Abstract
The CRISPR/Cas9 system provides a revolutionary genome editing tool for all areas of molecular biology. In long non-coding RNA (lncRNA) research, the Cas9 nuclease can delete lncRNA genes or introduce RNA-destabilizing elements into their locus. The nuclease-deficient dCas9 mutant retains its RNA-dependent DNA-binding activity and can modulate gene expression when fused to transcriptional repressor or activator domains. Here, we systematically analyze whether CRISPR approaches are suitable to target lncRNAs. Many lncRNAs are derived from bidirectional promoters or overlap with promoters or bodies of sense or antisense genes. In a genome-wide analysis, we find only 38% of 15929 lncRNA loci are safely amenable to CRISPR applications while almost two-thirds of lncRNA loci are at risk to inadvertently deregulate neighboring genes. CRISPR- but not siPOOL or Antisense Oligo (ASO)-mediated targeting of lncRNAs NOP14-AS1, LOC389641, MNX1-AS1 or HOTAIR also affects their respective neighboring genes. Frequently overlooked, the same restrictions may apply to mRNAs. For example, the tumor suppressor TP53 and its head-to-head neighbor WRAP53 are jointly affected by the same sgRNAs but not siPOOLs. Hence, despite the advantages of CRISPR/Cas9 to modulate expression bidirectionally and in cis, approaches based on ASOs or siPOOLs may be the better choice to target specifically the transcript from complex loci.
INTRODUCTION
Long non-coding RNAs (lncRNAs) represent a large subgroup of RNAs that are over 200 nucleotides long and have a limited protein-coding potential (1). They play an important role in diverse cellular processes by regulating the gene expression at transcriptional or post-transcriptional levels (2,3). This ability of lncRNAs to modulate gene expression renders them indispensable for normal development (4,5). Mutations or aberrations in lncRNA expression have been implicated in several diseases including cancer (6–10), making them an important class of molecules which demand interrogation.
RNA interference (RNAi), which involves short double-stranded RNA-based knockdown of target RNA (11), has been used extensively and successfully for loss-of-function studies of lncRNAs. However, there are several limitations of RNAi for lncRNAs. Firstly, unlike protein-coding genes, many lncRNAs are nuclear (12) and even though the RNAi machinery has been found to be active in the nucleus (13), siRNAs against nuclear lncRNAs have often proven to be less effective (14). Secondly, a few lncRNAs like MALAT1 are expressed at very high levels making it uncertain whether an RNAi-mediated knockdown would suffice to generate a complete loss of function (15). Lastly, the molecular functions of some lncRNAs can be transcript-independent, meaning that their functions are carried out by the act of transcription rather than by the transcripts (16). Although RNAi-mediated targeting of the promoter proximal region of a gene can be used for transcriptional gene silencing (17,18), there are only individual examples of RNAi-based targeting of lncRNA transcription (19,20). An alternative to RNAi is offered in antisense oligonucleotides (ASOs) which involve RNase-H-mediated cleavage of target RNA (21). ASOs can target nuclear lncRNAs with higher efficiency (14), and they can also deplete nascent transcripts (22,23). Hence, siRNAs, shRNAs, siPOOLs or ASOs can successfully be used for one fraction of lncRNAs, while a significant number require alternative approaches.
CRISPRs (Clustered Regularly Interspaced Palindromic Repeats) were first discovered in bacteria where they serve as an adaptive immune system against invading phages and plasmid DNAs (24). The type II CRISPR system from S. pyogenes is the most widely studied system. In its simplest form, this system consists of two components: the Cas9 nuclease enzyme and a single guide RNA (sgRNA) which directs Cas9 to its target DNA site (25). This ability of Cas9 to bind and cleave DNA in a sequence-specific manner makes it a very powerful tool for genome engineering that has been widely utilized across various genomic studies over the last four years (26–29). Wild-type Cas9 can be used to generate double-stranded breaks (DSB) in the open reading frame (ORF) of a protein coding gene (CRISPRn mutagenesis), which in turn can induce frameshift mutations via the Non-Homologous End Joining (NHEJ) repair pathway resulting in an effective knockout of the targeted coding gene (30). Alternatively, Homology-Directed Repair (HDR) of the Cas9-induced DSBs (CRISPRn HR) can be used for gene corrections or to knock-in DNA elements for gene overexpression, knockout as well as tagging (31,32). Cas9 can also be used to induce small or large genomic deletions by generating multiple DSBs (CRISPRn excision) (33,34). Finally, a nuclease-deficient version of Cas9 (dCas9) which still possesses its RNA-dependent DNA-binding activity can be fused to effector domains and thus generate custom transcription factors. dCas9, when recruited to the vicinity of the promoter of a gene can interfere with either transcription initiation or elongation, thereby resulting in reduced transcription (35,36). dCas9 fused to the KRAB (Krüppel-associated box) domain of ZNF10 results in an even more potent inhibitor of transcription (CRISPR interference or CRISPRi) (36). On the other hand, dCas9 fused to transcriptional activation domains like VP64, p65, or Rta (37–39) induces target gene expression in cis (CRISPR activation or CRISPRa). CRISPRi/a systems have two major advantages especially for lncRNA research: first, also effects in cis can be observed which would be undetectable by plasmid-based overexpression or RNA interference-mediated knockdown in trans. Second, the activation of the endogenous promoter may lead to the expression of splice variants - which are very frequently observed for lncRNAs - in their natural ratios. Given the importance and vast repertoire of uncharacterized lncRNAs, it is tempting to utilize CRISPR-based genomic manipulation tools for their molecular and functional characterization.
LncRNA loci are widely distributed throughout the genome, residing at both inter- and intragenic genomic regions (40). Intergenic lncRNAs, commonly referred to as long intergenic non-coding RNAs (lincRNAs), can be transcribed from independent promoters as well as bidirectional promoters shared with protein-coding or other non-coding genes as defined by the distance between the two transcriptional start sites (TSSs) (Supplementary Data). Out of the examined lncRNAs in a recent study, the majority of the bidirectional lncRNAs originated within 2000 bp from other TSSs (41). Hence, we selected this 2000 bp window as the starting point for our analysis of bidirectional promoters.
Intragenic lncRNAs are classified as ‘Sense’ if they are transcribed from the same DNA strand as the gene(s) they intersect (Supplementary Data). Vice versa, lncRNAs are termed ‘Antisense’ if they intersect genes transcribed in the opposite direction in a head-to-head or a tail-to-tail orientation (Supplementary Data), or if they are completely overlapping / overlapped by another gene (Supplementary Data).
Both, sense and antisense, lncRNAs that are fully located within a gene body of another gene are commonly referred to as ‘Internal’ lncRNAs (Supplementary Data, left), and the overlapping gene is then termed a ‘Host’ gene. ‘Intronic’ lncRNAs represent a subclass of internal lncRNAs where the whole lncRNA locus is located within an intron of the host gene (Supplementary Data, left panel). Inversely, when a gene is encoded within an intron of a lncRNA, such lncRNA is termed ‘Overlapping’ (Supplementary Data, right panel).
Promoters of intragenic lncRNAs can be positioned within gene bodies of other genes, and thus termed internal (Supplementary Data, left panel), or within intergenic regions. In the latter case, the promoters can also be bidirectional or independent in relation to the nearby genes, as described above. Notably, internal lncRNA promoters of head-to-head oriented antisense lncRNAs that arise less than 2000 basepairs (bp) downstream of the overlapping gene start are also considered as bidirectional (Supplementary Data) (23,41,42).
Given the complex architecture of genomic loci around lncRNA genes, we hypothesize that these intersections can greatly impair the specificity of the potential use of the CRISPR/Cas9 toolbox for the genetic manipulation of lncRNAs due to the possibility of perturbing the overlapping or neighboring genes. This may lead to the identification of phenotypes which in fact would be attributable to neighboring genes. In the present study, we set out to evaluate such risks and examine the utility of different CRISPR/Cas9-based systems for the purposes of lncRNA research.
MATERIALS AND METHODS
Genome-wide ‘CRISPRability’ analysis
The list of Gene IDs of all human lncRNAs annotated to date was downloaded from Gencode (Release 24, GRCh38.p5, released December 2015). This list was run through the Bedtools Intersect tool against the list of all human genes (coding and non-coding) annotated in the same Gencode release to identify all Intragenic Sense and Antisense lncRNAs and their position relative to the genes they intersected. Similarly, the Bedtools Closest tool was run to identify genes that neighbor, but do not overlap with the lncRNAs.
Bedtools is a publically available toolset for genomics analysis and can be found at http://bedtools.readthedocs.org (43). Based on the information retrieved with Bedtools, lncRNAs were categorized according to the adopted lncRNA and promoter classifications (see Results section) and assigned to different ‘CRISPRability’ subsets.
Construction of plasmids
PX458-2X-sgRNA
Individual sgRNAs (NOP14-AS1 US sgRNA and NOP14-AS1 DS sgRNA) for NOP14-AS1 deletion were cloned into PX458 (Addgene Plasmid 48138). The U6–NOP14-AS1 DS sgRNA expression cassette from the PX458–NOP14-AS1 DS sgRNA plasmid was PCR amplified and cloned into XbaI–KpnI restriction sites of the PX458–NOP14-AS1 US sgRNA plasmid. The resulting plasmid was named PX458-2X-sgRNA.
LentidCas9-blast
The Cas9 coding sequence in the lentiCas9-Blast plasmid (Addgene Plasmid 52962) was mutated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) to generate the nuclease-deficient dCas9 (D10A and H841A) (35). Primers can be found in Supplementary Data. The resulting plasmid was named lentidCas9-Blast.
LentidCas9-KRAB-blast
The cDNA for the repression domain of KRAB (amino acids 11–75) (44) fused with an XTEN linker (45) on its N-terminal end was synthesized (GeneArt / ThermoFisher Scientific). This was then cloned in frame downstream of dCas9. The resulting plasmid was named lentidCas9-KRAB-Blast.
LentidCas9-KRAB-PURO iv sgRNA
The dCas9-KRAB coding sequence was cut out from the lentidCas9-KRAB-Blast vector using FastDigest AgeI and BamHI (ThermoFisher Scientific) and cloned into the same sites in lentiCRISPR v2 (Addgene Plasmid #52961) replacing Cas9. An improved sgRNA scaffold (46) was synthesized (GeneArt) and cloned into the KpnI–NheI sites replacing the sgRNA. The resulting plasmid was named lentidCas9-KRAB-PURO iv sgRNA (iv = improved version). Plasmid sequences can be found in Supplementary Data.
All sgRNAs against the NOP14-AS1, LOC389641, MNX1-AS1, HOTAIR, LINC00441, HOXD1-AS1 and TP53 loci were designed using the online tool available at www.crispr.mit.edu. To control for specificity, only those guides were chosen which had two or more mismatches to any of the predicted off-targets in the genome. Sense and antisense oligonucleotides corresponding to the sgRNAs were annealed and cloned into PX458 / lentiGuide-Puro (Addgene Plasmid 52963) / lentidCas9-KRAB-PURO iv sgRNA as described in (31).
Cell culture, plasmid transfection, virus production and transduction, siPOOL / antisense LNA GapmeR knockdown
HEK293, HEK293T, Hela and HLE cells were cultured in Dulbecco's Modified Eagles Medium (DMEM, Sigma-Aldrich) supplemented with 10% FBS and 1% l-glutamine at 37°C in 5% CO2 in a humidified chamber. NCI-H460 cells were cultured in RPMI-1640 media supplemented with 10% FBS and 1% l-glutamine at 37°C in 5% CO2 in a humidified chamber.
For NOP14-AS1 deletion in HEK293, 3 × 105 cells in 2 ml media were seeded per well of a six-well plate (Greiner Bio-One) one day prior to transfection. Cells were transfected with 2.5 μg of PX458-2X-sgRNA plasmid using 15 μl Lipofectamine 2000 (ThermoFisher Scientific) in 200 μl Opti-MEM medium (ThermoFisher Scientific). Forty eight hours post transfection, EGFP expressing cells were single cell sorted into a 96-well plate. These were then allowed to proliferate for two weeks and each clone obtained was screened for NOP14-AS1 deletion.
For virus production, HEK293T cells (4 × 105 cells/well, 2 ml media) were seeded in poly-l-lysine coated 6-well plates (Greiner Bio-One) one day prior to transfection. Cells were co-transfected with 1.2 μg lentidCas9-Blast / lentidCas9-KRAB-Blast / lentiGuide-PURO (with the sgRNA) / lentidCas9-KRAB-PURO iv sgRNA (with the sgRNA), 0.9 μg packaging vector psPAX2 and 0.3 μg envelope plasmid pMD2.G using 6 μl Lipofectamine 2000 in 200 μl Opti-MEM medium. Forty eight hours post transfection, lentivirus-containing medium was filtered through a low protein binding 0.45 μm syringe filter (Millipore). NCI-H460 / HLE / HeLa cells were then infected with several dilutions of the virus along with Polybrene (final concentration: 8 μg/ml). Twenty four hours post-infection, the media was replaced with either 6 μg/ml Blasticidin (Invivogen) or 2 μg/ml Puromycin (ThermoFisher Scientific) containing RPMI-1640 / DMEM media. For Blasticidin selection (dCas9 / dCas9-KRAB expressing lentiviruses), cells were incubated in selection media for seven days, whereas for Puromycin selection (sgRNA expressing lentiviruses), cells were incubated for two days. Non-transduced cells were used as negative controls to monitor for a complete selection. The multiplicity of Infection (MOI) of the virus was estimated by comparing the number of surviving cells as compared to a non-transduced, non-selected control. Cells infected with viral particles with MOI <1 were lysed in 1 ml TRI reagent (Sigma-Aldrich) for RNA extraction or 1X RIPA buffer for protein extraction.
For NOP14-AS1 knockdown using Antisense LNA GapmeRs (Exiqon), NCI-H460 cells (2 × 105 cells per well) were reverse transfected with 50 nM (final concentration) of the control GapmeR or two independent GapmeRs against NOP14-AS1 using 2 μl Dharmafect1 reagent (Dharmacon GE Life Sciences) in 12-well plates. Twenty four hours post-transfection, cells were lysed in 1 ml TRI reagent. For MNX1-AS1 knockdown using ASOs (IDT DNA Technologies), NCI-H460 cells (4×105 cells per well) were reverse transfected with 30 nM (final concentration) of the control ASO or two independent ASOs against MNX1-AS1 using 4 μl Dharmafect1 reagent in six-well plates. For TP53 knockdown using siPOOLs (47), NCI-H460 cells (4 × 105 cells per well) were reverse transfected with 3 nM (final concentration) of siPOOL control / siPOOL TP53 (siTOOLs Biotech) using 6 μl RNAiMAX reagent (ThermoFisher Scientific) in six-well plates. Forty eight hours post transfection, cells were lysed in 1 ml TRI reagent for RNA extraction or 1× RIPA buffer for protein extraction.
RESULTS
Criteria of lncRNA availability for CRISPR/Cas9 targeting—‘CRISPRability’
To understand how the organization of lncRNA loci could limit the utility of CRISPR/Cas9, we established the criteria of lncRNA availability to existing CRISPR/Cas9-based genomic manipulation tools.
CRISPRn mutagenesis is generally not applicable for knocking out non-coding genomic loci due to the nature of ncRNA function. First, the sequences of the lncRNA transcripts responsible for carrying out transcript-dependent molecular functions remain generally uncharacterized, which makes their mutagenic targeting with Cas9 practically impossible. Predicting the active parts of a lncRNA for targeting is currently not possible and hence small mutations are unlikely to affect its activity. Secondly, for lncRNA loci which exert their phenotype by the act of their transcription per se, such functions will likely not be affected by Cas9-induced mutations, unless Cas9 is targeted to regulatory regions controlling lncRNA transcription, which remain unknown for the majority of lncRNAs. Hence, we did not consider this system applicable for lncRNA manipulation.
CRISPRn HR can be used to knockdown a lncRNA using Cas9 by homology-directed knock-in of a transcriptional termination signal or RNA destabilizing elements immediately downstream of the TSS of the lncRNA gene (48,49). We have previously utilized this method to achieve an efficient knockout of MALAT1, using zinc finger nucleases (ZFNs) instead of CRISPR/Cas9 (15). However, such strategy cannot be used for lncRNAs that arise from internal promoters or whose promoter-proximal regions span over other genes without disturbing their sequences.
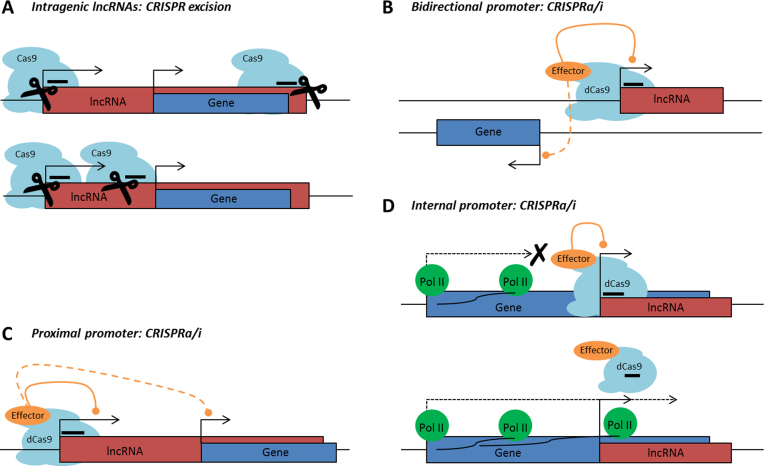
CRISPRn excision can be used to generate lncRNA knockouts by either deleting their promoters (48,50–53) or the entire lncRNA genes (48,54). Selective excision of full-length lncRNA loci is impossible for lncRNAs that intersect any other genes as this will inevitably alter the sequences of those genes (Figure 1A top panel). Deletion of lncRNA promoter regions can be performed for lncRNAs that arise from non-internal promoters (Figure 1A, bottom panel). For lncRNAs arising from internal / bidirectional promoters, it might be possible to generate a functional knockout by deleting a part of the lncRNA distant from its promoter and not overlapping with any other gene (55). However, there is a possibility that the functional domain of the lncRNA lies in the overlapping exon and in such a case, the lncRNA would still be functional. Also, deleting a part of the lncRNA without removing its TSS could lead to the generation of a new gene body. In any case, genomic excisions can lead to the deletion of regulatory DNA elements, which might affect transcription of other genes and give rise to phenotypes which are originally not attributable to the lncRNA (48,49,56,57). However, it is impossible to consider the position of all potential genomic regulatory elements, as they are largely uncharacterized. Thus, to minimize such risks, CRISPRn excision should be preceded by a detailed examination of the genomic region of interest.
Figure 1.
Overview of ‘non-CRISPRability’ cases. (A) Top: full-length excision of Intragenic lncRNAs leads to full or partial deletion of the genes they intersect, and therefore cannot be utilized. Bottom: partial deletion of 5΄-proximal genomic regions of lncRNAs arising from non-internal promoters represents a feasible tool for lncRNA manipulation, however bears certain major limitations (see text). (B) CRISPRa/i is not usable for targeting bidirectionally transcribed lncRNAs since the transcription of the adjacent gene might be affected. (C) CRISPRa/i efficiently represses transcription from any promoters located in the vicinity of the desired target site (up to 1.5 – 2 kb), therefore promoters of lncRNAs located in proximity to promoters of other genes cannot be selectively targeted. (D) Top: LncRNAs transcribed from internal promoters cannot be manipulated with CRISPRa/i, as the dCas9 complex binding might affect RNA-Pol II processivity, and thus hinder the transcription of the gene that embeds the internal promoter. Bottom: On the other hand, RNA-Pol II-mediated transcription of the overlapping gene might impair the binding of the dCas9-complex to its target site and therefore reduce the efficiency of CRISPRa/i. CRISPRa or CRISPR activation: dCas9 tagged to a transcriptional activation domain (e.g. VP64); CRISPRi or CRISPR interference: dCas9 alone or dCas9 tagged to a transcriptional inhibitory domain (e.g. KRAB).
CRISPRa / CRISPRi with dCas9-based transcription factors have been used for genome-wide gain- as well as loss-of-function screens for protein-coding genes (58,59). A few examples of lncRNA knockdown using CRISPRi also exist (58,60,61). Since both CRISPRa and CRISPRi involve recruitment of dCas9 to the promoter proximal region of the target gene, it is highly likely that they affect the expression of nearby genes. Therefore, this technique should not be used for modulation of lncRNAs arising from bidirectional promoters or promoters that are positioned in close proximity to the start sites of other genes (Figure 1B and C). Lastly, targeting dCas9 to internal promoters might also interfere with the transcription of the host genes (Figure 1D).
In the end, we subsumed all the aforementioned locus architectures to formulate the lncRNA ‘CRISPRability’ rules based on potential effects on neighboring genes:
LncRNAs (Sense, Antisense and Intergenic) were considered ‘non-CRISPRable’ when transcribed from bidirectional promoters, defined by the presence of another promoter 2000 bp upstream or downstream of the lncRNA start;
LncRNAs (Sense, Antisense and Intergenic) were considered ‘non-CRISPRable’ when the start of a lncRNA was located closer than 2000 bp to the start of the neighboring gene, excluding lncRNAs transcribed from bidirectional promoters. We termed such cases as ‘proximal promoters’;
LncRNAs (Sense and Antisense) were considered ‘non-CRISPRable’ when transcribed from internal promoters, i.e. the start of the lncRNA fell within the gene body of another transcript (coding or non-coding, Sense or Antisense), excluding lncRNAs transcribed from bidirectional or proximal promoters.
Next, we applied these rules to lncRNAs to determine the extent of their ‘non-CRISPRability’.
Genome-wide analysis reveals that the majority of lncRNAs cannot be targeted by CRISPR/Cas9
We first applied the ‘CRISPRability’ criteria to a group of lncRNAs deregulated in four major human cancers, namely lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), breast carcinoma (BRCA) and liver hepatocellular carcinoma (LIHC). Per cancer entity, we selected 25 lncRNAs that exhibited the highest significance level of differential expression, calculated from the RNA sequencing data of patient samples available in ‘The Atlas of non-coding RNA in Cancer’ (TANRIC) database (62). We then examined a total of 100 lncRNAs in the UCSC Genome Browser (63) in order to determine their ‘CRISPRability’ (Supplementary Data).
The investigated lncRNA sets exhibited varying ‘CRISPRability’, ranging from 20% in LIHC to 68% in LUSC, in all cases leaving a large proportion of the lncRNAs ‘non-CRISPRable’ (Supplementary Data). In total, targeting lncRNAs with CRISPR/Cas9-based tools appeared to be problematic for at least 59% of the examined lncRNAs, mainly as a result of the bidirectional nature of their promoters (38% of total) (Supplementary Data). However, since this study was limited to 100 lncRNAs, we further performed a similar analysis on a genome-wide scale to confirm our preliminary observations.
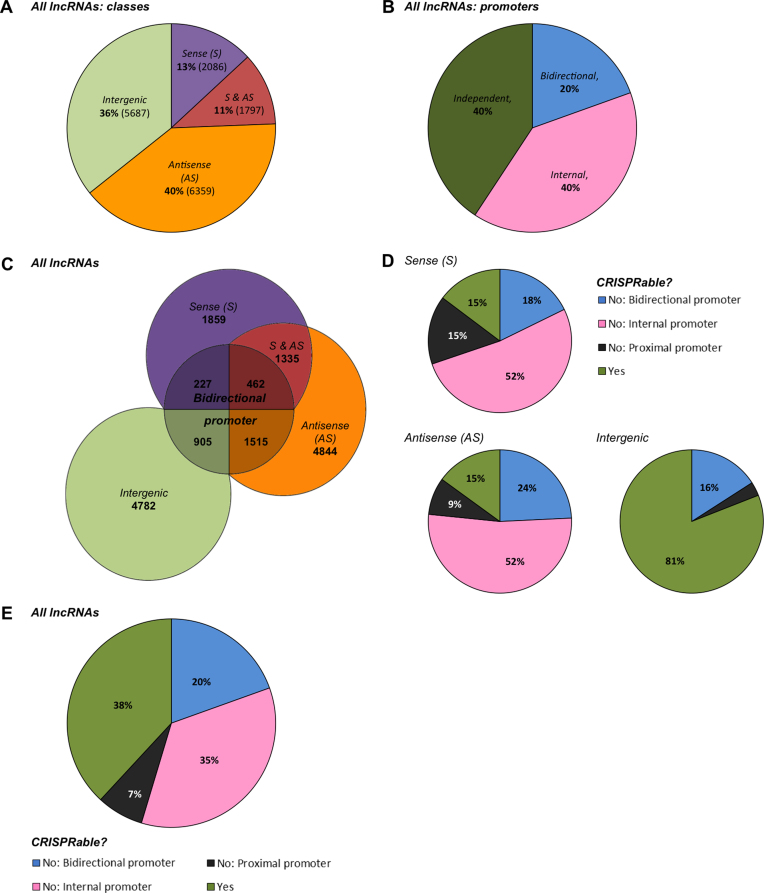
First, we categorized all lncRNAs annotated by Gencode based on the defined lncRNA and promoter classifications (Figure 2A–C, Supplementary Data). Intragenic lncRNAs constituted 64% of all lncRNAs, with the majority (40%) classified as Antisense lncRNAs. Sense lncRNAs comprised a fraction of 13% and 11% fell in both Sense and Antisense categories due to multiple neighboring genes. Intergenic lncRNAs made up the remaining 36% (Figure 2A and C). LncRNAs were transcribed from independent and internal promoters in equal shares (40%). Additionally, 20% of all lncRNAs turned out to be transcribed in a bidirectional fashion (Figure 2B and C).
Figure 2.
Genome-wide lncRNA classification and ‘CRISPRability’. (A) Genome-wide lncRNA distribution by classes: Antisense (AS) lncRNAs comprised the most abundantly represented class of lncRNAs, encompassing 51% of all lncRNAs annotated in the human genome. Sense (S) lncRNAs made up 24% of lncRNAs with 11% of lncRNAs falling into both Sense and Antisense categories (S & AS). Intergenic lncRNAs included 36% of all lncRNAs and thus comprised the second largest lncRNA group. (B) Genome-wide lncRNA promoter distribution: 40% of lncRNAs were transcribed from independent promoters. The same fraction (40%) of lncRNAs was transcribed from promoters located within other genes and are termed internal. Furthermore, 20% of lncRNAs were transcribed in a divergent manner from bidirectional promoters shared with other genes (coding and non-coding). (C) Genome-wide lncRNA representation: Out of a total of 15929 lncRNAs annotated by Gencode Release 24 (GRCh38p.5, released December 2015), Antisense (AS) lncRNAs represented the largest class consisting of 8156 lncRNAs with 1797 of them falling into both Sense and Antisense (S & AS) categories. Sense (S) lncRNAs represented a smaller class consisting of 3883 lncRNAs. In total, 1515 AS lncRNAs, 1335 S & AS lncRNAs, and 227 S lncRNAs were transcribed in a bidirectional fashion. The Intergenic lncRNA class comprised 5687 lncRNAs, with 905 transcribed from bidirectional promoters. (D) LncRNA ‘CRISPRability’—accessibility for CRISPR/Cas9-based tools—by classes: 85% of all Sense as well as Antisense and 19% of all Intergenic lncRNAs appeared ‘non-CRISPRable’, mostly due to the internal or bidirectional nature of the lncRNA promoters. (E) Genome-wide lncRNA ‘CRISPRability’: In total, 38% of all lncRNAs fulfilled the set of ‘CRISPRability’ criteria, while 62% of lncRNAs fell into the ‘non-CRISPRable’ category. Transcription from internal and bidirectional promoters accounted for the ‘non-CRISPRability’ of 35% and 20% of lncRNAs, respectively. 9% of lncRNAs were considered ‘non-CRISPRable’ due to proximal promoters.
Based on this characterization, we performed the genome-wide ‘CRISPRability’ analysis, employing the aforementioned criteria for targeting lncRNAs with CRISPRs. As much as 62% of lncRNAs did not fulfill the ‘CRISPRability’ requirements: 85% of Sense and Antisense lncRNAs as well as 19% of Intergenic lncRNAs (Figure 2D and E). In total, 62% of all lncRNAs were determined ‘non-CRISPRable’ by our analysis, mostly due to their internal (35%) or bidirectional (20%) promoters (Figure 2E, Supplementary Data). Narrowing down the bidirectionality window to 1000 bp or 500 bp around the TSS did not result in a significant drop in the number of ‘non-CRISPRable’ lncRNAs. The most stringent 500 bp criterion still resulted in 58% ‘non-CRISPRable’ lncRNAs (11% of total due to bidirectional promoters) (Supplementary Data).
Interestingly, only 23 out of 15929 lncRNAs were annotated by GENCODE as pseudogenes, and the distribution of these pseudogene-derived lncRNAs by ‘CRISPRability’ classes followed the general trend (bidirectional—5 (21.7%), internal—8 (34.8%), proximal—1 (4.4%), independent—9 (39.1%)). Given that the current GENCODE release contains 14517 pseudogenes, they comprise a separate entity beyond the scope of our study.
These results suggested that the experimental possibilities to safely and specifically apply CRISPR/Cas9-based lncRNA manipulations were severely limited by the complex genomic organization of lncRNA loci. Importantly, the same caution needs to be exercised when complex loci of coding genes or pseudogenes are targeted. Hence, we further set out to experimentally confirm multiple lncRNAs classified as ‘non-CRISPRable’ to corroborate the relevance of the classification.
NOP14-AS1 deletion using Cas9 or knockdown using dCas9 / dCas9-KRAB affects MFSD10 expression
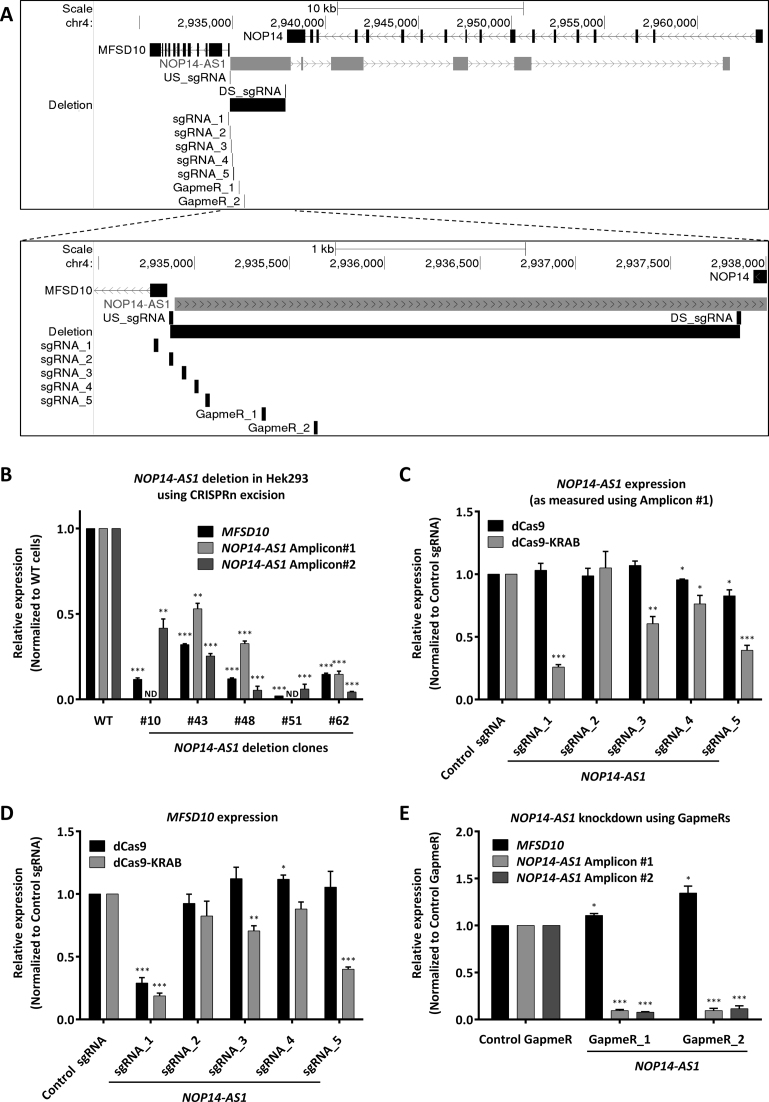
As an example of a lncRNA arising from a bidirectional promoter, we selected NOP14-AS1 which is transcribed from a promoter upstream region of the MFSD10 gene. We first deleted NOP14-AS1 in HEK293 cells using the CRISPRn excision approach by co-transfecting Cas9 along with two sgRNAs targeting a part of the NOP14-AS1 gene that did not overlap with any other gene. The upstream sgRNA (US_sgRNA) was designed immediately upstream of the NOP14-AS1 TSS without affecting the MFSD10 TSS, whereas the downstream sgRNA (DS_sgRNA) was designed in the NOP14-AS1 gene body immediately downstream of the overlapping NOP14 gene (Figure 3A). Clonal expansion and genotyping of the deletion clones revealed that 4 clones were heterozygous and one clone (clone #51) was homozygous for NOP14-AS1 deletion (Supplementary Data). Sanger sequencing of the region around the deletion uncovered deletions extending beyond the sgRNA sites by varying lengths (±40 bp) (Supplementary Data). RT-qPCR analysis of these clones showed that all of them had reduced NOP14-AS1 expression with two independent RT-qPCR amplicons to measure NOP14-AS1 expression. Notably, we observed a strong decrease in MFSD10 expression in all clones indicating that the promoter of this gene was also affected by the deletion (Figure 3B).
Figure 3.
NOP14-AS1 gene modulation using CRISPRn / CRISPRi and antisense LNA GapmeRs. (A) Schematic representation of the MFSD10 / NOP14-AS1 genomic locus depicting the sgRNAs and ASOs used in this study to target NOP14-AS1. (B) Expression analysis of NOP14-AS1 deletion clones in HEK293 cells. RT-qPCR results for MFSD10 and NOP14-AS1 (amplicon #1 and amplicon #2) normalized to cyclophilin A and Wild Type (WT) cells. ND = not detected. Error bars represent SEM (n = 3). * P < 0.05; ** P < 0.01; *** P < 0.001 compared to WT, unpaired two-sided t-test. (C, D) NCI-H460 cells expressing either dCas9 or dCas9-KRAB were transduced with either Control sgRNA or one of the five indicated sgRNAs targeting NOP14-AS1. RT-qPCR results for NOP14-AS1 amplicon #1 (C) and MFSD10 (D) normalized to cyclophilin A and control sgRNA. Error bars represent SEM (n ≥ 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to dCas9 / dCas9-KRAB + control sgRNA, unpaired two-sided t-test. (E) NCI-H460 cells were transfected with either a control or two independent antisense LNA GapmeRs against NOP14-AS1. RT-qPCR results for NOP14-AS1 amplicon#1, amplicon#2 and MFSD10 normalized to cyclophilin A and control antisense LNA GapmeR. Error bars represent SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control GapmeR, unpaired two-sided t-test.
Next, we used a CRISPRi approach to knockdown NOP14-AS1. We designed five sgRNAs spanning this locus (Figure 3A). sgRNA_1 and sgRNA_2 (US_sgRNA) were designed upstream of the NOP14-AS1 TSS whereas sgRNA_3 to sgRNA_5 were designed at the 5΄-end of the gene body of NOP14-AS1, at increasing distance from the NOP14-AS1 TSS with the aim of identifying at least one such sgRNA which could target only the NOP14-AS1 and not its neighboring gene. When these sgRNAs were expressed in NCI-H460 cells expressing dCas9 (without an inhibitory domain), only sgRNA_4 and sgRNA_5 showed a minor repression of NOP14-AS1 Amplicon #1 but not Amplicon #2 (Figure 3C, Supplementary Data). However, sgRNA_1 strongly repressed MFSD10 expression indicating that this sgRNA targeted the core promoter of MFSD10 (Figure 3D). These results showed that dCas9 alone was not a potent inhibitor of transcription unless it was targeted to the core promoter.
On the other hand, introduction of these sgRNAs in dCas9-KRAB expressing NCI-H460 cells resulted in strong repression of the intended target NOP14-AS1 for the sgRNAs except sgRNA_2 and sgRNA_4 (Figure 3C, Supplementary Data). Importantly, also the neighboring MFSD10 coding gene was affected by all sgRNAs which repressed NOP14-AS1 (Figure 3D). We could not find a sgRNA which reduced NOP14-AS1 expression without impacting the expression of the neighboring gene MFSD10.
To determine whether the simultaneous knockdown of MFSD10 and NOP14-AS1 expression using dCas9-KRAB could be due to an endogenous regulatory mechanism or was an artifact of this technique, we used antisense LNA GapmeRs to knockdown expression of the lncRNA NOP14-AS1. Knockdown of NOP14-AS1 using two independent antisense LNA GapmeRs reduced the expression of NOP14-AS1. In stark contrast to the results from a dCas9-KRAB-mediated knockdown, MFSD10 expression was unchanged or even slightly induced upon NOP14-AS1 knockdown using antisense LNA GapmeRs (Figure 3E). To validate these results in a second cell line, we also expressed sgRNA_1 and sgRNA_3 in A549 cells along with dCas9-KRAB and observed the same reduction in NOP14-AS1 as well as MFSD10 expression (Supplementary Data). We also transfected an antisense LNA GapmeR into A549 cells and observed strong repression of NOP14-AS1 without any effect on MFSD10 expression (Supplementary Data). Hence, MFSD10 and NOP14-AS1 expression were independent, but effective knockdown of NOP14-AS1 without affecting MFSD10 was not achievable with dCas9 or dCas9-KRAB.
CRISPRi, but not RNAi- / ASO-mediated knockdown of different lncRNAs affects expression of their divergent neighbors
As additional examples of ‘non-CRISPRable’ lncRNAs, we chose LOC389641, MNX1-AS1, HOTAIR, LINC00441, and HOXD-AS1.
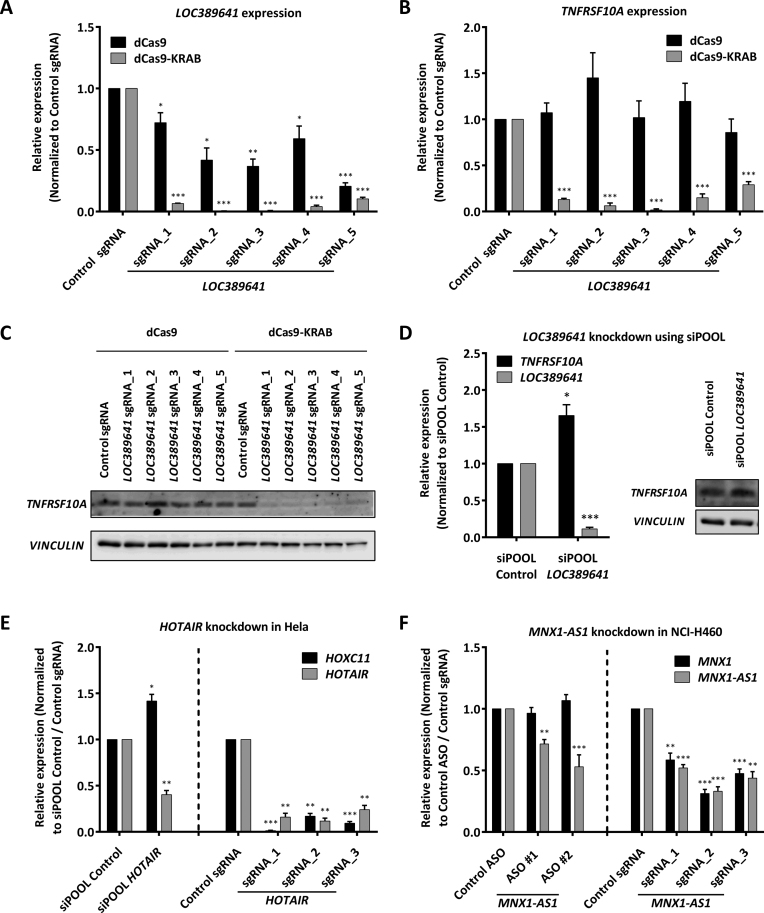
LOC389641 arises from the bidirectional promoter of the TRAIL receptor TNFRSF10A. We designed five sgRNAs spanning the LOC389641 promoter (Supplementary Data). Combined with dCas9-KRAB, all five sgRNAs strongly inhibited the expression of LOC389641 (Figure 4A), but at the same time also decreased TNFRSF10A mRNA as well as protein expression (Figure 4B and C, Supplementary Data). In this case, dCas9 alone had a modest inhibitory effect on the lncRNA LOC389641, but not on the mRNA. Hence, dCas9 displayed a slightly better specificity here not observed for NOP14-AS1, but its potency was much lower than for dCas9-KRAB preventing its alternative use. RNAi-mediated silencing of LOC389641 using an siPOOL of 30 defined siRNAs with non-identical seed regions to minimize any potential off-target effects (47) resulted in strong repression of LOC389641 without affecting TNFRSF10A expression (Figure 4D, Supplementary Data).
Figure 4.
CRISPRi and siPOOL- / ASO-mediated knockdown of LOC389641, MNX1-AS1 and HOTAIR. (A–C) HLE cells expressing either dCas9 or dCas9-KRAB were transduced with either control sgRNA or one of five indicated sgRNAs targeting LOC389641. RT-qPCR results for LOC389641 (A) and TNFRSF10A (B) normalized to cyclophilin A and control sgRNA. Error bars represent SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to dCas9 / dCas9-KRAB + control sgRNA, unpaired two-sided t-test. (C) Western blot results for TNFRSF10A. VINCULIN was used as loading control. (D) HLE cells were transfected with either siPOOL control or siPOOL LOC389641. RT-qPCR results for TNFRSF10A and LOC389641 normalized to cyclophilin A and siPOOL control. Error bars represent SEM (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control siPOOL, unpaired two-sided t-test. Western blot results for TNFRSF10A. VINCULIN was used as loading control. (E) NCI-H460 cells were transfected with either a control ASO or two independent ASOs against MNX1-AS1. Also, NCI-H460 cells expressing dCas9-KRAB were transduced with either control sgRNA or one of the three indicated sgRNAs targeting MNX1-AS1. RT-qPCR results for MNX1 and MNX1-AS1 normalized to cyclophilin A and control ASO / control sgRNA. Error bars represent SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control ASO / sgRNA, unpaired two-sided t-test. (F) Hela cells were transfected with either siPOOL control or siPOOL HOTAIR. Also, Hela cells were transduced with lenti dCas9-KRAB-PURO iv sgRNA containing either control sgRNA or one of the three indicated sgRNAs targeting HOTAIR. RT-qPCR results for HOXC11 and HOTAIR normalized to cyclophilin A and siPOOL Control or control sgRNA. Error bars represent SEM (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control ASO / sgRNA, unpaired two-sided t-test.
HOTAIR is a well-studied lncRNA which arises from the HOXC locus and regulates transcription of several genes by mediating chromatin modifications (64–66). One of the variants of HOTAIR arises from the HOXC11 intron, ∼1.8 kb downstream of its TSS (Supplementary Data). dCas9-KRAB-mediated targeting using three independent sgRNAs but not siPOOL-mediated knockdown of HOTAIR resulted in simultaneous knockdown of HOXC11 (Figure 4E).
MNX1-AS1 arises from the bidirectional promoter of MNX1 (Supplementary Data). In NCI-H460 lung cancer cells, dCas9-KRAB-mediated knockdown of MNX1-AS1 using three independent sgRNAs led to knockdown of both MNX1-AS1 as well as its neighboring mRNA MNX1, while MNX1-AS1 knockdown using two independent ASOs reduced MNX1-AS1 expression without affecting MNX1 expression (Figure 4F).
LINC00441 arises from the bidirectional promoter of the tumor suppressor gene RB1 and its knockdown using siRNAs in A549 cells did not impact RB1 expression (67). However, dCas9-KRAB mediated knockdown of LINC00441 using three independent sgRNAs targeting its promoter strongly reduced RB1 mRNA as well as protein expression in these cells (Supplementary Data). Similarly, knockdown of the lncRNA HOXD-AS1 using siRNAs did not affect the expression of its neighboring gene (HOXD1) (68). However, dCas9-KRAB-mediated targeting of HOXD-AS1 resulted in strong repression of HOXD1, as well (Supplementary Data).
TP53 knockdown using dCas9 / dCas9-KRAB affects WRAP53α expression
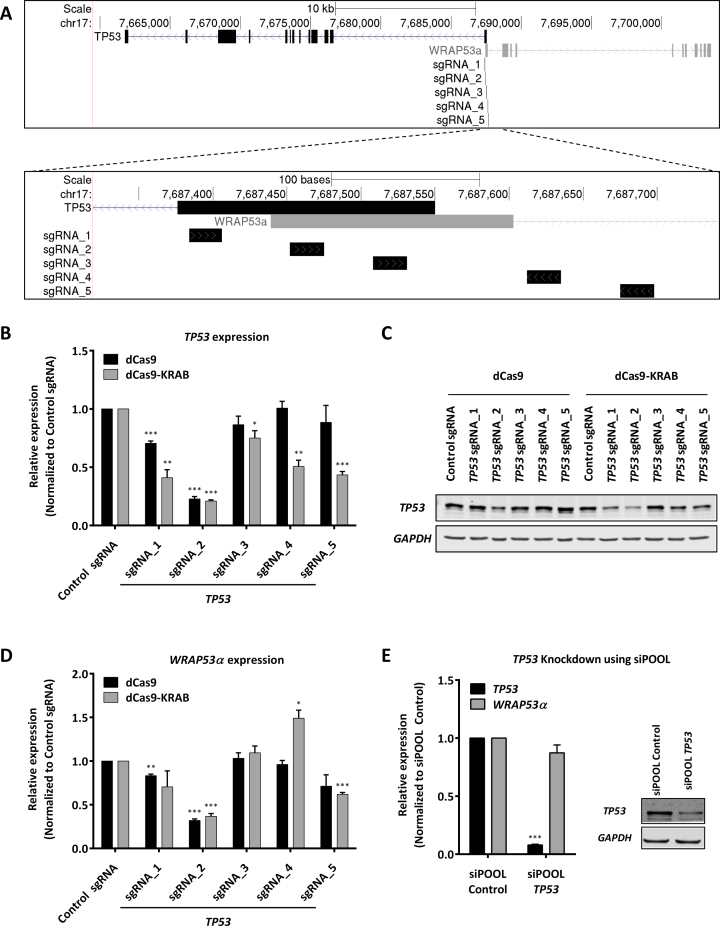
The well-known tumor suppressor gene TP53 is also transcribed in a bidirectional fashion, partially overlapping with another gene (WRAP53) in a head-to-head antisense orientation. WRAP53 is expressed from three different TSSs, namely α, β and γ. The α-TSS lies roughly 100 bp downstream of the TP53 TSS overlapping with its first exon (Figure 5A). The transcript arising from this TSS (WRAP53α) binds to TP53 mRNA via sense-antisense base-pairing and regulates TP53 mRNA stability, expression and is also required for TP53 induction upon DNA damage (69). Since TP53 is one of the most widely characterized genes and this locus is another example of a bidirectional promoter, we knocked down TP53 expression using dCas9 / dCas9-KRAB as well as siPOOLs to compare the effects on WRAP53α expression. We designed five different sgRNAs distributed across the TP53 promoter (Figure 5A). When these sgRNAs were expressed in dCas9 expressing NCI-H460 cells, sgRNA_1 and sgRNA_2 resulted in TP53 knockdown (Figure 5B and C, Supplementary Data). Both of these sgRNAs also resulted in WRAP53α knockdown (Figure 5D). When these sgRNAs were expressed in dCas9-KRAB expressing NCI-H460 cells, all sgRNAs led to a significant repression of TP53 expression to varying extents (Figure 5B, and C, Supplementary Data). We also tested one of these cell lines (dCas9-KRAB + sgRNA_2) for a TP53 functional knockdown. Doxorubicin treatment of these cells followed by PARP cleavage assay showed a strong reduction in apoptosis as compared to control sgRNA expressing cells, indicating a functional knockdown of TP53 using dCas9-KRAB (Supplementary Data). All sgRNAs strongly affecting TP53 expression also had a strong impact on WRAP53α expression except sgRNA_1 with only a non-significant trend (Figure 5D).
Figure 5.
TP53 knockdown using CRISPRi and siPOOL. (A) Schematic representation of the TP53 / WRAP53α genomic locus and the sgRNAs used in this study to target TP53. (B–D) NCI-H460 cells expressing either dCas9 or dCas9-KRAB were transduced with either Control sgRNA or one of five indicated sgRNAs targeting TP53. RT-qPCR results for TP53 (B) and WRAP53α (D) normalized to cyclophilin A and control sgRNA. Error bars represent SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to dCas9 / dCas9-KRAB + control sgRNA, unpaired two-sided t-test. (C) Western blot results for TP53. GAPDH was used as loading control. (E) NCI-H460 cells were transfected with either siPOOL Control or siPOOL TP53. RT-qPCR results for TP53 and WRAP53α normalized to cyclophilin A and siPOOL Control. Error bars represent SEM (n = 6). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control siPOOL, unpaired two-sided t-test. Western blot results for TP53. GAPDH was used as loading control.
In contrast, RNAi-mediated silencing of TP53 using a siPOOL resulted in a strong downregulation of TP53 mRNA as well as protein, but had no effect on WRAP53α expression (Figure 5E, Supplementary Data). Thus, dCas9- / dCas9-KRAB-mediated targeting of bidirectional promoters again led to an artifactual repression of both genes in the locus. A superior strategy to knockout such a gene could be to either use CRISPRn to disrupt the ORF or CRISPRn HR-mediated introduction of transcriptional / translational termination sequences in the non-overlapping part of the gene (e.g. exon2 in this case).
DISCUSSION
The CRISPR/Cas9 system has proven to be revolutionary in all fields of molecular biology allowing genome editing as well as transcriptional modulation in cis. However, several important challenges remain. Besides the question of specificity of the Cas9-sgRNA complex in DNA targeting and the efficacy of dCas9-mediated effects, we emphasize that Cas9-mediated approaches can have another major limitation: they can affect the neighboring or overlapping genes in loci harboring multiple genes as frequently found in the human genome. As examples, we show that targeting the lncRNAs NOP14-AS1, LOC389641, MNX1-AS1, LINC00441 and HOXD-AS1, as well as the protein-coding mRNA TP53 with the CRISPR/Cas9 system significantly affects their neighboring genes, while an RNAi- / ASO-mediated knockdown did not have these effects.
Notably, discrepancies between RNAi- and CRISPR-based screens can already be found in the literature although only very few studies involving CRISPRi screens have been published so far: SEC24C was found to protect cells from ricin-mediated cell death in a dCas9-KRAB-mediated CRISPRi screen. However, the resulting phenotype was much stronger as compared to a shRNA-based knockdown of SEC24C (58,70). Analyzing the locus in more detail, we noticed that the SEC24C promoter is also a bidirectional promoter and gives rise to an unannotated lncRNA, as well (Supplementary Data). This underlines the fact that special attention needs to be paid to genes sharing promoters or overlapping with other genes in the design of sgRNAs.
In summary, the CRISPR/Cas9 system provides an important and essential tool that also comparatively easily allows genome-wide screens. However, the limitations of this system need to be taken into account in order to obtain relevant and biologically significant results for the genes targeted. The complexity of the targeted gene locus needs to be evaluated for sgRNA design and data analysis. Also, to avoid false positives, validation using orthogonal techniques like RNAi-based methods using siRNAs, siPOOLs or shRNAs or RNase H-mediated RNA degradation using ASOs or GapmeRs should complement the CRISPR-based experiments.
As a practical guideline, we suggest the following three simple steps for CRISPR-mediated genome-editing or transcriptional modulation approaches to ensure the correct attribution of a phenotype to the desired target: (i) the locus of the target gene should be carefully studied for neighboring or overlapping genes before the design of the sgRNAs; (ii) the expression of neighboring or overlapping genes should be monitored in parallel to the intended target gene; (iii) phenotypes—if not mediated in cis—should be reproducible with RNAi- or RNase H-mediated approaches or rescued by exogenous expression of the intended target gene.
Supplementary Material
ACKNOWLEDGEMENTS
The PX458, lentiCas9-Blast, lentiCas9-Puro and lentiCRISPR v2 plasmids were kindly provided by Feng Zhang via Addgene. psPAX2 and pMD2.G were a gift from Prof. Didier Trono. We would like to thank Dr. Maïwen Caudron-Herger for introducing the Bedtools toolset and executing the scripts. We would like to thank Dr Kai Breuhan and Prof. Dr Peter Schirmacher for providing us with the antibodies and laboratory facilities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in the Diederichs labs is supported by the German Research Foundation [DFG Di 1421/7-1, SFB 850, CellNetworks EXC81 EcTop 5]; RNA@DKFZ Cross Program Topic; Helmholtz International Graduate School for Cancer Research (to K.M.); Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (to M.K.); Parts of this work are part of the Ph.D. theses of A.G., K.M. and M. K., respectively. Funding for open access charge: Core funding.
Conflict of interest statement. Sven Diederichs is co-owner of the siTOOLs Biotech GmbH.
REFERENCES
- 1. Rinn J.L., Chang H.Y.. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012; 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilusz J.E., Sunwoo H., Spector D.L.. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kung J.T., Colognori D., Lee J.T.. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geisler S., Coller J.. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013; 14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fatica A., Bozzoni I.. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014; 15:7–21. [DOI] [PubMed] [Google Scholar]
- 6. Gutschner T., Diederichs S.. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012; 9:703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 8. Schmitt A.M., Chang H.Y.. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016; 29:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmitz S.U., Grote P., Herrmann B.G.. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016; 73:2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 12. Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A.. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015; 16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R.. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014; 6:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lennox K.A., Behlke M.A.. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016; 44:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutschner T., Baas M., Diederichs S.. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011; 21:1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M.. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013; 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J.. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004; 305:1289–1292. [DOI] [PubMed] [Google Scholar]
- 18. Weinberg M.S., Morris K.V.. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016; 44:6505–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Golding M.C., Magri L.S., Zhang L., Lalone S.A., Higgins M.J., Mann M.R.. Depletion of Kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development. 2011; 138:3667–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stojic L., Niemczyk M., Orjalo A., Ito Y., Ruijter A.E., Uribe-Lewis S., Joseph N., Weston S., Menon S., Odom D.T. et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016; 7:10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bennett C.F., Swayze E.E.. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010; 50:259–293. [DOI] [PubMed] [Google Scholar]
- 22. Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F.. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003; 278:7108–7118. [DOI] [PubMed] [Google Scholar]
- 23. Luo S., Lu J.Y., Liu L., Yin Y., Chen C., Han X., Wu B., Xu R., Liu W., Yan P. et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016; 18:637–652. [DOI] [PubMed] [Google Scholar]
- 24. Bhaya D., Davison M., Barrangou R.. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011; 45:273–297. [DOI] [PubMed] [Google Scholar]
- 25. Wang H., La Russa M., Qi L.S.. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016; 85:227–264. [DOI] [PubMed] [Google Scholar]
- 26. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R.. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016; 34:192–198. [DOI] [PubMed] [Google Scholar]
- 30. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Proto. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R.. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013; 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiao A., Wang Z., Hu Y., Wu Y., Luo Z., Yang Z., Zu Y., Li W., Huang P., Tong X. et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013; 41:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B.V., Kralovics R., Brummelkamp T.R., Nijman S.M., Burckstummer T.. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014; 24:2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K.. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013; 10:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013; 10:973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., Iyer E. P R, Lin S., Kiani S., Guzman C.D., Wiegand D.J. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methds. 2015; 12:326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu H.Y., He L., Khaitovich P.. Deep sequencing reveals a novel class of bidirectional promoters associated with neuronal genes. BMC Genomics. 2014; 15:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cong L., Zhou R., Kuo Y.C., Cunniff M., Zhang F.. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012; 3:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guilinger J.P., Thompson D.B., Liu D.R.. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014; 32:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013; 155:1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hannus M., Beitzinger M., Engelmann J.C., Weickert M.T., Spang R., Hannus S., Meister G.. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014; 42:8049–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H. et al. Opposing roles for the lncRNA Haunt and Its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015; 16:504–516. [DOI] [PubMed] [Google Scholar]
- 49. Paralkar V.R., Taborda C.C., Huang P., Yao Y., Kossenkov A.V., Prasad R., Luan J., Davies J.O., Hughes J.R., Hardison R.C. et al. Unlinking an lncRNA from Its Associated cis Element. Mol. Cell. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han J., Zhang J., Chen L., Shen B., Zhou J., Hu B., Du Y., Tate P.H., Huang X., Zhang W.. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014; 11:829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aparicio-Prat E., Arnan C., Sala I., Bosch N., Guigo R., Johnson R.. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015; 16:846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deng C., Li Y., Zhou L., Cho J., Patel B., Terada N., Bungert J., Qiu Y., Huang S.. HoxBlinc RNA recruits Set1/MLL complexes to activate hox gene expression patterns and mesoderm lineage development. Cell Rep. 2016; 14:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welsh I.C., Kwak H., Chen F.L., Werner M., Shopland L.S., Danko C.G., Lis J.T., Zhang M., Martin J.F., Kurpios N.A.. Chromatin architecture of the Pitx2 locus requires CTCF- and Pitx2-dependent asymmetry that mirrors embryonic gut laterality. Cell Rep. 2015; 13:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Durruthy-Durruthy J., Sebastiano V., Wossidlo M., Cepeda D., Cui J., Grow E.J., Davila J., Mall M., Wong W.H., Wysocka J. et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 2016; 48:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013; 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Groff A.F., Sanchez-Gomez D.B., Soruco M.M., Gerhardinger C., Barutcu A.R., Li E., Elcavage L., Plana O., Sanchez L.V., Lee J.C. et al. In Vivo characterization of linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 2016; 16:2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bassett A.R., Akhtar A., Barlow D.P., Bird A.P., Brockdorff N., Duboule D., Ephrussi A., Ferguson-Smith A.C., Gingeras T.R., Haerty W. et al. Considerations when investigating lncRNA function in vivo. eLife. 2014; 3:e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. et al. Genome-Scale CRISPR-mediated control of gene repression and activation. Cell. 2014; 159:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lenstra T.L., Coulon A., Chow C.C., Larson D.R.. Single-Molecule imaging reveals a switch between spurious and functional ncRNA Transcription. Molecular Cell. 2015; 60:597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ghosh S., Tibbit C., Liu J.L.. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 2016; 44:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H.. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015; 75:3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Re. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007; 129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010; 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Musahl A.S., Huang X., Rusakiewicz S., Ntini E., Marsico A., Kroemer G., Kepp O., Orom U.A.. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene. 2015; 34:5046–5054. [DOI] [PubMed] [Google Scholar]
- 68. Yarmishyn A.A., Batagov A.O., Tan J.Z., Sundaram G.M., Sampath P., Kuznetsov V.A., Kurochkin I.V.. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014; 15:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mahmoudi S., Henriksson S., Corcoran M., Mendez-Vidal C., Wiman K.G., Farnebo M.. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009; 33:462–471. [DOI] [PubMed] [Google Scholar]
- 70. Bassik M.C., Kampmann M., Lebbink R.J., Wang S., Hein M.Y., Poser I., Weibezahn J., Horlbeck M.A., Chen S., Mann M. et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013; 152:909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.