Data from all aspects of bacteriology, from genomics through to single-protein localization, have been accumulating at an astonishing rate over the past few years. How then to make sense of all this information and how to try and assemble it into a rational, robust model that might go some way to explaining “a bacterium”? How does such a cell sense, respond, and adapt as it interacts with its local environment? These are the long-term goals of many bacteriologists, and 120 assembled in May 2004 in San Feliu in northern Spain to tackle aspects of this problem from a very wide range of perspectives. This was the fourth of the conferences funded by the European Union and now organized by the European Science Foundation under the title “Bacterial Neural Networks.” The aim of this conference series is to bring together scientists investigating different aspects of bacterial behavior and using very different approaches to understanding bacterial complexity. In this way it is hoped that new insights or interpretations will be developed and new collaborations established. It has been very exciting to see the subject of “bacterial systems biology” develop over the years of the conferences, from a vague hope to a solid achievement. With the increasing number of sequenced genomes, we now have some idea of the core components that make up a bacterium. We may not know what all these components do (a great many may still have the wrong labels), and we may not know where they are all located or who their partners are, but probably sooner than any of us could have anticipated 8 years ago, we shall have resolved many of these uncertainties.
A few years ago most researchers working on, for example, transcriptional regulation would focus, in vitro, on a single promoter and one or two regulators; now we need to analyze complex interactive pathways with multiple levels of regulation, from phosphoproteins to small RNAs (sRNAs). Technological developments have allowed us to move from immunoelectron microscopy of fixed cells to the capacity to pinpoint a protein in a living cell and follow the dynamics of that protein as the cell develops. While quantitation is still limited, the idea of following the movement, behavior, and interaction of a specific protein in a 2-μm cell is still truly amazing. Given the mass of data now being generated, it should not be surprising that this is now seen as a subject ripe for computational modelling, a point well illustrated in San Feliu. However, the talks and posters served to reveal not only how far we have come but also how far we are from understanding the complexity of what most scientists still see as “simple” organisms. Bacteria have more complexity, more organization, and more control than we ever imagined.
What follows is a summary of the plenary sessions and some of the short talks; unfortunately there is not enough space to discuss the high-quality data presented in the very large number of posters.
REGULATORY NETWORKS
The first session of the meeting dealt with approaches for modelling aspects of the control circuits of bacterial cells. By considering the cell as a chemical system, with very specific system properties, several researchers have used “systems theory” to try to comprehend metabolic and regulatory pathways in bacteria as a step towards producing generic models for use not only for bacteria but also for higher organisms. Hans Westerhoff (Free University of Amsterdam) opened the series of plenary lectures with a mathematical description of the cell as a typical nonlinear system. He emphasized that life depends on multiple interactions and molecular communications, and since in a cell when something changes then “everything” changes, mathematical modelling and simulations are going to be essential. In living organisms, elements or components evolve their properties through interactions with other components, thereby giving even the simplest living cell a very high level of complexity. To develop robust models, Westerhoff described two pathways in enterobacteria regulated at both a genetic and physiological level: the phosphoenolpyruvate:carbohydrate phosphotransferase systems (PTS) and ammonia assimilation. The PTS pathways combine transport, metabolism, and signal transduction, and an accurate model would have to integrate these different processes in a single “system”. PTS-mediated carbohydrate uptake has been analyzed experimentally and by simulation to suggest that bacteria, unlike mammalian cells, may be just small enough to escape diffusion limitation when using a group translocation system, but possibly only if the individual components are separate and not part of a complex (8). The nitrogen assimilation system (or glutamine synthase cascade) and its regulation are also ideally suited for modelling of responses to complex patterns of stimulation. The combination of modelling and experimentation suggests that this cascade is more complex than would appear necessary for the simple regulation of ammonia assimilation. While the ammonia uptake flux is extremely robust, the flux through the assimilation pathways is not. This suggests that the regulatory system can adjust the precise pathways used, and there may be regulatory mechanisms other than the well-characterized adenylylation of glutamine synthetase regulating that flux. Work is in progress, using system approaches combined with biochemistry, to identify the different pathways, their regulatory mechanisms, and their relative importance.
Joseph Lengeler (MPI “Dynamics of Complex Technical Systems, ” Magdeburg, Germany) also uses the PTS pathways as a paradigm of the bacterial system, but uses a “box-in-box” model approach to elucidate the nature and importance of the different circuit diagrams of the cell. His approach describes cellular activities as defined by their metabolic activities, genetic regulatory networks, and global physiological networks, as units of increasing complexity, with larger units formed by aggregation of units of a lower hierarchical level (22). Therefore, a cell can be described at different levels of detail. Lengeler argues that most extant computer models rely on qualitative and in vitro data and reflect the nonliving state. However, to understand a living cell, we need to understand the fluxes through complex networks, the dynamics of the system, and its variability under natural conditions in vivo. As yet, he maintains, for even a system as apparently well studied as carbohydrate metabolism in Escherichia coli, the quantity and quality of biochemical data are insufficient to accurately describe the pathways. To begin to describe these pathways with precision, it is necessary to identify intermediates that accurately reflect the flux through the pathway, eliminating the need to measure the kinetics of each enzyme in vivo, and then to measure these in a single strain in defined conditions. Lengeler noted that the past history of a bacterium alters these functions and emphasized that measurements must use chemostat-grown isogenic strains, and where expression relies on the use of plasmids, these must be single copy with well-characterized expression activities. Thus, measurement of the transport rate of the carbohydrates, the PEP/pyruvate ratio (reflecting the flux through gluconeogenesis and glycolysis), and the intracellular concentration of cyclic AMP (also controlled by the PTS) together circumscribe a functional unit reflecting the state of carbohydrate metabolism. Taking this approach further, Lengeler suggested that all the activity of the tricarboxylic acid cycle might be measured with a few essential parameters, such as cyclic AMP concentration and either the oxidized-to-reduced ratio of the quinone pool or the electrochemical proton gradient (20). Applying this approach to model, for example, chemotaxis to PTS carbohydrates, he found that only the protein kinase EI of the PTS pathways has the necessary kinetic properties that enable E. coli to mount a chemotactic response in 10 ms to PTS substrates.
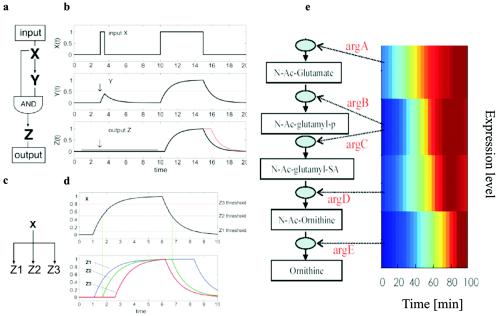
Rather than looking at protein interactions, Uri Alon (Weizmann Institute of Science, Rehovot, Israel) is trying to identify features in the transcriptional regulation network of E. coli that will allow patterns to be identified, what he terms the “design principles” of the transcriptional network (2). Using a comprehensive library of gfp-promoter fusions in E. coli and automated multiwell fluorimetry, Alon's group identified three abundant network motifs; feed-forward loops, single input modules, and dense overlapping regulons. Each network motif has a specific biological function in determining gene expression (Fig. 1). The coherent feed-forward loop turns out to be the most common (27). This filters out signals which are (too) transient, whereas the incoherent feed-forward loop allows a rapid response to the loss of a signal. The single input module is important for generating temporal expression, as in flagellar synthesis and accompanying assembly. Very little organizational hierarchy is present in these transcriptional networks, possibly because the cells need a fast response to external stimuli. There is also little overlap between regulons and rarely more than three steps in a cascade. The temporal expression of the flagellar regulon has the largest number of steps at five. Where there are multiple steps in an E. coli cascade, different sigma factors seem to be involved.
FIG. 1.
Transcriptional network motifs. Complex transcription networks are made out of simple network motifs (31, 36): patterns that recur throughout the network much more frequently than in randomized networks. Each of the network motif circuits has a defined information processing role. The feed-forward loop circuit (a), which appears in hundreds of systems from bacteria to humans, acts as a persistence detector (27) (b): it filters out short input pulses and responds only to persistent signals. The single-input module (c) can generate temporal expression programs. It allows serial expression of genes by means of differential activation thresholds by the master regulator (d). The single-input module in the arginine biosynthesis system of E. coli (e) was experimentally found to generate a temporal order of genes by means of high-resolution expression measurement employing fluorescent reporter strains (49). Strikingly, the temporal order corresponds to the order of the gene products in the metabolic pathway: the closer the gene to the beginning of the pathway, the earlier its expression. Thus, evolution uses a just in time production strategy, similar to engineering principles of production pipelines. (Figure kindly provided by Uri Alon.)
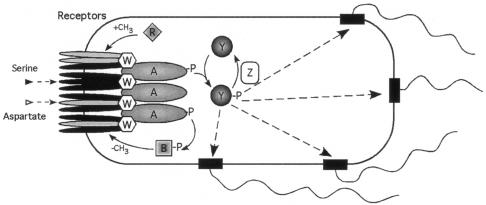
Probably the most tractable system for systems biology is E. coli chemotaxis (Fig. 2). This is a relatively simple system composed of a limited number of well-characterized proteins and with decades of behavioral and genetic data underpinning current models. Consequently, therefore, it is not surprising that predictive modelling has a firm foundation in this area. Denis Bray (University of Cambridge) has developed a program to predict the swimming trajectories of a model cell under conditions where a wide range of parameters are varied. This program has a graphical output allowing the observation of multiple bacteria swimming in defined gradients of attractants and repellents. Another algorithm developed by this group allows the diffusive trajectory and the chemical reactions of the individual chemosensory molecules to be followed in three dimensions. These algorithms have been used to look at the behavior of chemosensory proteins within a model E. coli cell and the effect on its behavior of varying different parameters (37). These include the response thresholds of the chemoreceptors, the occupancy of the flagellar motor by CheY-P, the rate of CheY-P diffusion from the kinase at the receptors to the motor, and the effects of changing the location of the CheY phosphatase, CheZ, in the E. coli cell. Experimental analysis has shown that the chemoreceptors form clusters at the poles of all bacterial cells. Modelling cooperative signaling through these arrays provides an in silico threshold value for chemotaxis that is now close to that observed in vivo. The use of stochastic models suggests that the spatial positioning of CheZ may regulate the spatial distribution of CheY-P in the cells. If CheZ is close to the receptor cluster together with the kinase, CheA, an almost even distribution of CheY-P in the cells is achieved and motors receive the same concentration of CheY-P wherever they are relative to the chemoreceptors (24).
FIG. 2.
Chemosensory pathway of E. coli. Shown are the abundant transmembrane chemoreceptors, Tar (grey) and Tsr (Black), which respond to aspartate and serine, respectively, localized at the pole of the cell as trimers of dimers. These are held in a lattice by CheW and the histidine protein kinase, CheA. On a reduction in receptor binding CheP is autophosphorylated and the phosphoryl residue is transferred to one of two competing response regulators, CheY and CheB. CheY when phosphorylated diffuses to the flagellar motor to increase switching of rotation. CheZ increases the rate of CheY∼P dephosphorylation to terminate the signal. The methylesterase activity of CheB increases when phosphorylated and modifies the cytoplasmic domain of the chemoreceptors by removing methyl groups added to glutamate by the methyl transferase, CheR. (Figure kindly provided by Victor Soujik.)
Despite the improved precision of the models described above and their closeness to some experimental measurements, we are still a long way from understanding many of the details of signal transduction, and this was illustrated in the talk by Jeff Stock (Princeton University). His group has been looking at the molecular architecture and the activity of the chemoreceptor clusters at the cell poles. The receptor patch is about 250 nm across and is composed of the chemoreceptors methyl-accepting chemotaxis proteins (MCPs), the kinase (CheA), and CheW, a protein with no known enzymatic activity. CheW probably helps couple CheA to the receptors within the patches and is important in bringing the components of the cluster together. The maximum kinase activity is seen when these proteins are in the ratio 6 MCPs to 4 CheWs to 1 CheA, with CheW increasing the activity of the kinase 200 fold (47). Many current models of receptor activity, based on the crystal structure of the cytoplasmic domain, suggest that the receptors are present as trimers of dimers. However, Stock presented data suggesting that the dimers may interact both through their periplasmic domains and through their cytoplasmic domains by strand exchange between monomers of different dimers, swapping antiparallel strands (present within the cytoplasmic “tails” of each monomer) within the lattice. He also suggested that, given the large molecular sizes of CheW and CheA relative to the helical loops of the MCPs and the relative copy numbers of the proteins, CheW and CheA could individually span more than one dimer, linking the dimers into the lattice. Supporting these views, electron microscopy of active cytoplasmic domains held in the correct conformation by terminal leucine zippers formed tetramers of dimers, rather than trimers. Methylation of the MCPs, which is the biochemical basis for adaptation (or “desensitization”) in chemotaxis, controls the gain of the stimulus-response coupling, but the mechanism controlling the level of sensitivity and gain is unclear. Stock argued that no relationship has yet been found between receptor occupancy and kinase activity. He therefore suggested that methylation of the glutamates on the receptors alters the lateral interaction between MCP receptors, by modulating strand exchange, through a sliding interaction between the heptad repeat sequences that are identifiable in the coiled-coil region of the cytoplasmic domain of the MCPs.
A complementary approach to the analysis of E. coli chemotaxis as a model system for complex circuit regulation was discussed by Victor Sourjik (University of Heidelberg). He reported the first in vivo experimental data which support models assuming long-range cooperation between the different chemoreceptors in the polar clusters resulting from attractant binding. By using fluorescent fusions to the chemosensory proteins he indirectly measured the kinase activity of CheA in living cells through the interaction of CheY-P (yellow fluorescent protein [YFP] tagged) with its cognate phosphatase CheZ (cyan fluorescent protein [CFP] tagged), using fluorescence resonance energy transfer. By measuring the dose-response curves over a wide range of attractant concentrations and by the controlled variation of the number of different receptors in the cluster, he provided clear evidence for functional interactions between homologous and heterologous receptors in receptor activation (38). Chemoreceptor clusters followed the behavior of multisubunit allosteric proteins proposed by Monod et al. (32), with functional units of at least 10 receptor homodimers.
RESPONSES TO ENVIRONMENTAL CHANGE
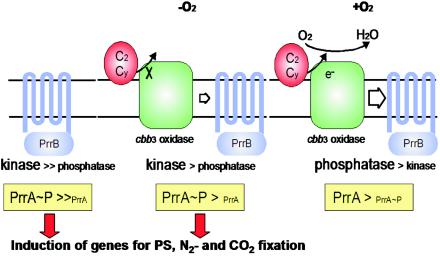
Rhodobacter sphaeroides (a purple nonsulfur bacterium) has long been a model system for photosynthesis. However, this organism is highly versatile, and in oxygenated environments can use one of at least three different respiratory electron transport chains, the terminal oxidase used depending on the oxygen tension. Under aerobic conditions the genes encoding the photosynthetic apparatus are repressed, but with falling oxygen levels derepression takes place. Sam Kaplan (University of Texas, Houston) described the key role in this control switch played by the interplay between one of these terminal oxidases (the cytochrome c oxidase cbb3) and the signal transduction histidine protein kinase-response regulator pair PrrBA (Fig. 3) (33). cbb3 cytochrome c oxidase monitors the flow of reductant, possibly with high levels of electron flow through the oxidase generating an “inhibitory signal” that favors the phosphatase activity of the histidine autokinase PrrB and hence repression of the photosynthetic genes (34). While the mechanism of communication between cbb3 and PrrB remains unclear, from microarray experiments the two-component Prr system appears directly or indirectly to control an amazing 20% of the organism's genes under anaerobic conditions and 40% under aerobic conditions. The neatness of the system allows fine homeostatic control as well as a capacity for major, rapid, but reversible, adaptation through the deployment of completely different modes of energy generation.
FIG. 3.
Regulation of PrrAB activity by electron flow through cytochrome c oxidase cbb3 in R. sphaeroides. PrrB is a transmembrane histidine protein kinase which acts as both a kinase and phosphatase for its cognate response regulator PrrA. In the absence of cytochrome c oxidase cbb3, PrrB functions predominantly as a kinase, phosphorylating PrrA, which is a direct or indirect transcriptional regulator for 20% of the genome, including the photosynthesis, nitrogenase, and carbon dioxide fixation genes. Under anaerobic conditions there is little electron flow from cytochrome c2 or cy and PrrB functions primarily as a kinase. As electron flow increases with increasing oxygen, the phosphatase activity of PrrB increases, decreasing the cytoplasmic concentration of PrrA∼P and reducing expression of target genes. (Figure reproduced from reference 34 with permission.)
Ian Booth (University of Aberdeen) emphasized the vital importance of water balance in maintaining the correct high positive internal pressures necessary for growth of bacteria. In E. coli at low external osmolarity the potentially lethal influx of water must be immediately (in milliseconds) countered by releasing solutes (such as K+, glutamate, or trehalose) through mechanosensitive channels, such as MscS, in order to restore the correct osmotic gradient. Such channels (a homoheptamer in this case) must be capable of extremely fast and efficient opening and closing. The crystal structure reported in 2002 (16a) elucidated the open state, and Ian Booth reported experiments involving cross-linking and mutagenesis studies, leading to a convincing model of channel closing (30). This involves rotation of the seven TM3 helices lining the pore, as specific Ala and Gly residues in TM3 on adjacent subunits slide past each other. The resulting narrowing of the pore is further restricted by the consequent closer packing of the large cytoplasmic domain. Excellent support for the proposed mechanism was obtained from molecular modelling (7).
The Aberdeen group has also recently reported that expression of MscS (and MscL) is induced by entry into stationary phase (39). This is dependent upon RpoS, the general stress and stationary-phase regulator in E. coli, whose own regulation was the subject of a detailed analysis described by Regine Hengge (Free University of Berlin). Impressively, regulation of the functional activity of this sigma factor, affecting up to 10% of all E. coli genes, is controlled at the levels of transcription, translation, and stability by an exceedingly complex multiple signal-integrating regulatory network. A key player in this network is the response regulator RssB. When phosphorylated, RssB targets RpoS for degradation by the ClpXP protease (41). It turns out that RssB can probably be phosphorylated by several histidine kinases, including ArcB, which regulates the aerobic-anaerobic transition. In parallel, the ArcA response regulator also represses rpoS transcription. Adding further complexity to the story, RpoS itself controls the expression of rssB. This negative feedback loop confers a highly nonlinear input-output behavior to the RpoS control systems. Thus, E. coli can rapidly adjust the availability of sufficient RpoS when conditions change by employing the control circuit, from the several interrelated regulatory mechanisms available, best suited to those particular conditions. These findings were again a timely reminder that cells are much more than an amalgamation of linear reactions whose significance can be figured out relatively intuitively but rather, regulation involves control networks whose overall understanding requires mathematical modelling and quantitative measurements in order to fully comprehend their significance.
The important role of sigma factors as global controllers of gene expression was portrayed by Thomas Nystrom (University of Göteborg) in terms of competition between sigma factors for limiting amounts of RNA polymerase, balancing the need for reproduction on the one hand and maintenance/repair on the other. In addition to the availability of RNA polymerase, nutrient availability and the consequent levels of the vitally important alarmone ppGpp (which itself participates in the control of rpoS expression) also play a part in the fate of the cell (21). Thus, new evidence shows that higher ppGpp levels, for example, as the cell moves towards stationary phase, determine the ability of RpoS to compete with the housekeeping transcription factor σ70. This also nicely explains why any RpoS lingering in cells during balanced exponential phase is not going to be effective as a transcription factor. This general idea of competition was confirmed by modulating the level of σ70 itself, with the result that reducing σ70 levels induces a stringent-like response, reflecting increased activity of RpoS (26).
The importance of RpoS was reiterated by Rita Horak (University of Tartu and Estonian Biocentre) who reported surprisingly (or perhaps not?) that a transposase gene in Pseudomonas putida is under the control of the stationary-phase sigma factor (15). Moreover, transpositional activity of this Tn4652, found in the TOL plasmid, is regulated directly or indirectly by the two-component signal transduction system ColRS, whose targets so far are unknown (14). In this case therefore, transposition is not a chance phenomenon but is related to changes in the host's external environment, and it is tempting to conclude that this may be advantageous to the host, promoting mutagenesis under conditions of stress. Thus, we may have to consider some transposons at least in terms of symbiosis rather than as parasites.
The recurring importance of using mathematical techniques to simulate complex regulatory circuits was again emphasized by Alon Zaslaver (Weizmann Institute of Science, Rehovot, Israel) (10, 19), in this case also relating to transcription regulation in E. coli. Importantly, this study was based on improved methods, developed in the Rehovot laboratory, for measuring transcript output with time. These allow detailed comparative time course analyses of many genes encoding one or more metabolic pathways by monitoring the green fluorescent protein (GFP) output signals from live cells in response to a variety of stimuli. The results so far, combined with mathematical modelling to identify most probable interpretations, reveal interesting design principles based on a “just in time” production of pathway intermediates, very much in the manner of the assembly line production of motor vehicles (36, 49).
David Clarke (University of Bath) reported on an overlap in the genes and regulatory circuits involved in the symbiotic and pathogenic properties of the entomopathogenic gram-negative bacterium Photorhabdus (17, 3). In addition to being pathogenic, Photorhabdus also has a mutualistic relationship with a nematode, Heterorhabditis. While Photorhabdus can be isolated as two distinct phenotypic variants—primary and secondary—that are equally virulent to the larva, only the primary variant can maintain the mutualistic relationship with the nematode. Screening of a bank of transposon mutants revealed a gene whose product, HexA, is required to repress the expression of factors linked to the mutualistic activity of Photorhabdus (17). In addition, the hexA mutant not only derepresses the expression of mutualism-related factors but also attenuates the pathogenic nature of Photorhabdus. Thus, it appears that HexA plays a central role in regulating the temporally separated pathogenic and mutualistic interactions, though further studies are required to delineate the mechanism by which HexA exerts its control and identify other factors involved in this process.
CELL-CELL INTERACTIONS
One area of cell-cell communication that has developed impressively over the past decade is that of N-acyl-homoserine lactone (HSL)-dependent quorum sensing, and three talks described different aspects of the process in three bacterial species. Quorum-sensing mechanisms have been a target for development of anti-infective drugs for several years. However, it has become apparent that many species have multiple quorum-sensing pathways, some which are expressed only under specific conditions and many of which interact. If effective drugs are ever to be developed targeting quorum sensing, it is essential that the interplay between the different quorum-sensing mechanisms is understood in situ. Peter Greenberg (University of Iowa) described the HSL signaling pathways of the opportunist pathogen Pseudomonas aeruginosa, which produces two HSL signals, one with a C4 chain and the other with a C12 chain. There are two receptors which respond to the oxo-C12-HSL, LasR and QscR, and one which responds to C4-HSL, RhlR. The two systems control the expression of hundreds of P. aeruginosa genes, either by repression or induction. This is very different from other species, e.g., Vibrio fischeri, where only a couple of operons are regulated by HSL. The complexity of this gene control and the interaction of the different pathways are being picked apart by microarray analysis. Interestingly, even when a saturating signal is added at low cell density there is a built-in delay in the response of most but not all HSL-controlled genes. What controls the delay? Greenberg presented several possible models for the delay and described provocative data indicating that several mechanisms were in play (22a). In addition, the C4 and C12 signals do not regulate specific subsets of genes, but there is a continuum, with some responding to both, or giving an increased response when both are present. This leads to the suggestion that specific promoters could be identified, but this proved not to be the case, possibly indicating that some genes might be indirectly regulated. Finally, Greenberg discussed some of the clinical studies of the HSLs. Cytokines respond to HSLs, stimulating inflammation. In in vitro lung tissue studies, C12-HSL was found to be inactivated by lung epithelial cells within 2 h, while C4 inactivation took over 8 h. However, this response was found to be cell line specific. These studies illustrate the complexity of both the pathogen and the host and how the observed responses reflect their interrelationship. Microarrays will help provide a framework, but also reveal how different conditions will produce very different results.
Vibrio anguillarum is a fish pathogen, causing a disease in at least 50 different species, and is capable of surviving in seawater for more than 50 months. Debra Milton (University of Umea) described the very complex quorum-sensing system of V. anguillarum, which consists of two HSL systems and a Vibrio harveyi-like AI-2 system (5). Unlike other Vibrio species, the V. harveyi LuxR homologue, VanT, is expressed at low cell population densities and expression levels do not change with increasing cell numbers; indeed VanT negatively regulates its own expression. VanT also regulates expression of a number of enzymes, including metalloprotease, peroxidase, and enzymes for exopolysaccharide production. Deletion of RpoS decreases both VanT and metalloprotease expression, indicating that RpoS is a positive regulator of vanT and protease genes. V. anguillarum utilizes the surface mucus of fish as a sole carbon source for growth and forms biofilms on fish scales. Since VanT regulates exopolysaccharide production, a complex interaction of signals likely occurs on the fish surface involving RpoS, VanT, and other virulence proteins, but this remains to be elucidated.
Rhizobium leguminosarum, on the other hand, is a free-living soil bacterium which forms a symbiotic relationship with legumes to produce nitrogen-fixing nodules. Allan Downie (John Innes Centre, Norwich, United Kingdom) described how quorum sensing influences interactions of the bacteria with legumes. The Downie group has identified four HSL synthesis genes and six LuxR-type regulators (46). The HSLs operate as a complex network with cross regulation involving different HSLs at different levels. Mutations in three chromosomal genes, cinI, cinR, and expR, cause enhanced biofilm formation, and this may be significant because R. leguminosarum forms a biofilm on root hair tips. There are two HSL regulatory systems on the Sym plasmid, one influencing nodulation (rhi) and one controlling plasmid transfer (tra). A novel mechanism of recipient-induced plasmid transfer was described (6). The HSL made by CinI acts to induce plasmid transfer because it activates plasmid-encoded BisR to induce traR expression and TraR then induces the plasmid transfer operons. However, BisR represses cinI expression, thereby preventing production of the HSL needed for traR induction. CinI-made C14 HSL is produced extracellularly by potential recipients, and this HSL is recognized by BisR, which then induces traR expression and plasmid transfer by the donor cells (6).
Cyanobacteria also play a global role in nitrogen fixation. As in all species, nitrogen fixation is essentially an anaerobic process, but many cyanobacteria live diazotrophically using oxygenic photosynthesis in oxic environments. Some filamentous cyanobacteria have evolved a complex intercellular communication system that controls development of periodic heterocysts, specialized for nitrogen fixation in an oxic background environment. Antonia Herrero (CSIC, Seville, Spain) described the complex interplay between nitrogen metabolism and heterocyst development (11). The crucial transcriptional regulator is NtcA, which belongs to the catabolite gene activator protein family of transcriptional regulators. With the regulatory protein PII, NtcA senses the C-to-N balance in the cyanobacterial cells. NtcA and PII respond to 2-oxoglutarate, probably increasing NtcA promoter binding under conditions of N deficiency. The NtcA protein is also required for heterocyst differentiation, controlling the sequential transcriptional activation of genes during the differentiation, initially activating hetR. HetR is a positive autoregulator and also a positive regulator of ntcA expression. Increased expression activates the next stages of heterocyst differentiation (12). Obviously, for the heterocysts to function as nitrogen fixation factories, there needs to be a system for carbon to get into the cells and amino acids to get out. The mechanism is unclear, but data were presented suggesting that the amino acids are exported into the periplasmic space, which is continuous along the filament, and taken up into vegetative cells by amino acid permeases.
The theme of cyanobacterial differentiation was continued by Cheng-Cai Zhang (CNRS, Marseille, France). Only 5 to 10% of cells along a cyanobacterial filament become heterocysts, and these are spaced at equal intervals along the filament. What induces a specific cell to deviate from the normal cell cycle and differentiate? As described by Herrero, HetR is the positive signal, but there is an additional negative regulator, PatS, which diffuses from the developing heterocyst, preventing neighboring cells from differentiating. Using GFP fusions and monitoring cell differentiation in vivo, with cell size as the marker for the stage of the cell cycle, Zhang showed that older cells were more likely to differentiate on N removal (23). Indeed, 90% of cells which became heterocysts had two nucleoids, and if the cell cycle was blocked, so was differentiation. Zhang also reported a study of the role of Ser/Thr and His kinases in development. Anabaena has 53 Ser/Thr kinases and 128 His protein kinases. Data suggest that a member of the HstK family may be involved in the later, oxic regulation of nitrogenase expression within the heterocysts.
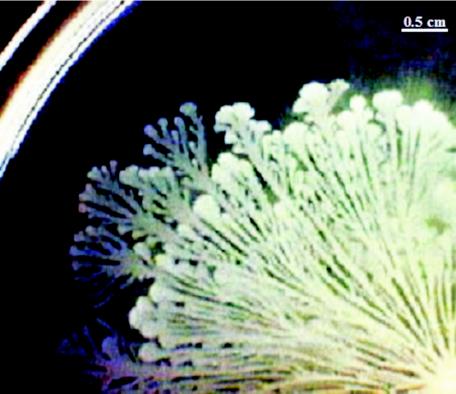
Many bacterial species have been identified that differentiate into multiflagellate forms and swarm extremely rapidly over agar surfaces. However, the developmental cycle shown by Bacillus subtilis when swarming, producing beautiful dendritic patterns, has been largely ignored (18). Simone Séror (Université Paris-Sud) reported on the many novel features of this process, including swarming by individual cells as well as by groups of cells at the tips of the dendrites (Fig. 4). Séror also described the properties of a signaling pathway, including PrkC (receptor kinase) and PrpC (phosphatase). Mass spectrometry identified seven Thr and one Ser autophosphorylated residues on PrkC. Phosphorylation of four Thr residues and one Ser residue was found to be essential for kinase activity (25). PrpC specifically dephosphorylates PrkC, and both enzymes are implicated in sporulation, biofilm formation, and swarming. The migrating cells produce surfactin, which precedes the swarm front and is involved in coordinated expansion of the complex community and the formation of dendrites. Future studies will have to show how cells communicate with each other during swarm development.
FIG. 4.
Swarming by B. subtilis. Section of a swarm community (B. subtilis 3610) formed from the point inoculum on a glucose minimal medium plate over a period of 20 h (bottom right hand corner). Swarms advance at more than 1 cm/h, forming waves of dendritic patterns. Formation of dendrites depends upon secreted surfactin, diffusing outwards ahead of the swarm front. (Figure kindly provided by Simone Seror.)
REGULATION OF CELLULAR DEVELOPMENT
How are bacterial cells organized, and how do these cells know where to put their DNA and proteins? It has become clear that bacteria are not simply bags of diffusing enzymes but instead show an astonishing degree of three-dimensional organization, which allows them to accurately perform basic functions of cell proliferation and behavior. Urs Jenal (Biozentrum, Basel, Switzerland) described the differentiation of Caulobacter crescentus. C. crescentus has long been a model organism for the bacterial cell cycle and development. Cells are able to attach to surfaces via an adhesive organelle, the holdfast, located at the tip of a stalk at one pole. On division these attached cells produce a motile, nondividing swarmer daughter cell, which swims to a new surface, attaches by its pole, loses flagella, and initiates a new round of growth and division. Polar development in C. crescentus is regulated by members of the two-component system signal transduction family. The histidine kinases PleC and DivJ were previously implicated in spatial and temporal control of development, but the role of the response regulator PleD, which is directly controlled by DivJ and PleC, has been uncertain (1). PleD contains a DUF1 domain and was shown to be a specific diguanylate cyclase whose enzymatic activity was increased upon phosphorylation by DivJ. A GFP-PleD fusion was shown to localize to the stalked pole in a phosphorylation-dependent manner (Fig. 5). Thus, diguanylate cyclase activity and polar localization of PleD seem to be coupled. This suggests that a local increase in cyclic di-GMP acts as specific signal to change the nature of the cell pole from one that is involved in cell motility to one specialized in adhesion (35). High levels of the secondary messenger cyclic di-GMP have been associated with community behavior such as biofilm formation, while low levels of cyclic di-GMP seem to favor cell dispersal. By spatial regulation of diguanylate cyclase the functions of the two cell poles, and hence of the daughter cells, can be regulated (16).
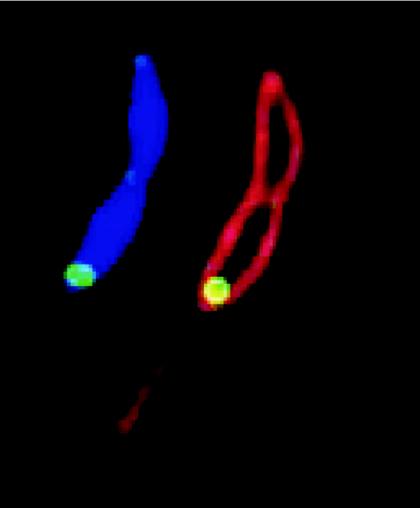
FIG. 5.
Localization of PleD in C. crescentus. Subcellular localization of PleD* (a constitutive active form of PleD) at the stalked pole of a Caulobacter predivisional cell. Green, PleD*-GFP; red, membrane (stained with FM4-64); blue, DNA (stained with DAPI [4′,6′-diamidino-2-phenylindole]). (Figure kindly provided by Urs Jenal.)
Another protein-targeting mechanism was described by Stephen Thompson (University of Oxford). Many bacterial species have several homologues of the components of chemosensory pathways, usually expressed from distinct operons, each containing genes for a complete pathway. Many species, including the photosynthetic bacterium Rhodobacter sphaeroides, with such multiple pathways also have genes encoding putative receptors without membrane-spanning domains. The Armitage group has already shown that the components of one pathway cluster with the membrane-spanning chemoreceptors at the cell poles, while the proteins for a second chemosensory pathway, expressed from a different operon, are targeted to the cytoplasmic chemoreceptors that form a cluster, surprisingly, in the center of the cell (43). This cluster duplicates, and the new clusters move to the daughter cell on cell division. Interestingly, encoded in the middle of the operon is a gene encoding a homologue of the chromosome-partitioning factor ParA. Stephen Thompson showed, using GFP fusions to proteins in the cluster, that this homologue (PpfA) is essential for the segregation of the protein cluster into the daughter cell and also regulates expression of the operon, very much as found for ParA itself. Whether other protein complexes are segregated in a similar controlled manner is an important question for the future and again illustrates the unexpected complexity of bacterial cells.
Mark Buttner (John Innes Centre) described how the filamentous soil bacterium Streptomyces coelicolor can remodel its cell wall to resist the antibiotic vancomycin, revealing a novel sensory pathway. Vancomycin binds the cell wall precursor d-Ala-d-Ala and prevents peptidoglycan cross-linking. In S. coelicolor a cluster of seven genes, vanSRJKHAX, confers vancomycin resistance, with vanRJKH promoters all vancomycin inducible. Expression of vanHAX (homologous to genes in enterococci), reprograms peptidoglycan synthesis to use d-Ala-d-Lac. VanK, with no counterpart in enterococci, is also essential for resistance and is a member of the Fem protein family, involved in forming cross bridges in Staphylococcus aureus (13). S. coelicolor turns out to have an additional gene encoding FemX, a protein involved in completing a single glycine cross bridge after addition of d-Ala-d-Ala rather than the d-Ala-d-Lac recognized by VanK. By examining the effect of mutating genes for VanK and FemX and isolating suppressors, an idea of the mechanisms involved in peptidoglycan synthesis and remodelling in response to glycopeptide antibiotics was developed. A VanSR sensor kinase/response regulator pair was identified which responds to glycopeptides.
In another gram-positive bacterium, Streptococcus pneumoniae, the CiaR/CiaH two-component system both increases resistance to β-lactams and interferes with competence. Regine Hakenbeck (University of Kaiserslautern) described the identification of the Cia regulon, which includes the entire competence regulon of 188 early/late and delayed genes (29). As with S. coelicolor, resistance involves reprogramming cell wall synthesis. The membrane-spanning histidine kinase CiaH probably senses the integrity of the cell wall and controls the phosphorylation state of the response regulator CiaR. CiaR is also implicated in penicillin binding protein-mediated β-lactam resistance.
The systems producing very exciting results from in vivo imaging technologies are cell division and chromosome segregation. Leendert Hameon (University of Oxford) described some elegant protein fusion studies showing how B. subtilis finds the midcell. The tubulin homologue FtsZ is crucial for cell division in all bacteria, forming the Z ring, but how do cells select the division site? One idea is that DivIVA-dependent MinCD localization at the poles, independent of FtsZ, prevents division close to the poles (9, 28). A second hypothesis is that of nucleoid occlusion, thought to prevent division near segregating chromosomes (48). Hameon brought these two hypotheses together, showing that a DNA binding protein, YyaA (renamed Noc), acts as a division inhibitor. In Noc mutants, FtsZ rings are misplaced and overlap the nucleoid, with the resulting septation going through the nucleoid. This suggests that a gradient of MinCD prevents division at the poles while Noc binds DNA in some way that prevents FtsZ forming a ring over the nucleoid. Together these events dictate that septation occurs only at midcell between two segregated nucleoids.
Earlier talks at the meeting had already emphasized that regulatory pathways are highly complex, interactive systems in most bacteria. However, Jörg Vogel (MPI for Infection Biology, Berlin, Germany) added yet another layer of complexity to the control of bacterial physiology, the role of regulatory RNA. A regulatory role for sRNA has been well established and exploited in mammalian systems. However, a corresponding process in bacteria was not widely appreciated until recently, even though sRNAs were first identified in bacteria in the early 1970s. Indeed, the capacity of bacteria and archaeal genomes to encode a plethora of sRNAs within the intergenic regions is now well established (Fig. 6 and 7) (44, 45). While a recent estimate of the number of sRNAs in E. coli suggests the existence of ∼55 sRNA genes, with another 1,000 or so possible candidates, the likely number is probably going to be in the range of 100 to 200 (4, 42). One problem in identifying sRNAs is the range of sizes, the range of targets, and the different interaction mechanisms. For example, some sRNAs interact with target mRNAs to affect stability or translation by a “kissing mechanism” whereby only a few nucleotides of the RNAs interact while other sRNAs bind to proteins to affect activity. Moreover, the number of sRNAs may be further underestimated by escaping detection when encoded within gene sequences (40). While sRNAs undoubtedly constitute a significant fraction of the transcriptional output of bacteria, the future challenge is to understand their role in regulating bacterial physiology whether they are involved in housekeeping or in environmental adaptation processes such as pathogenicity.
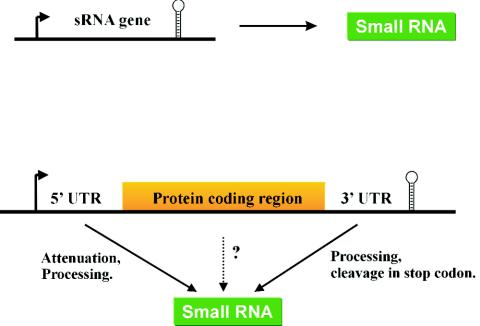
FIG. 6.
sRNA in bacteria. Definitions of sRNAs in bacteria and criteria for sRNA searches. sRNAs may be encoded by free-standing, independent genes flanked by a promoter and terminator (upper part) or could be generated through parallel transcriptional output by 5′ or 3′ processing of an mRNA transcript (lower part). This may even include sRNAs derived from coding regions; some of the mRNA-derived fragments observed in an E. coli RNomics screen (42) represent such candidates and are currently being tested. (Figure adapted from reference 42 with permission.)
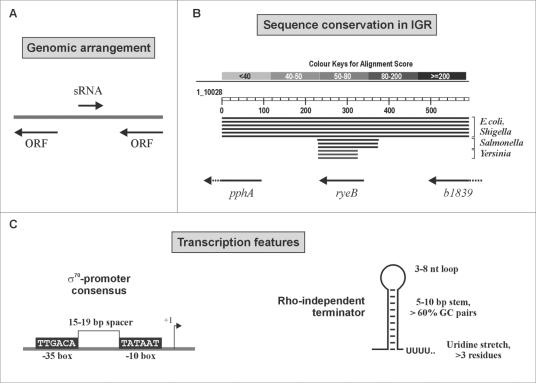
FIG. 7.
Biocomputational screens have employed individual sRNA features for predictions of these genes in bacteria. The predictive scheme used by Argaman et al. was used to search the “empty” intergenic regions of E. coli for new sRNA genes. (A) sRNA candidates in intergenic regions that were orientated oppositely to both neighboring genes were considered particularly “safe”; that is, they could not correspond to an mRNA leader or trailers. (B) Since many sRNA genes are conserved among closely related bacteria, comparative analysis of intergenic sequences has been used with great success for sRNA identification. Shown is a BLASTN search result for the intergenic region, including 100 bp of adjoining coding sequence, that harbors the sRNA gene ryeB (45). Conservation of the entire region from E. coli MG1655 is limited to the genomes of pathogenic E. coli and Shigella strains. However, the ryeB sequence itself displays high homology values in related enterobacteria such as Salmonella and Yersinia species. (C) Transcription features shared by many E. coli sRNAs that served as input to search for “orphan” promoter and terminator sequences in intergenic regions.
The importance of identifying and understanding those regulatory networks controlling the pathogenic nature of bacteria was emphasized by Aileen Allsop (AstraZeneca), who also stressed that, although interfering with the functioning of such pathways was no longer an aim for big pharmaceutical companies it was still ripe for exploitation by smaller companies. Nevertheless, the complexities of identifying new drugs was underscored. This requires the identification of chemicals that not only work on a variable human population (with individual health profiles) specifically to kill active pathogens, leaving unharmed host cells or commensal bacteria, but also have a broad spectrum activity and are capable of killing slow-growing or intracellular “hidden” pathogens. As a final reminder, if needed at such a meeting, the participants were left with the thought that approaches to drug discovery today need to employ more imaginative and directed strategies, based on fundamental knowledge, rather than the “random drug screening” approaches of old.
SUMMARY
This series of conferences beautifully illustrates and reflects the dramatic pace of development within the field of bacteriology. In the initial meeting the talks tended to be about the molecular details of single systems. Only 5 years on the talks centered on the complex integrative nature of signaling networks, their sensitivity to the changing external environment, and, in the case of pathogens and symbionts, their dialogue with their hosts. What is becoming very clear from the rapid increase in data on sensory networks in increasing numbers of bacterial species is that no two species are alike. Indeed, no cell within a population is identical to its neighbor, and each is influenced by its stage in the cell cycle, its recent and historical past, and its local environment. We need to make sense of this increasing volume of information in order to develop a deeper understanding of particular species and to create generic models that will allow some insight into how individual cells, dispersed populations, and communities of bacteria (single and mixed species) will respond to change. This is the challenge of the next series of meetings. Taking on this challenge requires the true integration of different disciplines, combining the skills of the microbial physiologist and biochemist, the physicist, the computational and mathematical modelers, the engineers, and the technologists. Each needs to understand the other's language and then mobilize their different skills to address a common biologically driven problem: the design and operation of bacterial sensory networks. We must not however neglect past experience, the roots of bacterial physiology, but combine this with new knowledge of the molecular details of the component parts of these complex circuits to develop (or revive?) a broad holistic approach. This will involve combining the accumulated wealth of biochemical and metabolic data with the current flood of data from genomics and proteomics, brought into a coherent whole through modelling, in order to produce useful guides on the way to the “total” understanding of the complex nature that makes up a bacterium.
Acknowledgments
We thank the European Science Foundation for organizational assistance with this Euresco Conference and the European Commission, Elsevier, AstraZeneca, the University of Barcelona, the Generalitat de Catalunya, and EMBO for funding.
We also specifically thank Corinne Le Moal and Anne-Sophie Gablin for their organizational skills and patience.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Alon, U. 2003. Biological networks: the tinkerer as an engineer. Science 301:1866-1867. [DOI] [PubMed] [Google Scholar]
- 2a.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, H. P. J., and D. J. Clarke. 2004. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J. Bacteriol. 187:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 5.Croxatto, A., J. Pride, A. Hardman, P. Williams, M. Cámara, and D. L. Milton. 2004. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol. Microbiol. 52:1677-1689. [DOI] [PubMed] [Google Scholar]
- 6.Danino, V. E., A. Wilkinson, A. Edwards, and J. A. Downie. 2003. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 50:511-525. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, M. D., I. R. Booth, and S. Miller. 2004. Gating the bacterial mechanosensitive channels: MscS a new paradigm? Curr. Opin. Microbiol. 7:163-167. [DOI] [PubMed] [Google Scholar]
- 8.Francke, C., P. W. Postma, H. V. Westerhoff, J. G. Blom, and M. A. Peletier. 2003. Why the phosphotransferase system of Escherichia coli escapes diffusion limitation. Biophys. J. 85:612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamoen, L. W., and J. Errington. 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J. Bacteriol. 185:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinrich, R., S. Schuster, and H. G. Holzhütter. 1991. Mathematical analysis of enzymic reaction systems using optimization principles. Eur. J. Biochem. 201:1-21. [DOI] [PubMed] [Google Scholar]
- 11.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero, A., A. M. Muro-Pastor, M. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 13.Hong, H.-J., M. I. Hutchings, J. M. Neu, G. D. Wright, M. S. B. Paget, and M. J. Buttner. 2004. Characterisation of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107-1121. [DOI] [PubMed] [Google Scholar]
- 14.Hõrak, R., H. Ilves, P. Pruunsild, M. Kuljus, and M. Kivisaar. The ColR-ColS two-component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol. Microbiol., in press. [DOI] [PubMed]
- 15.Ilves, H., R. Hõrak, and M. Kivisaar. 2001. Involvement of σS in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J. Bacteriol. 183:5445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 16a.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. The open pore conformation o potassium channels. Nature 417:523-526. [DOI] [PubMed] [Google Scholar]
- 17.Joyce, S. A., and D. J. Clarke. 2003. The hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic regulation. Mol. Microbiol. 47:1445-1457. [DOI] [PubMed] [Google Scholar]
- 18.Julkowska, D., M. Obuchowski, I. B. Holland, and S. J. Séror. 2004. Branched swarming patterns on a synthetic medium formed by wild type Bacillus subtilis: detection of different cellular morphologies and constellations of cells as the complex architecture develops. Microbiology 150:1839-1849. [DOI] [PubMed] [Google Scholar]
- 19.Klipp, E., R. Heinrich, and H-G. Holzhütter. 2002. Prediction of temporal gene expression. Metabolic optimization by redistribution of enzyme activities. Eur. J. Biochem. 269:5406-5413. [DOI] [PubMed] [Google Scholar]
- 20.Kremling, A., K. Bettenbrock, B. Laube, K. Jahreis, J. W. Lengeler, and E. D. Gilles. 2001. The organization of metabolic reaction networks. III. Application for diauxic growth on glucose and lactose. Metab. Eng. 3:362-379. [DOI] [PubMed] [Google Scholar]
- 21.Laurie, A. D., L. M. Bernardo, C. C. Sze, E. Skarfstad, A. Szalaewska-Palasz, T. Nystrom, and V. Shingler. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 278:1494-1503. [DOI] [PubMed] [Google Scholar]
- 22.Lengeler, J. W. 2000. Metabolic networks: a signal-oriented approach to cellular models. Biol. Chem. 381:911-920. [DOI] [PubMed] [Google Scholar]
- 22a.Lequette, Y., and E. P. Greenberg. 2004. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J. H., S. Laurent, V. Konde, S. Bedu, and C.-C. Zhang. 2003. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 149:3257-3263. [DOI] [PubMed] [Google Scholar]
- 24.Lipkow, K., S. S. Andrews, and D. Bray. 2004. Simulated diffusion of CheYp through the cytoplasm of Escherichia coli. J. Bacteriol. 187:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madec, E., A. Stensballe, S. J. Kjellström, L. Cladière, M. Obuchowski, O. N. Jensen, and S. J. Séror. 2003. Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis. J. Mol. Biol. 330:459-472. [DOI] [PubMed] [Google Scholar]
- 26.Magnusson, L. U., T. Nystrom, and A. Farewell. 2003. Underproduction of sigma 70 mimics a stringent response. A proteome approach. J. Biol. Chem. 278:968-973. [DOI] [PubMed] [Google Scholar]
- 27.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100:11980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcrptional profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, S., M. Edwards, C. Ozdemir, and I. R. Booth. 2003. The mechanosensitive channel, MscS, of Escherichia coli forms a compact closed state. J. Biol. Chem. 278:32246-32250. [DOI] [PubMed] [Google Scholar]
- 31.Milo, R., S. Shen-Orr, S. Itzkovitz, N. Kashtan, D. Chklovskii, and U. Alon. 2002. Network motifs: simple building blocks of complex networks. Science 298:824-827. [DOI] [PubMed] [Google Scholar]
- 32.Monod, J., J. Wyman, and J.-P. Changeux. 1965. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12:88-118. [DOI] [PubMed] [Google Scholar]
- 33.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 19:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J. I., I. J. Ko, and S. Kaplan. 2004. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 43:7915-7923. [DOI] [PubMed] [Google Scholar]
- 35.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64-68. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu, T. S., S. V. Aksenov, and D. Bray. 2003. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J. Mol. Biol. 329:291-309. [DOI] [PubMed] [Google Scholar]
- 38.Sourjik, V., and H. C. Berg. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437-441. [DOI] [PubMed] [Google Scholar]
- 39.Stokes, N. R., H. D. Murray, C. Subramaniam, R. L. Gourse, P. Louis, W. Bartlett, S. Miller, and I. R. Booth. 2003. A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc. Natl. Acad. Sci. USA 100:15959-15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 41.Studemann, A., M. Noirclerc-Savoye, E. Klauck, G. Becker, D. Schneider, and R. Hengge. 2003. Sequential recognition of two distinct sites in sigma(S) by the proteoyltic targeting factor RssB and ClpX. EMBO J. 22:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Hüttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadhams, G. H., A. C. Martin, A. Warren, and J. P. Armitage. 2003. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol. Microbiol. 50:763-770. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, E. G. H., and J. Vogel. 2003. Noncoding RNAs encoded by bacterial chromosomes, p. 243-259. In J. Barciszewski and V. Erdmann (ed.), Noncoding RNAs. Landes Bioscience, Georgetown, Tex.
- 45.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wisniewski-Dye, F., and J. A. Downie. 2002. Quorum sensing in Rhizobium. Antonie Leeuwenhoek 81:397-407. [DOI] [PubMed] [Google Scholar]
- 47.Wolanin, P. M., and J. B. Stock. 2004. Bacterial chemosensing: cooperative molecular logic. Curr. Biol. 14:486-487. [DOI] [PubMed] [Google Scholar]
- 48.Wu, L. J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915-925. [DOI] [PubMed] [Google Scholar]
- 49.Zaslaver, A., A. E. Mayo, R. Rosenberg, P. Bashkin, H. Sberro, M. Tsalyuk, M. G. Surette, and U. Alon. 2004. Just-in-time transcriptional program in metabolic pathways. Nat. Genet. 36:486-491. [DOI] [PubMed] [Google Scholar]