In this issue of the Journal of Bacteriology, van Beilen and colleagues (20) propose an elegant model that explains how members of the family of integral membrane alkane hydroxylases can distinguish the lengths of their substrate alkanes. Identification of the active-site pocket of this enzyme, and understanding the basis for its specificity, has been a long-standing problem that is relevant not only from the point of view of fundamental science but from a biotechnological perspective considering the applications of this enzyme.
Alkanes are saturated, linear hydrocarbons. Their chain lengths can vary from 1 carbon atom (in methane) to more than 50 carbon atoms in length. Well-known examples are octane and dodecane, etc. Alkanes constitute about 20 to 50% of crude oil, depending on the source of the oil. However, many living organisms, such as bacteria, plants, and some animals, also produce them. As a result, alkanes are widespread in nature, and many microorganisms have evolved enzymes to use them as a carbon source. Alkanes, however, are chemically inert and must be activated before they can be metabolized. In the presence of oxygen, activation is usually achieved by oxidation of one of the terminal methyl groups to generate the corresponding primary alcohol, which is further oxidized by dehydrogenases to render fatty acids.
Several unrelated enzymes can catalyze the terminal activation of alkanes. The best known of these enzymes is the Pseudomonas putida GPo1 alkane hydroxylase complex, which consists of an integral membrane hydroxylase (a monooxygenase named AlkB) and two soluble proteins (named rubredoxin [AlkG] and rubredoxin reductase [AlkT]). Rubredoxin reductase transfers electrons from NADH to rubredoxin, which in turn transfers the electrons to the membrane-bound alkane hydroxylase (for reviews, see references 15 and 22). AlkB transfers one oxygen atom from O2 to one of the terminal methyl groups of the alkane molecule, rendering an alcohol, while the other oxygen atom is reduced to H2O by the electrons transferred by rubredoxin. P. putida GPo1 AlkB oxidizes alkanes containing between 5 and 12 carbon atoms (C5 to C12 alkanes). However, the substrate specificity of this enzyme is rather relaxed, and it can oxidize many other molecules as well. Besides hydroxylating terminal methyl groups in aliphatic and alicyclic hydrocarbons, AlkB generates epoxides from alkenes and other chemicals with a terminal double bond, oxidizes alcohols to aldehydes, and catalyzes demethylation and sulfoxidation reactions (21, 23). Oxidation is regio- and stereospecific, which opens doors for many applications in fine chemistry. For example, when acting on a compound with a terminal double bond, AlkB produces an (R)-epoxide with a high enantiomeric excess. Optically active epoxides can be used to generate a number of chemicals that are useful precursors from which to derive several value-added products. The setup of a cost-effective high-scale process based on this enzyme is no simple task, since many practical issues hamper its productivity. Some of these problems are substrate uptake, toxicity of the substrate and/or the product generated, uncoupling, oxygen mass transfer, low turnover with some compounds, and product recovery. B. Witholt's research group at the Eidgenössische Technische Hochschule (Zürich, Switzerland) is dedicated to finding solutions to these problems (14, 21). One direction, though not the only one, has been to gain a deeper understanding of the enzyme itself. As can be expected, the highest efficiency of AlkB is achieved with some of its natural substrates (octane and nonane), but its activity towards other compounds of biotechnological interest is often significantly lower. There are several ways to improve its catalytic properties towards substrates of interest. One way is to investigate natural AlkB variants that may have different catalytic properties (10). Other alternatives are mutagenesis and directed evolution, strategies that would strongly benefit from the identification of the substrate-binding pocket of AlkB. However, the three-dimensional structure of AlkB is still unknown, since all attempts to obtain crystals have failed thus far. A combination of the analysis of AlkB-related proteins with the selection of substrate range mutants has finally allowed the proposal of an AlkB topology model that locates the catalytic site and explains substrate discrimination.
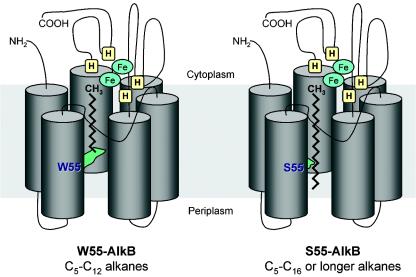
While P. putida GPo1 oxidizes only C5 to C12 alkanes, most alkane-degrading bacteria assimilate alkanes containing more than 10 carbon atoms (C12 to C20 or even C30 alkanes). The majority of these strains contain alkane hydroxylases that are clearly related to P. putida GPo1 AlkB (10, 11, 15, 16, 19). Although sequence comparisons between these AlkB homologs have not allowed the identification of amino acids involved in substrate binding, these comparisons have been useful in strengthening previous conclusions about AlkB topology based on analyses of hydrophobicity and of gene fusions with reporter genes. These results led to the proposal that AlkB is an integral membrane protein with six transmembrane segments that leave some amino acid stretches facing the periplasm and others facing the cytoplasm (Fig. 1) (18). AlkB is a non-heme diiron monooxygenase that shares sequence similarity with a large class of integral membrane fatty acid desaturases and with the P. putida xylene monooxygenase, all of which contain a number of highly conserved histidine residues that interact with two iron atoms that are thought to participate in the O2 activation required for the alkane oxidation reaction (6, 8). The conserved histidines are essential for AlkB activity (7), and in the topology model, all face the cytoplasm (18). These findings tell us how alkanes are oxidized but not how they are recognized.
FIG. 1.
Model for the AlkB alkane hydroxylase. The enzyme is proposed to have six transmembrane helices arranged in a hexagonal distribution that would define a long, hydrophobic pocket into which the linear alkane molecule can slip. The four histidine clusters (H) believed to bind the two iron atoms (Fe) would lie on the cytoplasmic side. In P. putida GPo1 AlkB, residue W55 lies in transmembrane helix 2 and extends its bulky arm towards the hydrophobic pocket (left). This hampers the proper insertion of alkanes longer than C13. The replacement of W55 by a serine residue (S55) (right), which has a shorter arm, allows longer alkanes to enter the pocket without impeding the proper alignment of the terminal methyl group relative to the histidine clusters. Sizes are not to scale. The soluble components of the alkane hydroxylase complex, rubredoxin and rubredoxin reductase, are not shown. The model is modified from reference 20 with permission.
To tackle this problem, van Beilen and colleagues decided to isolate mutant derivatives with altered substrate specificities. They introduced the alkB gene from P. putida GPo1, which oxidizes C5 to C12 alkanes, into an engineered Pseudomonas fluorescens strain that is unable to grow at the expense of C10 to C16 alkanes unless it is provided with an alkane hydroxylase that can oxidize them. As expected, P. putida GPo1 AlkB did not allow this P. fluorescens strain to grow on C14 or C16 alkanes, since its substrate specificity is restricted to C5 to C12 alkanes. However, after prolonged incubation, culture growth started, indicating the appearance of mutant AlkBs with an altered substrate specificity. Sequencing of individual alkB genes from seven independent cultures showed that they all contained point mutations in codon 55 so that the wild-type tryptophan was replaced by serine or cysteine. A similar strategy with the Alcanivorax borkumensis AP1 AlkB1 alkane hydroxylase, which also oxidizes C5 to C12 alkanes, allowed the isolation of AlkB1 mutants in which the corresponding tryptophan (position 58 in this protein) was replaced by serine, cysteine, glycine, or leucine. It is worth noting that the expansion of the substrate range to longer alkanes did not impair the original ability of the enzyme to oxidize C5 to C12 alkanes. Inspection of the topology model of P. putida GPo1 AlkB showed that W55 lies close to the middle of transmembrane helix 2 (Fig. 1). Other AlkB homologs oxidizing C5 to C12 alkanes also have a W residue at a similar position. However, all AlkB-related proteins that oxidize alkanes longer than C12 do not have a tryptophan at this position but rather have other, less bulky residues. If we assume that the six AlkB transmembrane helices are assembled in a hexagonal structure, the protein would have a deep hydrophobic pocket in the center of the hexagon, with the conserved histidine residues located on the cytoplasmic surface (Fig. 1). In this model, the bulky side chain of W55 protrudes into the hydrophobic pocket, about midway between the cytoplasmic end with the conserved histidines and the opposite end of the pocket. The estimated distance between W55 and the histidine residues is similar to the length of a linear C13 molecule. This length suggests that the tryptophan side chain prevents alkanes longer than C12 to C13 from binding to the hydrophobic pocket in such a way that the terminal CH3 group of the alkane is properly oriented relative to the iron liganded to the histidines. The presence of a less bulky side chain at position 55 may then allow larger alkanes to enter deeper into the hydrophobic pocket so that the terminal CH3 group can be properly positioned. This hypothesis was experimentally tested. The AlkB proteins encoded by the Mycobacterium tuberculosis H37Rv genome include a functional alkane hydroxylase that can oxidize C10 to C16 alkanes and has a leucine residue at the position corresponding to W55 of the AlkB protein of strain GPo1 (10). Modification of this leucine residue to a more bulky amino acid, phenylalanine or tryptophan, rendered alkane hydroxylase variants that could no longer oxidize C12 to C16 alkanes, although these variants were still active on C10 and C11 alkanes. Furthermore, if strains containing these AlkB variants were forced to grow on C16, which cannot be oxidized by these enzymes, after prolonged incubation, new mutants could be isolated that could oxidize the alkane. As could have been predicted, these isolates had mutations changing the phenylalanine or tryptophan residues into less bulky residues, such as leucine or serine. These results strongly support the proposed model and therefore give a suggestive clue to the nature and position of the substrate-binding pockets of AlkB-related alkane hydroxylases. These findings open a door to obtain AlkB variants with improved catalytic properties by directed evolution.
The search for the catalytic pocket of AlkB enzymes has raised many interesting observations. The observation regarding the diversity and distribution of this class of enzymes is perhaps one of them. Some bacterial species have only one alkane hydroxylase, while other ones have two, three, or even more alkane hydroxylases. This diversity can be expected if the enzymes differ, for example, in their substrate specificities, as occurs with Acinetobacter sp. strain M1 (13). However, the exact nature of the difference may not be straightforward. In Pseudomonas aeruginosa PAO1, the two AlkB-related alkane hydroxylases have very similar substrate ranges (9), but they are induced at different moments of the growth phase (exponential or early stationary phase) (5), which suggests that the two enzymes are different in unknown aspects other than their substrate ranges.
Close to 60 AlkB homologs are known to date, and they show high sequence diversity (15). Interestingly, there is no clear linkage between the diversity of these genes and that of the bacterial species from which they were isolated. For example, AlkB homologs from fluorescent pseudomonads are almost as diverse as the entire collection of AlkB-related genes (15). This diversity is indicative of a wide horizontal transfer of genes coding for this class of enzymes. An interesting case is that of A. borkumensis AP1, a hydrocarbonoclastic bacterium that assimilates very few substrates besides alkanes and makes up a large part of the biomass in oil-polluted marine environments (3). This strain has two alkane hydroxylases, one of them being very similar to the P. aeruginosa PAO1 AlkB2 enzyme and the other being highly related to the P. putida GPo1 AlkB enzyme (16). The gene encoding the latter enzyme is flanked by other genes involved in alkane metabolism, genes forming a cluster that appears to have been mobilized to other bacterial strains and that may well be the origin of the alkane degradation genes present in the OCT plasmid originally isolated from P. putida GPo1 (16, 17). Finally, AlkB-related alkane hydroxylases are found not only in soil and water bacteria but also in some unexpected hosts. The finding of a functional alkane hydroxylase in M. tuberculosis H37Rv was not anticipated (10), since this bacterium is believed to grow only inside its mammalian hosts.
The wide distribution of alkane degradation genes suggests that alkane degradation is a useful trait. However, in most cases, alkanes have been found to be nonpreferred growth substrates, and the induction of the alkane degradation pathways is subject to a strong catabolite repression effect by many other growth substrates (2, 4, 5, 12, 24). Furthermore, at least for P. putida GPo1, growth at the expense of alkanes leads to high expression of the alkB gene, which is detrimental to cell growth (1). Perhaps alkane degradation genes are useful to scavenge the low hydrocarbon concentrations that bacteria are likely to find in different environments but may pose some problems when they are induced to high levels. We should not forget that alkane degraders, even hydrocarbonoclastic bacteria, such as A. borkumensis, normally thrive in unpolluted environments where alkanes synthesized by algae, plants, and other organisms are present at low concentrations. In summary, recent progress has clarified many aspects of alkane degradation by bacteria but has raised many new questions, as well. Clearly, there is still much to be learned about the biodegradation of hydrocarbons.
Acknowledgments
Work at my laboratory was funded by grants BMC2003-00063 and GEN2001-4698-C05-01 from the MInisterio de Educacion y Ciencia.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Chen, Q., D. B. Janssen, and B. Witholt. 1996. Physiological changes and alk gene instability in Pseudomonas oleovorans during induction and expression of alk genes. J. Bacteriol. 178:5508-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinamarca, M. A., I. Aranda-Olmedo, A. Puyet, and F. Rojo. 2003. Expression of the Pseudomonas putida OCT plasmid alkane degradation pathway is modulated by two different global control signals: evidence from continuous cultures. J. Bacteriol. 185:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harayama, S., Y. Kasai, and A. Hara. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205-214. [DOI] [PubMed] [Google Scholar]
- 4.Marín, M. M., T. H. M. Smits, J. B. van Beilen, and F. Rojo. 2001. The alkane hydroxylase gene of Burkholderia cepacia RR10 is under catabolite repression control. J. Bacteriol. 183:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marín, M. M., L. Yuste, and F. Rojo. 2003. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 185:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanklin, J., C. Achim, H. Schmidt, B. G. Fox, and E. Munck. 1997. Mossbauer studies of alkane omega-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. Proc. Natl. Acad. Sci. USA 94:2981-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shanklin, J., and E. Whittle. 2003. Evidence linking the Pseudomonas oleovorans alkane omega-hydroxylase, an integral membrane diiron enzyme, and the fatty acid desaturase family. FEBS Lett. 545:188-192. [DOI] [PubMed] [Google Scholar]
- 8.Shanklin, J., E. Whittle, and B. G. Fox. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787-12794. [DOI] [PubMed] [Google Scholar]
- 9.Smits, T. H., B. Witholt, and J. B. van Beilen. 2003. Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Leeuwenhoek 84:193-200. [DOI] [PubMed] [Google Scholar]
- 10.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits, T. H. M., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ. Microbiol. 1:307-317. [DOI] [PubMed] [Google Scholar]
- 12.Staijen, I. E., R. Marcionelli, and B. Witholt. 1999. The PalkBFGHJKL promoter is under carbon catabolite repression control in Pseudomonas oleovorans but not in Escherichia coli alk+ recombinants. J. Bacteriol. 181:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Beilen, J. B., W. A. Duetz, A. Schmid, and B. Witholt. 2003. Practical issues in the application of oxygenases. Trends Biotechnol. 21:170-177. [DOI] [PubMed] [Google Scholar]
- 15.van Beilen, J. B., Z. Li, W. A. Duetz, T. H. M. Smits, and B. Witholt. 2003. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci. Technol. 58:427-440. [Google Scholar]
- 16.van Beilen, J. B., M. M. Marín, T. H. M. Smits, M. Röthlisberger, A. G. Franchini, B. Witholt, and F. Rojo. 2004. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 6:264-273. [DOI] [PubMed] [Google Scholar]
- 17.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk-genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 18.van Beilen, J. B., D. Penninga, and B. Witholt. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:9194-9201. [PubMed] [Google Scholar]
- 19.van Beilen, J. B., T. H. Smits, L. G. Whyte, S. Schorcht, M. Rothlisberger, T. Plaggemeier, K. H. Engesser, and B. Witholt. 2002. Alkane hydroxylase homologues in Gram-positive strains. Environ. Microbiol. 4:676-682. [DOI] [PubMed] [Google Scholar]
- 20.van Beilen, J. B., T. H. M. Smits, F. Roos, T. Brunner, S. B. Balada, M. Röthlisberger, and B. Witholt. 2005. Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Beilen, J. B., M. Wubbolts, Q. Chen, M. Nieboer, and B. Witholt. 1996. Effects of two-liquid-phase systems and expression of alk genes on the physiology of alkane-oxidizing strains, p. 35-47. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonads. ASM Press, Washington, D.C.
- 22.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 23.Witholt, B., M. J. de Smet, J. Kingma, J. B. van Beilen, M. Kok, R. G. Lageveen, and G. Eggink. 1990. Bioconversions of aliphatic compounds by Pseudomonas oleovorans in multiphase bioreactors: background and economic potential. Trends Biotechnol. 8:46-52. [DOI] [PubMed] [Google Scholar]
- 24.Yuste, L., I. Canosa, and F. Rojo. 1998. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 180:5218-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]