Abstract
Thousands of long non-coding RNAs (lncRNAs) have been identified in mammals, many of which represent important regulators of gene expression. However, the mechanisms used by lncRNAs to control transcription remain largely uncharacterized. Here, we report on HOTAIRM1, a promising lncRNA biomarker in leukemia and solid tumors. We find that HOTAIRM1 contributes to three-dimensional chromatin organization changes required for the temporal collinear activation of HOXA genes. We show that distinct HOTAIRM1 variants preferentially associate with either UTX/MLL or PRC2 complexes to modulate the levels of activating and silencing marks at the bivalent domain. HOTAIRM1 contributes to physical dissociation of chromatin loops at the cluster proximal end, which delays recruitment of the histone demethylase UTX and transcription of central HOXA genes. Interestingly, we find overall proximal HOXA gene activation without chromatin conformation changes by HOTAIRM1 in a different cell type. Our results reveal a previously unappreciated relationship between chromatin structure, architecture and lncRNA function.
INTRODUCTION
Transcriptome mapping has revealed the existence of thousands of long non-coding RNA (lncRNA) transcripts expressed throughout mammalian genomes (1–6). Although the physiological role of most lncRNAs remains unknown, different types of mechanisms have emerged from studies investigating functionally-validated transcripts in transcriptional regulation (7). Many of these act through the recruitment of histone-modifying complexes such as the polycomb repressive complex 2 (PRC2) to modulate silencing marks at target genes. This form of regulation can occur either in cis as seen with XIST (8,9) or in trans as with the lncRNA HOTAIR (10). Interestingly, certain lncRNAs such as HOTAIR and steroid receptor RNA activator have been found to interact with different chromatin-modifying complexes (11,12). Consequently, transcription may be activated or repressed depending on the type(s) of complexes recruited to the chromatin. Although the methods used by lncRNAs to direct targeting of these complexes remain elusive, three-dimensional chromatin organization has recently been suggested, but strictly as a means to distribute transcripts over target genes (13).
With a growing list of lncRNAs validated as bona fide transcriptional regulators, many studies are looking for these transcripts as potential human disease biomarkers and therapeutic targets (14). One such lncRNA is HOTAIRM1, described first as a myeloid-specific regulator of HOXA genes (15). HOTAIRM1 has since been identified as a promising molecular signature in leukemia and several solid tumors, yet little is known about how it regulates the expression of genes (16–19). Also, its expression has been found in various cell types and it is unclear whether it maintains its regulatory function on the HOXA gene cluster in different tissues.
Here, we investigate the regulation of HOXA genes by HOTAIRM1 in the NT2-D1 and NB4 cellular differentiation models. Surprisingly, we find that whereas HOTAIRM1 acts solely as an activator of the proximal HOXA genes in NB4, it has a repressor role for the more distant HOXA4/5/6 genes in NT2-D1. Further investigation on the mechanism of this lncRNA in NT2-D1 reveals that it recruits histone-modifying complexes and contributes to temporal collinear HOXA gene activation by altering chromatin organization at the cluster. Our study shows for the first time that a lncRNA can deliver opposing activities within and between different cell types. We suggest that lncRNAs can activate or repress transcription based on several criteria including the transcript variants expressed, the chromatin landscape surrounding both lncRNA and target genes, and their physical proximity in three-dimensional space.
MATERIALS AND METHODS
Cell culture
The NT2-D1 cell line (NTERA-2 clone D1) was obtained from the American Type Culture Collection (ATCC; CRL-1973) and cultured as previously described (20). To induce HOXA gene expression in NT2-D1, cells were seeded at 2.4 × 106 per 10 cm plate in complete Dulbecco's modified Eagle's medium (DMEM) containing 10 μM RA (Sigma-Aldrich) or 1% ethanol control.
The NB4 cell line was kindly provided by Dr J. Teodoro (McGill) (21). NB4 were derived from the bone marrow of a 20-year old female suffering from typical acute promyelocytic leukemia (22). The cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco®) supplemented with 10% fetal bovine serum (Wisent Inc.), in the presence of 1% Penicillin/Streptomycin (‘complete’ RPMI). To induce HOXA genes in NB4, cells were seeded at 2 × 105 per ml in complete RPMI containing 1 μM RA (Sigma-Aldrich) or 1% ethanol control. Inductions were for 72 h unless otherwise indicated. NT2-D1 and NB4 cell authentication was by shape, size, morphology, gene expression of HOXA and differentiation marker genes before and after RA-induced differentiation. Cell lines were tested negative for mycoplasma contamination using the Mycoplasma Plus PCR Primer Set as recommended by the manufacturer (Agilent Technologies; cat. no. 302008).
RNA interference
HOTAIRM1 knockdown was performed by reverse transfection in NT2-D1 using the Lipofectamine® RNAiMAX transfection reagent in the presence of 30 nM small interfering RNA (siRNA) as recommended by the manufacturer (Thermo Fisher Scientific). The control siRNA (siGFP: 5΄-GCAAGCTGACCCTGAAGTTC-3΄) was purchased from GE Healthcare Dharmacon Inc. (cat. no. P-002048-01-20). HOTAIRM1 siRNAs (siM1-1: 5΄-GAAAGGCGAGCTTGGTTACGCTTAA-3΄, siM1-2: 5΄-GACTTCGAAGCATTAACGATC-3΄ and siE2/3: 5΄-ACATGCTGCGTTTTCTCACGGTCTGT-3΄) were purchased from Invitrogen™ (Stealth RNAi™ siRNA; Thermo Fisher Scientific). For experiments where cells are differentiated with RA, complete DMEM containing 10 μM RA was added 4 h after transfection and the cells were allowed to continue growing for 3 days.
Knockdown of HOTAIRM1 was also conducted with a short hairpin RNA (shRNA) using a construct kindly provided by Dr P.E. Newburger (University of Massachusetts Medical School). An shRNA vector containing a non-target sequence was used as negative control (Sigma-Aldrich; SHC002). Lentivirus production and knockdowns were performed following a procedure described previously (23). For NT2-D1, the cells were plated at 8 × 105 cells per 60-mm dish 24 h before infection. The cells were infected in the presence of 4 μg/ml Polybrene (Millipore Inc.) for 24 h before the media was changed for selection with 2 μg/ml puromycin (Wisent Inc.) over 3 days. After selection, the cells were passaged to 10-cm plates and processed for RA differentiation.
For NB4 infections, the cells were seeded at 1 × 106 cells/ml in 6-well plates in complete RPMI containing 4 μg/ml Polybrene. After 24 h, the cells were pelleted at 1000 rpm for 5 min in a clinical centrifuge and the media was changed for selection with 2 μg/ml puromycin over 3 days. Following selection, NB4 cells were passaged to T75 flasks and processed for RA differentiation.
RNA extraction
Total RNA was extracted from cells with the TRIzol® RNA isolation reagent according to the manufacturer's instructions (Thermo Fisher Scientific). The RNA was resuspended in RNase-free water, quantified by NanoDrop™ and treated with DNaseI (NEB) for 15 min at 37°C before re-extraction with TRIzol®.
Quantitative real-time polymerase chain reaction (RT-qPCR)
For gene expression measurements, 1 μg of total RNA was used to generate cDNA with reverse transcriptase SuperScript® III and either oligo(dT)20 or random hexamers (Thermo Fisher Scientific). Real-time polymerase chain reaction (PCR) quantification was performed as described in (24). The primer sequences used to quantify gene expression in this study are found in Supplementary Table S1.
Northern blotting
DNaseI-treated total RNA (10 μg per lane) from either NT2-D1 or NB4 cells was resolved on 1% agarose/formaldehyde denaturing gels. The RNA was transferred overnight by upward capillary transfer onto a Hybond™-N+ nylon membrane (GE Healthcare) using 20× saline sodium citrate (SSC) transfer buffer (3M NaCl, 0.3M sodium citrate, pH 7.0). Membranes were air-dried and the RNA was cross-linked with UV light (Stratalinker). The membranes were then re-hydrated in 6× SSC and transferred to pre-hybridization buffer (5× SSC, 1× Denhardt's buffer, 50% formamide, 2% sodium dodecyl sulfate (SDS), 10% dextran sulfate, 100 μg/ml denatured salmon sperm DNA) for 4 h at 42°C.
A purified PCR product amplified from NT2-D1 oligo(dT)20 cDNA with two primers against Exon 3 of HOTAIRM1 (Exon3 Fwd2: 5΄-GTCTGTTTTGCCTGAACCCATCAAC-3΄; Exon3 Rev1: 5΄-TCAGTGCACAGGTTCAAGCC-3΄) was used to produce the radiolabelled probes (Supplementary Table S1). The probes were generated with Amersham Ready-To-Go™ DNA labeling beads (GE Healthcare), 50 ng of PCR product and 5 μl of 32P-dCTP (3000 Ci/mmol; PerkinElmer). Unincorporated 32P-dCTP was removed with CHROMA SPIN™ columns (Clontech). Hybridization was conducted with 1 × 106 cpm radiolabeled probe per 1 ml of hybridization buffer containing 100 μg/ml of sheared salmon testes DNA overnight at 42°C. Membranes were washed three times with 2× SSC/0.1% SDS at room temperature followed by two washes with 1× SSC/0.1% SDS at 50°C before exposure onto KODAK BioMax XAR film for 24–72 h at −80°C.
Western blotting
Protein samples from cell fractionation and RNA immunoprecipitation (RIP) analysis were denatured in SDS sample buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 7.5% glycerol, 5% β-mercaptoethanol, 0.04% bromophenol blue) for 10 min at 95°C, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto 0.45 μm nitrocellulose membrane (Whatman Protran®) using a Hoefer TE77X semi-dry transfer unit at 20 V for 45 min. Immunoblotting was performed with antibodies against Hsp90 (Santa Cruz (at-115), gift from Dr S. Huang, McGill), Lamin A/C (Santa Cruz, sc-20681), UTX (Bethyl Laboratories, A302-374A), SUZ12 (Abcam, ab12073) and ASH2L (Bethyl Laboratories, A300-489A). Horseradish peroxidase-conjugated AffiniPure goat α-rabbit IgG (cat. no. 111-035-003) and rabbit α-mouse IgG (cat. no. 315-035-003) secondary antibodies were purchased from Jackson ImmunoResearch Laboratories Inc. Signals were visualized by chemiluminescence using Western Lightning® Plus-ECL (PerkinElmer Inc.).
Cell fractionation
Ten million cells were lysed in 200 μl of hypotonic lysis buffer (10 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1× protease inhibitor cocktail (Sigma Aldrich), and 300 U/ml RNasin Plus (Promega Corp.)) for 6 min on ice. Lysate was centrifuged at 1300 g for 5 min to separate the nuclei from the crude cytoplasmic fraction. The supernatant was re-centrifuged 5 min at 20 000 g, and the purified cytoplasmic fraction was transferred to a new eppendorf tube before three volumes of TRIzol® was added for cytoplasmic RNA extraction. Pelleted nuclei were washed once with lysis buffer and 300 μl of TRIzol® was added for RNA extraction. RNA pellets were resuspended in RNase-free water and quantified by NanoDrop™. Following DNaseI treatment, 1 μg of RNA from each fraction was used to generate cDNA using random hexamer priming and the reverse transcriptase SuperScript® III (Thermo Fisher Scientific). The amount of transcripts present in either the cytoplasm or the nucleus is shown as a percentage of the total quantity of RNA extracted from each fraction.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) experiments were conducted as described previously (24). Samples equivalent to 2 million cells were used for each ChIP. A total of 5 μg of antibody against either H3K4me3 (Abcam, ab8580), H3K27me3 (Abcam, ab6002), UTX (Bethyl Laboratories, A302-374A) or a control IgG (Abcam, ab37415) was added to each sample and magnetic Dynabeads® Protein G were used for pull down according to the manufacturer's instructions (Thermo Fisher Scientific).
RNA immunoprecipitation (RIP)
NT2-D1 and NB4 cells were induced with RA for 3 days and collected as 20 million cell pellets. Each pellet was lysed in 500 μl of hypotonic lysis buffer (see ‘Cell fractionation’ above) for 6 min on ice, before nuclei were pelleted by centrifugation at 1300 g for 5 min at 4°C. Nuclei were washed once with lysis buffer and resuspended in 1 ml of RIP buffer (10 mM Hepes pH 7.5, 250 mM KCl, 1 mM ethylenediaminetetraacetic acid, 1% NP-40, 0.5 mM DTT, 1× protease inhibitor cocktail, 1 mM PMSF, 100 U/ml RNasin Plus (Promega Corp.)). Nuclear material was then sheared by 20 strokes though a 23G1 needle on ice, followed by four 15-s sonication bursts with a Branson 450D cup-horn system at 80% amplitude. Debris was removed by centrifugation at 13 000 rpm for 10 min at 4°C. Each RIP used the lysate equivalent of 5 million cells in a final volume of 500 μl (supplemented with RIP buffer), and the volume equivalent of 10% input was set aside. Cell lysates were incubated with 5 μg of antibodies against UTX (Bethyl Laboratories, A302-374A), SUZ12 (Abcam, ab12073), or a control IgG (Abcam, ab12073) on an end-over-end overnight at 4°C. Magnetic Dynabeads® were used to pull down protein–RNA complexes, which were then washed three times with RIP buffer. Aliquots were set aside for western blotting, and the RNA was extracted from the rest with TRIzol®. Reverse transcription was done with random hexamer priming and the reverse transcriptase SuperScript® III (Thermo Fisher Scientific).
Chromosome conformation capture carbon copy (5C)
Aliquots of 2 million cells for chromosome conformation capture (3C)/chromosome conformation capture carbon copy (5C) libraries were fixed exactly as for ChIP experiments. The 3C libraries were generated with the BglII restriction enzyme as previously published (25). 5C libraries were produced and sequenced using a protocol and experimental design already described (26).
Databases URLs
The NT2-D1 ChIP-seq datasets for H3K27me3 (wgEncodeEH000908), H3K4me3 (wgEncodeEH000909), H3K36me3 (wgEncodeEH000915) and input (wgEncodeEH000654) were obtained and visualized from the UCSC genome browser (http://http://genome.ucsc.edu). The NB4 ChIP-seq data for H3K4me3/input (wgEncodeEH000970/wgEncodeEH000969), and RNAP2/input (wgEncodeEH000617/whEncodeEH000618) were also from UCSC. The ‘my5C-heatmap’ bioinformatics tool can be found at http://3dg.umassmed.edu/my5Cheatmap/heatmap.php.
RESULTS
The lncRNA gene HOTAIRM1 is highly induced upon RA stimulation of NT2-D1 cells
The HOXA gene cluster (Figure 1A) encodes a tightly-regulated group of homeotic transcription factors with important roles in embryonic development and hematopoietic lineage regulation (27). The HOX genes are evolutionarily conserved and mammalian loci have many embedded noncoding RNAs antisense to the HOX genes proper, including at least four intergenic lncRNAs at the human HOXA locus (Figure 1B) (3,10,28–30). To understand how HOTAIRM1 regulates the expression of HOXA genes, we began characterizing its expression in the NT2-D1 cellular differentiation model. This cell model has been extensively used to study various aspects of HOX regulation as RA treatment recapitulates their induction patterns in developing axial systems (20,30–32).
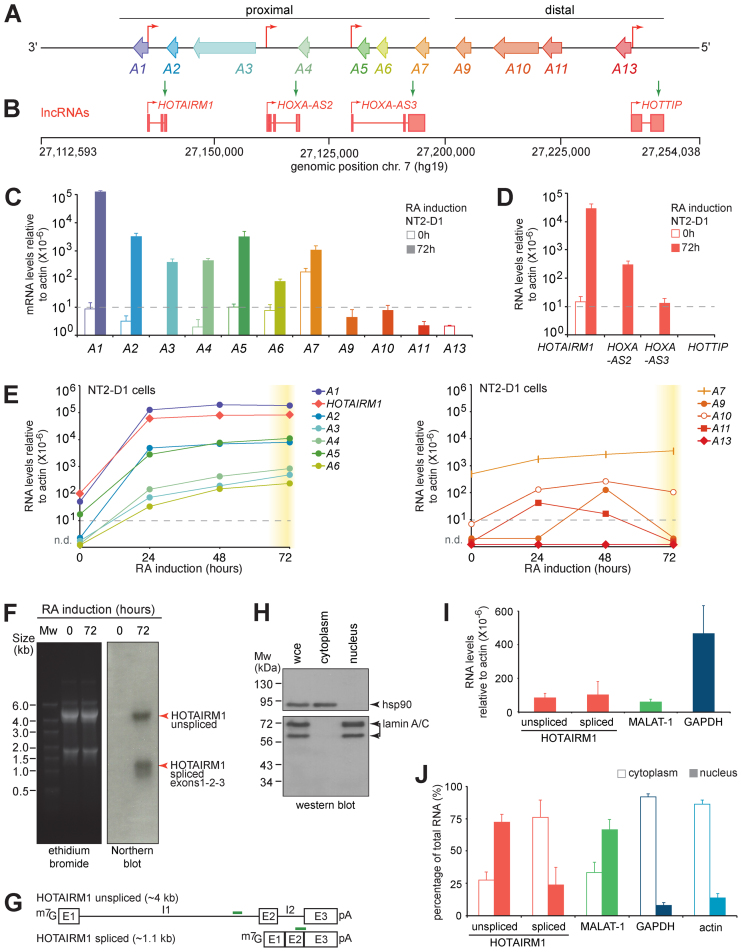
Figure 1.
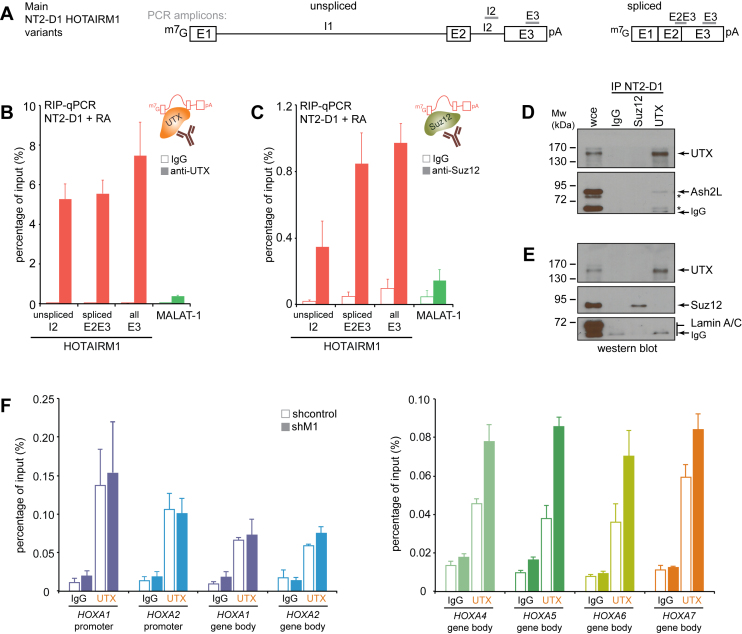
The HOTAIRM1 lncRNA gene is highly induced during RA treatment of NT2-D1 cells. (A) Linear diagram of the HOXA gene cluster region examined in this study. The HOXA genes are presented as left-facing arrows to indicate transcription direction, and are color-coded to facilitate data interpretation throughout the study. Red arrows pointing right identify the TSS of previously reported intergenic lncRNAs. The gene structures and genomic positions of lncRNAs along human chromosome 7 are shown in (B). The HOXA (C) and lncRNA (D) genes in the 3΄ (proximal) part of the cluster are induced upon 10 μM RA treatment in NT2-D1 cells. Gene expression was measured by RT-qPCR and is presented relative to actin. The primer sequences used for quantification are found in Supplementary Table S1. (E) The expression of 3΄ (left) and 5΄ (right) HOXA cluster genes was measured as in (C). The RT-qPCR values and standard deviations are found in Supplementary Table S2. (F) Northern blot analysis reveals the stable expression of unspliced and spliced HOTAIRM1 in NT2-D1 upon RA induction. Diagrams of the predicted HOTAIRM1 transcript variants are shown in (G). (H) Western blotting of the cytoplasmic hsp90 and nuclear lamin A/C shows the quality of the cell fractionation protocol. (I) The total unspliced and spliced HOTAIRM1 transcripts were measured as in (C), and are shown next to the nuclear MALAT-1 and cytoplasmic GAPDH transcripts for comparison. The percentage of unspliced and spliced HOTAIRM1 in the nucleus and cytoplasm is seen in (J), next to MALAT-1, GAPDH and actin included for quality control. The regions used to quantify the HOTAIRM1 variants are indicated with green lines in (G). E; exon. I; intron. All RNA expression measurements in this figure are from at least three PCRs in three biological replicates, and error bars represent standard deviations.
We first verified that HOTAIRM1 was induced upon RA addition in NT2-D1 cells (Figure 1C and D; Supplementary Table S1). As expected (30,33), the proximal HOXA genes—particularly HOXA1 to A5—were dramatically induced upon RA addition while distal HOXA9 to A13 remained silent (Figure 1C). RA treatment also potently induced HOTAIRM1, and to lesser extent HOXA-AS2 and HOXA-AS3 encoded within the cluster proximal region, but not HOTTIP found 5΄ of HOXA13 (Figure 1D). Over a 3-day RA differentiation time course, we found HOTAIRM1 induced at a similar rate and level as HOXA1 (Figure 1E and Supplementary Table S2). RA induction proceeded in a collinear fashion 3΄ to 5΄ along the cluster with HOXA1, A2 and HOTAIRM1 reaching their maximum expression at 24 h, while levels of HOXA3, A4, A5 and A6 continued to rise until 72 h. HOXA7 was the only gene that showed noticeable expression prior to RA induction and gradually increased over the time course. Meanwhile, the distal genes beyond HOXA9 remained nominal throughout the treatment (Figure 1E, right). These results were consistent with the known collinear induction of HOXA genes in this cell model (33,34).
Northern blot analysis of RA-treated NB4 cells originally identified HOTAIRM1 as ∼500 nt alternatively spliced and polyadenylated transcript lacking the middle exon of the annotated HOTAIRM1 gene (15). As alternative splicing is often tissue-specific and given that NT2-D1 cells are pluripotent rather than of the myeloid lineage, we probed uninduced and RA-induced NT2-D1 by northern blotting to identify the transcript variant expressed. Surprisingly, we found that RA induces the stable expression of an unspliced transcript measuring over 4 kb in length as well as a spliced form of ∼1.1 kb (Figure 1F). Molecular cloning and sequencing of the spliced transcript revealed HOTAIRM1 containing all three annotated exons in NT2-D1 (Figure 1G and Supplementary Table S3).
Comparable levels of both HOTAIRM1 forms were detected on northern blot (Figure 1F), and we sought to better quantify this observation and determine if they were similarly distributed within cells (Figure 1H–J). We designed RT-qPCR primers to detect the unspliced and spliced NT2-D1 forms (Figure 1G, green lines), and verified the amplification efficiencies of primer pairs (Supplementary Table S4). We then prepared samples from whole cell extracts (wce), cytoplasm and nuclei that were controlled for quality by western blotting (Figure 1H). We used total RNA extracted from each sample to measure the expression and cellular localization of the transcripts (Figure 1I and J), and confirmed that both transcripts were expressed at comparable levels in induced NT2-D1, and within range of the nuclear lncRNA MALAT-1 (Figure 1I). Interestingly, unspliced HOTAIRM1 preferentially localized to the nucleus whereas most of the spliced form was observed in the cytoplasm (Figure 1J). Together, our results show that two forms of HOTAIRM1 are induced upon RA treatment of NT2-D1 cells.
Opposite HOXA gene regulation by HOTAIRM1 within a given cell type
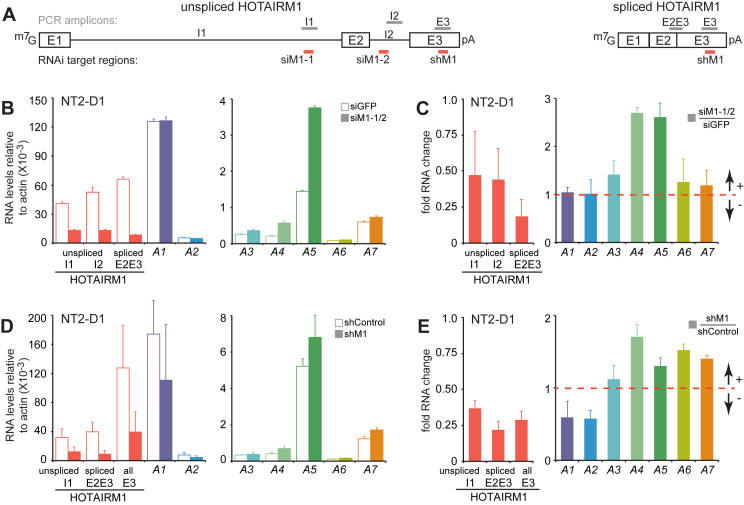
HOTAIRM1 depletion prior to RA-induced NB4 granulocyte differentiation was shown to reduce the expression of several proximal HOXA genes (15). Given that the transcripts induced by RA from the HOTAIRM1 gene differed in NT2-D1, we wondered if this gene retained its activating function across cell types. We designed several siRNAs targeting HOTAIRM1 and retained the most effective combination consisting of two siRNAs targeting its introns (Figure 2A, siM1-1 and siM1-2). Knockdown with these siRNAs during RA induction considerably reduced the accumulation of both forms of HOTAIRM1 transcripts at 72 h post-induction (Figure 2B and C) despite the fact that introns were targeted to deplete the transcripts. Surprisingly, we observed increased levels of HOXA4 and A5 in NT2-D1 instead of the suppression reported in NB4 under similar conditions. Expression of HOXA6 and A7 also increased slightly, but the results were more variable between the three biological replicates.
Figure 2.
HOTAIRM1 mediates opposite HOXA gene regulation in a given cell type. (A) Schematic representation of the unspliced (left) and spliced (right) HOTAIRM1 variants in NT2-D1. The gray (top) and salmon (bottom) lines indicate regions used for RT-qPCR quantification and for targeting transcripts by RNAi, respectively. (B and C) RNAi knockdown of HOTAIRM1 using siRNAs. RT-qPCR analysis of steady-state HOTAIRM1 and 3΄ HOXA transcripts in control (siGFP) and HOTAIRM1 (siM1-1/2) knockdown cells induced 72 h with 10 μM RA. Expression levels relative to actin in (B) is shown to display differences in steady-state expression levels between HOXA genes. Fold RNA change (C) is relative to the siGFP control set to 1. (D and E) Same as in (B and C), except that knockdowns were conducted with a short hairpin RNA control (shControl) or HOTAIRM1 (shM1). All RNA expression measurements are from at least three PCRs in three biological replicates, and error bars represent standard deviations.
We wondered if targeting the intronic region might have influenced the inferred regulatory role of HOTAIRM1 on the HOXA cluster in NT2-D1. To explore this question, we used a previously described shRNA against the final exon of HOTAIRM1 (Figure 2A, shM1 (15)). We found that shM1 curbed the accumulation of both unspliced and spliced HOTAIRM1 during RA treatment, and still led to an increase of HOXA4, A5, A6 and A7 expression (Figure 2D and E). Interestingly, the expression of HOXA1 and A2 decreased considerably when using the shRNA. We noticed that the shRNA was more effective at reducing unspliced HOTAIRM1 levels compared to when siRNAs were used (Figure 2C and E), suggesting that the full length transcript may be acting more as an activator than a repressor of HOXA genes (see below).
As our results in NT2-D1 revealed an opposing regulatory role for HOTAIRM1 on proximal HOXA genes, we verified its overall activating property in NB4 cells during RA induction (Supplementary Figure S1A and B). As previously found, the expression of HOXA1, A4 and A5 were specifically decreased when HOTAIRM1 was depleted in this cell line. In fact, we observed reduced levels for all proximal HOXA genes except for A3, and no noticeable changes for any of the distal genes. We also verified that the main HOTAIRM1 transcript expressed in RA-treated NB4 cells was ∼500 nt in length by northern blotting and RT-qPCR (Supplementary Figure S1A, C and D). Together, these results show that lncRNAs derived from the HOTAIRM1 gene can activate and/or repress HOXA gene expression within and in different cell types.
HOTAIRM1 is required for RA-induced three-dimensional chromatin organization changes at the HOXA cluster in NT2-D1
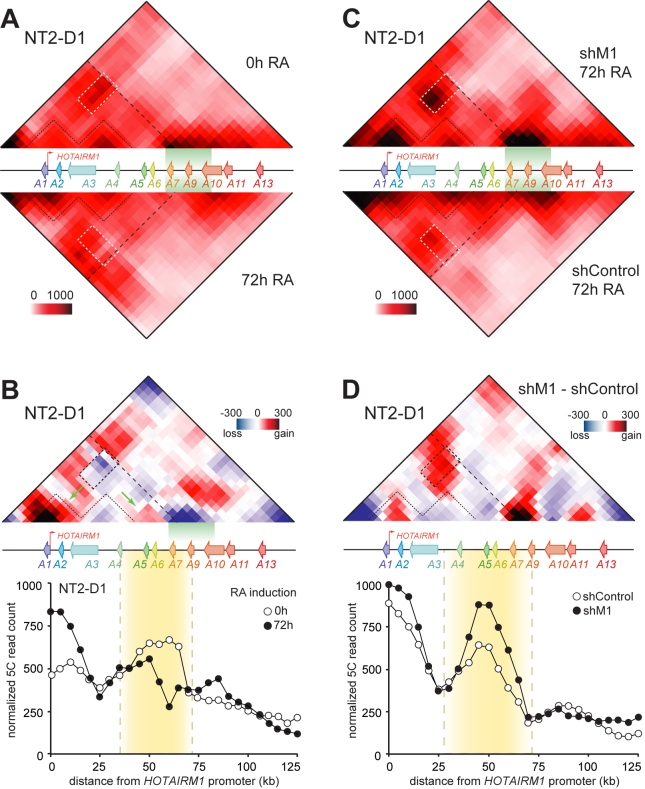
If a lncRNA can either activate or silence gene expression, then what guides which genes will be targeted and how their expression will be affected? A well-recognized mechanism by which regulatory elements such as enhancers find their target genes is through three-dimensional chromatin organization (35). This mechanism was suggested for the lncRNA HOTTIP to explain how it regulates gene expression at the HOXA cluster distal end (13). To determine whether chromatin architecture plays a role in targeting HOTAIRM1 activity, we started by defining the chromatin conformation changes induced by RA at the HOXA cluster in NT2-D1 cells. We used 5C-seq to measure the frequency of pairwise interactions across the entire locus in untreated (0 h RA) and RA-induced (72 h RA) cells (Figure 3A and B). In both conditions, we found the cluster organized into several consecutive and interacting subdomains or ‘subTADs’, characterized by their chromatin contact enrichment (Figure 3A, top). HOXA1, HOTAIRM1 and HOXA2 were all contained within a subdomain, while HOXA3 and HOXA4 resided in another subTAD adjacent to it (outlined with dashed lines). We observed a high degree of interactions between subTADs in untreated cells, especially between those containing HOXA1/2 and HOXA5/6/7 (Figure 3A, top; white rectangle).
Figure 3.
HOTAIRM1 is required for RA-induced three-dimensional chromatin organization changes at the HOXA cluster in NT2-D1 cells. (A and B) RA induces three-dimensional chromatin organization changes at the HOXA cluster in NT2-D1 cells. (A) Physical contacts along the HOXA cluster were measured by 5C-seq in uninduced (0h RA; Supplementary Table S7), and RA-induced (72 h RA; Supplementary Table S8) cells. Pairwise interaction frequencies (IFs) are shown in heatmap form (10 kb bins, 10 kb smoothing, 2× steps) according to the color scale displayed at the bottom. The HOXA region is shown to scale between the two conditions. (B) Chromatin contact changes associated with RA treatment (top) are shown in heatmap form according to the scale on the right. Heatmap values represent the difference of IFs between 72 h RA and 0h RA treatment. Blue values indicate a loss of contact after induction, and red a gain of interactions. The dashed black line dividing the cluster in two parts highlights the position of a TAD boundary. 5C chromatin interaction profile of HOTAIRM1 along the HOXA cluster (bottom) in uninduced (0 h) and RA-induced (72 h) cells. Interaction frequency (y-axis; normalized 5C read count) is correlated with the distance from the HOTAIRM1 promoter (x-axis). The looping region is highlighted in yellow. (C and D) Loss of looping contacts between HOTAIRM1-regulated genes requires the lncRNA in NT2-D1. (C) 5C-seq analysis of the HOXA cluster in control (shControl 72 h RA; Supplementary Table S11) and HOTAIRM1-depleted cells (shM1 72 h RA; Supplementary Table S12). (D) Chromatin contact changes associated with HOTAIRM1 depletion with shRNAs (top) and the HOTAIRM1 chromatin interaction profile with the HOXA cluster (bottom) are as described in panel B.
Certain sharp differences in the cluster subTAD organization were present amid the two states, including a considerable decrease in contact frequency between the HOXA1/2 and HOXA5/6/7 subTADs after RA treatment (Figure 3A, bottom; white rectangle). This result suggested that transcription activation here associated with unfolding of the cluster as observed for the mouse HoxB and D clusters (36,37), and is consistent with our cursory analysis of the HOXA locus in undifferentiated and fully differentiated NT2-D1 with the 3C technique (20). For instance, we recapitulated the general loss of contacts around HOXA9 even after 72 h of RA treatment (Figure 3B, top; green highlighted area). Our 5C analysis also further revealed higher local contact frequencies around HOXA1/2, and HOXA4/5/6 when the cells were induced (green arrows). Therefore, RA induction of the proximal HOXA genes in NT2-D1 associates with higher local interaction frequencies and a loss of long-range contacts bridging HOXA1/2 with HOXA5/6/7 (Figure 3B, top).
Having established that proximal HOXA gene expression in NT2-D1 associates with physical uncoupling of HOTAIRM1-regulated genes, we wondered if the lncRNA itself contributes to this loss of long-range contacts. We measured chromatin contacts across the cluster with 5C-seq in RA-induced HOTAIRM1-depleted NT2-D1 cells. Surprisingly, depletion of the lncRNA resulted in stronger contacts between the HOXA1/2 and HOXA5/6/7 regions (Figure 3C and D). The effect on chromatin architecture was recapitulated in an shRNA biological replicate and in cells where HOTAIRM1 was depleted by siRNAs targeting both isoforms of the lncRNA (Supplementary Figure S2). These results indicate that HOTAIRM1 transcripts are required to physically uncouple these two subTADs during RA induction in this cell model.
Given that regulation by HOTAIRM1 is so much different in NB4, we examined whether RA treatment induced a distinct physical shift at the HOXA cluster and if the lncRNA was required for such a change. As in NT2-D1, we found the cluster organized into a series of subTADs in NB4 cells, both in untreated and RA-treated states (Supplementary Figure S3A). Interestingly, the contacts between HOXA1/2 and HOXA5/6/7 were weaker in this cell type and RA did not considerably change their interaction frequency (Supplementary Figure S3A and B), suggesting an open structure even prior induction. Furthermore, HOTAIRM1 depletion in NB4 did not lead to considerable changes in the cluster's organization (Supplementary Figure S3C–F), indicating that HOXA gene regulation by the lncRNA does not involve alteration of chromatin architecture in this cell system.
Together, these findings show that the HOTAIRM1 gene resides in close physical proximity to its targets both in NT2-D1 and NB4. Furthermore, in NT2-D1 where RA triggers the collinear activation of proximal HOXA genes, we find that physical dissociation of the HOXA1/2 and HOXA5/6/7 subTAD regions accompanies gene induction. This result suggests that uncoupling of the two domains might represent an important component to HOXA collinearity. Given that in this cell system RNAi depletion of HOTAIRM1 prevents uncoupling of the subTADs and leads to higher expression of the central HOXA genes, we suggest that HOTAIRM1 could play a pivotal role in the temporal collinear induction of HOXA genes by RA. In contrast, in a cellular context like in NB4 cells where RA does not trigger the collinear activation of HOXA genes, HOTAIRM1 appears to regulate HOXA gene expression differently and without altering the spatial organization of chromatin.
HOTAIRM1 depletion alters RA-induced changes in activating H3K4me3 and silencing H3K27me3 marks
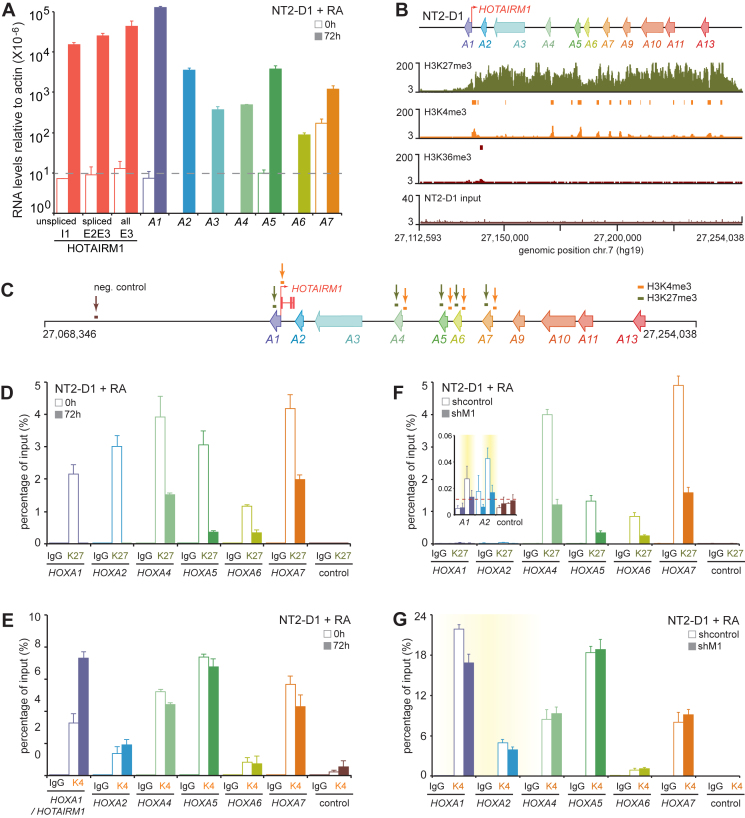
We next considered why the NT2-D1 transcriptional output of HOTAIRM1 might activate HOXA1/2 located in one contacting subTAD, and inhibit HOXA4/5/6 located in the other. To this end, we first examined the basal and RA-induced expression levels of HOXA genes in this cell model. We observed that the HOXA cluster was virtually silent prior induction and that RA led to a very strong induction of the HOXA1 to A6 genes (Figure 4A). Based on available ChIP-seq datasets from uninduced NT2-D1 (Figure 4B), we found the HOXA cluster covered with the silencing H3K27me3 mark. Consistent with its known bivalent nature (38,39), the HOXA gene promoters were also marked with the activating H3K4me3 modification, but not transcribed as seen by the absence of the H3K36me3 elongation mark along the gene bodies.
Figure 4.
Depletion of HOTAIRM1 in NT2-D1 cells alters the changes in H3K4me3 and H3K27me3 levels triggered by RA. (A) The 3΄ HOXA cluster genes are strongly induced when NT2-D1 cells are treated with RA. Measurements with values below the horizontal dashed lines represent those for which quantification was deemed unreliable. Gene expression was measured by RT-qPCR, and measurements are from at least three PCRs in three biological replicates. Error bars represent standard deviations. (B) The HOXA cluster is a transcriptionally silent bivalent domain in untreated NT2-D1 cells. The H3K27me3, H3K4me3, H3K36me3 marks were measured by ChIP-seq. Significant peaks are shown as colored rectangles above the tracks. These datasets and their matching inputs are from the UCSC browser as indicated in ‘Materials and Methods’ section. (C) Position of PCR amplicons used to profile the abundance of H3K4me3 or H3K27me3 by RT-qPCR in panels D–G. The ChIP results shown here are from the same three biological replicates analyzed for gene expression in Figure 2D and E. (D) RA induction in NT2-D1 cells is accompanied by a sharp decrease of the silencing H3K27me3 mark in the gene bodies. (E) H3K4me3 promoter levels upon RA treatment of NT2-D1 increase at HOXA1 and A2 genes. (F) 3΄ HOXA genes that are more highly expressed in HOTAIRM1 knockdowns show less H3K27me3 in their gene bodies. (G) The 3΄ HOXA1 and A2 genes that are less expressed in HOTAIRM1 knockdowns have less H3K4me3 at their promoters. Measurements are presented as a percentage of input and are from at least three PCRs each in three biological replicates. Error bars represent the standard error of the mean (sem). The primer sequences used for quantification are found in Supplementary Table S5.
We then measured the levels of H3K4me3 at the promoters and H3K27me3 in the gene bodies before and after RA induction to assess the changes caused by the morphogen (Figure 4C–E and Supplementary Table S5). As expected, we detected high levels of both H3K4me3 and H3K27me3 at the HOXA1 to A7 genes before induction (Figure 4D and E). We found much lower levels of H3K27me3 in the gene bodies after induction—particularly at HOXA1 and A2 where they were undetectable (Figure 4D). Interestingly, RA treatment increased H3K4me3 strictly at the HOXA1 and A2 promoters (Figure 4E), demonstrating that although HOXA induction in NT2-D1 occurs primarily through the removal of H3K27me3, clearing the path for transcription at the poised genes, activation of HOXA1 and A2 additionally involves enhancement of H3K4me3 at their promoters.
The fact that RA triggers distinct histone modification changes at regulated HOXA genes suggests that HOTAIRM1 depletion in NT2-D1 may activate or repress genes by differentially affecting these changes. We tested this hypothesis by examining if loss of HOTAIRM1 affects the level of histone marks in RA-induced cells (Figure 4F and G). Depletion of the transcript caused a dramatic loss of H3K27me3 at HOXA4, A5, A6 and A7 (Figure 4F), which correlates with their higher expression as measured by RT-qPCR in the same samples (Figure 2D and E). Given that we observed no change in H3K4me3 at these promoters (Figure 4G), upregulation of HOXA4 to A7 in the absence of HOTAIRM1 occurred via regulation of H3K27me3 levels.
As shown in Figure 4D, H3K27me3 was virtually absent at HOXA1 and A2 after RA induction such that HOTAIRM1 could no longer play a role in modulating the silencing mark at these genes (Figure 4F, inset). However, we detected less H3K4me3 at HOXA1 and A2 (Figure 4G)—the only two gene promoters for which higher levels of H3K4me3 are observed upon RA treatment (Figure 4E). Thus, it appears that HOTAIRM1 depletion leads to lower expression of HOXA1 and A2 by preventing further activation of their promoters. Together, our results show that during RA induction of NT2-D1 cells, HOTAIRM1 contributes to the activation of HOXA1/2 by increasing H3K4me3 levels, while it helps suppress the expression of HOXA4/5/6/7 by curbing the removal of H3K27me3 silencing marks.
The situation is different in NB4, where HOTAIRM1 seems to promote the overall activation of proximal HOXA genes. Indeed, compared to NT2-D1, the basal expression levels of proximal HOXA genes and of HOTAIRM1 was very high, such that a 72-h RA treatment only led to modest inductions of some genes (Supplementary Figure S4A). We verified this result in an RA-induction time course where the genes were either slightly induced (3-fold) by RA or not at all (Supplementary Figure S4C and Table S5). Also, we found that gene promoters were already marked with high levels of H3K4me3 and RNAPII signals even before induction in NB4 cells (Supplementary Figure S4C).
We reasoned that these basal conditions might explain the different activities of HOTAIRM1 in the two cell lines and provide two distinct chromatin states from which different mechanisms of regulation from a single lncRNA gene could be explored. Given the HOXA chromatin landscape of each cell line prior to induction, we expected RA to have a different impact on chromatin structure in NB4 compared to NT2-D1 cells. We found that H3K27me3 was virtually absent across the cluster in both uninduced and RA-treated NB4 cells (Supplementary Figure S4D and E), indicating that RA did not induce HOXA expression by reducing this silencing mark. However, H3K4me3 levels were increased after RA induction, particularly at the HOXA1 and HOXA4 promoters (Supplementary Figure S4F). These results were consistent with the ChIP-seq data shown in Supplementary Figure S4C, and with the observation that HOXA1 and A4 are the two most highly expressed genes at the cluster in NB4 (Supplementary Figure S1A). Therefore, HOXA expression changes upon RA treatment in NB4 cells associate solely with higher activating marks at promoters.
Accordingly, HOTAIRM1 depletion did not alter the very low levels of H3K27me3 across the cluster (Supplementary Figure S4G). Instead, we found less activating H3K4me3 at the promoters of HOXA1 and A4 (Supplementary Figure S4H), the two most downregulated genes in knockdown cells treated with RA (Supplementary Figure S1A and (15)) and the promoters for which the greatest increase in activating marks could be observed after RA treatment (Supplementary Figure S4F). Thus, HOTAIRM1 depletion leads to the downregulation of certain HOXA genes by curbing promoter activation signals in both NT2-D1 and NB4 cells. Together, these results show that the chromatin landscape from which HOTAIRM1 is transcribed differs significantly in NT2-D1 and NB4, and that RA induces distinct histone modification changes in the two cell models. These differences may in fact explain why HOTAIRM1 expression can lead to more or less HOXA gene expression in different cell types.
HOTAIRM1 associates with activating UTX/MLL and silencing PRC2 complexes
The results presented above show that HOTAIRM1 depletion interferes with RA-induced histone modification changes at regulated HOXA genes in both NT2-D1 and NB4. Loss of HOTAIRM1 resulted either in a failure to enhance H3K4me3 at promoters or a more rapid displacement of H3K27me3 along gene bodies, suggesting that the lncRNA has a role in balancing the rate of change of both activating and silencing marks. Whether HOTAIRM1 expression leads to more or less transcription may therefore depend on the types of chromatin-modifying complexes it can interact with, guided at least in part by the chromatin landscape from which it is transcribed. Given that either H3K4me3 or H3K27me3 levels were altered in knockdown samples and that other lncRNAs have been shown to bind complexes involved in modulating these marks (10,12–13), we wondered if HOTAIRM1 regulates HOXA genes by interacting with such complexes.
We addressed this question by performing RIPs with nuclear extracts from RA-treated NT2-D1 and NB4 cells (Figure 5 and Supplementary Figure S5) where we strategically tested its interaction with the H3K27me3 demethylase UTX and the PRC2 component Suz12. PRC2 is the only complex known to trimethylate histone H3 on lysine 27 in these cells (40), and has been shown to interact with numerous lncRNAs. RA induction is known to recruit UTX/MLL at HOX genes to promote body demethylation of H3K27me3 with concomitant methylation of H3K4 (41). UTX was also shown to interact with both retinoic acid receptor α (RARα) in vivo (42), and ASH2L, a mandatory component of the mixed-lineage leukemia (MLL) complex for H3K4 methylation (43–45). Furthermore, we could detect comparable expression of UTX and ASH2L in both cell lines (Supplementary Figure S5).
Figure 5.
Preferential association of unspliced HOTAIRM1 with activating UTX/ASH2L and of silencing PRC2 complexes with the spliced form. (A) Linear diagrams of the unspliced (left) and spliced (right) HOTAIRM1 variants in NT2-D1. The gray lines (top) indicate regions used for RT-qPCR quantification in (B and C). E; exon. I; intron. (B) UTX RIP in RA-induced NT2-D1 nuclear extracts. (C) Suz12 RIP in RA-induced NT2-D1 nuclear extracts. RIP measurements are presented as a percentage of input and are from at least three PCRs each in three biological replicates. (D and E) Western blot analysis of control (IgG), UTX and Suz12 RIP samples show the specific isolation of UTX/MLL complexes distinct from PRC2 in NT2-D1 cells. (F) Higher UTX binding to HOXA4-7 in HOTAIRM1-depleted NT2-D1 cells induced with RA. The ChIP-RT-qPCR results are from three biological replicates and error bars represent the standard error of the mean (sem). The primer sequences used for ChIP are found in Supplementary Table S5.
In NT2-D1 where two HOTAIRM1 variants were expressed (Figure 5A), we found a similar fraction of nuclear spliced and unspliced HOTAIRM1 transcripts associated with UTX (Figure 5B). Since nuclear extracts contain approximately three times more unspliced transcripts (Figure 1J), we can therefore reasonably assume that nuclear UTX is more frequently bound to unspliced HOTAIRM1 than the spliced form. The preferential association of unspliced HOTAIRM1 with UTX implies that it can act through both H3K27me3 removal and addition of H3K4me3, which supports our initial finding that this form acts more like an activator—particularly at neighboring HOXA1 and A2 genes where higher concentration of this variant can be expected (Figure 2C and E). Interestingly, we were also able to detect HOTAIRM1 binding to Suz12 (Figure 5C), which unlike UTX, seemed to favor the spliced lncRNA, especially when considering that this form is three times less abundant in the nucleus of NT2-D1 cells (Figure 1J). Though binding to different chromatin-modifying complexes has been reported previously for a small number of lncRNAs, we were surprised to find HOTAIRM1 interacting with two functionally opposing complexes. We verified the specificity of our RIP conditions by western blot (Figure 5D and E), and confirmed that the pull-down of UTX could indeed co-immunoprecipitate ASH2L but not Lamin A/C used as control. Suz12 was not co-immunoprecipitated with the UTX antibody and vice versa validating that two distinct types of HOTAIRM1 ribonucleoproteins were probed by RIP.
Since the higher HOXA4-A7 gene expression in HOTAIRM1-depleted samples was accompanied by a loss of silencing histone marks (Figure 4F), we wondered if stronger contacts between the two subTADs might influence the distribution of UTX. We measured occupancy of the demethylase by ChIP in RA-induced HOTAIRM1 knockdown samples and found that binding of UTX at the HOXA4-7 genes increased considerably when the lncRNA was depleted (Figure 5F). This finding provides a potential mechanistical explanation for the more rapid H3K27me3 demethylation observed at these genes upon HOTAIRM1 depletion. As could be expected, we found no substantial change in UTX occupancy at the HOXA1/2 genes since this region was already devoid of silencing marks at 72 hours of RA treatment (Figure 4F). Together with our ChIP data, our results suggest that HOTAIRM1 regulates the expression of proximal HOXA genes in NT2-D1 by affecting both three-dimensional chromatin organization and the activity of histone-modifying complexes at the cluster.
In NB4, where a shorter spliced variant is expressed (Supplementary Figure S5A) we found that HOTAIRM1 can also associate with both UTX and Suz12 in the nucleoplasm (Supplementary Figure S5B and C) using RIP conditions that precipitate distinct UTX/MLL and PRC2 complexes (Supplementary Figure S5D and E). Given the much lower relative abundance of unspliced and fully-spliced HOTAIRM1 in NB4 cells (Supplementary Figure S1A and C), it is reasonable to expect that UTX and Suz12 complexes containing HOTAIRM1 will mainly feature the shorter ∼500 nt form. However, we suggest that HOTAIRM1 will mainly associate with the activating MLL complex at the cluster in these cells since PRC2 was not found at the HOXA genes, which potentially explains why HOTAIRM1 does not inhibit HOXA genes in this cell system. Taken altogether, we propose that the type(s) of lncRNP(s) formed will depend on the transcript variant(s) expressed and the protein complexes in its vicinity.
DISCUSSION
We know very little about the mechanisms by which lncRNAs regulate gene expression despite the fact that many of them have now been functionally validated (46–48). Several studies demonstrate that lncRNA function requires their association with chromatin-modifying complexes but how and why they achieve specific regulation at given target genes has remained mostly uncharacterized. Here, we investigate the role of HOTAIRM1 in regulating the expression of HOXA genes in NT2-D1 and NB4, two well-known cellular differentiation systems. Our study provides evidence that a lncRNA gene can have distinct regulatory functions in different cell types. We propose that lncRNA function is guided by the type of transcript produced, which may favor binding to particular protein complexes, the unique chromatin state around the lncRNA and target genes that will direct which lncRNPs can form and to the manner in which the chromatin is folded to create spatial neighborhoods.
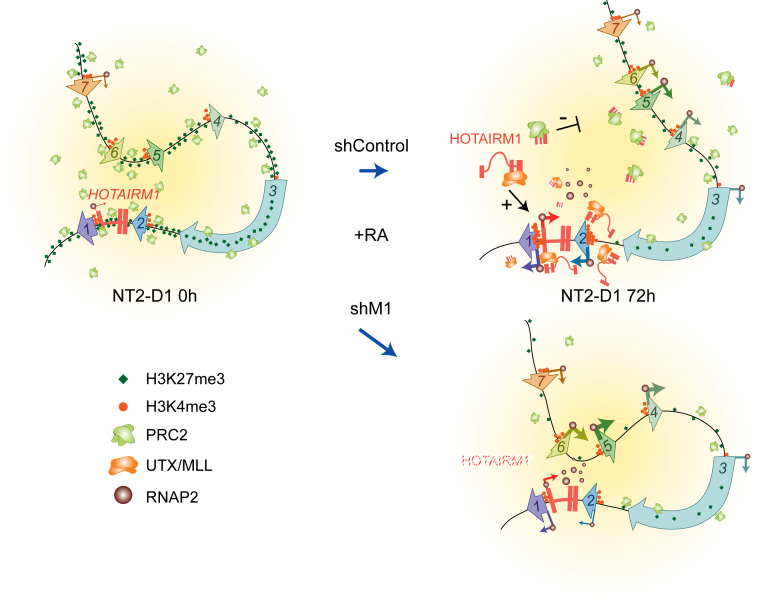
In a previous study (20), we speculated that ‘looping’ contacts may be required to regulate the temporal collinearity of HOX genes in vivo as both are conserved features of the clusters. Based on our findings here, we propose that HOTAIRM1 uses chromatin looping to promote the temporal collinear activation of HOXA genes by RA in NT2-D1. We suggest that looping between the HOXA1/2 and HOXA5/6/7 genes initially contributes to the maintenance of their silent state, and later helps coordinate their regulation by HOTAIRM1. In our model (Figure 6), we propose that when a morphogen such as RA activates the locus in a collinear manner, the local accumulation of unspliced HOTAIRM1 help enhance early activation of HOXA1/2 through recruitment of UTX/MLL complexes. This is supported by our lower H3K4me3 ChIP signals at the HOXA1/2 promoters when HOTAIRM1 is depleted during RA stimulation (Figure 4E and G). Simultaneously, interactions between the HOXA1/2 and HOXA5/6/7 subTADs are relinquished in order to prevent the rapid demethylation and expression of the downstream genes.
Figure 6.
Diagram illustrating how HOTAIRM1 regulates the expression of HOXA genes in NT2-D1 cells. The model suggests how HOTAIRM1 contributes to the collinear activation of proximal HOXA genes through modulation of both spatial chromatin organization and the distribution of histone-modifying complexes.
Although not yet fully tested experimentally, we propose two processes that contribute to this physical uncoupling and that are consistent with our data. First, early removal of polycomb proteins around HOXA1/2 (Figure 3A and B, top), which are known to mediate long-range contacts (49–51), will facilitate physical uncoupling of the two regions. Second, the initial spatial proximity between the HOTAIRM1 gene and HOXA5/6/7 permits rapid delivery of the spliced lncRNAs by diffusion where they will maintain PRC2 residence to delay transcription. We propose that combined, these events could altogether permit sequential HOXA induction.
Failure to dissociate the HOXA1/2 and HOXA5/6/7 subTADs as seen in HOTAIRM1 knockdowns might contribute to premature expression of the downstream genes. Indeed, their physically proximity to the highly expressed HOXA1/2 genes might, for instance, allow UTX recruitment to pervade throughout the proximal region and accelerate H3K27me3 demethylation (Figure 5F). This is supported by the observation that RNAi knockdown of both HOTAIRM1 isoforms leads to higher expression of the middle HOXA genes as compared to when only the spliced HOTAIRM1 variant is specifically targeted (Supplementary Figure S6). We found that depletion of spliced HOTAIRM1 does not change chromatin organization at the locus (Supplementary Figure S6C) and that de-repression of HOXA4/5/6/7 is less prominent under these conditions (Supplementary Figure S6F). This result suggests that spliced HOTAIRM1 does contribute to the repression of the HOXA genes. In contrast, unspliced HOTAIRM1—not the spliced form—is required to change the cluster conformation during RA induction, and the physical proximity resulting from its depletion also contributes to premature gene expression at the cluster.
Whether additional elements contribute to the observed HOXA deregulation when HOTAIRM1 is depleted remains unknown as other features, including a potential deregulation of other lncRNAs encoded within the proximal region, might add to this effect. However, our data shows that uncoupling of the two domains represents an important event for proper collinear HOXA gene activation by RA. This result is particularly interesting as it provides an example where regulation of gene expression by a lncRNA is not solely due to the direct recruitment of a chromatin-modifying complex, but rather to a combination of this interaction, its impact on chromatin architecture and how the resultant change in chromatin conformation affects delivery of protein complexes that can regulate transcription. Thus, our analysis in NT2-D1 provides the first evidence that expression of a lncRNA can induce a conformational shift in the chromatin that might be required for timely gene expression. To our knowledge, this is the first functionally validated demonstration of the suggested self-enforcing feedback between chromatin activity and spatial organization (52).
Our study also shows how the behavior of a lncRNA can differ greatly when expressed in an entirely different chromatin context. In NB4 cells, the actively transcribed HOTAIRM1 gene is surrounded by a much more open chromatin devoid of silencing marks. Although still capable of associating with both UTX/MLL and PRC2 complexes in the nucleoplasm (Supplementary Figure S5A–E), the splice variant appears to regulate HOXA gene expression only by modulating H3K4me3 at promoters, likely because UTX/MLL complexes are present around HOTAIRM1 whereas PRC2 is not. Furthermore, chromatin architecture at the HOXA cluster in NB4 cells remains largely unchanged during RA induction and HOTAIRM1 does not appear to regulate proximal HOXA gene expression by changing this organization (Supplementary Figure S3C–F). Instead, HOTAIRM1 appears to act more like HOTTIP in this cellular context, where static chromatin architecture simply guides the lncRNA to its target genes. Taken together, these two cellular models show how different chromatin landscapes and conformations at the HOXA locus might impact the manner in which HOTAIRM1—and perhaps other lncRNAs—can regulate gene expression.
Much effort has been directed at annotating the wide range of lncRNAs expressed across cell types (53), and many studies searching for highly expressed lncRNAs—like HOTAIRM1—as potential biomarkers have started to emerge (14). However, these projects generally do not consider the potential cell-type specificity of lncRNAs, which may yield confounding data and halt progress in this area. Our study provides new insights on parameters that can influence gene regulation by lncRNAs. It shows that predicting how a transcript may impact transcription can require far more than simply measuring its expression, and highlights how chromatin state and architecture might be important in guiding lncRNA gene function.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our laboratory for insightful discussions, and Dr J. Teodoro (McGill University) for critical reading of this manuscript. We are indebted to D. Paquette for excellent technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CHIR) [MOP-142451 to J.D.]; Canadian Cancer Society [702834 to J.D.]; Cole Foundation Scholarships (to X.Q.D.W.); FRSQ (Fonds de la Recherche en Santé du Québec); CIHR; FRSQ Research Scholar (Junior 2) (to J.D.). Funding for open access charge: CIHR [MOP-142451].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R.. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002; 296:916–919. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H. et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002; 420:563–573. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009; 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X.Q., Crutchley J.L., Dostie J.. Shaping the genome with non-coding RNAs. Curr. Genomics. 2011; 12:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessing D., Anguera M.C., Lee J.T.. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 2013; 14:85–110. [DOI] [PubMed] [Google Scholar]
- 9.Galupa R., Heard E.. X-chromosome inactivation: new insights into cis and trans regulation. Curr. Opin. Genet. Dev. 2015; 31:57–66. [DOI] [PubMed] [Google Scholar]
- 10.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007; 129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010; 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wongtrakoongate P., Riddick G., Fucharoen S., Felsenfeld G.. Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet. 2015; 11:e1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X., Lian Z., Padden C., Gerstein M.B., Rozowsky J., Snyder M., Gingeras T.R., Kapranov P., Weissman S.M., Newburger P.E.. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009; 113:2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y., Gong B., Jiang Z.L., Zhong S., Liu X.C., Dong K., Wu H.S., Yang H.J., Zhu S.K.. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int. J. Oncol. 2016; 48:670–680. [DOI] [PubMed] [Google Scholar]
- 17.Diaz-Beya M., Brunet S., Nomdedeu J., Pratcorona M., Cordeiro A., Gallardo D., Escoda L., Tormo M., Heras I., Ribera J.M. et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015; 6:31613–31627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Wu J.J., Lin X.B., Bao Y., Chen Z.H., Zhang C.R., Cai Z., Zhou J.Y., Ding M.H., Wu X.J. et al. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. Int. J. Clin. Exp. Med. 2015; 8:5033–5043. [PMC free article] [PubMed] [Google Scholar]
- 19.Su X., Malouf G.G., Chen Y., Zhang J., Yao H., Valero V., Weinstein J.N., Spano J.P., Meric-Bernstam F., Khayat D. et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014; 5:9864–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferraiuolo M.A., Rousseau M., Miyamoto C., Shenker S., Wang X.Q., Nadler M., Blanchette M., Dostie J.. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010; 38:7472–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zagani R., El-Assaad W., Gamache I., Teodoro J.G.. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015; 6:28282–28295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Berger R.. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood. 1991; 77:1080–1086. [PubMed] [Google Scholar]
- 23.Kucharski T.J., Gamache I., Gjoerup O., Teodoro J.G.. DNA damage response signaling triggers nuclear localization of the chicken anemia virus protein Apoptin. J. Virol. 2011; 85:12638–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseau M., Crutchley J.L., Miura H., Suderman M., Blanchette M., Dostie J.. Hox in motion: tracking HoxA cluster conformation during differentiation. Nucleic Acids Res. 2014; 42:1524–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferraiuolo M.A., Sanyal A., Naumova N., Dekker J., Dostie J.. From cells to chromatin: capturing snapshots of genome organization with 5C technology. Methods. 2012; 58:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser J., Ethier S.D., Miura H., Dostie J.. A torrent of data: mapping chromatin organization using 5C and high-throughput sequencing. Methods Enzymol. 2012; 513:113–141. [DOI] [PubMed] [Google Scholar]
- 27.Pineault N., Helgason C.D., Lawrence H.J., Humphries R.K.. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 2002; 30:49–57. [DOI] [PubMed] [Google Scholar]
- 28.Bae E., Calhoun V.C., Levine M., Lewis E.B., Drewell R.A.. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16847–16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J. 3rd, Gingeras T.R. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005; 120:169–181. [DOI] [PubMed] [Google Scholar]
- 30.Sessa L., Breiling A., Lavorgna G., Silvestri L., Casari G., Orlando V.. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007; 13:223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews P.W. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev. Biol. 1984; 103:285–293. [DOI] [PubMed] [Google Scholar]
- 32.Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K.. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006; 20:1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boncinelli E., Simeone A., Acampora D., Mavilio F.. HOX gene activation by retinoic acid. Trends Genet. 1991; 7:329–334. [DOI] [PubMed] [Google Scholar]
- 34.Mavilio F., Simeone A., Boncinelli E., Andrews P.W.. Activation of four homeobox gene clusters in human embryonal carcinoma cells induced to differentiate by retinoic acid. Differentiation. 1988; 37:73–79. [DOI] [PubMed] [Google Scholar]
- 35.Fraser J., Williamson I., Bickmore W.A., Dostie J.. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. 2015; 79:347–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambeyron S., Bickmore W.A.. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004; 18:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morey C., Da Silva N.R., Perry P., Bickmore W.A.. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007; 134:909–919. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 39.Voigt P., Tee W.W., Reinberg D.. A double take on bivalent promoters. Genes Dev. 2013; 27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D.. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002; 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Di Croce L., Shiekhattar R.. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007; 318:447–450. [DOI] [PubMed] [Google Scholar]
- 42.Rocha-Viegas L., Villa R., Gutierrez A., Iriondo O., Shiekhattar R., Di Croce L.. Role of UTX in retinoic acid receptor-mediated gene regulation in leukemia. Mol. Cell. Biol. 2014; 34:3765–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M. et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007; 282:20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G.. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006; 13:713–719. [DOI] [PubMed] [Google Scholar]
- 45.Steward M.M., Lee J.S., O'Donovan A., Wyatt M., Bernstein B.E., Shilatifard A.. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006; 13:852–854. [DOI] [PubMed] [Google Scholar]
- 46.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010; 42:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gendrel A.V., Heard E.. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu. Rev. Cell Dev. Biol. 2014; 30:561–580. [DOI] [PubMed] [Google Scholar]
- 48.Goff L.A., Rinn J.L.. Linking RNA biology to lncRNAs. Genome Res. 2015; 25:1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiwari V., Cope L., McGarvey K., Ohm J., Baylin S.. A novel 6C assay uncovers polycomb-mediated higher order chromatin conformations. Genome Res. 2008; 18:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eskeland R., Leeb M., Grimes G.R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A.I., Wutz A. et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010; 38:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francis N.J., Kingston R.E., Woodcock C.L.. Chromatin compaction by a polycomb group protein complex. Science. 2004; 306:1574–1577. [DOI] [PubMed] [Google Scholar]
- 52.Cavalli G., Misteli T.. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013; 20:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.