Abstract
Functional annotation of metagenomic and metatranscriptomic data sets relies on similarity searches based on e-value thresholds resulting in an unknown number of false positive and negative matches. To overcome these limitations, we introduce ROCker, aimed at identifying position-specific, most-discriminant thresholds in sliding windows along the sequence of a target protein, accounting for non-discriminative domains shared by unrelated proteins. ROCker employs the receiver operating characteristic (ROC) curve to minimize false discovery rate (FDR) and calculate the best thresholds based on how simulated shotgun metagenomic reads of known composition map onto well-curated reference protein sequences and thus, differs from HMM profiles and related methods. We showcase ROCker using ammonia monooxygenase (amoA) and nitrous oxide reductase (nosZ) genes, mediating oxidation of ammonia and the reduction of the potent greenhouse gas, N2O, to inert N2, respectively. ROCker typically showed 60-fold lower FDR when compared to the common practice of using fixed e-values. Previously uncounted ‘atypical’ nosZ genes were found to be two times more abundant, on average, than their typical counterparts in most soil metagenomes and the abundance of bacterial amoA was quantified against the highly-related particulate methane monooxygenase (pmoA). Therefore, ROCker can reliably detect and quantify target genes in short-read metagenomes.
INTRODUCTION
Omics approaches are commonly applied to the study of microbial communities in a variety of clinical and environmental settings, but numerous technical challenges remain for accurately analyzing short gene sequences recovered from metagenomes or metatranscriptomes (1). Most importantly, several standard bioinformatic tasks rely on widely used similarity search algorithms (e.g. BLAST) that, through the comparison of nucleic or protein sequences to reference databases, allow for the identification of homologous genetic features among millions of unrelated sequences. However, in short-read metagenomes or metatranscriptomes representing diverse microbial communities (e.g. those of soils, oceans or the human gut), the rate of false positive (i.e. incorrectly identified, FP) or false negative (i.e. incorrectly rejected, FN) matches obtained from similarity searches are rarely addressed or quantified. An important underlying cause for FP and FN matches is the use of thresholds for a match based on a fixed e-value, a statistical parameter that reflects the number of expected matches by chance but not necessarily true homology. Although the use of e-values represents an efficient strategy for selecting matches, it can result in a substantial number of false positives, especially for protein sequences that share functional domains or motifs. Only lately, these limitations have received adequate attention but mostly for taxonomic assignment purposes (2,3).
Recently, we employed the receiver operating characteristic curve (ROC) approach to refine the results of similarity searches and calculate a reliable, fixed bitscore value across the sequence of the target gene that maximizes the sensitivity (true positive rate) and specificity (true negative rate) for detecting short-gene fragments encoding nitrous oxide reductase (nosZ) in soil metagenomes (4). This approach was clearly advantageous compared to the use of an arbitrary e-value threshold by decreasing both the false discovery rate [FDR = FP/(TP + FP)] to ∼1% and the false negative rate [FNR = FN/(TP+FN)] to ∼2%. Accordingly, our approach resulted in a small fraction of false positive metagenomic reads recruited by (or annotated as) reference nosZ sequences, i.e. metagenomic reads encoding non-nosZ gene fragments but showing a significant score due to the presence of shared domains and/or motifs with nosZ. Unlike nosZ, other genes sharing highly conserved domains and motifs such as metal binding or ATP-hydrolyzing domains can retrieve a higher fraction of false positive matches when analyzing short-read sequences, therefore, representing more challenging cases. Such genes require comparatively higher thresholds in similarity searches in order to achieve a low rate of false positives matches. However, the latter typically comes at the expense of increased frequency of false negatives. Therefore, a variable bitscore threshold across the sequence of the target gene, which would be stringent in highly conserved, non-discriminative regions in order to minimize false positives but can be lowered in less conserved regions in order to avoid false negatives, should be advantageous compared to the common practice of using arbitrary fixed e-value thresholds. To the best of our knowledge, the idea of a variable threshold across the sequence of a target protein/gene has not yet been implemented in an automated bioinformatic tool.
Here, we introduce an automated bioinformatic pipeline, called ROCker, which uses the ROC curve to estimate the most-discriminating bitscore thresholds in sliding windows across the sequences of a protein family of interest and evaluates non-discriminative domains shared with unrelated proteins. The pipeline takes as input a list of identifiers for proteins of interest (e.g. beta subunit of RNA polymerase, RpoB) and generates a simulated shotgun data set using sequenced microbial genomes encoding these proteins (i.e. simulated reads from genomes that encode the reference proteins together with reads from non-target regions of the genome). This data set of known composition is then used as a training data set for generating a ROCker profile of most discriminating, position-specific, bitscore values across the target protein alignment, which maximize the recovery of true positive and minimize false positive matches. Therefore, a ROCker profile essentially represents an adaptable filter for minimizing FDR and FNR in similarity search results to accurately detect metagenomic reads related to a single function of interest. We further tested the effectiveness of ROCker with available short-read metagenomes and assessed the diversity of nitrogen cycle genes in terrestrial soils and marine sediments.
MATERIALS AND METHODS
Implementation
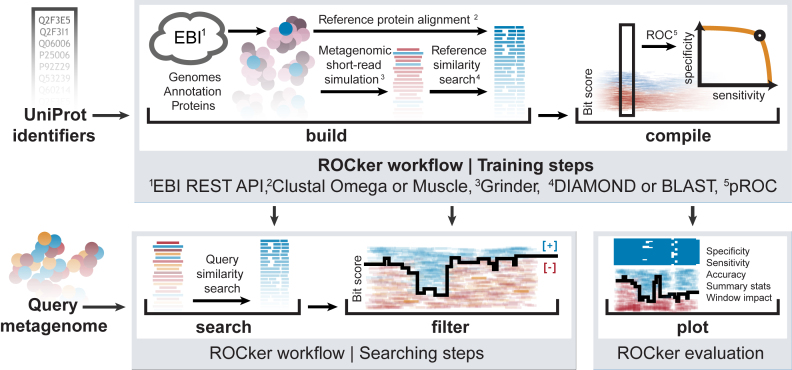
ROCker is implemented in the Ruby programming language and its workflow consists of five tasks. (i) Build: Reads a user-provided list of UniProt (Universal Protein Resource) protein identifiers and downloads the corresponding whole genome sequences encoding these proteins for generating data sets that simulate shotgun, short-read, Illumina metagenomes using GRINDER (5). A second list of known negative references, i.e. closely related proteins that should not be considered as true matches can also be given at this step in order to increase the performance of ROCker (see amoA example below). The training reference sequences are downloaded and annotated using the European Bioinformatics Institute REST API (6) and aligned using ClustalΩ (7). Subsequently, ROCker queries the reference protein sequences provided against the simulated shotgun data sets using BLASTx (8) or DIAMOND (9). (ii)Compile: Translates search results to alignment columns, and identifies the most discriminant bitscore per alignment in a 20 amino acid window (or another, user-defined length) in a set of sequences using pROC (10). The latter algorithm calculates sensitivity and specificity using the number of true and false positive matches in each window. The bitscore thresholds are calculated as the value in the ROC curve that maximizes the distance to the identity line (i.e. the non-discriminatory diagonal line in the ROC curve) according to the Youden method. Windows are iteratively refined to reduce low-accuracy regions (<95% estimated accuracy), for all windows with sufficient data (≥5 amino acid positions and ≥3 true positives available). Thresholds in regions with insufficient data are inferred by linear interpolation of surrounding windows. (iii)Filter: Uses the calculated set of bitscore thresholds (as estimated by the compile task) to filter the result of a preexisting search. (iv)Search: Executes a search of metagenomic sequences against target protein sequences (i.e. single protein function) using BLASTx or DIAMOND, and filters the output according to the most-discriminating bitscores calculated in the Compile step. (v)Plot: Generates a graphical representation of the alignment, the thresholds and the matches obtained, together with summary statistics (See Supplementary Data).
Target gene sequences
Protein sequences for nitrogen cycle reference genes were obtained from the National Center for Biotechnology Information (NCBI) (downloaded in March 2014) and Uniprot (downloaded in June 2015). In order to avoid mis-annotated references, all protein sequences were aligned and visually inspected for the presence of characteristic amino acids or protein motifs and their phylogenetic relationships. Having a list of well-curated reference sequences is key for accurate ROCker results. All reference protein sequences used in the analysis for NirK (n = 147), NosZ (n = 173), PmoA (n = 9), archaeal AmoA (n = 5), bacterial AmoA (n = 7) and RpoB (n = 757) are available through http://enve-omics.gatech.edu.
Simulated data sets and benchmark analyses
Generation of simulated shotgun data sets
Simulated data sets were constructed using the ‘Build’ function in ROCker based on an input list of UniProt identifiers for each protein sequence (-P option). GRINDER's parameters differed from their default options as follows: sequencing depth of 3 (for NosZ and NirK, 10 for bacterial and archaea AmoA simulated data sets), remove ‘-∼*NnKkMmRrYySsWwBbVvHhDdXx’ characters, sequencing error ‘uniform 0.1’, mutation ratio ‘95 5’ and read length distribution ‘L uniform 5’, where L is the average read length of the simulated data set. Simulated data sets ranged from 1 to 43 million reads in size (Supplementary Data). The CPU time (cput) in hours required for generating simulated data sets can be approximated by using a power law regression as follows: cput = 3.0672*D1.096 (r2 = 0.948), where D is the number of protein reference sequences used. Calculated ROCker profiles can be re-used in following similarity searches. The processing of a similarity search output (i.e. ROCker-based filtering) typically takes from a few seconds to a couple of minutes on a personal computer, depending on the number of matching sequences.
Similarity search analysis
The simulated shotgun data sets were used as query sequences for BLASTx (BLAST+2.2.8) and DIAMOND (v0.7.9.58) searches against the reference protein sequences that corresponded to the input UniProt IDs. Default settings were used for BLASTx except that e-value was set to 0.01. For DIAMOND, the settings used were ‘min score’ of 20 and ‘sensitive’. These settings were used to make DIAMOND comparable to BLASTx in terms of sensitivity, albeit at the expense of speed; users that want faster DIAMOND searches should opt for the default settings instead. In all cases, only best matches were considered by using the script BlastTab.best_hit_sorted.pl from the enveomics collection (11). The BLASTx searches were used for generating ROCker profiles for NosZ, NirK and RpoB protein references (profiles available through http://enve-omics.ce.gatech.edu/rocker). Hidden Markov models for each set of proteins were built using full-length alignments with HMMer (12). For hidden Markov model (HMM)-based searches, the read sequences were first translated to amino acids using FragGeneScan (13), and subsequently used as query sequences in the hmmsearch algorithm implemented in HMMer (12) (Supplementary Data).
Ten-fold cross-validation calculations
Both NosZ and NirK ROCker profiles were further evaluated by performing a tenfold cross-validation test. To ensure that multi-copy references encoded in the same genome were grouped together in cross-validation sets, we randomly separated the genomes into ten subsets (rather than using protein UniProt identifiers). For each subset, a simulated data set was generated as a query (Test) to challenge a ROCker profile built with the remaining nine subsets (Model). Similarity searches were performed using BLASTx with the parameters described above. FNR and FDR were calculated for each subset and for 100, 150, 200, 250 and 300 bp read length simulated data sets. All generated data sets are available through http://enve-omics.ce.gatech.edu/data/rocker.
Shotgun metagenomes
Publicly available shotgun metagenomes were downloaded from the Sequence Read Archive, Metagenomics RAST or other web resources (see Supplementary Data for details). The data sets included two representative Midwest USA agricultural sites (Havana and Urbana, Illinois, USA) (4), two prairie soils that underwent infrared heating for 10 years (warming and control; Oklahoma, USA) (14), tropical (Misiones, Argentina) and boreal forests (Alaska, USA) (15), Alaskan permafrost active layer (Alaska, USA) (16), two beach sands (17) and a deep marine sediment (18) related to the Deepwater Horizon oil spill (Florida, USA), human stool (19) and a waste water enrichment sample (20).
Sequence processing of shot-gun metagenomes
SolexaQA (21) was used for quality trimming of raw Illumina metagenomic reads to extract the longest continuous segment with a Phred score ≥ 20. All paired-end or single reads (when only one read was available) longer than 50 bp were used for further analysis.
Fraction of genomes encoding nitrogen cycle genes
RpoB (RNA polymerase beta subunit) sequences were obtained from reviewed proteins in UniProt/Swiss-Prot. A total of 757 sequences were visually inspected for conservation of functional domains and complete alignment and were used to construct a simulated data set and ROCker profile (similar options as above for nitrogen cycle genes but using the ‘–per-genus’ option in the building step in order to reduce redundancy caused by sampling individual species with many representative sequences). Short-reads from soil metagenomes were used as query sequences for independent BLASTx searches (same settings as above) against the NosZ, NirK, AmoA or RpoB protein references. The ROCker-filtered or e-value-filtered counts were normalized by the median length of the sequences of each protein reference. The fraction of microbial genomes encoding either nosZ, nirK or amoA (i.e. genome equivalent) was calculated as the ratio of nirK, nosZ or amoA read counts to rpoB read counts using ROCker profiles or e-values.
Phylogenetic placement of amoA and nosZ reads
Protein reference sequences for NosZ or Amoa/PmoA were aligned using ClustalΩ (7) with default parameters. The alignment was used to build a phylogenetic tree in RAxML (22) v8.0.19 (LG model). nosZ- or amoA-reads were extracted from soil metagenomes using ROCker (BLASTx option), and their protein-coding sequences were predicted using FragGeneScan. The latter sequences were added to the NosZ or Amoa/PmoA protein alignment using MAFFT (‘addfragments’) (23) and were placed in the corresponding phylogenetic tree using RAxML EPA (24) (-f v option). An in house script (‘JPlace.to_iToL.rb’ available through http://enve-omics.gatech.edu) was used to prepare the visualization of the generated jplace file (25) in iTOL (26).
Availability and dependencies of ROCker
The ROCker package, documentation and pre-computed profiles are available through http://enve-omics.ce.gatech.edu/rocker. ROCker is distributed both as a packaged Ruby gem (https://rubygems.org/gems/bio-rocker) and source code (https://github.com/lmrodriguezr/rocker) under the terms of the Artistic License 2.0. Complete ROCker execution requires the rest-client and json Ruby gems, as well as R (including the pROC package), NCBI-BLAST+ or DIAMOND, GRINDER and ClustalΩ or MUSCLE (27). In addition, ROCker models can be built online through http://enve-omics.ce.gatech.edu/rocker-build/.
RESULTS
ROCker benchmark
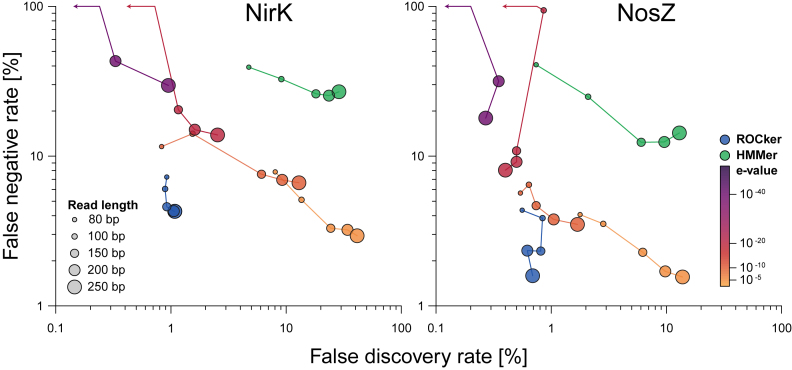
We applied ROCker to identify short-reads in simulated data sets of known composition encoding two denitrification genes, namely nitrite reductase (nirK) and nitrous oxide reductase (nosZ), and compared the results to other strategies for filtering the output of similarity searches. For this, two manually verified lists of NirK and NosZ protein identifiers were provided to ROCker (as positive references) to generate simulated data sets of known composition resembling short-read metagenomes of different lengths (see Figure 1 and Supplementary Data). The data sets were subsequently searched against NirK and NosZ reference sequences to provide the similarity search outputs for comparisons. The coupling of BLASTx with ROCker yielded substantially better performance compared to using fixed e-values, e.g. ∼3 and 15 fold-decrease in FDR when compared to the use of a low stringency e-value of 10−5 for NosZ and NirK, respectively (100 bp simulated data sets; see Figure 2 and Supplementary Data). However, the use of high e-values (i.e. low stringency) provided similar FNR results to ROCker. In fact, for NirK simulated data sets of longer read lengths, the FNR was slightly lower by ∼0.6% to 1.3% when an e-value of 10−5 was used compared to ROCker (Figure 2). Nevertheless, the high FDR observed for the same searches (at least 24 times higher, on average, compared to ROCker) makes the use of fixed e-values a less accurate approach. In other words, even though using lower e-values (higher stringency, e.g. 10−10) decreased FDR values, this was at the expense of much higher FNR values. In contrast, ROCker's FDR and FNR values were consistently low for all evaluated data sets (Figure 2).
Figure 1.
ROCker workflow for generating simulated shotgun data sets and calculating position-specific and most-discriminant bitscores. (Upper panel) ROCker can be used to perform five independent tasks: (i) Build: Using a user-provided list of unique UniProt protein identifiers for a target protein of interest, ROCker downloads the reference sequences, their corresponding whole genomes and annotation from the European Bioinformatics Institute (EBI) using the REST API. The protein references are aligned and the whole genomes used for the simulation of short-read Illumina metagenomes and then are searched against the protein reference sequences. The outputs of these searches are then provided to the (ii) Compile function, where the results are translated to alignment windows where it identifies the most discriminant bitscore that minimizes false positives but maximizes true positive matches. These results are compiled in ‘ROCker profiles’ that essentially represent an adaptable and reusable filter for the output of similarity searches increasing the accuracy of finding a true match compared to the most common practice of using fixed e-value thresholds. (Lower panel) (iii) Search: Short-read metagenomes are used as query in a similarity search using the target protein sequences as database (iv) Filter: This tool filters similarity searches using pre-calculated ROCker profiles. Finally, (v) Plot generates a graphical representation of the ROCker profiles along with the reference sequence alignments and summary statistics (see Supplementary Data for an extract of this feature).
Figure 2.
Comparison of false negative and false positive rates for simulated shotgun data sets of different read lengths using ROCker profiles and e-value thresholds. Simulated shotgun data sets of 80, 100, 150, 200 and 250 bp read length (figure legend) were generated using ROCker and searched against reference NirK and NosZ protein sequences using BLASTx. The outputs were filtered using the calculated ROCker profiles (circles in blue) and fixed e-value thresholds (circles in orange to purple gradient). Results from hidden Markov models search of the references NirK and NosZ sequences against the simulated reads are also shown (circles in green).
In all searches, the recently developed DIAMOND algorithm (using sensitive settings) showed low FNR and FDR when coupled with ROCker, similar to BLASTx (Supplementary Data and Supplementary Data), and was up to ∼13-fold faster than BLASTx, consistent with the results reported previously (9). Nonetheless, in every simulation, DIAMOND required more RAM than BLASTx (e.g. 9.6 Gb compared to 0.45 Gb for the 80 bp NirK simulated data set, respectively). Therefore, the choice of DIAMOND or BLASTx coupled with ROCker would depend on the number of sequences analyzed (e.g. size of metagenomic data sets) and the computational resources available. We also evaluated HMM as implemented in HMMer (12). Searches of both NirK and NosZ simulated data sets showed higher FNR values (about 5-fold higher, on average) compared to ROCker when the same simulated shotgun data sets and reference sequences were used. A better FDR was obtained in HMMer searches compared to the use of a fixed e-value threshold in BLASTx searches, but not as low as those obtained with ROCker (Figure 2). Moreover, HMMer required the least amount of memory and was ∼860 and 5700-fold faster, on average, compared to DIAMOND and BLASTx, respectively, consistent with previous results (12). Finally, we compared the results of BLASTx to those of other high-speed protein classification tools such as UproC (28) or GRASP (29), which showed similar FDR but much higher FNR values (Supplementary Data). Accordingly, the latter tools were not pursued further.
The evaluation of the performance of ROCker in 10-fold cross-validation tests showed low FDR values for both NosZ and NirK ROCker profiles (0.48% and 1.62%, on average, respectively) in 100, 150, 200, 250 and 300 bp simulated data sets (Supplementary Data). However, higher FNR values (5.33% and 17.33%, on average, for NosZ and NirK, respectively) were observed compared to when all references were used for generating ROCker profiles. These results showed that the more reference sequences used when building a ROCker profile and/or the higher the diversity of the reference sequences represented, a better recovery of reads encoding the target gene can be expected. Compared to the use of fixed e-values, ROCker showed lower FDR values in all simulations, consistent with the result reported above. For instance, up to 48- and 35-fold decrease in FDR were observed when compared to the use of low (10−5) and high (10−15) stringency e-values for the NirK simulated data sets, respectively.
Targeting a specific group of proteins using negative references
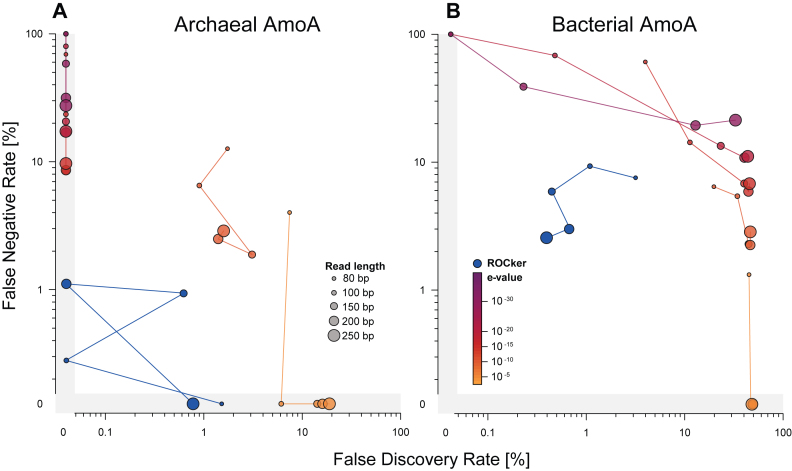
It is important to realize that ROCker attempts to optimize the number of matching (simulated) sequences originating from a target gene (true positives) against those originating from the remaining, non-target genes encoded in the same genomes (false positives). If a closely related, yet distinct, protein is encoded by other genomes than those corresponding to the input, simulated sequences from the former genes will not be included in ROCker analyses. To account for such cases and further improve the robustness of the calculated ROCker profile, a second list of non-target, negative references can also be provided to ROCker in order to obtain a filter that can exclude sequences originating from the provided non-target genes, in addition to the other non-target genes encoded in the genomes that correspond to the input. Under this configuration, ROCker simulates data sets generated from both positive (target) and negative references (non-target), and uses them as queries for similarity searches against positive (target) references. However, only matches derived from positive references are considered for determining the position-specific thresholds of the ROCker profile. Using this setup, ROCker was applied to analyze two highly-similar proteins, the bacterial and archaeal ammonia monooxygenase (amoA) and the particulate methane monooxygenase (pmoA), which are not typically encoded on the same genome and are often challenging to distinguish from each other based on sequence similarity searches. Archaeal AmoA ROCker profiles using bacterial AmoA and PmoA sequences as negative references (Supplementary Data), showed a moderate decrease of 23-fold and 5-fold in FNR and FDR compared to the use of 10−5 and 10−10 e-values, respectively (Figure 3A). Only low score matches from negative references (considered as false positives) were observed in the similarity search output (Supplementary Data), consistent with the higher divergence of archaeal amoA from bacterial amoA or pmoA relative to the divergence between bacterial amoA or pmoA. In contrast, the performance of the bacterial AmoA ROCker profile using archaeal AmoA and PmoA as negative references was decreased by 66- and 59-fold, on average, for FDR compared to the use of fixed e-values of 10−5 and 10−10, respectively (Figure 3B). Slightly higher FNR values were observed for bacterial AmoA ROCker profile compared to the archaeal AmoA profile (Figure 3B), as expected based on the high sequence similarity between bacterial amoA and pmoA. The increased FNR values obtained in all searches were attributed to the higher bitscore values calculated for each ROCker profile in order to efficiently discard high-scoring matches derived from negative references (Supplementary Data). Therefore, bacterial AmoA ROCker profiles including negative references showed low FDR at the cost of a slightly higher FNR. In summary, having a well-curated set of positive, and, if necessary, negative references is an essential prerequisite for achieving low FDR and FDR values with ROCker.
Figure 3.
Effect of including negative references in AmoA ROCker profiles for simulated shotgun data sets of different read lengths. Simulated shotgun data sets of 80, 100, 150, 200 and 250 bp read length were searched against (target) AmoA reference sequences using BLASTx. Panel A shows the results of using ROCker archaeal AmoA profiles, including bacterial AmoA and PmoA as negative references, and e-values for filtering the simulated data sets. Panel B shows the results of using ROCker bacterial AmoA profiles, including archaeal AmoA and PmoA as negative references, and e-values.
Using ROCker on shotgun metagenomes from marine and soil habitats
nosZ gene abundance in soil metagenomes
In order to assess the abundance and diversity of nosZ genes in different habitats, we analyzed the phylogenetic classification of nosZ gene fragments detected by ROCker (BLASTx search) in 10 short-read metagenomes representing agricultural, forest, permafrost and marine sediments (no planktonic samples were analyzed). A maximum likelihood method for the phylogenetic placement of these short reads into a NosZ tree revealed a consistent placement of the recovered fragments according to their habitat of origin (Supplementary Data), further supporting that the reads identified by ROCker are indeed NosZ-encoding reads. For instance, the marine genera Rhodothermus, Maribacter and Caldilinea, independently recruited ∼11- to 320-fold more nosZ reads from marine (beach and marine sediments) than terrestrial environments. On the other hand, the Anaeromyxobacter, Opitutus and Gemmatimonas genera, all commonly found in terrestrial soils, recruited between ∼2- and 33-fold more nosZ reads from terrestrial than marine environments. The analysis also revealed that atypical or clade II NosZ (4,30,31) reads were 2 times more abundant, on average, than the typical or clade I counterparts, which was consistent with our previous analysis using a fixed bitscore threshold across the sequence of NosZ and a smaller set of samples from Midwestern agricultural soils (4). However, typical nosZ gene fragments were relatively more abundant in marine sediments than soils, since marine sequences comprised almost 80% of the total typical gene fragments found in all samples.
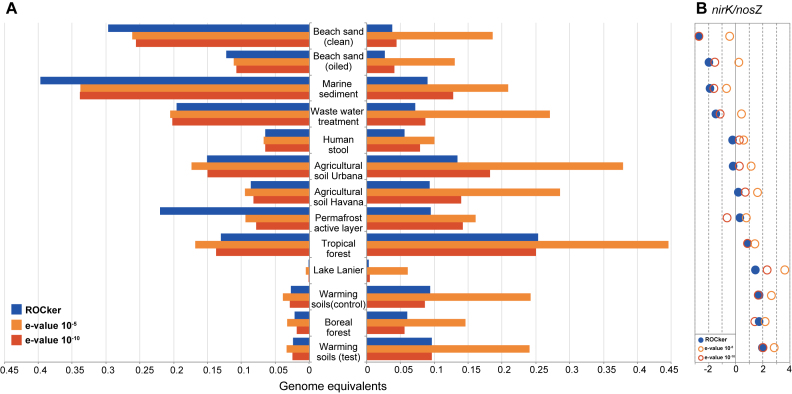
Quantifying nirK/nosZ ratio in terrestrial and marine habitats
The abundance of nirK and nosZ genes in publicly available short-read metagenomes was quantified based on position-specific bitscore thresholds calculated by ROCker (Figure 4A). The use of fixed e-value thresholds (e.g. 10−5 or 10−10) generally provided higher abundance estimates compared to those of ROCker, consistent with our expectations from the FDR results reported for simulated data sets. For instance, when a 10−5 e-value was used to estimate nirK genome equivalents (using universal RpoB protein to normalize abundances), these values exceeded four times, on average, the estimations of ROCker. A similar trend was observed for nosZ, albeit ROCker and e-value-based estimates for genome equivalents were closer to each other compared to those calculated for nirK, reflecting the less problematic conserved functional domains of NosZ. Further, a higher ratio of nirK/nosZ was observed for most terrestrial soil metagenomes compared to metagenomes from sand beaches and sediments when ROCker values were used (Figure 4B).
Figure 4.
Abundance for nirK and nosZ genes in short-read metagenomes calculated using ROCker or fixed e-value thresholds. Panel A shows the abundance, calculated as the fraction of the microbial community encoding nirK or nosZ, based on searching short-read metagenomes against NirK (a) and NosZ (b) reference protein sequences. BLASTx searches were filtered using the calculated ROCker profiles or fixed e-values (10−5 and 10−10). Panel B shows the log2 ratio of nirK/nosZ gene abundances using ROCker.
Recovering amoA gene fragments from soil metagenomes
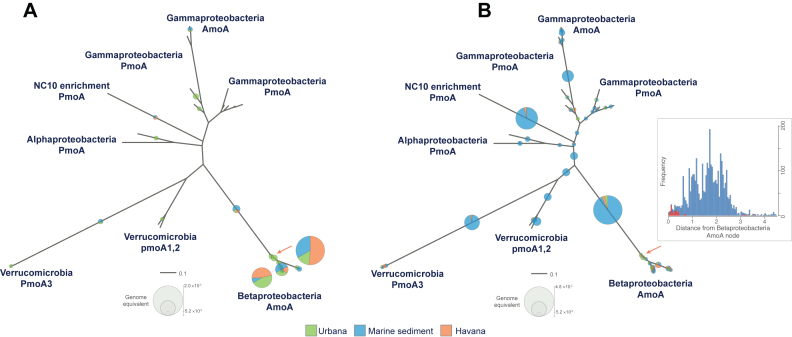
We tested the performance of ROCker for extracting bacterial amoA reads from soil and sediment shotgun metagenomes (Havana and Urbana soils, and Florida marine sediments) and assessed their phylogenetic placement. Even though more than 30-fold amoA reads were extracted when a ROCker profile not including negative references was used (Figure 5, inset), only ∼10% of these reads were placed in the correct (target) bacterial AmoA clade; the majority of the remaining reads were likely related to PmoA or represented deep-branching members of the membrane-bound monooxygenase (CuMMO) protein family (Figure 5B). Conversely, when a bacterial AmoA ROCker profile including negative references (i.e. archaeal AmoA and PmoA) was used to filter the similarity searches, 81% of the amoA reads were placed in the expected nodes and branches containing AmoA references (Figure 5A).
Figure 5.
Placement of amoA reads recovered from terrestrial and marine metagenomes in an AmoA and PmoA phylogenetic tree. A total of 27 bacterial AmoA and PmoA sequences available in the public databases were used to build a reference phylogenetic tree. ROCker bacterial AmoA profiles including (left panel) and not including negative bacterial PmoA references (right panel) were used to identify amoA reads from three metagenomes (see figure key). Reads were placed in the phylogenetic tree using RAxML EPA. The radii of the pie charts represent the abundance for each node (calculated as genome equivalents). Note that most reads in the left tree were placed in the betaproteobacterial AmoA clade. However, the reads in the right tree were placed in more deep-branching nodes of the tree or PmoA clades. The inset shows the distribution of the evolutionary distances of the reads from the (target) betaproteobacterial AmoA node (orange arrow), obtained when a ROCker profile including (red bars) and not including (blue bars) negative references was used.
Comparison of ROCker to alternative approaches
While several approaches have been recently developed to functionally annotate metagenomic reads (e.g. functional profilers), these tools are based on competitive matches against a large database of functions (28) or they attempt to reconstruct gene variants present in the metagenomes (29,32), and thus, have different objectives and underlying ideas than ROCker. However, ROCker can be used complementary with these approaches, especially in low sequencing depth metagenomes or with tools that are prone to detect or assemble non-target references (false positives). For instance, in simulated data sets with low sequencing depth for NosZ and NirK (e.g. 1 and 5X), ROCker showed less than 3.33% and 6.6% FNR, respectively, whereas Xander (32) failed to detect and reconstruct more than half of the target sequences (Supplementary Data). While Xander's performance was better with target sequences showing 10X coverage (e.g. 70–90% of target sequences reconstructed), consistent with results of the earlier study (32), it was still missing target sequences recovered by ROCker (Supplementary Data and Supplementary Data). Furthermore, in cases where the target references showed high identity to non-related references and also have a different biological role (e.g. AmoA versus PmoA), ROCker effectively recovered bacterial amoA-encoding reads instead of pmoA ones (maximum of 3.45% FDR), at the cost of a slightly higher FNR (>9.7%, Supplementary Data). In contrast, Xander showed increased values of FDR (above 30.1%) and FNR (above 10%) due the assembly of false positive non-target references (Supplementary Data and Supplementary Data). However, when the reads identified by ROCker were provided as input to Xander, there were no false positive sequences reconstructed by Xander, and Xander's processing time decreased by several orders of magnitude due to the lower sequence complexity of the input. Hence, ROCker can be used complementary to assemblers of target sequences such as Xander in order to increase the accuracy of the reconstructed targets.
DISCUSSION
The results presented here using ROCker underscore the advantages of using calculated position-specific versus fixed thresholds when analyzing short-read metagenomes. E-values depend on the size of the database used and the length of the query sequences, making the determination of the optimal e-value threshold to use a challenging task for short-length queries against different databases. For instance, a closer agreement between ROCker and fixed e-value approaches was observed for NirK abundances in metagenomes when a more stringent 10−10 e-value was used (Figure 2), but it remains challenging to decide what optimal e-value should be used for other references. In addition, our simulations showed that even considering the bitscore values from the 10% of the best matching reads as thresholds, it is not as robust as ROCker, since such bitscores can represent false positive matches instead. Further, the estimated abundance of proteins with several conserved functional domains such as NirK was frequently overestimated, by at least 2- to 3-fold, when using fixed e-values (Figure 4). Notably, ROCker overcomes these limitations, providing consistent results, independent of the frequency of shared functional domains in the reference of interest.
Two denitrification proteins were chosen to showcase ROCker because they encode a different number of conserved domains, which can increase FDR in similarity searches by recruiting reads encoding similar motifs but originating from non-target (and not related) proteins. NirK is a copper nitrite reductase that contains type-1 and -2 copper centers, commonly found in multicopper oxidases (33). Even though NosZ contains two copper centers, CuZ and CuA, short-reads of 100 bp or longer have sufficient length in this case to prevent false positive matches from non-nosZ-containing reads. Consistent with these characteristics, a 3- to ∼5-fold increase in FDR was observed for NirK versus NosZ when the e-value strategy (10−5) and different read lengths were used. In contrast, ROCker showed less than 1.5-fold increase in FDR and FNR for NirK versus NosZ, for the same data sets (Figure 2), consistent with ROCker's ability to robustly deal with genes containing different numbers of conserved domains and/or domains with different degrees of conservation and phylogenetic distribution. Even though low FDR were observed in a 10-fold cross validation test, the slightly higher FNR observed was attributable to the reduced sequence diversity in the reference subsets used to generate the ROCker profiles. These findings revealed that users should try to maximize the number of (trusted) reference sequences for building ROCker profiles, and especially the phylogenetic/sequence diversity encompassed by these references for more accurate results. The results presented here for NirK and NosZ illustrate a useful guide for building ROCker profiles and analyzing additional proteins, depending mostly on the number of conserved domains and motifs encoded by the target protein of interest and their degree of sequence conservation.
It is also important to note that a ROCker profile, while computationally demanding to create (e.g. building in silico data sets) and labor intensive (e.g. manual checking of reference sequences) at the building step (but not for filtering a similarity search output), needs to be built only once and can be subsequently used multiple times, such as in similarity searches for different metagenomic data sets.
We also evaluated popular, alternative algorithms to BLASTx for the similarity search step, including the recently described DIAMOND (9), and HMM as implemented in HMMer (12). ROCker results using DIAMOND (Supplementary Data) were faster and comparable in terms of FDR and FNR with BLASTx and thus, the former configuration is recommended for studies with limited computational time available without compromising sensitivity (Supplementary Data).
ROCker is intended to accurately detect short metagenomic fragments related to a single gene function rather than performing a complete gene functional profile or reconstructing full target sequences from metagenomes. Nonetheless, ROCker can be used complementary to the latter approaches and thus, leads to more accurate analyses of abundance and diversity of target genes in metagenomes. For instance, ROCker showed to be advantageous compared to tools for reconstructing target sequences such as Xander, especially when the target gene sequences had low sequencing depth (e.g. below 5X), or they were prone to be mistakenly identified as their highly-related but functionally distinct (non-target) gene families (e.g. AmoA versus PmoA; see Supplementary Data). Having full-length sequences reconstructed from metagenomes enables downstream analyses of the naturally occurring diversity (e.g. diversity surveys, design improved PCR primers); hence, an approach that combines ROCker with tools like Xander could strengthen future studies.
Copper-containing membrane-bound monooxygenase (CuMMO) enzymes catalyze the oxidation of ammonia (AMO), methane (pMMO) and other hydrocarbons, and are encoded in the genomes of methanotrophs and nitrifiers (34–38). Subunit ‘A’ is typically used as a diagnostic marker of the specific substrate of the enzyme (39). Even though PCR primers can effectively distinguish between bacterial and archaeal amoA (40,41), differences in sensitivity and performance have been identified for primers intended to discriminate between pmoA and amoA genes (42). These difficulties are mostly due to the high similarity at the nucleotide level because of their recognized evolutionary relatedness (43). To deal with such cases of high sequence identity between target versus non-target genes, especially when the latter are encoded by different genomes than those encoding the former, we implemented the use of negative references for generating ROCker profiles. Remarkably, bacterial AmoA ROCker profiles including PmoA sequences as negative references showed 60-fold improvement in FDR compared to the use of a fixed e-value (e.g. 10−5) (Figure 3B), and almost all reads identified were placed in the target bacterial AmoA tree clade, unlike reads extracted using a ROCker profile without negative references (Figure 5A versus B panels). The use of negative references is also recommended when discrimination between different variants or clades of the same gene family is intended. However, it is important to point out that the decrease in FDR when including negative references was at the expense of a slightly increased FNR, by about 8%, on average, according to our simulated AmoA data sets of different read lengths. Therefore, unless discrimination between closely related protein sequences encoded by the same or different genomes is required, the use of negative sequences should be avoided in order to maximize the number of reads detected that encode the target gene (true positives).
Interestingly, the analysis of soil metagenomes showed a higher ratio of nirK/nosZ for terrestrial samples relative to marine sediments (Figure 4B), in agreement with previous results based on quantitative real-time PCR (44,45). These findings are consistent with the hypothesis that in some environments a high fraction of denitrifiers does not possess the genetic potential to reduce N2O, a potent greenhouse gas. Assuming that gene abundance can be used as a proxy for gene activity (46), these results imply that microbial-mediated reduction of N2O might be higher (and hence, emissions might be lower) in marine sediments than on land, which remains to be experimentally verified.
Recent studies have shown that previous efforts to determine the abundance of nosZ genes have missed a group of divergent sequences, the so-called atypical sequences or clade II, which are functional as N2O reductases and are frequently more abundant than their more studied, typical counterparts (4,30,31). Consistently, ROCker identified twice as many reads, on average, encoding atypical versus typical nosZ gene fragments in ten short-read metagenomes representing terrestrial and marine environments. Phylogenetic placement of these short-reads into a NosZ tree revealed that typical nosZ reads were mostly derived from marine sediments (Supplementary Data), probably reflecting differences in nitrogen cycle pathways and/or regulation between these environments. For instance, typical nosZ genes are frequently associated with complete denitrifiers (30), which might account for the higher N2O reduction potential detected in marine sediments compared to soils. Many atypical nosZ reads found in the terrestrial metagenomes were affiliated with the Anaeromyxobacter, Opitutus and Gemmatimonas genera, and accordingly nosZ sequences assigned to these taxa have been frequently recovered from soils based on PCR and/or cloning approaches (30,47,48). The high consistency observed between the results of the phylogenetic placement of nosZ reads and the habitats of origin of the reads are also in agreement with previous literature and further corroborates the robustness of ROCker.
The only input required to generate simulated data sets and calculate position-specific, most-discriminant bitscores, is a list of UniProt protein sequence identifier numbers for the protein of interest. It should be pointed out, however, that these reference sequences should be carefully selected to represent the protein family of interest (target), as opposed to closely-related homologs of distinct function (when available), in order to obtain accurate ROCker results. Sequences of related, yet distinct, protein families (negative sequences), which could provide false-positives during similarity searches, can be also given to ROCker in order to increase the performance of the profiles during the ‘build’ stage. Therefore, careful, manual curation of the reference sequences is typically the most time-consuming step of ROCker, and the only step that is not currently fully automated. In our experience, using protein families generated automatically or unsupervised commonly brings error/noise to the generated ROCker models, and thus, is not recommended. A few manually curated repositories such as the Functional Gene Pipeline and Repository (FUNGENE) (49) have started to become available, although they are still limited in the number of protein families they encompass.
Finally, finding reads distantly related to the target references might be challenging for ROCker (as is the case for any similarity search-based approach) since ROCker's thresholds (bitscores) are often high, reflecting close similarity to the reference set (particularly in conserved domains present in reference sequences). Using high e-value cutoffs might be advantageous for the latter purpose, albeit at the cost of an unknown (and probably high) number of false positive matches.
In summary, ROCker expands the molecular toolbox for clinical and environmental surveys in the prokaryotic and eukaryotic domain, providing a pipeline to efficiently detect and quantify the abundance of gene fragments of interest in short-read metagenomes. The idea underlying ROCker can also be extended beyond metagenomics to (full-length) protein-protein searches and have broad applications in bioinformatic sequence analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alissa Hooker and Janet Hatt for their helpful discussions regarding the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program [award DE-SC0006662]; US National Science Foundation [awards 1241046 and 1356288]; Chilean Fulbright-Conicyt doctoral scholarship [L.H.O.]. Funding for open access charge: US National Science Foundation [awards 1241046 and 1356288].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P.. A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008; 72:557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huson D.H., Auch A.F., Qi J., Schuster S.C.. MEGAN analysis of metagenomic data. Genome Res. 2007; 17:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gerlach W., Stoye J.. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res. 2011; 39:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orellana L.H., Rodriguez-R L.M., Higgins S., Chee-Sanford J.C., Sanford R.A., Ritalahti K.M., Löffler F.E., Konstantinidis K.T.. Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio. 2014; 5, doi:10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Angly F.E., Willner D., Rohwer F., Hugenholtz P., Tyson G.W.. Grinder: A versatile amplicon and shotgun sequence simulator. Nucleic Acids Res. 2012; 40:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R.. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013; 41:W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al. . Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: Architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 10. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M.. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodriguez-R L.M., Konstantinidis K.T.. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints. 2016; 4:e1900v1. [Google Scholar]
- 12. Eddy S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rho M., Tang H., Ye Y.. FragGeneScan: Predicting genes in short and error-prone reads. Nucleic Acids Res. 2010; 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo C., Rodriguez-R L.M., Johnston E.R., Wu L., Cheng L., Xue K., Tu Q., Deng Y., He Z., Shi J.Z. et al. . Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl. Environ. Microbiol. 2014; 80:1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fierer N., Leff J.W., Adams B.J., Nielsen U.N., Bates S.T., Lauber C.L., Owens S., Gilbert J.A., Wall D.H., Caporaso J.G.. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mackelprang R., Waldrop M.P., DeAngelis K.M., David M.M., Chavarria K.L., Blazewicz S.J., Rubin E.M., Jansson J.K.. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011; 480:368–371. [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez-R L.M., Overholt W.A., Hagan C., Huettel M., Kostka J.E., Konstantinidis K.T.. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015; 9:1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mason O.U., Scott N.M., Gonzalez A., Robbins-Pianka A., Bælum J., Kimbrel J., Bouskill N.J., Prestat E., Borglin S., Joyner D.C. et al. . Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 2014; 8:1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Consortium T.H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McIlroy S.J., Albertsen M., Andresen E.K., Saunders A.M., Kristiansen R., Stokholm-Bjerregaard M., Nielsen K.L., Nielsen P.H.. ‘Candidatus Competibacter’-lineage genomes retrieved from metagenomes reveal functional metabolic diversity. ISME J. 2014; 8:613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox M.P., Peterson D.A., Biggs P.J.. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010; 11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006; 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 23. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berger S.A., Krompass D., Stamatakis A.. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 2011; 60:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsen F.A., Hoffman N.G., Gallagher A., Stamatakis A.. A format for phylogenetic placements. PLoS One. 2012; 7:e31009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Letunic I., Bork P.. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011; 39:W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2015; 31:1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong C., Yang Y., Yooseph S.. GRASP: Guided reference-based assembly of short peptides. Nucleic Acids Res. 2015; 43:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanford R.A., Wagner D.D., Wu Q., Chee-Sanford J.C., Thomas S.H., Cruz-García C., Rodríguez G., Massol-Deyá A., Krishnani K.K., Ritalahti K.M. et al. . Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19709–19714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones C.M., Graf D.R., Bru D., Philippot L., Hallin S.. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013; 7:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q., Fish J.A., Gilman M., Sun Y., Brown C.T., Tiedje J.M., Cole J.R.. Xander: employing a novel method for efficient gene-targeted metagenomic assembly. Microbiome. 2015; 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. MacPherson I.S., Murphy M.E.P.. Type-2 copper-containing enzymes. Cell. Mol. Life Sci. 2007; 64:2887–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hooper A.B., Vannelli T., Bergmann D.J., Arciero D.M.. Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie Van Leeuwenhoek. 1997; 71:59–67. [DOI] [PubMed] [Google Scholar]
- 35. Lieberman R.L., Rosenzweig A.C.. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005; 434:177–182. [DOI] [PubMed] [Google Scholar]
- 36. Könneke M., Bernhard A.E., de la Torre J.R., Walker C.B., Waterbury J.B., Stahl D.A.. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005; 437:543–546. [DOI] [PubMed] [Google Scholar]
- 37. Tavormina P.L., Orphan V.J., Kalyuzhnaya M.G., Jetten M.S.M., Klotz M.G.. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ. Microbiol. Rep. 2011; 3:91–100. [DOI] [PubMed] [Google Scholar]
- 38. Lawton T.J., Ham J., Sun T., Rosenzweig A.C.. Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins. 2014; 82:2263–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rotthauwe J.H., Witzel K.P., Liesack W.. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997; 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G.W., Prosser J.I., Schuster S.C., Schleper C.. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006; 442:806–809. [DOI] [PubMed] [Google Scholar]
- 41. Jia Z., Conrad R.. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 2009; 11:1658–1671. [DOI] [PubMed] [Google Scholar]
- 42. Junier P., Kim O.-S., Molina V., Limburg P., Junier T., Imhoff J.F., Witzel K.-P.. Comparative in silico analysis of PCR primers suited for diagnostics and cloning of ammonia monooxygenase genes from ammonia-oxidizing bacteria. FEMS Microbiol. Ecol. 2008; 64:141–152. [DOI] [PubMed] [Google Scholar]
- 43. Holmes A.J., Costello A., Lidstrom M.E., Murrell J.C.. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995; 132:203–208. [DOI] [PubMed] [Google Scholar]
- 44. Henry S., Bru D., Stres B., Hallet S., Philippot L.. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006; 72:5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Čuhel J., Šimek M., Laughlin R.J., Bru D., Chèneby D., Watson C.J., Philippot L.. Insights into the effect of soil pH on N(2)O and N(2) emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 2010; 76:1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petersen D.G., Blazewicz S.J., Firestone M., Herman D.J., Turetsky M., Waldrop M.. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 2012; 14:993–1008. [DOI] [PubMed] [Google Scholar]
- 47. Sanford R.A., Cole J.R., Tiedje J.M.. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 2002; 68:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chin K.J., Liesack W., Janssen P.H.. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int. J. Syst. Evol. Microbiol. 2001; 51:1965–1968. [DOI] [PubMed] [Google Scholar]
- 49. Fish J.A., Chai B., Wang Q., Sun Y., Brown C.T., Tiedje J.M., Cole J.R.. FunGene: The functional gene pipeline and repository. Front. Microbiol. 2013; 4:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.