cell lysate was centrifuged at 15 000g for 10 min. at 4°C and the soluble fraction was collected. For each IP, 2 mg of the extract was pre-cleared with protein G beads, and then incubated with 20 μl of GFP-Trap beads (Chromotech) at 4°C for 2 hours with end-over-end rotation. The beads were subsequently washed five times with lysis buffer and dissolved in 2× Laemmli loading buffer for immunoblotting analysis. To study the effect of CD2BP2 down-regulation by siRNA, 2 × 105 HeLa-CCL2 cells were transfected with either a universal scrambled negative control siRNA duplex or CD2BP2-C siRNA duplex (5΄-CUCAGGCAGCGAGGAAAUUGGAGGC-3΄ (ORIGENE)) at 10 nM using Lipofectamin 2000 according to the manufacturer's instructions (Life Technologies). Cells were harvested for knock-down efficiency and co-IP analyses at 48 hours post-transfection.
RESULTS
teg-1 mutants display heterochronic defects
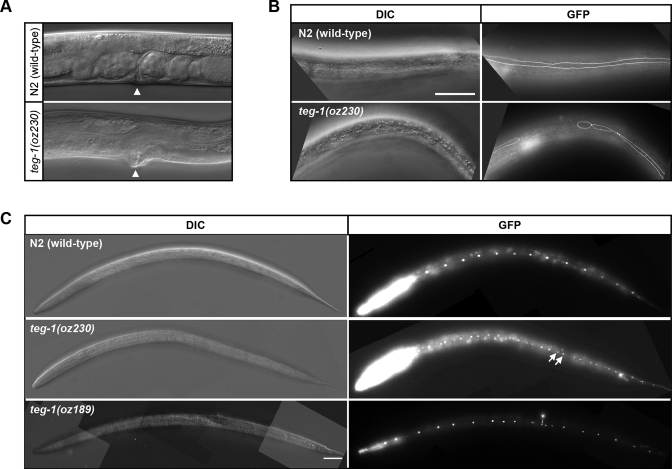
Although we previously reported TEG-1's roles in regulating germline development via its interaction with the spliceosome (13), teg-1 mutants also display somatic defects that are not observed in other splicing factor mutants. Similarly, CD2BP2, the mammalian ortholog of TEG-1, interacts with other non-spliceosomal proteins, and has non-splicing functions (24,25). Thus, it is likely that the somatic phenotypes observed in teg-1 mutants are due to non-splicing functions of TEG-1. We found that teg-1 mutants develop slower than wild-type animals, with teg-1(oz230) animals taking ∼67.5 hours to progress from the first larval stage (L1 arrest) to the fourth larval stage (L4, vulval Christmas tree stage) at 20°C (n = 30), as compared to ∼45.5 h for wild-type animals (n = 22). In addition, a mild protruding vulva phenotype is present in teg-1(oz230) animals (Figure 1A).
Figure 1.
teg-1 mutants display heterochronic phenotypes. Mutations in teg-1 lead to somatic phenotypes, such as abnormal vulva morphology (A), defective seam cell syncytium formation (B) and over-replication of seam cells (C). (A) 62% of teg-1(oz230) mutants have a mild protruding vulva phenotype (n = 88), whereas 100% of the N2 (wild-type) animals do not (n = 50). Arrow head indicates the location of vulva. (B) Incomplete fusion of lateral seam cells in teg-1 mutants results in gaps in the seam cell syncytium. Scale bar = 50 μm. (C) Over-replication of hypodermal seam cells results in formation of paired nuclei marked by arrows in teg-1 mutants. Each control animal has exactly 16 seam cells, whereas an average of 18 and 20 seam cells were found in teg-1(oz189) and teg-1(oz230) mutants, respectively. Seam cell membranes (B) and nuclei (C) are marked by AJM-1::GFP and SCM::GFP, respectively.
Since these somatic phenotypes are often associated with developmental timing (heterochronic) defects (26,27), we sought to determine if teg-1 mutants have heterochronic defects by analyzing three well-characterized heterochronic phenotypes: seam cell number, seam cell fusion and cuticular alae formation. There are 16 seam cells in wild-type animals at the L2 stage, whereas the stage is reiterated in certain heterochronic mutants, such as in the let-7(n2853) mutants, which were reported to have 23 seam cell nuclei (28). An increase in the seam cell number was also observed in loss-of-function let-7 miRNA family member mutants (miR-48, miR-84 and miR-241) (29). We found that teg-1 mutants have more seam cells than wild-type animals (between 18 and 20, depending on the allele), indicating that the L2 stage program is reiterated in teg-1 mutants (Figure 1C, Table 1).
Table 1. Analysis of developmental timing progression in teg-1 mutants.
| Genotypea | Seam cell nucleib | Seam cell fusionf | Alae at L4-to-adult transition | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean; rangec | = 16d | > 16e | n | WTg | Gappedh | n | WT | None | n | |
| teg-1(+);wIs78i | 16; 16 - 16 | 100% | 0% | 51 | – | – | – | – | – | – |
| teg-1(oz189);wIs78 | 18; 16 - 23 | 41% | 58% | 41 | – | – | – | – | – | – |
| teg-1(oz230);wIs78 | 20; 16 - 30 | 34% | 66% | 50 | – | – | – | – | – | – |
| teg-1(+);jcIs1j | – | – | – | – | 100% | 0% | 81 | – | – | – |
| teg-1(oz230);jcIs1 | – | – | – | – | 42% | 58% | 117 | – | – | – |
| N2 (wild-type) | – | – | – | – | – | – | – | 100% | 0% | 22 |
| teg-1(oz230)/hT2g | – | – | – | – | – | – | – | 100% | 0% | 34 |
| teg-1(oz230) | – | – | – | – | – | – | – | 51% | 49% | 75 |
| teg-1(oz230);jcIs1; pL4440(RNAi) 20°C | – | – | – | – | 37% | 63% | 54 | – | – | – |
| teg-1(oz230);jcIs1;lin-41(RNAi) 20°C | – | – | – | – | 67% | 33% | 58 | – | – | – |
aAll the stains were maintained at 15°C other than the ones indicated.
bSeam cell nuclei were counted in L4-stage worms.
cThe average number of seam cell nuclei and the range of seam cell nuclei.
dThe percentage of animals with seam cell nuclei = 16.
eThe percentage of animals with seam cell nuclei > 16.
fSeam cell fusions were determined by observing between vulva and tail of the worms.
gThe percentage of animals with WT seam cell fusion phenotype.
hThe percentage of animals with gapped seam cell fusion defects.
iwIs78 [ajm-1::gfp; scm-1::gfp; unc-119(+); F58E10(+)] IV.
jjcIs1 [Pajm-1::gfp; unc-29] IV, a gift from Dr. Jay Kormish at University of Manitoba.
Next, we examined seam cell fusion in teg-1 mutants at the L4-to-adult transition, when the 16 seam cells fuse into a single syncytium and form specialized cuticular alae structures (30). While all animals in the control group have a continuous seam cell syncytium at the late L4 stage, 58% of teg-1(oz230) mutants, like other heterochronic mutants (31,32), display a variety of seam cell fusion defects, including gaps along the seam cell syncytium and single cells separated from the rest of the syncytium (Figure 1B and Supplementary Figure S1B, Table 1). These defects in seam cell fusion suggest that teg-1 mutants fail to properly progress through the L4-to-adult transition.
We also examined if late L4 stage alae formation occurs properly in teg-1 mutants. We found both wild-type animals and teg-1(oz230)/+ heterozygotes form alae properly at the late L4 stage. However, 48% of teg-1(oz230) homozygotes do not form alae at this stage. These homozygous mutants, however, do form alae as older adults, suggesting the teg-1(oz230) mutants have retarded alae formation (Table 1). Consistent with teg-1 functioning in these somatic tissues, we found that TEG-1 protein is expressed in the seam cells, vulva, and uterus (Supplementary Figure S3), and our previous work demonstrates that while TEG-1 is predominantly found in the nucleus, TEG-1 is also found in the cytoplasm (13).
miRNA levels are reduced in teg-1 mutants
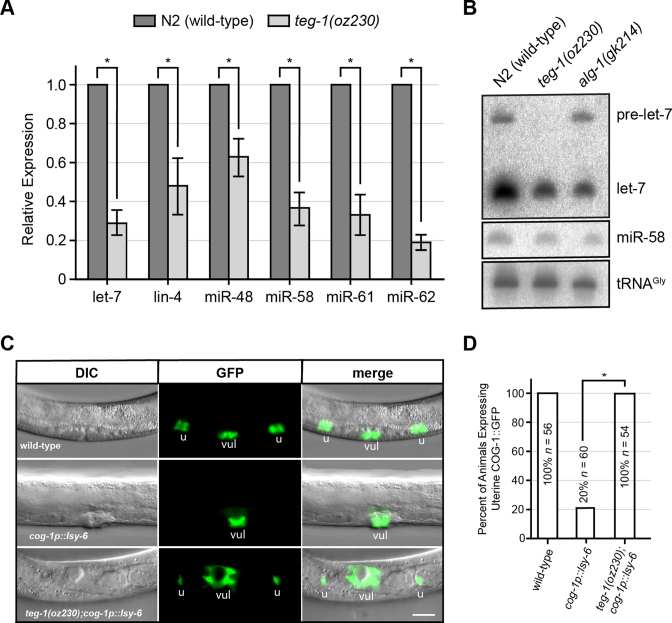
Genetic analysis of C. elegans heterochronic mutants led to the identification of miRNAs as key regulators in controlling developmental timing (1,2). Therefore, to determine if the heterochronic phenotypes observed in teg-1 mutants are due to abnormal miRNA function, we first measured the levels of the mature form of let-7. In C. elegans, let-7 helps to regulate terminal differentiation of the lateral hypodermal cells, and accumulation of let-7 in the L4 stage decreases LIN-41 expression at the L4-to-adult transition (2,31). Loss-of-function mutations in let-7 result in abnormal vulval formation and heterochronic phenotypes, including protruding vulva, additional seam cell divisions, and retarded alae formation (2,33), which are similar to the somatic phenotypes observed in teg-1 mutants. We found that mature let-7 levels are reduced in two loss-of-function teg-1 alleles, teg-1(oz230) and teg-1(oz189), suggesting that the reduction in let-7 levels is specifically due to a loss of TEG-1 activity (Figure 2A – B, Supplementary Figures S1A, and S2A). To test if this lower level of let-7 could explain the heterochronic defects in teg-1 mutants, we reduced the activity of lin-41 in teg-1(oz230) mutants by RNAi. let-7 represses lin-41 translation; therefore, LIN-41 levels are higher in let-7 mutants, contributing to the heterochronic defects (2,31). We found an almost two fold reduction in the seam cell fusion defect in teg-1(oz230);lin-41(RNAi) worms as compared to teg-1(oz230) mutants (Table 1). Together, the lower let-7 levels and the ability of lin-41(RNAi) to partially suppress the seam cell fusion defects suggest that the abnormal timing defects observed in teg-1 mutants are due, at least in part, to a lower level of mature let-7 miRNA. To further test if teg-1 functions with let-7 in regulating developmental timing, we analyzed teg-1(oz230);let-7(mg279) double mutants. let-7(mg279) is a partial loss-of-function allele; therefore, if teg-1 mutants were to lower let-7 abundance, we would expect teg-1 mutants to enhance let-7(mg279) phenotypes. Indeed, we observed synthetic lethality in both teg-1(oz230);let-7(mg279) males and hermaphrodites (Table 2), as well as enhancement of the ruptured vulva and adult lethal phenotypes. Therefore, the enhancement of let-7(mg279) is consistent with TEG-1 affecting let-7 function.
Figure 2.
TEG-1 function is required for maintaining mature miRNA levels and function. (A) Reduced mature let-7, lin-4, miR-48, miR-58, miR-61 and miR-62 levels are found in teg-1(oz230) mutants by quantitative real time PCR using TaqMan probes. (B) Northern blot detection of let-7 and miR-58 RNAs in various genetic backgrounds of late L4-stage animals. The detection of tRNAGly was served as a loading control. (C) Ectopic expression of lsy-6 miRNA from the cog-1 promoter suppresses COG-1::GFP expression in the uterus. However, COG-1::GFP expression is partially restored upon removal of TEG-1 in lsy-6 ectopic expression background (vul = vulva and u = uterus). (D) Quantification of uterine COG-1::GFP expression in various genetic backgrounds. In (A) and (D), the error bars indicate standard deviation, and two-tailed Student's t-tests were used to determine the statistical significance: *P < 0.0005.
Table 2. teg-1 genetically interacts with let-7 in males and hermaphrodites.
| Genotypea | % Adult Male Lethalb | % Hermaphrodite Mutantc | n |
|---|---|---|---|
| let-7(mg279) | 0 | – | 30 |
| teg-1(oz230) | 0 | – | 30 |
| teg-1(oz230)/+;let-7(mg279) | 80 | – | 30 |
| let-7(mg279) | – | 2 | 50 |
| teg-1(oz230) | – | 0 | >50 |
| teg-1(oz230)/+; let-7(mg279)d | – | 0 | >50 |
| teg-1(oz230)/teg-1(oz230);let-7(mg279) | – | 91 | 56 |
aAll the stains were maintained at 18°C.
bAnimals die within 24 h after reaching adult stage.
cPhenotypes analyzed at 24–48 h. past L4 stage; Mutant animals were adult lethal with or without ruptured vulva.
dActual genotype is teg-1(oz230)/hT2g;let-7(mg279)
We next asked whether a reduction in TEG-1 function lowers let-7 miRNA levels specifically, or if teg-1 mutations also affect the levels of other miRNAs. To address this question, we analyzed the levels of mature lin-4, miR-48, miR-58, miR-61 and miR-62 in teg-1 mutants. The activity of lin-4 regulates the timing of fate specification in neuronal and hypodermal lineages during post-embryonic development (1,27). miR-58 is a highly abundant miRNA (34) and is a member of a miRNA family orthologous to the bantam miRNA in Drosophila, which controls apoptosis and cell proliferation (35). miR-62 is a mirtron that displays Drosha-independent, yet splicing-mediated, biogenesis (36). miR-48 is a member of let-7 family, while miR-61 functions in promoting the secondary vulval cell fate via LIN-12/Notch signaling (37). Therefore, lin-4, let-7, miR-58, miR-61 and miR-62 belong to different families of miRNAs, differing in expression, biogenesis and biological function, providing a representative sample of mature miRNAs. We found that lin-4, miR-48, miR-58, miR-61, and miR-62 levels are reduced in teg-1 mutants as compared to wild-type levels (Figure 2A). Consistent with the qPCR results, northern blotting also showed a reduction of mature let-7, lin-4, miR-58, and miR-62 levels in teg-1(oz230) mutants (Figure 2B and Supplementary Figure S2B). In contrast to alg-1(gk214) animals, no accumulation of the precursor levels in let-7, lin-4, or miR-62 was found in teg-1(oz230) mutants (Figure 2B and Supplementary Figure S2B). The detection of accumulated precursor levels in alg-1(gk214) mutants is thought to be attributed to the role of ALG-1 in processing precursor miRNAs into mature forms during miRNA biogenesis (6). It is intriguing that let-7, lin-4 and miR-62 precursor miRNA levels are lower in teg-1(oz230) animals than in wild-type. This could be due to a disruption of precursor miRNA biogenesis, or an increased amount of precursor miRNA being converted to mature miRNA, as compared to wild-type, in an attempt to maintain mature miRNA levels (see Discussion). However, since teg-1 mutants do not show a buildup of miRNA precursor levels, TEG-1 is unlikely to be required for processing precursor miRNAs into mature forms during miRNA biogenesis, unlike ALG-1.
We have demonstrated that teg-1 affects the abundance of miRNAs from a number of miRNA families, and that a lowering of let-7 levels in teg-1 mutants leads to heterochronic phenotypes. To test if teg-1 affects not only the abundance, but also the function of other miRNAs, we examined the effects of teg-1 mutations on the activity of lsy-6 miRNA. lsy-6 functions in determining the lateralization of two chemosensory ASE neurons, ASEL and ASER (38). Expression of lsy-6 miRNA in the ASEL neuron results in suppression of the ASER cell fate by down regulating the transcription factor COG-1 (38). cog-1, unlike lsy-6, also functions in other tissues, such as the vulva and uterus, that do not normally express lsy-6 miRNA. However, ectopic expression of lsy-6 under the control of the cog-1 promoter (cog-1p::lsy-6) results in a significant reduction of cog-1 expression in uterine cells (10,38). Therefore, we reasoned that if teg-1 regulates the levels of miRNAs, removing teg-1 activity should reduce lsy-6 repression of the COG-1::GFP reporter. We found that COG-1::GFP expression was significantly restored in the uterus of teg-1 mutants that ectopically express cog-1p::lsy-6 (Figure 2C – D), suggesting that lsy-6 function is reduced in teg-1 mutants.
To further test the hypothesis that TEG-1 regulates the activities of miRNAs from a number of miRNA families, we tested for potential synthetic interactions between teg-1 and the miR-35 family of miRNAs. This miRNA family consists of eight members (miR-35-42) that reside in two genomic loci, miR-35-41 and miR-42, separated by 350 kb on LGII (34). In general, miRNAs of the miR-35 family work redundantly as deletion of seven of the eight members in the miR-35(gk262) mutant results in a partially penetrant embryonic lethality phenotype at the permissive temperature, while deletion of all eight members results in fully penetrant embryonic or L1-stage lethality (34,39). We found that reducing the activity of teg-1 by half (by using a teg-1 heterozygote), strongly enhanced miR-35(gk262) lethality, further suggesting that teg-1 affects the function of miRNAs from multiple families (Supplemental Table S1).
TEG-1 directly interacts with VIG-1, a component of miRISC
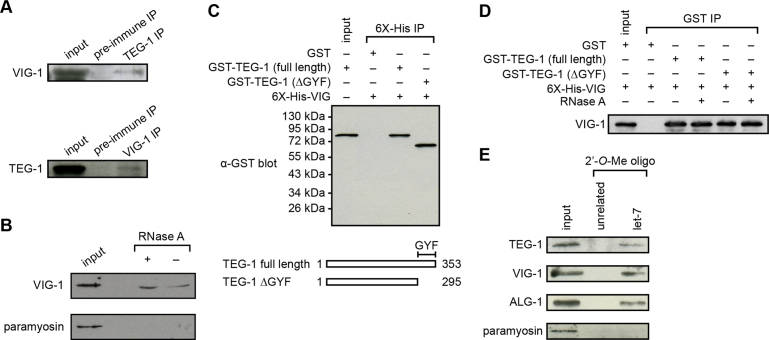
To understand how TEG-1 could affect miRNA levels, we analyzed a list of proteins that co-immunoprecipitate (co-IP) with TEG-1, which we described previously (13), and found that VIG-1 co-IP'ed with TEG-1 in two separate experiments (Supplemental Table S2). VIG-1 has been reported to be a component of the miRISC in both C. elegans and Drosophila (8,9,40), and its function is necessary for let-7-mediated translational repression of a lacZ-lin-41 reporter (9). Thus, the interaction between TEG-1 and VIG-1 could help to explain TEG-1's role in miRNA function. To verify the TEG-1-VIG-1 interaction, we raised VIG-1 specific antibodies and performed reciprocal IPs using whole worm lysates (Supplementary Figure S4, Figure 3A). VIG-1 co-IP's with TEG-1 using TEG-1 specific antibodies, and TEG-1 co-IP's with VIG-1, suggesting that at least a subset of TEG-1 and VIG-1 exist in a complex in worms (Figure 3A). Also, this interaction can be detected in the presence of RNase A, indicating that the binding between TEG-1 and VIG-1 is independent of RNA (Figure 3B).
Figure 3.
TEG-1 associates with VIG-1 and miRISC. (A) A subset of TEG-1 is detected in association with VIG-1 by reciprocal co-IP. In the TEG-1 IP, VIG-1 is detected with VIG-1-specific antibodies, but not with the pre-immune serum. Conversely, in the VIG-1 IP, TEG-1 is co-IP'd, whereas TEG-1 is not detected in an IP using pre-immune rabbit serum. (B) A subset of VIG-1 is detected in association with TAP-tagged TEG-1 using TEG-1 IP followed by detection with VIG-1-specific antibodies in both the presence and absence of RNase A. (C and D) Physical interaction of TEG-1 with VIG-1 using bacterial pull-down experiments followed by immunoblotting. 6X-His-VIG-1 was co-expressed in E. coli with GST, GST-TEG-1 (full length), or GST-TEG-1 with the GYF domain removed (GST-TEG-1(ΔGYF)). When a Ni column was used to pull-down the 6X-His-VIG-1 fusion protein, both full-length and GYF-truncated GST-TEG-1, but not GST alone, were co-precipitated (C). Similarly, VIG-1 was pulled-down from extracts containing full-length and GYF-truncated TEG-1 in the presence and absence of RNase A, using GST beads to pull down GST-TEG-1 fusion proteins (D). (E) TEG-1 associates with a complex containing the mature let-7 miRNA. Immunoblotting analysis using TEG-1-specific antibodies detects the presence of TEG-1 only in the let-7 complementary, but not in the unrelated, 2΄-O-methylated oligonucleotide IPs.
To test if the interaction between TEG-1 and VIG-1 is direct, rather than being part of a larger complex, we performed bacterial pull-down experiments using E. coli co-expressing GST-tagged TEG-1 and 6X-His-tagged VIG-1. We found that bacterially expressed TEG-1 and VIG-1 co-precipitated, even in the presence of RNase A, indicating the two proteins directly associate with each other in an RNA independent manner (Figure 3C and D). Furthermore, the physical interaction remains uninterrupted in bacterial extracts co-expressing full-length VIG-1 and a C-terminal truncated version of TEG-1, which lacks a potential GYF protein-binding motif, suggesting that the GYF domain is not essential for the TEG-1-VIG-1 interaction (Figure 3C and D).
To determine if the TEG-1-VIG-1 interaction is functionally relevant to our understanding of TEG-1 function, we tested if miRNA levels are also reduced in vig-1 mutants. We found that, like teg-1 mutants, the levels of mature let-7, miR-58, and miR-62 miRNAs are decreased in vig-1(tm3383) mutants, suggesting that the interaction between TEG-1 and VIG-1 may be relevant to our understanding of the role of TEG-1 in miRNA function (Supplementary Figure S5, S2A, and Figure 2A and B).
The observations that mutations in teg-1 result in reduced mature miRNA levels, and that the TEG-1 protein interacts with VIG-1, suggest that TEG-1 interacts with the miRISC. To test this hypothesis, we first asked if ALG-1, an Argonaute protein of the miRISC (41), could be IP'ed in a complex with TEG-1. For this, we IP'ed a TAP-tagged version of TEG-1 and probed for ALG-1 in the immunoprecipitate. We found that ALG-1 does co-IP with TAP-TEG-1 (Supplementary Figure S6), suggesting that TEG-1 interacts with the miRISC. To further confirm TEG-1's interaction with miRISC, we tested if TEG-1 associates with mature let-7, because mature miRNAs are components of the miRISC (42,43). We used an affinity pull-down assay to purify mature let-7 and the associated miRISC (22). We found that TEG-1 associates with biotinylated 2΄-O-methylated let-7, but not with the control affinity column (Figure 3E). The association of TEG-1 with ALG-1 and the let-7-associated ribonucleoprotein complex suggest that TEG-1 interacts with the miRISC.
VIG-1 levels are lower in teg-1 mutants
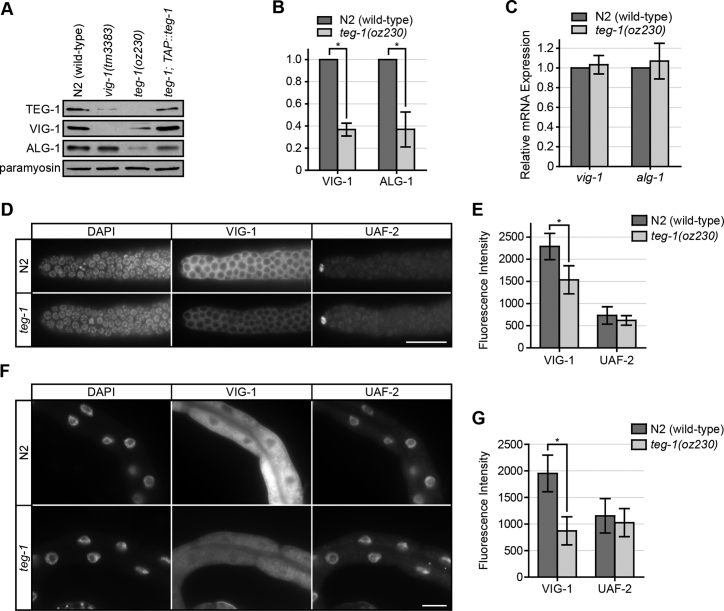
The association of TEG-1 with the miRISC, together with the teg-1 heterochronic mutant phenotypes, suggests that TEG-1 could affect miRISC function. MiRNAs are destabilized when Argonaute levels are lowered (44,45); therefore, lower levels of miRNAs in teg-1 mutants could be due to a lowering in the level of ALG-1 and/or miRISC components. We first asked if TEG-1 affects the abundance of miRISC proteins. We found that VIG-1 protein levels are reduced approximately three fold in teg-1 mutants (Figure 4A – B). This reduction in VIG-1 levels is found throughout the animal, including the intestine and germ line (Figure 4D – G). Interestingly, we found that TEG-1 protein levels are also lowered in vig-1(tm3383) mutants (Figure 4A), suggesting that the association between TEG-1 and VIG-1 plays an important role in maintaining each other's protein levels.
Figure 4.
VIG-1 and ALG-1 protein levels are reduced in teg-1 mutants. (A) A representative immunoblot showing lower TEG-1 protein levels in whole worm extracts from vig-1(tm3383) animals, and lower VIG-1 and ALG-1 protein levels in teg-1(oz230) extracts, with paramyosin as a loading control. The reduction of VIG-1 and ALG-1 levels can be restored in teg-1 mutants expressing exogenous TAP-tagged TEG-1 fusion protein. Approximately 100 one day past L4-staged animals were loaded per lane. (B) A roughly three fold reduction of VIG-1 and ALG-1 protein levels are detected in teg-1(oz230) extracts in comparison to the wild-type N2 extracts by intensity measurement using data collected from three independent experiments. Using paramyosin as an internal control, intensities of the paramyosin bands in N2 (wild-type) samples were set to 100%, whereas the intensities of the corresponding experimental samples were normalized to this value. A two-tailed student t-test was used to determine the statistical significance: * P < 0.002. (C) vig-1 and alg-1 mRNA levels in teg-1(oz230) mutants are similar to wild-type levels using relative quantitative real time PCR. (D–G) Reduced VIG-1 protein levels in both germ line (D and E) and somatic (F and G) tissues of teg-1(oz230) mutants. Indirect immunofluoresence staining of VIG-1 was performed on dissected gonads (D and E) and intestine (F and G) of N2 (wild-type) and teg-1(oz230) animals. UAF-2 was used as control. Images were collected with identical exposure times and the intensity was measured by ImageJ software. Fluorescence intensities of UAF-2 in N2 samples were set to 100%, whereas fluorescence intensities of the experimental samples were normalized to this value. Error bars = standard deviation. *P < 0.001. Scale bar = 25 μm.
To determine if this influence of TEG-1 is specific to VIG-1, or if it also influences other miRISC components, we analyzed ALG-1 protein levels in teg-1 mutants. We found that ALG-1 protein levels are also reduced to approximately 35% of wild-type levels in teg-1 mutants (Figure 4A and B), suggesting that TEG-1 may be generally involved in stabilizing miRISC components. Intriguingly, ALG-1 levels are not reduced in vig-1(tm3383) mutants, suggesting that TEG-1 may be more closely involved, than VIG-1, in regulating miRISC protein abundance. Furthermore, the amount of TEG-1 still present in the vig-1(tm3383) must be sufficient to maintain ALG-1 levels.
Many mechanisms can be envisioned as to how TEG-1 could affect VIG-1 and ALG-1 levels, including regulation at the transcriptional, translational, and post-translational levels. To distinguish between these possibilities, we compared the mRNA levels of vig-1 and alg-1 in teg-1(oz230) animals with those in wild-type animals. In contrast to the protein levels, vig-1 and alg-1 mRNA levels remain unchanged in teg-1 mutants (Figure 4C), suggesting that the reduction in VIG-1 and ALG-1 protein levels is not due to differences in transcriptional activity and/or mRNA stability.
Since VIG-1 and TEG-1 physically interact, we reasoned that their binding could increase their stability. Cellular proteolysis is mediated largely by the ubiquitin-proteasome system, with polyubiquitinated proteins being degraded by the 26S proteasome (46). To determine if VIG-1 and ALG-1 levels are controlled, at least in part, by ubiquitin/proteasome-mediated degradation, we examined their protein levels in wild-type animals treated with the selective irreversible proteasome inhibitor, lactacystin. We found that treatment with lactacystin increased VIG-1 and ALG-1 protein levels by ∼1.5–2 fold (Figure 5A–C). Furthermore, we identified ubiquitinated forms of VIG-1 when we immunoprecipitated it from wild-type extracts and probed with ubiquitin-specific antibodies (Figure 5D). Therefore, proteasomal degradation appears to be utilized for the turnover of these miRISC proteins.
Figure 5.
VIG-1 protein levels are regulated by proteasome ubiquitin degradation pathway. (A) Increased VIG-1 and ALG-1 protein levels are detected in animals treated with lactacystin, a specific inhibitor for the 26S proteasome. (B and C) Intensity measurements of VIG-1 and ALG-1 protein levels were averaged from three independent experiments. Both VIG-1 and ALG-1 protein levels are increased >1.5-fold upon lactacystin treatment in N2 (B) and teg-1(oz230) (C). A two-tailed student t-test was used to determine the statistical significance: *P < 0.01. Error bars in B and C = standard deviation. (D) VIG-1 is ubiquitylated. VIG-1 was immunoprecipitated from wild-type extracts in the presence or absence of lactacystin followed by immunoblotting with ubiquitin-specific antibodies. Bands consistent in size with ubiquitylated VIG-1 are detected and become more intense with lactacystin treatment. The two bands labeled as Ub-VIG-1 at ∼60 kDa and ∼70 kDa (arrow heads) are consistent in size with mono- and di-ubiquitylated VIG-1, respectively; whereas the poly-ubiquitylated VIG-1 is detected in the upper smear region. The lower panel represents the results of re-probing the membrane with anti-VIG-1 antibodies.
Role of TEG-1 in association with miRISC is conserved
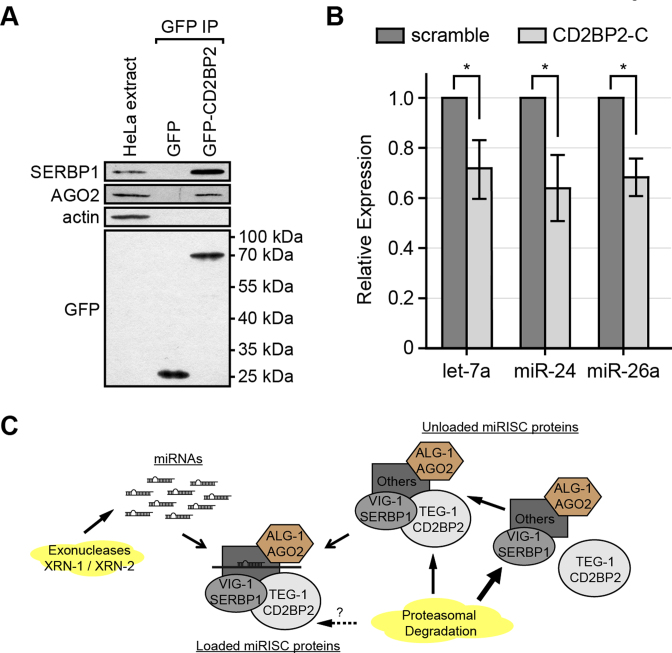
Given that many of the miRISC components are conserved between nematodes and higher eukaryotes (3), it is possible that the association of TEG-1 with miRISC proteins is also conserved. To test this, we transiently expressed GFP-tagged CD2BP2, the human ortholog of TEG-1 (24), in HeLa cells and IP'ed the protein with GFP-Trap beads (Figure 6A). As a negative control, a construct expressing GFP alone was transfected and IP'ed. Using antibodies specific to SERBP1/PAI-RBP1, the human VIG-1 ortholog (40), and AGO2, the human ALG-1 ortholog (6), we found that SERBP1/PAI-RBP1 and AGO2 co-IP'ed with GFP::CD2BP2, but not with the GFP-negative control (Figure 6A), suggesting that CD2BP2 is part of a complex with these miRISC components in HeLa cells. This finding is consistent with a recent report identifying SERBP1/PAI-RBP1 as a potential CD2BP2 binding partner by differential pull-down and quantitative mass spectrometric analyses using extracts obtained from bone marrow-derived macrophages (47). SERBP1/PAI-RBP1 has also been identified in proteomic experiments aimed to identify proteins that interact with Ago2 in a Dicer-independent manner (48). In addition, consistent with the subcellular localization in C. elegans, CD2BP2 (TEG-1 ortholog) is more abundant in the nucleus in HeLa cells (49), whereas SERBP1 (VIG-1 orthologue) is predominantly found in the cytoplasm (50).
Figure 6.
Association of TEG-1 with miRISC proteins is conserved in human tissue culture cells. (A) SERBP1/PAI-RBP1 (VIG-1 ortholog) and AGO2 (ALG-1 ortholog) are co-IP'd with CD2BP2 (TEG-1 ortholog). GFP or GFP-CD2BP2 was transiently expressed in HeLa cells and proteins were co-IP'd using anti-GFP antibodies. SERBP1/PAI-RBP1 and AGO2 are detected when IP'd with anti-GFP antibodies followed by immunoblotting, while the negative control, actin, was not detected. (B) Mature let-7a, miR-24, and miR-26a miRNAs are significantly reduced in HeLa cells treated with CD2BP2 siRNA in comparison with a scrambled siRNA treatment by quantitative real time PCR using TaqMan probes. A two-tailed Student's t-test was used to determine the statistical significance: *P < 0.0003. Error bar = standard deviation. (C) Proposed model. Free miRNAs are degraded by exonucleases (XRN-1/XRN-2), while miRISC proteins are subjected to proteasomal degradation. Our model suggests that TEG-1 CD2BP2 interacts with miRISC. TEG-1 CD2BP2 helps stabilize miRISC protein components. In the absence of TEG-1 CD2BP2, miRISC protein components are more susceptible to proteasomal degradation (thick arrow), which in turn results in decreased stability of miRNAs.
Together, our immunoprecipitation data, in conjunction with the previously reported proteomic results, suggest that, similar to C. elegans, a complex is formed between CD2BP2, SERBP1/PAI-RBP1, and Ago2 in human tissue culture cells.
Our current model is that TEG-1 stabilizes the activity of the miRISC, perhaps through interaction with one or more of its components, and destabilization of the complex in teg-1 mutants results in a reduction in miRNA levels (Figure 6C). To determine the extent to which this function may be conserved, we tested if miRNA levels are lowered in HeLa cells when CD2BP2 levels are reduced. We found that mature let-7a, miR-24 and miR-26a levels are reduced in HeLa cells treated with CD2BP2 siRNA, but not in cells treated with control siRNA (Figure 6B, Supplementary Figure S7). These data suggest that the association between TEG-1 and the miRISC, which we initially identified in C. elegans, is also present in human tissue culture cells, and that the effect of this association on miRNA abundance is also conserved.
DISCUSSION
In C. elegans, mis-regulation of miRNA levels often results in defective temporal developmental events (27). Here, we show that loss of teg-1 function leads to a range of developmental defects, and the levels of miRNAs from diverse miRNA families are lowered in teg-1 mutants. In addition, TEG-1 physically interacts with the miRISC protein VIG-1, and TEG-1 complexes with mature let-7 miRNA and the core miRISC Argonaute protein, ALG-1. Importantly, the protein levels, but not the mRNA levels, of VIG-1 and ALG-1 are reduced in teg-1 mutants. These data provide the first evidence of TEG-1 as a miRISC interacting protein and a regulator of miRNA function. The involvement of TEG-1 in miRNA function appears to be conserved, as we also identified similar interactions between CD2BP2, AGO2, and SERBP1/PAI-RBP1, and a reduction of let-7a, miR-24 and miR-26a levels when we reduced CD2BP2 levels in HeLa cells. Thus, our results demonstrate that TEG-1 CD2BP2 is an interacting partner of miRISC, affecting both miRISC stability and miRNA function in nematodes and humans.
Homeostatic control of miRNAs and miRISC stability
In teg-1 mutants, we observe decreased levels of certain miRNAs and two miRISC proteins. We envision two basic models by which TEG-1 could affect the abundance of both miRNAs and miRISC proteins. In the first, TEG-1 could be directly involved in regulating the abundance of miRNAs, and in its absence, miRNA levels are decreased, which decreases the stability of the miRISC and its associated proteins. In the second model, TEG-1's interaction with the miRISC proteins could assist in their assembly, or stability of miRISC proteins, by protecting them from proteasomal degradation. In the absence of teg-1 function, the miRISC would become destabilized, which in turn would lead to the destabilization of miRNAs.
With respect to the first model, Ago2 in mammalian cells has been observed to possess endonuclease activity, suggesting that it can catalyze the maturation of miRNAs in the absence of Dicer (51,52). In fact, it has been shown that Ago2 cleaves the 3΄ hairpin loop of pre-miR-451 to produce the mature miR-451 in a Dicer-independent manner (53,54). Thus, like Ago2, TEG-1 could also have miRNA biogenesis activity, with loss of TEG-1 resulting in lower miRNA levels. Indeed, lower miRNA levels have been demonstrated to decrease Argonaute stability in both Drosophila and mammals (55,56). However, our northern analysis did not show a build-up of let-7, lin-4 and miR-62 precursors in teg-1 mutants, as observed in alg-1 mutants, suggesting that TEG-1 is unlikely to have a role in miRNA biogenesis, or at least must be involved at a different stage of biogenesis than ALG-1.
In the second model, TEG-1 could directly affect the stability of miRISC proteins. In human cells, over-expression of Argonaute leads to a global increase of mature miRNAs (44). Additionally, the C. elegans 5΄ to 3΄ exonuclease XRN-1 and XRN-2 are capable of catalyzing the degradation of mature miRNAs that are not in association with the miRISC (57,58). Recent studies also revealed that the decapping scavenger enzyme, DCS-1, interacts with XRN-1 to promote miRNA degradation independent of its decapping scavenger activity (20). Therefore, in teg-1 mutants, ALG-1 and VIG-1 may be destabilized, which would reduce the protection of mature miRNAs from exonucleolytic degradation.
We suggest that our results are more consistent with the second model in which TEG-1 affects the stability of the miRISC proteins. First, the physical interaction between TEG-1 and VIG-1, and the reduced VIG-1 and ALG-1 protein levels in teg-1 mutants, suggest a direct effect of TEG-1 on the miRISC. A recent study has reported that the DDX-23, a DEAD box helicase with similar germline phenotypes as teg-1 mutants, is involved in processing the primary lin-4 and let-7 family members in C. elegans (59). However, unlike TEG-1, no interaction between DDX-23 and miRISC was observed, and the protein levels of ALG-1 and other miRNA pathway components remained unchanged in DDX-23 depleted animals (59). Second, TEG-1's association with a complex containing the mature let-7 miRNA suggests a role with the miRISC rather than with the biogenesis of miRNAs. Third, unlike in alg-1 mutants, there is no build-up of pre-let-7, pre-lin-4 or pre-miR-62 levels in teg-1 mutants, suggesting that the lower levels of mature miRNAs observed in teg-1 mutants are likely due to degradation of the mature form rather than abnormal biogenesis. However, we cannot rule out the possibility that in addition to a role in stabilizing miRISC protein components, TEG-1 may have an additional role in miRNA biogenesis, perhaps in association with other protein partners, such as splicing factors (13). Whether TEG-1 has this additional function or not, there appear to be multiple mechanisms to fine-tune the optimum levels of miRNAs and miRISC proteins for proper function, and additional inputs from interacting proteins, including TEG-1 (20,52,60,61).
TEG-1 CD2BP2 fine tunes miRNAs and miRISC homeostasis
Recent studies have shown that proper maintenance of miRISC stability is a key limiting factor for controlling miRNA levels in regulating T cell differentiation, mouse stem cell potency, and Xenopus development (38,62,63). In these studies, the turnover of the Argonaute protein by ubiquitin-mediated protein degradation has emerged as a common theme in controlling miRNA levels, supporting our findings that loss of TEG-1 reduces the levels of the miRISC proteins, VIG-1 and ALG-1, and miRNAs from a wide range of miRNA families in C. elegans. Therefore, the regulation of mature miRNA levels via miRISC stability could be a conserved mechanism found in many systems; however, as we have not analyzed level changes for all miRNAs, we cannot conclusively state that miRISC stability affects all miRNAs. Importantly, our work has identified TEG-1 CD2BP2 as a molecular regulator of miRISC stability. Since the somatic phenotypes of teg-1 mutants are relatively mild as compared to when the activities of both of the core components, ALG-1 and ALG-2, are removed (6,45), and the loss of TEG-1 reduces the levels of miRISC proteins and miRNAs, but does not eliminate them all together, TEG-1 is not essential for miRNA function. Rather, we propose that TEG-1 CD2BP2 fine-tunes the levels of miRISC proteins and miRNAs in order to optimize their activities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tom Blumenthal for UAF-2 antibodies, Christian Freund for providing the GFP::CD2BP2 construct, and Carrie Shemanko for using her tissue culture facility. We thank Priscilla Van Wynsberghe and Amy Pasquinelli for providing detailed protocols and helpful discussions. We thank Jim McGhee, Paul Mains, and members of the Hansen lab for helpful advice. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Present address: Chris Wang, St. Mary's University, Calgary, Alberta T2X 1Z4 Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) [477786-2015 and 06647-2015 to D.H.]; Canadian Institutes of Health Research (CIHR) [MOP-97815 to D.H., MOP-81186 to M.J.S.]; G.D.B. is a NSERC Graham Bell Scholar; M.J.S. is a Senior scholar from Fonds de Recherche du Québec en Santé. Funding for open access charge: NSERC [477786-2015 and 06647-2015 to D.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G.. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 3.Kozomara A., Griffiths-Jones S.. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayed D., Abdellatif M.. MicroRNAs in development and disease. Physiol. Rev. 2011; 91:827–887. [DOI] [PubMed] [Google Scholar]
- 6.Vasquez-Rifo A., Jannot G., Armisen J., Labouesse M., Bukhari S.I., Rondeau E.L., Miska E.A., Simard M.J.. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One. 2012; 7:e33750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., Liu X., Yates J.R. 3rd, Han M.. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007; 28:598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan S.P., Ramaswamy G., Choi E.Y., Slack F.J.. Identification of specific let-7 microRNA binding complexes in Caenorhabditis elegans. RNA. 2008; 14:2104–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B., Silva J.M., Myers M.M., Hannon G.J., Plasterk R.H.. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003; 425:411–414. [DOI] [PubMed] [Google Scholar]
- 10.Hammell C.M., Lubin I., Boag P.R., Blackwell T.K., Ambros V.. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009; 136:926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akay A., Craig A., Lehrbach N., Larance M., Pourkarimi E., Wright J.E., Lamond A., Miska E., Gartner A.. RNA-binding protein GLD-1/quaking genetically interacts with the mir-35 and the let-7 miRNA pathways in Caenorhabditis elegans. Open Biol. 2013; 3:130151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin K.H., Choi B., Park Y.S., Cho N.J.. Analysis of C. elegans VIG-1 expression. Mol. Cells. 2008; 26:554–557. [PubMed] [Google Scholar]
- 13.Wang C., Wilson-Berry L., Schedl T., Hansen D.. TEG-1 CD2BP2 regulates stem cell proliferation and sex determination in the C. elegans germ line and physically interacts with the UAF-1 U2AF65 splicing factor. Dev. Dyn. 2012; 241:505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmons L., Court D.L., Fire A.. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001; 263:103–112. [DOI] [PubMed] [Google Scholar]
- 16.Jones A.R., Schedl T.. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995; 9:1491–1504. [DOI] [PubMed] [Google Scholar]
- 17.Zorio D.A., Blumenthal T.. Both subunits of U2AF recognize the 3΄ splice site in Caenorhabditis elegans. Nature. 1999; 402:835–838. [DOI] [PubMed] [Google Scholar]
- 18.Van Wynsberghe P.M., Kai Z.S., Massirer K.B., Burton V.H., Yeo G.W., Pasquinelli A.E.. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2011; 18:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 20.Bosse G.D., Ruegger S., Ow M.C., Vasquez-Rifo A., Rondeau E.L., Ambros V.R., Grosshans H., Simard M.J.. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell. 2013; 50:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasquez-Rifo A., Bosse G.D., Rondeau E.L., Jannot G., Dallaire A., Simard M.J.. A new role for the GARP complex in microRNA-mediated gene regulation. PLoS Genet. 2013; 9:e1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jannot G., Vasquez-Rifo A., Simard M.J.. Argonaute pull-down and RISC analysis using 2΄-O-methylated oligonucleotides affinity matrices. Methods Mol. Biol. 2011; 725:233–249. [DOI] [PubMed] [Google Scholar]
- 23.Kofler M., Heuer K., Zech T., Freund C.. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J. Biol. Chem. 2004; 279:28292–28297. [DOI] [PubMed] [Google Scholar]
- 24.Freund C., Kuhne R., Yang H., Park S., Reinherz E.L., Wagner G.. Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules. EMBO J. 2002; 21:5985–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kofler M., Motzny K., Beyermann M., Freund C.. Novel interaction partners of the CD2BP2-GYF domain. J. Biol. Chem. 2005; 280:33397–33402. [DOI] [PubMed] [Google Scholar]
- 26.Mondol V., Pasquinelli A.E.. Let's make it happen: the role of let-7 microRNA in development. Curr. Top. Dev. Biol. 2012; 99:1–30. [DOI] [PubMed] [Google Scholar]
- 27.Ambros V. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 2011; 21:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes G.D., Frand A.R., Ruvkun G.. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006; 133:4631–4641. [DOI] [PubMed] [Google Scholar]
- 29.Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V.. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005; 9:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page A.P., Johnstone I.L.. The cuticle. WormBook. 2007; 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slack F.J., Basson M., Liu Z., Ambros V., Horvitz H.R., Ruvkun G.. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000; 5:659–669. [DOI] [PubMed] [Google Scholar]
- 32.Brenner J.L., Kemp B.J., Abbott A.L.. The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One. 2012; 7:e37185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meneely P.M., Herman R.K.. Lethals, steriles and deficiencies in a region of the X chromosome of Caenorhabditis elegans. Genetics. 1979; 92:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Saavedra E., Horvitz H.R.. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010; 20:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M.. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003; 113:25–36. [DOI] [PubMed] [Google Scholar]
- 36.Ruby J.G., Jan C.H., Bartel D.P.. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007; 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo A.S., Greenwald I.. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005; 310:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston R.J., Hobert O.. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003; 426:845–849. [DOI] [PubMed] [Google Scholar]
- 39.McJunkin K., Ambros V.. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3. 2014; 4:1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caudy A.A., Myers M., Hannon G.J., Hammond S.M.. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002; 16:2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutvagner G., Simard M.J., Mello C.C., Zamore P.D.. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004; 2:E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D.. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007; 130:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell. Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 44.Diederichs S., Haber D.A.. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007; 131:1097–1108. [DOI] [PubMed] [Google Scholar]
- 45.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C.. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001; 106:23–34. [DOI] [PubMed] [Google Scholar]
- 46.Navon A., Ciechanover A.. The 26 S proteasome: from basic mechanisms to drug targeting. J. Biol. Chem. 2009; 284:33713–33718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert G.I., Schell C., Kirschner K.M., Schafer S., Naumann R., Muller A., Kretz O., Kuropka B., Girbig M., Hubner N. et al. The GYF domain protein CD2BP2 is critical for embryogenesis and podocyte function. J. Mol. Cell Bio. 2015; 7:402–414. [DOI] [PubMed] [Google Scholar]
- 48.Frohn A., Eberl H.C., Stohr J., Glasmacher E., Rudel S., Heissmeyer V., Mann M., Meister G.. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol. Cell. Proteomics : MCP. 2012; 11:1442–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinze M., Kofler M., Freund C.. Investigating the functional role of CD2BP2 in T cells. Int. Immunol. 2007; 19:1313–1318. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y.J., Wei H.M., Chen L.Y., Li C.. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2014; 281:352–364. [DOI] [PubMed] [Google Scholar]
- 51.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 52.Bouasker S., Simard M.J.. The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic Acids Res. 2012; 40:10452–10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O'Carroll D., Lai E.C.. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez N.J., Gregory R.I.. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013; 19:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smibert P., Yang J.S., Azzam G., Liu J.L., Lai E.C.. Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 2013; 20:789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterjee S., Grosshans H.. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009; 461:546–549. [DOI] [PubMed] [Google Scholar]
- 58.Chatterjee S., Fasler M., Bussing I., Grosshans H.. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011; 20:388–396. [DOI] [PubMed] [Google Scholar]
- 59.Chu Y.D., Chen H.K., Huang T., Chan S.P.. A novel function for the DEAD-box RNA helicase DDX-23 in primary microRNA processing in Caenorhabditis elegans. Dev. Biol. 2016; 409:459–472. [DOI] [PubMed] [Google Scholar]
- 60.Jannot G., Bajan S., Giguere N.J., Bouasker S., Banville I.H., Piquet S., Hutvagner G., Simard M.J.. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011; 12:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zisoulis D.G., Kai Z.S., Chang R.K., Pasquinelli A.E.. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012; 486:541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lund E., Sheets M.D., Imboden S.B., Dahlberg J.E.. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011; 25:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H. Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007; 27:6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.