SUMMARY
The emergence of ZIKV infection has prompted a global effort to develop safe and effective vaccines. We engineered a lipid nanoparticle (LNP) encapsulated modified mRNA vaccine encoding wild-type or variant ZIKV structural genes and tested immunogenicity and protection in mice. Two doses of modified mRNA LNPs encoding prM-E genes that produced virus-like particles resulted in high neutralizing antibody titers (~ 1/100,000) that protected against ZIKV infection and conferred sterilizing immunity. To offset a theoretical concern of ZIKV vaccines inducing antibodies that cross-react with the related Dengue virus (DENV), we designed modified prM-E RNA encoding mutations destroying the conserved fusion-loop epitope in the E protein. This variant protected against ZIKV and diminished production of antibodies enhancing DENV infection in cells or mice. A modified mRNA vaccine can prevent ZIKV disease and be adapted to reduce the risk of sensitizing individuals to subsequent exposure to DENV, should this become a clinically-relevant concern.
Keywords: flavivirus, Zika virus, Dengue virus, RNA vaccine, protection, pathogenesis, immunity, antibody neutralization
eTOC
A modified mRNA vaccine induces sterilizing immunity against Zika virus while avoiding the generation of cross-reactive antibodies that may enhance dengue infection.
INTRODUCTION
Zika virus (ZIKV) was identified in 1947 from a Rhesus monkey in the Zika Forest of Uganda (Dick, 1952; Dick et al., 1952). Historically, ZIKV circulated between Aedes species mosquitoes and non-human primates, and episodically spilled into human populations in parts of Africa and Asia. Prior to 2010, ZIKV Infection was described as a self-limiting febrile illness with headache, rash, conjunctivitis, and myalgia. More recently, and especially in the context of its spread in the Western Hemisphere, more severe clinical consequences have been observed (Lazear and Diamond, 2016). Infection of fetuses during pregnancy has been associated with placental insufficiency and congenital malformations including cerebral calcifications, microcephaly, and miscarriage (Brasil et al., 2016; Rasmussen et al., 2016; van der Eijk et al., 2016). In adults, ZIKV infection is linked to Guillain-Barré syndrome (GBS), an autoimmune disease characterized by paralysis and polyneuropathy (Cao-Lormeau et al., 2016; Oehler et al., 2014). Sexual transmission of ZIKV also has been described from men-to-women (Foy et al., 2011), men-to-men (Deckard et al., 2016), and women-to-men (Davidson et al., 2016). Persistent ZIKV has been detected in semen, sperm, and vaginal secretions up to 6 months following infection (Mansuy et al., 2016; Murray et al., 2017). ZIKV is now a global disease of the Americas, Africa, and Asia.
ZIKV is a member of the Flavivirus genus of the Flaviviridae family of enveloped RNA viruses. ZIKV has an ~11 kb positive sense RNA genome. Translation of viral RNA in the cytoplasm generates a polyprotein that is cleaved into three structural proteins (capsid (C), pre-membrane/membrane (prM/M), and envelope (E)) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). ZIKV buds into the lumen of the endoplasmic reticulum as an immature virion composed of 60 icosahedrally arranged prM-E heterotrimers (Prasad et al., 2017). As the virus transits through the secretory pathway, the acidic environment of the Golgi network triggers exposure of a furin protease cleavage site within prM. Cleavage of prM and release of the pr peptide in the extracellular space produces mature, infectious virions that display 90 antiparallel E homodimers on their surface. The ZIKV E protein is composed of three ectodomains (DI, DII, and DIII) and is the primary target of neutralizing antibodies. Potently inhibitory monoclonal antibodies (mAbs) against ZIKV target epitopes in all three E protein domains as well as quaternary structures composed of multiple domains within or across E dimers (Barba-Spaeth et al., 2016; Sapparapu et al., 2016; Stettler et al., 2016; Swanstrom et al., 2016; Wang et al., 2016; Zhao et al., 2016). In addition to induction by infectious or inactivated virus particles, neutralizing antibodies against ZIKV can be produced after immunization with DNA plasmids encoding M-E protein (Larocca et al., 2016) or prM-E, which in some cases generates secreted virus-like subviral particles (SVPs) (Dowd et al., 2016b; Muthumani et al., 2016).
The existence of two ZIKV lineages, African and Asian/American (Haddow et al., 2012) does not impact antibody neutralization substantively and thus, ZIKV is classified as a single serotype (Dowd et al., 2016a). ZIKV is related to several pathogens that cause disease globally including Dengue (DENV), yellow fever (YFV), West Nile (WNV), Japanese encephalitis (JEV), and tick-borne encephalitis (TBEV) viruses. Of these viruses, ZIKV is most closely related to the four serotypes of DENV as it shares 54 to 59% amino acid identity in the viral E protein (Dejnirattisai et al., 2016).
Because of its potential to infect and cause harm to developing fetuses and neonates (Huang et al., 2016), there is an urgent call to develop countermeasures (Marston et al., 2016). Several groups have developed subunit (prM-E or M-E DNA plasmid or adenovirus-vectored) or inactivated virus vaccine platforms capable of eliciting neutralizing antibodies that protect against ZIKV viremia in mice and non-human primates (Abbink et al., 2016; Dowd et al., 2016b; Larocca et al., 2016; Muthumani et al., 2016). Some of these vaccine candidates have initiated or are scheduled to begin recruitment of subjects for evaluation in humans (NCT02840487, NCT02887482, NCT02809443, NCT02937233, NCT02952833, and NCT02963909).
The sequence similarity between ZIKV and DENV poses issues for vaccine development due to cross-reactivity of the human anti-ZIKV antibody response (Dejnirattisai et al., 2016; Sapparapu et al., 2016; Stettler et al., 2016). Cross-reactive antibody responses may contribute minimally to protection, consistent with the limited neutralizing activity of many broadly-reactive anti-flavivirus mAbs in cell culture. Whereas primary infection with DENV generates an antibody response that protects against the homologous serotype, secondary infection with a heterologous DENV serotype can result in a potentially lethal shock syndrome. This disease is attributed in part to antibody-dependent enhancement of infection (ADE), whereby cross-reactive antibodies elicited by the first DENV serotype augment infection of the second DENV serotype in cells expressing Fc-γ receptors (Morens, 1994). This phenomenon could be relevant to ZIKV vaccination because DENV and ZIKV are related closely, and vaccinated subjects in or travelers to endemic areas could become exposed secondarily to DENV, which infects ~390 million people per year (Bhatt et al., 2013). Indeed, cross-reactive antibodies targeting the highly conserved fusion loop in DII of E (E-DII-FL) derived during natural ZIKV infection can augment infectivity of DENV in cell culture (Dejnirattisai et al., 2016; Kawiecki and Christofferson, 2016; Stettler et al., 2016) and in vivo in mice (Stettler et al., 2016). This laboratory-based data is not conclusive, as epidemiological studies, which may take years to perform, are required to establish the impact of ZIKV humoral immunity on DENV pathogenesis. Vaccine strategies that reduce induction of cross-reactive antibodies (Crill et al., 2012) might minimize the risk of sensitizing recipients to severe DENV infections, should the clinical relevance of ZIKV immunity on DENV pathogenesis be established.
We generated a versatile ZIKV vaccine platform in which lipid nanoparticles encapsulate modified mRNA encoding wild-type (WT) or variant ZIKV structural genes. Unlike DNA plasmid based vaccines, mRNA does not integrate into chromosomes, which can lead to insertional mutagenesis and potential oncogenesis (Pardi and Weissman, 2017). Non-amplifying or self-amplifying mRNA-based vaccines were used recently to generate humoral responses against influenza A virus in mice (Hekele et al., 2013; Petsch et al., 2012) or HIV in non-human primates (Bogers et al., 2015). Our modified non self-amplifying mRNA vaccines have an optimized mRNA containing an open reading frame that encodes the antigen of interest, and 5′ and 3′ untranslated regions that optimize translation efficiency and intracellular stability, and a proprietary nucleoside modification to minimize the indiscriminate activation of innate immunity. mRNA can stimulate innate immunity through Toll-like and RIG-I-like receptors (Desmet and Ishii, 2012; Kariko et al., 2004). Although the adjuvant effect of stimulating innate immunity might be advantageous for protein vaccines, indiscriminate immune activation can inhibit mRNA translation, thus reducing antigen expression and immunogenicity of an mRNA vaccine (Claudio et al., 2013; Coffman et al., 2010). This can be overcome by replacing uridine nucleosides with naturally-occurring base modifications, such as pseudouridine and 5-methylcytidine (Anderson et al., 2011). Here, a two-dose immunization of modified mRNA encoding ZIKV prM-E induced high levels of neutralizing antibodies that protected mice with genetic or acquired innate immune deficiencies against severe infection. Because mRNA vaccines can be synthesized with virtually any sequence, we created modified mRNA immunogens encoding mutations that abolished an immunodominant cross-reactive epitope in E-DII-FL. These mRNA induced protective antibody responses against ZIKV in mice and minimized the generation of cross-reactive antibodies that enhanced DENV infection in cell culture and pathogenicity in mice.
RESULTS
An mRNA vaccine platform for ZIKV
We developed a vaccine platform for generating optimized lipid nanoparticles that encapsulate modified mRNA for intramuscular delivery to induce high levels of protein expression in vivo (Fig 1A and Fig S1). As a proof-of-principle, we designed a modified mRNA encoding a type 1 (N7mGpppAm) cap, optimized 5′ and 3′ untranslated sequences (see STAR Methods), the signal sequence from human IgE, and the full-length prM and E genes from an Asian (Mirconesia 2007) ZIKV strain (Lanciotti et al., 2008), which has > 99% amino acid sequence identity relative to strains from the Americas (Fig 1B). The modified mRNA was synthesized chemically and packaged into lipid nanoparticles (LNPs). Incubation of LNPs containing IgE signal-prM-E mRNA (IgEsig-prM-E) with 293T or HeLa cells resulted in efficient expression and secretion of ~30 nm SVPs, as judged by electron microscopy (Fig 1C), Western blotting (Fig 1D) and mass spectrometry for ZIKV structural proteins in the cell supernatants (data not shown).
Figure 1. ZIKV mRNA LNP vaccine testing in AG129 mice.
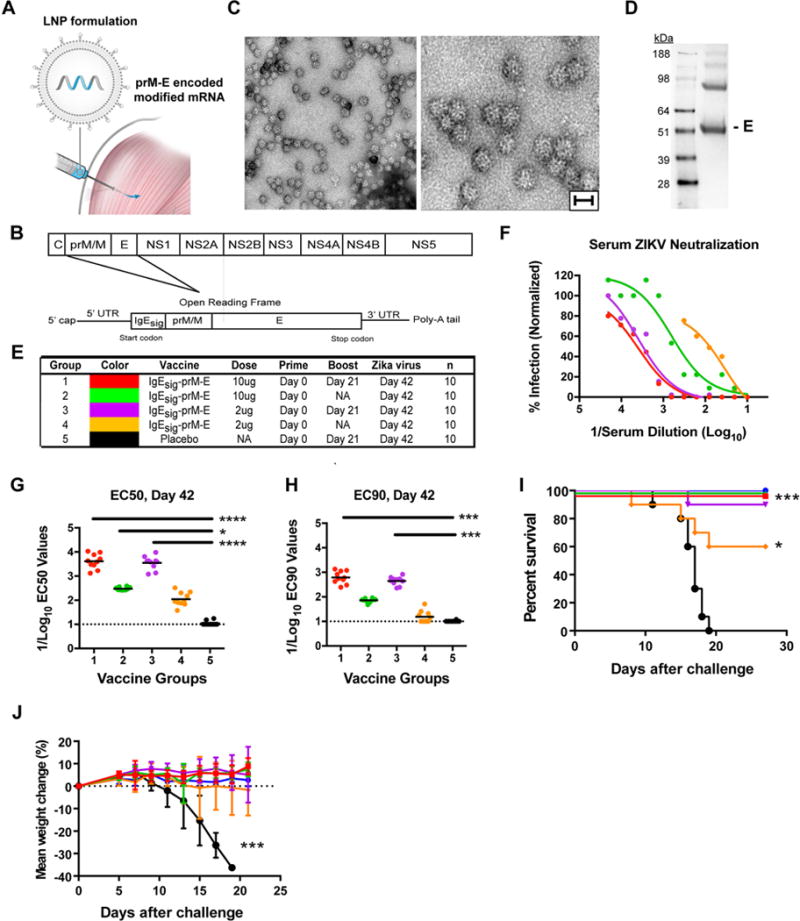
A. ZIKV prM-E modified mRNA is packaged into LNPs for intramuscular delivery. B. Schematic of ZIKV genome (top). An mRNA encoding the ZIKV prM/M and E structural genes was engineered (bottom). In this construct, prM is directed into the ER using a heterologous IgE signal sequence (IgEsig) at the amino-terminus. C. HeLa cells were transfected with prM-E mRNA, and SVPs in the supernatant were purified and concentrated by ultracentrifugation and then subjected to electron microscopy and negative staining. Low- and high-power images of purified SVPs are shown. Scale bar = 30 nm. One representative experiment of several is shown D. HeLa cell supernatants were collected for Western blotting under non-reducing conditions with a mAb against the ZIKV E protein. E. Scheme of immunization of AG129 mice with one (prime) or two (prime-boost) doses of 2 or 10 μg with IgEsig prM-E or placebo mRNA LNP vaccines. F–H. Serum was collected at 6 weeks after vaccination and analyzed for neutralization of ZIKV by PRNT assay. Representative curves are shown (F), and EC50 (G) and EC90 (H) values were calculated for individual animals in each group. Each point represents the mean of two independent experiments per animal. Data are a composite of two independent experiments with sera from each of the 10 animals per group. EC50 and EC90 data were analyzed by a Kruskal-Wallis test with a multiple comparisons correction and compared to the placebo LNP vaccine (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001). The dashed lines indicate the limit of detection of the assay. I–J. AG129 Mice were challenged at 6 weeks post vaccination with 104 PFU of ZIKV P6-740. Animals were monitored for survival (I) and weight loss (J). Error bars indicate standard error the mean (SEM). Survival data was analyzed by the log rank test (*, P < 0.05; ***, P < 0.001). Weight loss was analyzed by two-way ANOVA (***, P < 0.001). See also related figures S1 and S2.
Vaccine efficacy in AG129 mice
We first assessed the immunogenicity and protective activity of IgEsig-prM-E LNPs in immunocompromised mice lacking type I and II interferon (IFN) signaling responses. Eight week-old Ifnar1−/− Ifngr−/− AG129 male and female mice were divided into five groups, which received an intramuscular inoculation of 2 or 10 μg of IgEsig-prM-E LNPs or 10 μg of a non-translating RNA LNP. Groups 1, 3, and 5 were boosted with the same dose at 21 days after vaccination whereas Groups 2 and 4 were not boosted (Fig 1E). At day 42 after immunization, mice were phlebotomized for serum neutralizing antibody analysis (Fig 1F–H and Fig S2). Mice receiving the IgEsig-prM-E LNPs with a boost had the strongest serum neutralizing response against ZIKV, with reciprocal dilution EC50 (half-maximal inhibition of virus infection) and EC90 values of up to 1/10,000 and 1/1,000, respectively. Mice receiving only a single immunization had lower neutralizing titers.
At 42 days after vaccination, AG129 mice were challenged with the ZIKV strain P6-740 (Malaysia, 1966). As expected (Aliota et al., 2016), AG129 mice receiving the negative control LNP vaccine succumbed to ZIKV infection (Fig 1I). With the exception of a single animal, recipients of the 2 or 10 μg IgEsig-prM-E LNP vaccine with a boost as well as those receiving a single 10 μg dose all survived infection. In comparison, mice receiving a single 2 μg dose of the vaccine displayed an intermediate phenotype with a 60% survival rate. Weight measurements correlated with lethality, as mice receiving the IgEsig-prM-E LNP vaccine were protected whereas the negative controls lost weight beginning at approximately day 10 after challenge (Fig 1J).
Vaccine efficacy in C57BL/6 mice
To test the immunogenicity and efficacy of the IgEsig-prM-E LNP vaccine efficacy in an immunocompetent mouse strain, we inoculated via intramuscular injection 8 week-old male C57BL/6 WT mice with 10 μg of IgEsig-prM-E LNPs. These animals were phlebotomized prior to a single boost at day 28 (4 weeks) and before challenge at either day 56 (8 weeks) or day 126 (18 weeks). As expected, serum neutralization titers were relatively low prior to boosting; however, titers peaked at 4 weeks after boosting (EC50 of 1/10,000) and remained elevated 18 weeks post initial vaccination (Fig 2A–C and Fig S3). To create a lethal challenge model, we passively transferred 2 mg of a blocking anti-Ifnar1 antibody one day prior to infection with 106 focus-forming units (FFU) of a mouse-adapted African ZIKV strain (Dakar 41519) (Sapparapu et al., 2016; Zhao et al., 2016). All mice immunized with IgEsig-prM-E LNPs were protected against lethal ZIKV infection compared to the control group, which had a 30% survival rate (Fig 2D). Mice vaccinated with IgEsig-prM-E mRNA LNPs did not display any loss in weight (Fig 2E) nor had measurable viremia in serum at 5 days after challenge (Fig 2F).
Figure 2. ZIKV mRNA LNP vaccine protects C57BL/6 mice.
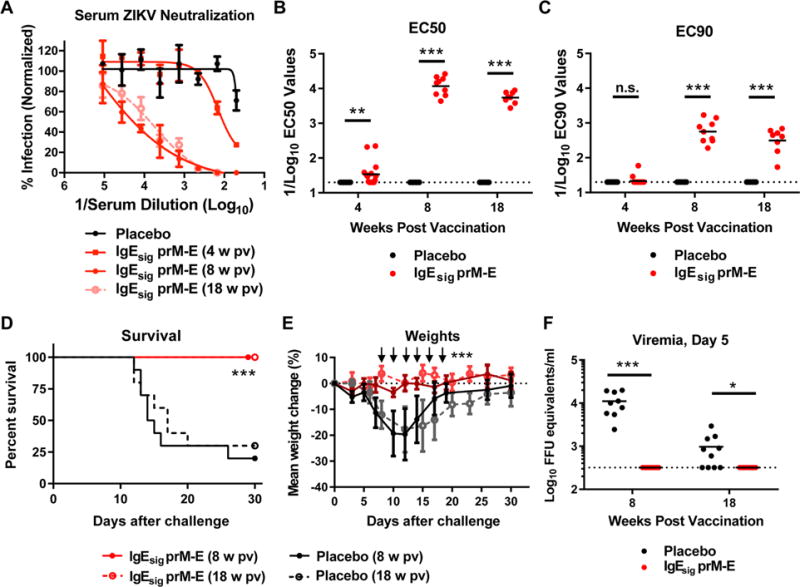
WT C57BL/6 mice (n = 10, pooled from two independent experiments) were immunized with 10 μg of placebo or IgEsig prM-E mRNA LNPs and boosted with an equivalent dose four weeks later. A. Serum was collected at 4, 8, and 18 weeks post initial vaccination and analyzed for ZIKV neutralization activity by FRNT assay. Representative neutralization curves are shown for each group. Error bars denote the standard deviation (SD) of triplicate technical replicates. B–C. EC50 (B) and EC90 (C) values were calculated for individual animals in each group. The dashed lines indicate the limit of detection of the assay. Data were analyzed using the Mann-Whitney test and compared to the placebo LNP vaccine at each time point (**, P < 0.01; ***, P < 0.001; n.s. indicates not significant). D–F. At week 8 or 18, vaccinated C57BL/6 mice were administered 2 mg of anti-Ifnar1 blocking antibody and one day later challenged with 105 FFU of mouse adapted ZIKV Dakar 41519. Animals were monitored for survival (D) and weight loss (E). At day 5 after viral challenge, serum was analyzed for levels of ZIKV RNA (F). The dashed line indicates the limit of detection of the assay. Survival data was analyzed by the log rank test (***, P < 0.001). Weight loss was analyzed by two-way ANOVA (***, P < 0.001) for surviving animals; arrows indicate days having statistically significant differences from placebo vaccine. Viremia data was analyzed by a Mann-Whitney test (*, P < 0.05; ***, P < 0.001). See also related figure S3.
Modified vaccines lacking the immunodominant E-DII-FL epitope generate a protective anti-ZIKV response
The highly conserved FL epitope in DII of the flavivirus E protein is immunodominant in humans (Beltramello et al., 2010; Crill et al., 2007; Dejnirattisai et al., 2010; Oliphant et al., 2007; Sapparapu et al., 2016; Stettler et al., 2016). As such, infection with ZIKV or vaccination with ZIKV structural proteins could induce cross-reactive antibodies, which might enhance DENV infection and disease through ADE (Morens, 1994; Stettler et al., 2016). To minimize this possibility, we generated modified mRNA vaccines by engineering four mutations (T76R, Q77E, W101R, and L107R) in or near the E-DII-FL (prM-E-FL) that abolish antibody reactivity of FL-specific antibodies (Chabierski et al., 2014; Crill et al., 2012; Oliphant et al., 2007). We also generated a separate series of mRNA LNPs by replacing the IgE leader sequence with one from Japanese encephalitis virus (JEVsig), a feature included in other flavivirus prM-E DNA vaccines to increase the efficiency of host signalase cleavage (Davis et al., 2001; Dowd et al., 2016b), and by further optimizing codon usage (Fig 3A). Western blotting analysis showed similar levels of SVP expression in HeLa cell supernatants after transfection of WT and FL mutant mRNA, and a loss of FL reactivity was confirmed for the prM-E-FL mRNA by an absence of binding of a mAb (WNV E60) that recognizes this epitope (Oliphant et al., 2006) (Fig 3B).
Figure 3. ZIKV mRNA LNP vaccines containing WT or mutant FL sequences induce neutralizing antibody responses and protect BALB/c mice A.
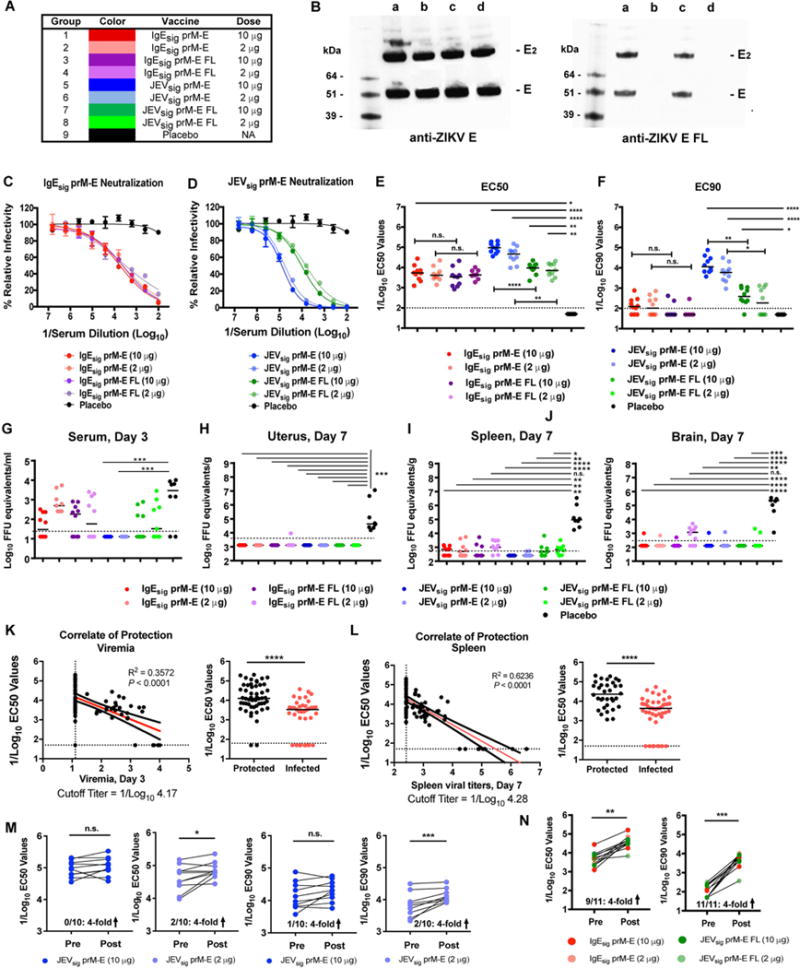
Immunization scheme. Female WT BALB/c mice (n = 10, pooled from two independent experiments) were immunized with 2 or 10 μg of prM-E mRNA LNP vaccines containing either IgE or JEV signal sequences at the N-terminus of prM and WT or mutant FL sequences in the E gene. Animals were boosted with the equivalent dose of the same vaccine 28 days later. B. HeLa cells were transfected with different modified mRNA (Lane a, IgEsig-prM-E; Lane b, IgEsig-prM-E FL mutant; Lane c, JEVsig-prM-E; and Lane d, JEVsig-prM-E FL mutant), and ZIKV E protein in the supernatant was detected by Western blotting with a type-specific anti-ZIKV E antibody (left panel) or a cross-reactive anti-ZIKV E antibody that binds the FL (right panel). Results are the representative of two independent experiments. E, monomer; E2, dimer. C–F. At week 8, serum was harvested from IgEsig-prM E (C) or JEVsig prM-E (D) mRNA LNP vaccinated mice and analyzed for neutralization capacity using ZIKV RVPs. Representative curves for each group (with EC50 values at or near the group median) are shown in (C) and (D). Error bars indicate the range of duplicate technical replicates. EC50 (E) and EC90 (F) values were calculated for individual animals in each group. Each point represents the results from a single experiment or the mean of two independent experiments. The dotted line indicates the limit of detection of the assay. G–J. At week 8, vaccinated BALB/c mice were administered 2 mg of anti-Ifnar1 blocking antibody and one day later challenged with 105 FFU of mouse adapted ZIKV Dakar 41519. At 3 (G) and 7 (H–J) days after viral challenge, serum (G), uterus (H), spleen (I), and brain (J) tissues were harvested and analyzed for ZIKV RNA. EC50, EC90, and viral titer data were analyzed by a Kruskal-Wallis test with a multiple comparisons correction and compared to the placebo LNP vaccine (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant). K–L. Correlates of day 3 viral load (K, left panel), day 7 spleen titers (L, left panel) and protective efficacy (K–L, right panels) are shown. Data from all JEVsig-prM-E and IgEsig-prM-E mRNA LNP vaccines was included in this analysis. P values and R2 values reflect Spearman rank-correlation tests. For correlate data, values at which the line would cross the limit of detection of the y-axis are indicated below the graphs. Red line represents the best fit linear regression. For protective efficacy data, bars indicate median values (****, P < 0.0001; Mann-Whitney test). M–N. Anamnestic neutralizing antibody response. Paired sera were collected from vaccinated animals (JEVsig-prM-E (M) or IgEsig-prM-E and JEVsig-prM-E FL (N)) immediately before (Pre) or 7 days after (Post) ZIKV challenge and analyzed for neutralizing activity using ZIKV RVPs. EC50 and EC90 values were analyzed for differences by a paired t-test (n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Indicated at the bottom of the graphs are the number of animals showing a 4-fold increase in neutralization titer (positive anamnestic response) at 7 days after ZIKV challenge. See also related figure S4.
Immunocompetent 8 week-old female BALB/c mice were immunized with 2 μg or 10 μg of IgEsig-prM-E or JEVsig-prM-E (WT or FL mutant) LNPs and boosted with the same LNPs 4 weeks later. At 8 weeks after initial vaccination, serum was analyzed for neutralizing activity using ZIKV reporter virus particles (RVPs) (Dowd et al., 2016a) (Fig 3 and Fig S4). Mice receiving 2 or 10 μg doses of the IgEsig-prM-E WT and FL mutant LNPs showed similar neutralization titers (Fig 3C), with EC50 values of ~1/5,000 (Fig 3E). The 2 and 10 μg dose of the WT JEVsig-prM-E LNPs induced stronger inhibitory responses with EC50 values of ~1/100,000 (Fig 3D–E) and EC90 values of ~1/10,000 (Fig 3F). The mutant JEVsig-prM-E-FL LNPs, however, induced antibody responses with lower EC50 and EC90 values, which still approached 1/10,000 and 1/500, respectively.
At ~13 weeks after initial vaccination, mice were challenged with ZIKV Dakar 41519 after pre-administration of anti-Ifnar1 blocking antibody. At day 3 after infection, serum was analyzed for viremia. Consistent with their high neutralizing titers, all mice immunized with 2 or 10 μg doses of JEVsig-prM-E LNPs lacked measurable viremia (Fig 3G). All other vaccine groups (IgEsig-prM-E LNPs (WT or FL) or JEVsig-prM-E FL LNPs) had breakthrough viremia in some animals, although levels were 10 to 100-fold lower than observed with placebo LNPs. Mice were euthanized at day 7 after infection and spleen, uterus, and brain were analyzed for ZIKV RNA levels (Fig 3H–J). Most mice vaccinated with prM-E LNPs showed markedly reduced levels of viral RNA in the spleen (>100-fold) and virtually no detectable viral RNA in the uterus or brain, whereas the placebo immunized animals had mean levels of 105 to 106 FFU equivalents per gram of tissue. Mice vaccinated with the 2 μg dose of IgEsig-prM-E FL or JEVsig-prM-E FL LNPs showed slightly less protection, as viral RNA was detectable in some animals in the spleen and brain, albeit at much lower levels than those immunized with placebo LNPs.
To establish an immune correlate of complete protection against ZIKV in this model, we compared the EC50 values from all mRNA LNP vaccines with the levels of viral RNA recovered in the serum and spleen from individual mice. This analysis revealed an expected inverse relationship between neutralizing titers and levels of ZIKV RNA in serum and tissues, and defined a cut-off EC50 value of ~1/10,000 to completely prevent viremia and tissue dissemination (Fig 3K–L). We next evaluated whether the JEVsig-prM-E mRNA LNP vaccines, which protected almost completely against viremia or infection in tissues, conferred sterilizing immunity. The vast majority of animals (80 to 90%) receiving 2 or 10 μg doses of the JEVsig-prM-E mRNA vaccines failed to boost neutralizing titers one week after challenge with infectious ZIKV, consistent with sterilizing immunity (Fig 3M). In comparison, most animals (82–100%) showing breakthrough viremia or tissue burden with the IgEsig-prM-E or JEVsig-prM-E FL mRNA vaccines sustained marked increases in EC50 and EC90 values after challenge (Fig 3N), consistent with the induction of an anamnestic response.
Mutation of the DII-fusion loop epitope diminishes ADE in cells and mice
To evaluate whether the FL mutant vaccines diminished induction of cross-reactive enhancing antibodies, we incubated dilutions of serum obtained at 8 weeks after immunization with DENV serotype 1 (DENV-1) RVPs (Dowd et al., 2015) and assessed infection in K562 cells, which express the activating human Fc-γ receptor IIA (CD32A). Whereas sera from mice vaccinated with WT IgEsig-prM-E or JEVsig-prM-E mRNA LNPs all showed bell-shaped, canonical antibody enhancement curves, many of the sera from mice immunized with serum from IgEsig-prM-E-FL or JEVsig-prM-E-FL LNPs supported enhancement at only high concentrations and with considerably reduced efficiency (Fig 4A–C and Fig S5). Indeed, the peak serum enhancement titer (PET (Boonnak et al., 2008)) was ~100-fold lower in mice immunized with mutant forms of the fusion loop (Fig 4D). A similar reduction in enhancing power, or the fraction of infected cells at the PET was observed (Fig 4A, B, and E). Thus, introduction of mutations in the FL of ZIKV E reduced the production of enhancing antibodies against DENV, as judged by cell culture assays.
Figure 4. ZIKV mRNA LNP vaccines containing mutant FL sequences showed reduced ADE against DENV in cell culture and in AG129 mice.
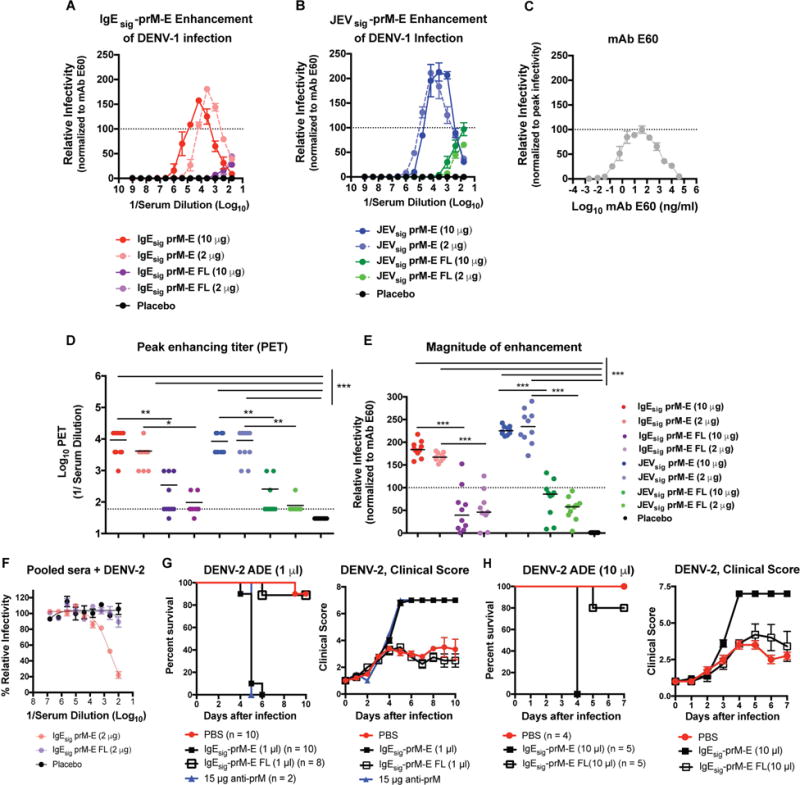
A–B. Serial dilutions of serum obtained at week 8 (see Fig 3) from BALB/c mice immunized with 2 or 10 μg of IgEsig-prM E (A) or JEVsig-prM-E (B) mRNA LNPs with WT or mutant FL sequences were mixed with DENV-1 RVPs and incubated with Fc-γ receptor expressing K562 cells. Infected cells were quantified by flow cytometry. Representative curves for each group (with peak enhancement titers (PET) at or near the group median) are shown in (A) and (B). Results are displayed relative to the maximum infectivity observed with the control cross-reactive WNV mAb E60 (FL-specific) run in parallel (C). The dotted line is provided as a reference for mAb E60 normalization. Error bars indicate the range of duplicate technical replicates. D. Peak enhancement titer (PET) for each mouse per group. Data were analyzed by a Kruskal-Wallis one-way ANOVA with a multiple comparisons correction and compared to the control LNP vaccine (**, P < 0.01; ***, P < 0.001). E. Magnitude of enhancement (extent of infection at the PET) for each mouse per group. F. Neutralization of DENV-2 RVPs by sera pooled from placebo or IgEsig-prM-E (2 μg dose of WT or FL mutant) vaccinated mice. Error bars indicate the range of duplicate technical replicates. G–H. Enhancing effects of ZIKV immune serum on DENV-2 infection in AG129 mice. Recipient AG129 mice were passively transferred PBS, 1 μl (G) or 10 μl (H) of pooled serum from BALB/c mice vaccinated with WT or FL mutant IgEsig-prM E LNPs, or 15 μg of anti-prM mAb (2H2, positive control). One day later, animals were challenged with 105 FFU of DENV-2 (strain S221) and followed for mortality (left panels) and clinical score (right panels) (1 (healthy) to 7 (deceased) scale; see STAR Methods). Results are pooled from two to three independent challenge experiments (numbers of animals indicated beneath graph) with the exception of the anti-prM mAb, which was administered in only one of the two experiments as a positive control. Survival curves between serum transfers from IgEsig-prM-E (WT and FL mutant LNPs) vaccinated mice were statistically different (****, P < 0.0001, log-rank test). See also related figures S5 and S6.
To determine the physiological significance of these results, we used an established passive transfer model of ADE for DENV in AG129 mice (Balsitis et al., 2010; Zellweger et al., 2010) with serum from the IgEsig-prM-E and IgEsig-prM-E-FL or JEVsig-prM-E and JEVsig-prM-E-FL vaccinated mice. We first evaluated the relative neutralizing activity of DENV-2 of pooled ZIKV serum using an established RVP assay in Raji-DCSIGNR cells (Dowd et al., 2015). Consistent with studies using FL-specific mAbs (Williams et al., 2013), the pooled sera from IgEsig-prM-E or or JEVsig-prM-E but not IgEsig-prM-E-FL or JEVsig-prM-E-FL vaccinated animals inhibited DENV-2 infection in cell culture (Fig 4F and Fig S6A), indicating a cross-reactive antibody (likely FL-specific) was produced only in mice receiving the WT but not FL mutant vaccines, and this antibody could neutralize infection in Raji-DCSIGNR cells lacking activating Fc-γ receptors. We next transferred pooled sera to AG129 mice one day prior to challenge with a non-lethal dose of DENV-2. Whereas administration of 1 or 10 μl of serum from mice vaccinated with WT IgEsig-prM-E or JEVsig-prM-E LNPs or a positive control anti-prM mAb (2H2) resulted in uniformly lethal infection and severe disease due to antibody enhancement, transfer of equivalent amounts of sera from mice vaccinated with IgEsig-prM-E FL or JEVsig-prM-E-FL mutant LNPs resulted in significantly less morbidity and mortality (Fig 4G–H and Fig S6B).
DISCUSSION
As ZIKV emerges, the urgency for development and deployment of counter-measures to control infection increases (Marston et al., 2016). Here, we developed a ZIKV vaccine platform using modified mRNA encoding the prM-E proteins packaged into LNPs. Although mRNA-based vaccines were used recently to generate humoral responses against influenza A virus in mice (Hekele et al., 2013; Petsch et al., 2012) or HIV in non-human primates (Bogers et al., 2015), these platforms differed from ours in their use of self-replicating RNA or protamine complexed mRNA. We selected LNPs for delivery of the modified mRNA as they have been validated in clinical trials for siRNA delivery and are well tolerated compared to other non-viral delivery systems (Coelho et al., 2013). In proof-of-principle studies, we showed that LNP-delivered modified mRNA induced neutralizing antibodies that protected several strains (129 Sv, BALB/c, or C57BL/6) of susceptible mice with genetic or acquired deficiencies in IFN signaling against lethal ZIKV challenge. The neutralization titers induced by the IgEsig-prM-E mRNA LNPs were uniformly high (50% neutralization titer of ~1/10,000) in all three mouse models. The protective responses conferred by mRNA LNP vaccines were durable; even 14 weeks after boosting, challenged mice showed no morbidity or mortality. Furthermore, by further optimizing the prM signal sequence and codon usage (JEVsig-prM-E), we induced even more potent responses (EC50 of ~1/100,000 and EC90 of 1/10,000) that resulted in sterilizing immunity in the majority of mice. The basis for the enhanced immunogenicity of the JEVsig-prM-E versus IgEsig-prM-E mRNA remains uncertain at this time.
While other candidate ZIKV vaccines have demonstrated protection in mice against ZIKV viremia with IgG titers of ~1/2,000 (Larocca et al., 2016), some of these studies used immunocompetent strains for challenge, which do not support as efficient ZIKV infection due to a species-specific lack of antagonism of IFN signaling (Grant et al., 2016; Kumar et al., 2016; Lazear et al., 2016). Passive transfer and challenge studies in non-human primates with DNA plasmid or inactivated vaccines suggested that a neutralization titer of 1/100 (measured by microneutralization assay) or 1/1,000 (measured by RVP assay) is required to prevent viremia in the majority of ZIKV-infected animals (Abbink et al., 2016; Dowd et al., 2016b). However, no prior study assessed sterilizing immunity, as judged by the combined absence of virus in target tissues and lack of evidence of an anamnestic humoral response. For vaccine development, a key question remains as to correlates of vaccine protection in pregnancy, and whether sterilizing immunity will be required to prevent seeding of the placenta and fetal infection and injury. In anti-Ifnar1-treated susceptible mice, the level of neutralizing antibodies required for conferring sterilizing immunity was substantially higher (>100 to 1,000-fold) than required to reduce viral burden or confer survival protection. Vaccination studies in pregnant animals are planned to establish the correlate of immune protection that completely prevents maternal-fetal transmission.
Because of the versatility of the platform, we engineered additional mRNA LNP vaccines with mutations in the conserved E-DII-FL, which abolish reactivity of monoclonal and polyclonal antibodies targeting this region (Chabierski et al., 2014; Crill et al., 2012; Hughes et al., 2012; Vogt et al., 2011). The FL mutant mRNA vaccines induced neutralizing antibody responses in non-pregnant female BALB/c mice that protected against virus dissemination to the uterus and brain. Moreover, the FL mutant ZIKV vaccines induced serum antibody responses that resulted in less ADE of DENV-1 infection in cell culture and immune enhancement of DENV-2 infection in AG129 mice. Our results are analogous to a prior study with a DENV-1 DNA plasmid vaccine in which the FL epitope was altered by introducing G106R and L107D mutations (Crill et al., 2012); in that study, mice immunized with cross-reactivity-reduced monovalent DENV-1 DNA vaccines had diminished immune enhancement of DENV-2 infection in mice. Although our FL mutant mRNA LNP vaccines minimized ADE, we did observe a reduction in neutralizing titer (~7-fold decrease in EC50) for JEVsig-prM-E FL LNPs compared to the respective WT LNPs; however, no significant difference in neutralization was observed between IgEsig-prM-E WT and FL mutant vaccines. Although further studies are warranted, mRNA LNPs with FL mutations may induce lower neutralizing responses because (a) they produce SVPs that are less stable; (b) the absence of the FL results in a change in the display of neutralizing epitopes; or (c) the mutation of the FL in ZIKV SVPs prevents induction of neutralizing antibodies that bind epitopes proximal to the FL, as observed with EDE antibodies (Barba-Spaeth et al., 2016; Dejnirattisai et al., 2016). Thus, there is a trade-off for using the FL mutant LNPs; although cross-reactivity and ADE of DENV infection is reduced, neutralizing activity and the likelihood of conferring sterilizing immunity also are reduced. Second generation cross-reactivity reduced constructs with a different array of mutations in conserved regions may prove even more immunogenic.
Our mRNA LNP ZIKV vaccine platform adds to a burgeoning pipeline that focuses on subunit-based or inactivated virion approaches. Three groups reported DNA plasmid vaccines encoding ZIKV M-E or prM-E genes that induced neutralizing antibodies in mice and non-human primates and protected against viremia or lethal virus challenge (Abbink et al., 2016; Dowd et al., 2016b; Larocca et al., 2016; Muthumani et al., 2016). A rhesus adenovirus-vectored vaccine encoding ZIKV M-E genes induced neutralizing antibodies after a single dose in four rhesus macaques (Abbink et al., 2016). Immunization of BALB/c mice or rhesus macaques with an alum-adjuvanted chemically inactivated ZIKV vaccine induced neutralizing antibodies and cellular immunity after two doses (Larocca et al., 2016) and protected against plasma viremia, or viral RNA in urine, cerebrospinal fluid, colorectal, and cervicovaginal secretions (Abbink et al., 2016). A human adenovirus vectored vaccine producing soluble E protein trimers induced neutralizing antibodies and protected one week-old mice from ZIKV challenge (Kim et al., 2016). Although all of these vaccine platforms show promise, none have been evaluated for induction of cross-reactive antibodies that in theory, could enhance DENV infection and disease. Nonetheless, it remains to be established whether the enhancement of DENV infection and disease, which can be demonstrated in vitro and in mice, becomes a clinically-relevant concern for ZIKV vaccines. In the absence of epidemiological proof, it may be difficult to justify the evaluation of cross-reactivity reduced variant vaccines in humans, in view of the decreased immunogenicity, especially when compared to the extraordinarily strong response generated with our WT JEVsig-prM-E construct. Nonetheless, the modified mRNA platform is well-positioned to respond rapidly to emerging epidemiology data.
In summary, we describe the generation of a ZIKV vaccine, utilizing modified mRNA encapsulated into LNPs that can be easily manipulated to optimize neutralization capacity and limit potentially undesired cross-reactivity. Our JEVsig-prM-E mRNA LNPs induced remarkable neutralizing titers that in most animals conferred sterilizing immunity. Consistent with our results, a recent study also reported the utility of LNP-encapsulated mRNA encoding the prM and E genes as a candidate vaccine for ZIKV infection (Pardi et al., 2017). The flexibility of this platform allows for the future inclusion of additional mRNA encoding other flavivirus proteins (e.g., NS1) that could augment protective responses (Costa et al., 2007; Costa et al., 2006) or immunodominant helper CD4 T cell epitopes (Hung et al., 2007). Future studies will be directed on evaluating the immunogenicity and protective efficacy of these mRNA vaccines in pregnant mice, non-human primates, and ultimately, humans.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests should be addressed and will be fulfilled by lead author Michael S. Diamond; diamond@wusm.wustl.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the Washington University School of Medicine (Assurance Number: A3381-01), the IACUC at the La Jolla Institute for Allergy & Immunology under protocol # AP028-SS1-0615, and the IACUC at Utah State University under protocol 2598. Dissections and footpad injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize suffering.
Mouse experiments
BALB/c and C57BL/6 mice were purchased from The Jackson Laboratory, and AG129 mice were bred in the animal facilities at Utah State University, Washington University, or the La Jolla Institute for Allergy and Immunology. All mice were housed in pathogen-free mouse facilities. For immunizations, mice were inoculated via an intramuscular route with 50 μl of the indicated vaccine LNP constructs. For challenge studies, mice were inoculated subcutaneously with 104 PFU of ZIKV P6-740 or 106 FFU of mouse-adapted ZIKV Dakar 41519 in 50 μl of HBSS + 0.1% FBS. For ZIKV challenge studies in BALB/c and C57BL/6 mice, 2 mg of anti-IFNAR1 blocking antibody (MAR1-5A3 (Sheehan et al., 2006)) was administered via intraperitoneal injection 24 hours prior to viral infection. For DENV challenge studies in AG129 mice, animals were passively transferred pooled vaccine immune sera or PBS one day prior to infection with ~105 FFU of DENV-2 S221. Animals were monitored for mortality and clinical score (1 = heathy; 2 = slightly ruffled fur; 3 = very ruffled fur; 4 = mild lethargy, decreased scurrying activity; 5 = very sick, slow to no movement; 6 = very sick, euthanize (in distress); 7 = deceased) as described previously (Tang et al., 2016).
METHOD DETAILS
Viruses and cells
ZIKV strain Dakar 41519 (Senegal, 1984) and P6-740 (Malaysia, 1966) were provided by the World Reference Center for Emerging Viruses and Arboviruses (R. Tesh and S. Weaver, University of Texas Medical Branch). To create a mouse-adapted more pathogenic variant of ZIKV Dakar 41519, it was passaged twice in Rag1−/− mice (Sapparapu et al., 2016; Zhao et al., 2016). ZIKV strain Paraiba 2015 (Brazil) was provided by S. Whitehead (NIH, Bethesda, MD) (Tsetsarkin et al., 2016). DENV-2 strain S221 is a mouse-adapted strain that has been described previously (Yauch et al., 2009). Virus stocks were propagated in mycoplasma-free Vero cells and titrated by focus-forming assay (FFA), as described previously (Lazear et al., 2016). Experiments with ZIKV and DENV were conducted under biosafety level 2 (BSL2) containment at Washington University School of Medicine or under BSL3 containment at Utah State University with Institutional Biosafety Committee approval.
Generation of modified mRNA and LNP
The mRNA was synthesized in vitro using T7 polymerase-mediated DNA-dependent RNA transcription where the UTP was substituted with 1-methylpseudoUTP, using a linearized DNA template, which incorporates 5′ and 3′ untranslated regions (UTRs) and includes a poly-A tail (5′-UTR: TCAAGCTTTTGGACCCTCGTACAGAAGCTAATACGACTCACTATAGGGAAATAAGAGAGAAAAGAAGAGTAAGAAGAAATATAAGAGCCACC; and 3′-UTR: TGATAATAGGCTGGAGCCTCGGTGGCCATGCTTCTTGCCCCTTGGGCCTCCCCCCAGCCCCTCCTCCCCTTCCTGCACCCGTACCCCCGTGGTCTTTGAATAAAGTCTGAGTGGGCGGC). A donor methyl group S-adenosylmethionine (SAM) was added to the methylated capped RNA (cap 0), resulting in a cap 1 structure to increase mRNA translation efficiency. The modified mRNAs encoded the signal sequences from human IgE (MDWTWILFLVAAATRVHS) or JEV prM (MWLVSLAIVTACAGA) and the prM and E genes from an Asian ZIKV strain (Micronesia 2007, GenBank accession number EU545988 (Lanciotti et al., 2008)), which is >99% identical to circulating American strains.
LNP formulations were prepared using a modified procedure of a method described for siRNA (Chen et al., 2016). Briefly, lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (ionizable lipid: DSPC: cholesterol: PEG-lipid). The lipid mixture was combined with a 50 mm citrate buffer (pH 4.0) containing mRNA at a ratio of 3:1 (aqueous:ethanol) using a microfluidic mixer (Precision Nanosystems, Vancouver, BC). Formulations were dialyzed against PBS (pH 7.4) in dialysis cassettes for at least 18 h. Formulations were concentrated using Amicon Ultra Centrifugal Filters (EMD Millipore, Billerica, MA), passed through a 0.22-μm filter and stored at 4°C until use. All formulations were tested for particle size, RNA encapsulation, and endotoxin and were found to be between 80 to 100 nm in size, with greater than 90% encapsulation and <1 EU/ml of endotoxin.
Viral protein analysis
SVPs were floated on a 10–50% sucrose gradient (50 mM MES pH 5.5, 120 mM NaCl) and ultracentrifuged at 32,000 rpm at 4°C for 4 h in a Beckman SW55 rotor. Fractions (1 ml) were collected and pelleted by a second ultracentrifugation step over a 20% sucrose cushion at 32,000 rpm at 4°C for 2 h. P rotein concentration of SVPs was measured using a spectrophotometer (280 nm wavelength). Western blotting was performed using NuPAGE MES Western blot system (Thermo Fisher) reagents as per the manufacturer’s instructions. Samples were boiled at 100°C for 10 min in the absence of reducing agent. Samples (100 or 500 ng total protein) were loaded on 4–12% NuPAGE gradient gel and electrophoresed. Proteins were transferred onto nitrocellulose membranes using iBlot 2 gel transfer system. Membranes were washed three times with deionized water and blocked in PBS with 5% Blotto (Thermo Fisher) overnight at 4°C. Primary mAbs (WNV E60 (Oliphant et al., 2006) or mouse anti-ZIKV E (Biofront, BF-1176-86)) were added at 1 μg/ml in PBS supplemented with 5% Blotto and 0.2% (v/v) Tween 20 (Sigma) and incubated for 2 h at room temperature with agitation. Membranes were washed three times with PBS supplemented with 0.2% Tween 20. Secondary antibody (horseradish peroxidase conjugated goat anti-mouse IgG (Ab97023, Abcam)) was added at 0.25 μg/ml in PBS supplemented with 5% Blotto and 0.2% Tween 20 for 2 h at room temperature with agitation. After final washing, blots were developed using Amersham ECL prime solution (GE Healthcare Life Sciences) for 2 min and imaged. SeeBlue plus 2 pre-stained protein ladders were included for molecular weight references.
To compare expression of SVPs from different mRNA vaccine constructs, HeLa cells were transfected with 1.25 μg of mRNA constructs using Lipofectamine 2000 (Thermo Fisher), and cells were harvested 24 h later. Cell lysates were prepared in RIPA lysis buffer (Thermo Fisher) with phosphatase and protease inhibitors (Millipore) added.
Electron microscopy of SVPs
The SVPs were imaged by electron microscopy and negative staining using a fee-for-service facility (University of California, Los Angeles). Briefly, purified SVPs (2.5 μl) were applied to a Glow-discharge carbon-coated grid (Ted Pella Inc.). Staining (2% uranyl acetate) was added in a drop-wise manner for 60 sec. After blotting of excess liquid and drying, the images were collected on a FEI Tecnai TF20 transmission electron microscope at an accelerating voltage of 200 kV using TVIPS EM-Menu program. The nominal magnifications used were 50,000, 29,000 and 14,500 with 2 binning.
Measurement of viral burden
At specified time points after ZIKV challenge, blood was collected and organs were recovered. Organs were weighed and homogenized using a bead-beater apparatus (MagNA Lyser, Roche), and serum was prepared after coagulation and centrifugation. Tissue samples and serum from ZIKV-infected mice were extracted with the RNeasy Mini Kit (Qiagen). ZIKV RNA levels were determined by TaqMan one-step quantitative reverse transcriptase PCR (qRT-PCR) on an ABI 7500 Fast Instrument using standard cycling conditions. Viral burden is expressed on a log10 scale as viral RNA equivalents per gram or per milliliter after comparison with a standard curve produced using serial 5-fold dilutions of ZIKV RNA from known quantities of infectious virus. For ZIKV, the following primer sets were used: 1183F: 5′-CCACCAATGTTCTCTTGCAGACATATTG-3′; 1268R: 5′-TTCGGACAGCCGTTGTCCAACACAAG-3′; and probes (1213F): 5′-56-FAM/AGCCTACCTTGACAAGCAGTC/3IABkFQ-3′.
Neutralization assays
(a) PRNT or FRNT assays. Serial dilutions of heat-inactivated sera obtained from AG129 or C57BL/6 mice were incubated with 50 to 100 FFU of ZIKV (Paraiba, Brazil 2015) for 1 h at 37 °C. The serum Ab-virus complexes were added to Vero cell monolayers in 96-well plates for 60 min at 37°C. Pl aque assays were performed as described previously (Brien et al., 2013; Lazear et al., 2016) For FRNT assays, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 4% heat-inactivated FBS. Plates were fixed 40 h later with 1% PFA in PBS for 1 h at room temperature. The plates were incubated sequentially with 500 ng/ml of humanized anti-WNV E60 (Oliphant et al) and horseradish-peroxidase-conjugated goat anti-human IgG in PBS supplemented with 0.1% (w/v) saponin (Sigma) and 0.1% BSA. ZIKV-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 macroanalyzer (Cellular Technologies). (b) RVP assays. RVPs incorporating the structural proteins of ZIKV or DENV were produced by complementation of a previously described sub-genomic GFP-expressing replicon derived from a lineage II strain of WNV (Dowd et al., 2016a; Dowd et al., 2015). Serial dilutions of heat-inactivated sera obtained from BALB/c mice were mixed with ZIKV (strain H/PF/2013; French Polynesia, 2013) or DENV-2 (strain 16681) reporter viral particles (RVPs) and incubated for 1 h at 37°C. Immune complexes wer e added in duplicate technical replicates to pre-plated Vero cells in a 96-well plate and incubated for two days. Cells were trypsinized, resuspended in 4% PFA in PBS, and RVP infection scored as a function of GFP expression by flow cytometry. All neutralization data were analyzed by non-linear regression to determine the dilution of sera required to inhibit 50% (EC50) and 90% (EC90) of infection. RVP studies were performed starting at an initial serum dilution of 1:100 (based on the final volume of cells, virus, and sera per well), which was designated as the limit of detection.
ADE assays
Serial dilutions of heat-inactivated sera obtained from BALB/c mice were mixed with DENV-1 RVPs (Western Pacific-74 strain) and incubated for 1 h at 37°C. Immune complexes were added in duplicate technical replicates to K562 cells that express the Fc-γ receptor CD32A and incubated for two days. Due to limited volumes of sera, a small number of samples (four) could not be performed in duplicate. Cells were fixed with 2% PFA, and RVP infection was scored as a function of GFP expression by flow cytometry. To normalize the magnitude of enhancement across independent experiments, results are displayed relative to the maximum infectivity observed with a control cross-reactive WNV mAb E60 (Oliphant et al., 2006) run in parallel. ADE studies were performed starting at an initial serum dilution of 1:60 (based on the final volume of cells, virus, and sera per well), which was designated as the limit of detection. For calculations of peak enhancing titer, samples for which no enhancement (infectivity) was observed are reported as a titer of 30 (one half the limit of detection).
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were analyzed with GraphPad Prism software. Kaplan-Meier survival curves were analyzed by the log rank test, and weight losses were compared using two-way ANOVA. For neutralization antibody titers and viral burden analysis, the log titers and levels of viral RNA were analyzed by a Kruskal-Wallis 2-way ANOVA with a multiple comparisons correction. A P value of < 0.05 indicated statistically significant differences.
DATA AND SOFTWARE AVAILABILITY
All data is available upon request to the lead contact author. No proprietary software was used in the data analysis.
ADDITIONAL RESOURCES
mRNA LNP vaccines are available from Valera/Moderna upon request and completion of appropriate Material Transfer Agreements.
Supplementary Material
Figure S1. Production of LNPs for vaccination, Related to Figure 1. A. Schematic representation of the process to encapsulate mRNA into LNPs. B. A representative cryo-electron microscopy image of an LNP solution, following mRNA encapsulation.
Figure S2. Serum neutralization curves from IgEsig-prM-E vaccinated AG129 mice, Related to Figure 1. Ten AG129 mice in each group (combined from two independent experiments) were immunized with 10 (Groups 1 and 2, panels A–B) or 2 (Groups 3 and 4, panels C–D) μgs of IgEsig prM-E mRNA LNPs. Some of the mice (Groups 1 and 3) were boosted with an equivalent dose 21 days later. Serum was collected at 6 weeks (day 42) post initial vaccination and analyzed for ZIKV neutralization activity by PRNT. Each line represents the neutralization curve from an individual mouse.
Figure S3. Serum neutralization curves from IgEsig prM-E vaccinated C57BL/6 mice, Related to Figure 2. Ten C57BL/6 mice in each group (combined from two independent experiments) were immunized with 10 μg of IgEsig-prM-E mRNA LNPs and boosted with an equivalent dose four weeks later. Serum was collected at 4 (B), 8 (C), and 18 (D) weeks post initial vaccination and analyzed for ZIKV neutralization activity by FRNT. Serum from mice immunized with placebo mRNA LNPs also were analyzed (A) Each line represents the neutralization curve from an individual mouse. Error bars indicate the standard deviation (SD) of triplicate technical replicates.
Figure S4. Serum neutralization curves from LNP vaccines containing WT or mutant FL sequences, Related to Figure 3. Ten BALB/c mice in each group (combined from two independent experiments) were immunized with 2 or 10 μg of prM-E mRNA LNP vaccines containing IgE or JEV signal sequences and WT or mutant fusion loop (FL) sequences or placebo mRNA LNPs (Groups 1–9, A–I). Animals were boosted with the equivalent dose of the same vaccine 4 weeks later. At week 8, serum was harvested and analyzed for ZIKV neutralization capacity by incubating serial dilutions of serum with GFP-expressing ZIKV RVPs, followed by infection of Vero cells. Infected cells were quantified 2 days later by flow cytometry. Each curve represents the data from an individual mouse analyzed by non-linear regression analysis. Error bars indicate the range of duplicate technical replicates.
Figure S5. ADE curves from LNP vaccines containing WT or FL mutant sequences, Related to Figure 4. Ten BALB/c mice in each group (combined from two independent experiments) were immunized with 2 or 10 μg of prM-E mRNA LNP vaccines containing IgE or JEV signal sequences and WT or mutant FL sequences or placebo mRNA LNPs (Groups 1–9, A–I). Animals were boosted with the equivalent dose of the same vaccine 4 weeks later. At week 8, serum was harvested from vaccinated mice. Serial dilutions of sera were mixed with GFP-expressing DENV-1 (strain West-Pac 74) RVPs and incubated with FcγR-expressing K562 cells. Infected cells were quantified by flow cytometry. In each experiment, the cross-reactive FL-specific mAb E60 was included as a control (J). To normalize the magnitude of enhancement across independent experiments, results are displayed relative to the maximum infectivity observed with mAb E60 run in parallel (designated by the dotted line at 100). Each line represents the enhancement curve from an individual mouse. Error bars indicate the range of duplicate technical replicates.
Figure S6. ZIKV mRNA LNP vaccines containing mutant FL sequences showed reduced ADE against DENV in AG129 mice, Related to Figure 4. A. Neutralization of DENV-2 RVPs by sera pooled from placebo or JEVsig-prM-E (2 μg dose of WT or FL mutant) vaccinated mice. Error bars indicate the range of duplicate technical replicates. B. Enhancing effects of ZIKV immune serum on DENV-2 infection in AG129 mice. Recipient AG129 mice were passively transferred PBS or 10 μl of pooled serum from BALB/c mice vaccinated with WT or FL mutant JEVsig-prM E LNPs. One day later, animals were challenged with 104 FFU of DENV-2 (strain S220) and followed for mortality. Results are pooled from two independent challenge experiments (numbers of animals indicated beneath graph). Survival curves between serum transfers from JEVsig-prM-E (WT and FL mutant LNPs) vaccinated mice were statistically different (***, P < 0.001, log-rank test).
HIGHLIGHTS.
A modified mRNA vaccine encoding prM-E protects in 3 different mouse strains
Extraordinarily high titers of neutralizing antibodies (>1/100,000 EC50) are produced
Sterilizing immunity was achieved with a single prime-boost strategy
A fusion loop mutant vaccine reduced production of enhancing anti-DENV antibodies
Acknowledgments
This work was supported by grants from the NIH-NIAID (R01 AI073755, R01 AI104972, and P01 AI106695 to M.S.D; R01 AI116813 to S.S), the intramural program of NIH-NIAID (T.C.P), and by a research grant from Moderna. We thank W. H. Hui (University of California, Los Angeles) for performing the negative stain imaging of the SVPs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
J.M.R, S.H., K.A.D, S.B., J.G.J., S.S, T.C.P, G.C., and M.S.D. designed the experiments. J.M.R., S.H., K.A.D., J.M.F., V.S., and W.T. performed the experiments. J.M.R, S.H., K.A.D, S.S, T.C.P, G.C., and M.S.D analyzed the data. J.M.R. and M.S.D. wrote the first draft of the paper; all authors edited the manuscript.
CONFLICT of INTERESTS STATEMENT
M.S.D. is a consultant for Inbios, Visterra, and Takeda Pharmaceuticals and on the Scientific Advisory Boards of Moderna and OvaGene. S.H., S.B., and G.C. are employees of Valera LLC, a Moderna Venture focusing on the development of therapeutic approaches for Infectious Diseases, including ZIKV mRNA vaccines.
References
- Abbink P, Larocca RA, De La Barrera RA, Bricault CA, Moseley ET, Boyd M, Kirilova M, Li Z, Ng’ang’a D, Nanayakkara O, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016;353:1129–1132. doi: 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. Characterization of Lethal Zika Virus Infection in AG129 Mice. PLoS neglected tropical diseases. 2016;10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′–5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 2010:e1000790. doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Loriere E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogers WM, Oostermeijer H, Mooij P, Koopman G, Verschoor EJ, Davis D, Ulmer JB, Brito LA, Cu Y, Banerjee K, et al. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J Infect Dis. 2015;211:947–955. doi: 10.1093/infdis/jiu522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonnak K, Slike BM, Burgess TH, Mason RM, Wu SJ, Sun P, Porter K, Rudiman IF, Yuwono D, Puthavathana P, et al. Role of dendritic cells in antibody-dependent enhancement of dengue virus infection. J Virol. 2008;82:3939–3951. doi: 10.1128/JVI.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro. N Engl J Med. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien JD, Lazear HM, Diamond MS. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol. 2013;31:15D 13 11–15D 13 18. doi: 10.1002/9780471729259.mc15d03s31. [DOI] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabierski S, Barzon L, Papa A, Niedrig M, Bramson JL, Richner JM, Palu G, Diamond MS, Ulbert S. Distinguishing West Nile virus infection using a recombinant envelope protein with mutations in the conserved fusion-loop. BMC Infect Dis. 2014;14:246. doi: 10.1186/1471-2334-14-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tam YY, Lin PJ, Sung MM, Tam YK, Cullis PR. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. Journal of controlled release : official journal of the Controlled Release Society. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- Claudio N, Dalet A, Gatti E, Pierre P. Mapping the crossroads of immune activation and cellular stress response pathways. Embo j. 2013;32:1214–1224. doi: 10.1038/emboj.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa SM, Azevedo AS, Paes MV, Sarges FS, Freire MS, Alves AM. DNA vaccines against dengue virus based on the ns1 gene: the influence of different signal sequences on the protein expression and its correlation to the immune response elicited in mice. Virology. 2007;358:413–423. doi: 10.1016/j.virol.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Costa SM, Freire MS, Alves AM. DNA vaccine against the non-structural 1 protein (NS1) of dengue 2 virus. Vaccine. 2006;24:4562–4564. doi: 10.1016/j.vaccine.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Crill WD, Hughes HR, Trainor NB, Davis BS, Whitney MT, Chang GJ. Sculpting humoral immunity through dengue vaccination to enhance protective immunity. Front Immunol. 2012;3:334. doi: 10.3389/fimmu.2012.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Trainor NB, Chang GJ. A detailed mutagenesis study of flavivirus cross-reactive epitopes using West Nile virus-like particles. J Gen Virol. 2007;88:1169–1174. doi: 10.1099/vir.0.82640-0. [DOI] [PubMed] [Google Scholar]
- Davidson A, Slavinski S, Komoto K, Rakeman J, Weiss D. Suspected Female-to-Male Sexual Transmission of Zika Virus - New York City, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:716–717. doi: 10.15585/mmwr.mm6528e2. [DOI] [PubMed] [Google Scholar]
- Davis BS, Chang GJ, Cropp B, Roehrig JT, Martin DA, Mitchell CJ, Bowen R, Bunning ML. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol. 2001;75:4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckard DT, Chung WM, Brooks JT, Smith JC, Woldai S, Hennessey M, Kwit N, Mead P. Male-to-Male Sexual Transmission of Zika Virus - Texas, January 2016. MMWR Morb Mortal Wkly Rep. 2016;65:372–374. doi: 10.15585/mmwr.mm6514a3. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet CJ, Ishii KJ. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46:521–534. doi: 10.1016/0035-9203(52)90043-6. [DOI] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- Dowd KA, DeMaso CR, Pelc RS, Speer SD, Smith AR, Goo L, Platt DJ, Mascola JR, Graham BS, Mulligan MJ, et al. Broadly Neutralizing Activity of Zika Virus-Immune Sera Identifies a Single Viral Serotype. Cell Rep. 2016a;16:1485–1491. doi: 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, DeMaso CR, Pierson TC. Genotypic Differences in Dengue Virus Neutralization Are Explained by a Single Amino Acid Mutation That Modulates Virus Breathing. MBio. 2015;6:e01559–01515. doi: 10.1128/mBio.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Ko SY, Morabito KM, Yang ES, Pelc RS, DeMaso CR, Castilho LR, Abbink P, Boyd M, Nityanandam R, et al. Rapid development of a DNA vaccine for Zika virus. Science. 2016b;354:237–240. doi: 10.1126/science.aai9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS neglected tropical diseases. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, Otten GR, Brazzoli M, Buccato S, Bonci A, et al. Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice. Emerging microbes & infections. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Abraham R, Shim BS, Choe H, Page DT. Zika virus infection during the period of maximal brain growth causes microcephaly and corticospinal neuron apoptosis in wild type mice. Sci Rep. 2016;6:34793. doi: 10.1038/srep34793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes HR, Crill WD, Chang GJ. Manipulation of immunodominant dengue virus E protein epitopes reduces potential antibody-dependent enhancement. Virol J. 2012;9:115. doi: 10.1186/1743-422X-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CF, Tsai YC, He L, Wu TC. DNA vaccines encoding Ii-PADRE generates potent PADRE-specific CD4+ T-cell immune responses and enhances vaccine potency. Mol Ther. 2007;15:1211–1219. doi: 10.1038/sj.mt.6300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Kawiecki AB, Christofferson RC. Zika Virus-Induced Antibody Response Enhances Dengue Virus Serotype 2 Replication In Vitro. J Infect Dis. 2016;214:1357–1360. doi: 10.1093/infdis/jiw377. [DOI] [PubMed] [Google Scholar]
- Kim E, Erdos G, Huang S, Kenniston T, Falo LD, Jr, Gambotto A. Preventative Vaccines for Zika Virus Outbreak: Preliminary Evaluation. EBioMedicine. 2016;13:315–320. doi: 10.1016/j.ebiom.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. Zika virus inhibits type-I interferon production and downstream signaling. EMBO reports. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca RA, Abbink P, Peron JP, Zanotto PM, Iampietro MJ, Badamchi-Zadeh A, Boyd M, Ng’ang’a D, Kirilova M, Nityanandam R, et al. Vaccine protection against Zika virus from Brazil. Nature. 2016;536:474–478. doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Diamond MS. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J Virol. 2016;90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy JM, Suberbielle E, Chapuy-Regaud S, Mengelle C, Bujan L, Marchou B, Delobel P, Gonzalez-Dunia D, Malnou CE, Izopet J, et al. Zika virus in semen and spermatozoa. Lancet Infect Dis. 2016;16:1106–1107. doi: 10.1016/S1473-3099(16)30336-X. [DOI] [PubMed] [Google Scholar]
- Marston HD, Lurie N, Borio LL, Fauci AS. Considerations for Developing a Zika Virus Vaccine. N Engl J Med. 2016;375:1209–1212. doi: 10.1056/NEJMp1607762. [DOI] [PubMed] [Google Scholar]
- Morens DM. Antibody-dependent of enhancement of infection and the pathogenesis of viral disease. Clin Inf Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- Murray KO, Gorchakov R, Carlson AR, Berry R, Lai L, Natrajan M, Garcia MN, Correa A, Patel SM, Aagaard K, et al. Prolonged Detection of Zika Virus in Vaginal Secretions and Whole Blood. Emerg Infect Dis. 2017;23:99–101. doi: 10.3201/eid2301.161394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K, Griffin BD, Agarwal S, Kudchodkar SB, Reuschel EL, Choi H, Kraynyak KA, Duperret EK, Keaton AA, Chung C, et al. In vivo protection against ZIKV infection and pathogenesis through passive antibody transfer and active immunisation with a prMEnv DNA vaccine. npj Vaccines. 2016 doi: 10.1038/npjvaccines.2016.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Euro Surveill. 2014;19:20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. The Induction of Epitope-Specific Neutralizing Antibodies against West Nile virus. J Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, et al. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, Dowd KA, Sutherland LL, Scearce RM, Parks R, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017 doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Weissman D. Nucleoside Modified mRNA Vaccines for Infectious Diseases. Methods Mol Biol. 2017;1499:109–121. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, Schlake T, Thess A, Kallen KJ, Stitz L, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- Prasad VM, Miller AS, Klose T, Sirohi D, Buda G, Jiang W, Kuhn RJ, Rossmann MG. Structure of the immature Zika virus at 9 A resolution. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects–Reviewing the Evidence for Causality. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, Baric RS. Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. MBio. 2016;7 doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Young MP, Mamidi A, Regla-Nava JA, Kim K, Shresta S. A Mouse Model of Zika Virus Sexual Transmission and Vaginal Viral Replication. Cell Rep. 2016;17:3091–3098. doi: 10.1016/j.celrep.2016.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Kenney H, Chen R, Liu G, Manukyan H, Whitehead SS, Laassri M, Chumakov K, Pletnev AG. A Full-Length Infectious cDNA Clone of Zika Virus from the 2015 Epidemic in Brazil as a Genetic Platform for Studies of Virus-Host Interactions and Vaccine Development. MBio. 2016;7:e01114–01116. doi: 10.1128/mBio.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eijk AA, van Genderen PJ, Verdijk RM, Reusken CB, Mogling R, van Kampen JJ, Widagdo W, Aron GI, GeurtsvanKessel CH, Pas SD, et al. Miscarriage Associated with Zika Virus Infection. N Engl J Med. 2016;375:1002–1004. doi: 10.1056/NEJMc1605898. [DOI] [PubMed] [Google Scholar]
- Vogt MR, Dowd KA, Engle M, Tesh RB, Johnson S, Pierson TC, Diamond MS. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fc-{gamma} receptor and complement-dependent effector mechanisms. J Virol. 2011;22:11567–11580. doi: 10.1128/JVI.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, Wong G, Peng R, Liu S, Li J, et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med. 2016;8:369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- Williams KL, Sukupolvi-Petty S, Beltramello M, Johnson S, Sallusto F, Lanzavecchia A, Diamond MS, Harris E. Therapeutic Efficacy of Antibodies Lacking FcgammaR against Lethal Dengue Virus Infection Is Due to Neutralizing Potency and Blocking of Enhancing Antibodies. PLoS Pathog. 2013;9:e1003157. doi: 10.1371/journal.ppat.1003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, et al. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Production of LNPs for vaccination, Related to Figure 1. A. Schematic representation of the process to encapsulate mRNA into LNPs. B. A representative cryo-electron microscopy image of an LNP solution, following mRNA encapsulation.
Figure S2. Serum neutralization curves from IgEsig-prM-E vaccinated AG129 mice, Related to Figure 1. Ten AG129 mice in each group (combined from two independent experiments) were immunized with 10 (Groups 1 and 2, panels A–B) or 2 (Groups 3 and 4, panels C–D) μgs of IgEsig prM-E mRNA LNPs. Some of the mice (Groups 1 and 3) were boosted with an equivalent dose 21 days later. Serum was collected at 6 weeks (day 42) post initial vaccination and analyzed for ZIKV neutralization activity by PRNT. Each line represents the neutralization curve from an individual mouse.
Figure S3. Serum neutralization curves from IgEsig prM-E vaccinated C57BL/6 mice, Related to Figure 2. Ten C57BL/6 mice in each group (combined from two independent experiments) were immunized with 10 μg of IgEsig-prM-E mRNA LNPs and boosted with an equivalent dose four weeks later. Serum was collected at 4 (B), 8 (C), and 18 (D) weeks post initial vaccination and analyzed for ZIKV neutralization activity by FRNT. Serum from mice immunized with placebo mRNA LNPs also were analyzed (A) Each line represents the neutralization curve from an individual mouse. Error bars indicate the standard deviation (SD) of triplicate technical replicates.
Figure S4. Serum neutralization curves from LNP vaccines containing WT or mutant FL sequences, Related to Figure 3. Ten BALB/c mice in each group (combined from two independent experiments) were immunized with 2 or 10 μg of prM-E mRNA LNP vaccines containing IgE or JEV signal sequences and WT or mutant fusion loop (FL) sequences or placebo mRNA LNPs (Groups 1–9, A–I). Animals were boosted with the equivalent dose of the same vaccine 4 weeks later. At week 8, serum was harvested and analyzed for ZIKV neutralization capacity by incubating serial dilutions of serum with GFP-expressing ZIKV RVPs, followed by infection of Vero cells. Infected cells were quantified 2 days later by flow cytometry. Each curve represents the data from an individual mouse analyzed by non-linear regression analysis. Error bars indicate the range of duplicate technical replicates.
Figure S5. ADE curves from LNP vaccines containing WT or FL mutant sequences, Related to Figure 4. Ten BALB/c mice in each group (combined from two independent experiments) were immunized with 2 or 10 μg of prM-E mRNA LNP vaccines containing IgE or JEV signal sequences and WT or mutant FL sequences or placebo mRNA LNPs (Groups 1–9, A–I). Animals were boosted with the equivalent dose of the same vaccine 4 weeks later. At week 8, serum was harvested from vaccinated mice. Serial dilutions of sera were mixed with GFP-expressing DENV-1 (strain West-Pac 74) RVPs and incubated with FcγR-expressing K562 cells. Infected cells were quantified by flow cytometry. In each experiment, the cross-reactive FL-specific mAb E60 was included as a control (J). To normalize the magnitude of enhancement across independent experiments, results are displayed relative to the maximum infectivity observed with mAb E60 run in parallel (designated by the dotted line at 100). Each line represents the enhancement curve from an individual mouse. Error bars indicate the range of duplicate technical replicates.
Figure S6. ZIKV mRNA LNP vaccines containing mutant FL sequences showed reduced ADE against DENV in AG129 mice, Related to Figure 4. A. Neutralization of DENV-2 RVPs by sera pooled from placebo or JEVsig-prM-E (2 μg dose of WT or FL mutant) vaccinated mice. Error bars indicate the range of duplicate technical replicates. B. Enhancing effects of ZIKV immune serum on DENV-2 infection in AG129 mice. Recipient AG129 mice were passively transferred PBS or 10 μl of pooled serum from BALB/c mice vaccinated with WT or FL mutant JEVsig-prM E LNPs. One day later, animals were challenged with 104 FFU of DENV-2 (strain S220) and followed for mortality. Results are pooled from two independent challenge experiments (numbers of animals indicated beneath graph). Survival curves between serum transfers from JEVsig-prM-E (WT and FL mutant LNPs) vaccinated mice were statistically different (***, P < 0.001, log-rank test).


