Abstract
Repeated adaptation to a new environment often leads to convergent phenotypic changes whose underlying genetic mechanisms are rarely known. Here, we study adaptation of color vision in threespine stickleback during the repeated postglacial colonization of clearwater and blackwater lakes in the Haida Gwaii archipelago. We use whole genomes from 16 clearwater and 12 blackwater populations, and a selection experiment, in which stickleback were transplanted from a blackwater lake into an uninhabited clearwater pond and resampled after 19 y to test for selection on cone opsin genes. Patterns of haplotype homozygosity, genetic diversity, site frequency spectra, and allele-frequency change support a selective sweep centered on the adjacent blue- and red-light sensitive opsins SWS2 and LWS. The haplotype under selection carries seven amino acid changes in SWS2, including two changes known to cause a red-shift in light absorption, and is favored in blackwater lakes but disfavored in the clearwater habitat of the transplant population. Remarkably, the same red-shifting amino acid changes occurred after the duplication of SWS2 198 million years ago, in the ancestor of most spiny-rayed fish. Two distantly related fish species, bluefin killifish and black bream, express these old paralogs divergently in black- and clearwater habitats, while sticklebacks lost one paralog. Our study thus shows that convergent adaptation to the same environment can involve the same genetic changes on very different evolutionary time scales by reevolving lost mutations and reusing them repeatedly from standing genetic variation.
Author summary
When organisms colonize a new environment in replicate, natural selection often leads to similar phenotypic adaptations. Such “convergent evolution” is known from both distant relatives, e.g., sea cows and whales adapting to an aquatic life, and from multiple populations within a species, but the causing genetic changes are rarely known. Here, we studied how a fish, the threespine stickleback, repeatedly adapted its color vision to living in red light–dominated blackwater lakes. Using multiple natural populations and a 19-y evolution experiment, we found selection on a blue light–sensitive visual pigment gene. One allele of this gene with a red-shifted light sensitivity facilitated repeated blackwater colonization. Surprisingly, two amino acid changes responsible for the red-shift have independently occurred 198 million years earlier, after the gene was duplicated in the ancestor of all spiny-rayed fish and modified into blue- and red-shifted gene copies. While other fish species today use these two gene copies to adapt to clear- and blackwater, stickleback have lost a copy and reevolved these mutations on different alleles of the same gene causing convergent adaptation to these habitats. Thus, we conclude that the same genetic changes can be responsible for convergent evolution on very different time scales.
Introduction
Successful colonization of a new habitat requires adaptation to a multitude of different selection pressures. When similar habitats are colonized in replicate by different populations or species, phenotypic adaptation is often convergent [1], and this is most striking in adaptive radiations in multiple lakes or on several islands [2]. Whether a similar phenotypic adaptation is caused by selection on variants present in a shared ancestor due to recurrent mutation or due to changes in different genes, however, is still poorly understood [3, 4]. Only recently, genetic and population genomic studies have started to unravel the evolutionary mechanisms of phenotypic convergence [5–12].
An adaptive radiation with replicate habitat colonization is found among threespine stickleback (Gasterosteus aculeatus) inhabiting the Haida Gwaii archipelago, British Columbia, Canada. Since the retreat of the ice sheets 12,000 years ago, and likely before that [13–15], marine stickleback have colonized hundreds of freshwater habitats independently in different watersheds and adapted in predictable ways to highly divergent “ecological theatres” [16]. One major predictor of natural selection in the Haida Gwaii stickleback radiation is the spectrum of visible light [16]. Most Haida Gwaii lakes are either oligotrophic clearwater lakes featuring full-spectrum light to blue-shifted light with increasing depth, or they are dystrophic blackwater lakes, stained by dissolved tannins leading to a red-shifted light spectrum [16–18]. Blackwater lakes are extreme, almost “nocturnal” visual environments, as both downwelling short-wavelength light and almost all up- or sidewelling light is absorbed, leaving only downwelling red light in a small cone above the focal animal.
Evolutionary adaptation to blackwater lakes in Haida Gwaii stickleback had consequences for multiple traits: stickleback have evolved larger body sizes and reduced lateral plates, both maximizing burst velocity and agility to escape from a predator on short reaction distance, and the former increasing postcapture resistance to predators [16]. Not only natural selection by predators, but also sexual selection interacts with light spectra: blackwater stickleback males have replaced red with black nuptial throat color, which maximizes contrast to the blue eye and against the background via reversed counter-shading [17]. And both traits, the black throat and blue eyes, are preferred by females choosing their mates [19]. Also, color vision was adapted to the blackwater light spectrum: double cones in the retina of stickleback from blackwater systems express only red light–sensitive photopigments instead of one red and one green light–sensitive photopigment, increasing the stickleback’s visual sensitivity to dominant red light [18]. Expression differences are heritable and replicated between independently colonized blackwater lakes [18], but the genetic mechanisms underlying these differences are still unknown.
Here, we study the evolutionary history of color vision genes during adaptation to blackwater environments. To perceive color, vertebrates use a combination of membrane-bound photosensitive proteins, called visual opsins, that are expressed in cone cells in the retina (i.e., cone opsins) and have peak sensitivities at different wavelengths [20]. Vertebrates have evolved large opsin repertoires via gene duplication and divergence [21, 22]. Previous research showed that opsins have evolved in response to the visual environment by sequence or expression divergence (i.e., “spectral tuning,” [18, 23–30]). Adaptation of color vision has been found both between [31–34] and within species [35–37], sometimes without gene duplication [38] or without sequence divergence [39]. Remarkably, spectral tuning of opsins has led to convergent adaptation by recurrent mutations, leading to the same amino acid sequences in distantly related species, families, orders, or phyla [31, 40–43]. These observations together with extensive biochemical and mutagenesis study of opsin proteins have led to the identification of several “key site” substitutions [44–46], from which genotypes the light absorption phenotype can be predicted. Both functional experiments and the evolutionary history of opsins thus show that there are many different, functionally tractable “molecular roads” to color vision adaptation.
Most ray-finned fish possess large repertoires of eight or more cone opsin genes, originating from a combination of ancient and recent lineage-specific gene duplication events, facilitating adaptation to a diversity of visual environments in aquatic systems [21, 22]. However, threespine stickleback have only four cone opsins: the UV sensitive SWS1, a single blue-sensitive SWS2, a single green-sensitive RH2, and a red-sensitive LWS [18, 22]. This is an impoverished repertoire compared to most other fish species, and when compared to relatives among spiny-rayed fish, two SWS2 paralogs and one RH2 paralog have been lost [22, 47].
Although we know of parallel and heritable expression differences between blackwater and clearwater habitats [18] and between marine and freshwater habitats [36], the targets of selection in the genome are unknown and it is unclear whether spectral tuning of amino acids is involved in adaptation to divergent freshwater habitats [6, 36]. Here, we assess whether and which cone opsin genes have experienced recent selection using two types of evidence. First, we use whole genomes from one oceanic and 27 freshwater fish from across the Haida Gwaii adaptive radiation and from coastal British Columbia, including 15 clearwater and 12 blackwater populations derived from 18 watersheds independently colonized by marine ancestors over the last 12,000 y [13]. Second, we use whole genomes from a selection experiment in which 100 adult stickleback from a blackwater lake were transferred to a barren clearwater pond, from which 11 individuals were resampled after 19 y [48]. Then, we screen cone opsin genes for amino acid variation with predictable effects on color vision and test whether selection on such coding changes or noncoding variation has facilitated the colonization of blackwater habitats. Finally, we compare the molecular mechanisms of adaptation to blackwater habitat among Haida Gwaii threespine stickleback to other blackwater-inhabiting fish species and therein uncover convergent evolution on vastly different time scales.
Results
Signature of selection around linked blue- and red-sensitive opsins
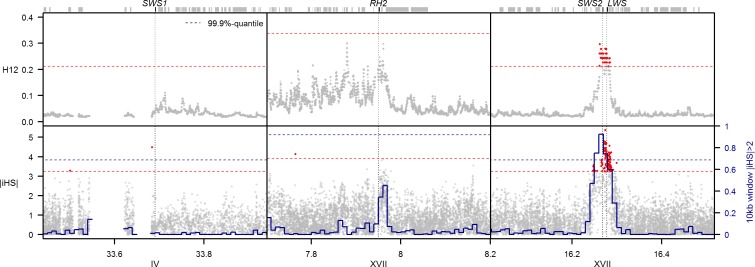
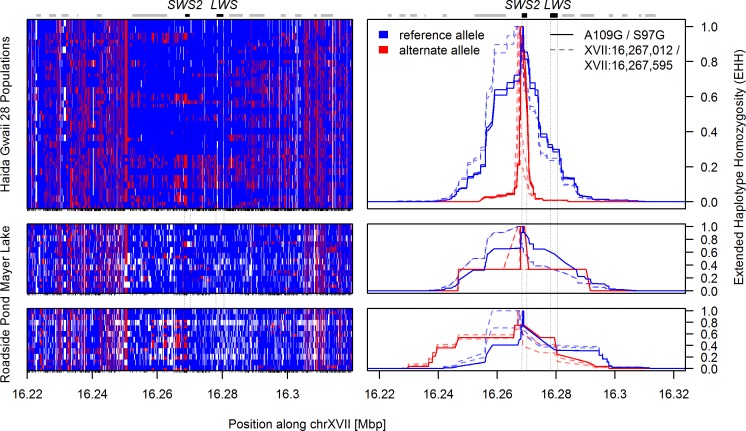
We sequenced the genomes of 58 threespine stickleback from 25 freshwater populations on Haida Gwaii, two freshwater sites from coastal British Columbia, and one marine site (Table 1), resulting in a dataset of 7,888,602 high-quality SNPs with transition to transversion ratio (Ts/Tv) 1.26 (see materials and methods). We split this dataset into an “adaptive radiation” partition, containing single individuals from each natural population on Haida Gwaii, two coastal British Columbia populations, and one mid-Pacific marine population (n = 28 individuals, 6,526,842 SNPs, Ts/Tv = 1.31), and a “selection experiment” partition, containing 12 individuals from the blackwater Mayer Lake source population and 11 individuals from the clearwater Roadside Pond transplant population (n = 23 individuals, 4,180,622 SNPs, Ts/Tv = 1.26). We scanned the adaptive radiation dataset for evidence of selective sweeps at the four cone opsin genes, using the haplotype-based statistics iHS and H12 [49, 50] and their variation across the genome to identify outlier regions. We identified a prominent outlier region for both iHS and H12 metrics in Haida Gwaii sticklebacks, suggesting a selective sweep centered on a genomic region containing both the blue- and red-sensitive opsin genes, SWS2 and LWS (Fig 1). In contrast, H12 and iHS around the green- and UV-sensitive opsins RH2 and SWS1 were not significantly different from the genome-wide expectation, although both statistics are elevated around RH2, which is in the vicinity of a more heterogeneous genomic background due to reduced recombination in this region. The selective sweep signature centered on SWS2 and LWS is caused by haplotypes with a long run of reference alleles, uninterrupted by recombination and thus leading to an extended haplotype homozygosity (EHH) across the 28 populations for the reference “sweep haplotype” (Fig 2). The same EHH signature for the sweep haplotype was found in the selection experiment within the blackwater population Mayer Lake. However, 13 generations after the transfer of Mayer Lake fish into the clearwater habitat of Roadside Pond, the alternate haplotype shows a stronger EHH signature (Fig 2), indicative of the alternate haplotype quickly rising to high frequency in the selection experiment.
Table 1. Population information.
| Abbreviation | Population | Habitat | Depth | Area | T400 | Blackwater | Watershed | Year | Number |
|---|---|---|---|---|---|---|---|---|---|
| ANSR | Anser | lake | 4.0 | 18.0 | 76.0 | no | Kumara | 2009 | 1 |
| BK70 | Banks 70 | lake | 3.0 | 5.0 | 91.0 | no | Banks 70 | 2010 | 1 |
| BKW2 | Banks W2 | lake | 2.0 | 2.0 | 85.0 | no | Banks W2 | 2010 | 1 |
| BOUL | Boulton | lake | 4.0 | 15.0 | 78.1 | no | Masset inlet | 2010 | 2 |
| BRNT | Branta | lake | 4.0 | 3.0 | 70.0 | yes | Sangan | 2009 | 1 |
| COAT | Coates | lake | 30.0 | 90.0 | 94.5 | no | Coates | 2009 | 1 |
| DARW | Darwin | lake | 15.0 | 13.9 | 90.0 | no | Darwin | 2009 | 1 |
| DAWS | Dawson | lake | 4.0 | 1.0 | 82.0 | no | Dawson | 2009 | 1 |
| DRIZ | Drizzle | lake | 16.0 | 97.0 | 67.0 | yes | Sangan | 2009 | 4 |
| EDEN | Eden | lake | 50.0 | 513.0 | 87.0 | no | Naden | 2010 | 1 |
| ESCP | Escarpment | lake | 50.0 | 97.0 | 93.6 | no | Barry | 1993 | 1 |
| GOLD | Gold Creek | stream | – | – | 50.2 | yes | Mayer | 2009 | 2 |
| LAUR | Laurel | lake | 2.0 | 2.0 | 70.0 | yes | Sangan | 1993 | 1 |
| LUTE | Lutea | lake | 2.0 | 3.0 | 93.9 | no | Burnaby | 2003 | 1 |
| MAYR | Mayer Lake | lake | 20.0 | 489.9 | 57.1 | yes | Mayer | 2004 | 12 |
| MNYN | Menyanthes | lake | 5.0 | 6.0 | 82.0 | no | Hughes | 2009 | 1 |
| MDPC | Mid-Pacific | marine | – | – | – | no | – | 1993 | 1 |
| POQU | Poque | lake | 25.0 | 17.0 | 90.8 | no | Poque | 2009 | 1 |
| RDSP | Roadside Pond | lake | 1.0 | 0.3 | 80.0 | no | – | 2012 | 11 |
| ROUG | Rouge | lake | 2.0 | 1.2 | 68.1 | yes | Kliki | 2000 | 1 |
| SDPY | Serendipity | lake | 2.0 | 3.0 | 70.5 | yes | Hiellen | 2006 | 1 |
| SKID | Skidegate | lake | 4.0 | 8.5 | 60.0 | yes | Copper | 2009 | 1 |
| SKON | Skonun | lake | 20.0 | 734.0 | 94.4 | no | Sangan | 2009 | 1 |
| SLTC | Solstice | lake | 15.0 | 51.0 | 68.0 | yes | Sangan | 2009 | 1 |
| SLVR | Silver | lake | 5.0 | 11.7 | 72.2 | yes | Kumara | 2009 | 1 |
| SPNC | Spence | lake | 30.0 | 95.0 | 75.0 | no | Hiellen | 2009 | 4 |
| STIU | Stiu | lake | 30.0 | 24.0 | 92.8 | no | Stiu | 2009 | 1 |
| WATT | Watt | lake | 2.0 | 4.0 | 60.0 | yes | Mayer | 2009 | 1 |
| WDPL | Woodpile | lake | 2.0 | 4.0 | 60.8 | yes | Mayer | 2009 | 1 |
Sample sizes (number) indicate sequenced individuals, lake area is given in hectares, lake depth in meters, and light transmission at 400 nm in percent (T400).
Fig 1. Scan for selective sweeps around cone opsins in Haida Gwaii threespine stickleback.
One of the strongest outlier regions for H12 and iHS in the genome of Haida Gwaii stickleback from 28 populations is centered on SWS2 and LWS, indicating a past selective sweep in this region. No such signature is found around the other two cone opsins. Grey dots show H12 (upper panel) and absolute iHS (lower panel) values for each SNP with a minor allele frequency (MAF) greater 5%, with top 0.1% outlier SNPs highlighted as red dots. The proportion of iHS-SNPs with a value greater than 2 per 10 kb window is shown as a blue line. The 99.9%-quantile boundaries are shown as red- and blue-dashed lines for SNPs and windows, respectively. Genomic coordinates in Mb based on an improved version of the stickleback reference genome [51] are given on the x-axis. The position of the four cone opsin genes is highlighted with black boxes and vertical dashed lines; grey boxes indicate other genes. Depicted values for H12, iHS, window-iHS, and 99.9%-quantile boundaries can be found in S1 Data.
Fig 2. Haplotype structure and extended haplotype homozygosity around SWS2 and LWS.
Left panels show phased haplotypes, right panels the decay of haplotype homozygosity around selected SNPs, each for the adaptive radiation (upper panels) and the selection experiment dataset (middle and lower panels). The selective sweep signature seen in Fig 1 is caused by a long run of reference alleles (blue, upper left panel). This led to an extended haplotype homozygosity (EHH) signal at SNPs in the top H12-window (dashed lines in top right panel): haplotype homozygosity decays slowly around the reference allele (blue) for these SNPs, but rapidly around their alternate alleles (red). Decay is similar for SWS2 key amino acid substitutions A109G and S97G (full lines). In the selection experiment (middle/lower panels), the same EHH decay is found for the reference haplotype, but the alternate haplotype shows reduced decay, in particular in the transplant population Roadside Pond. Rows in the left panel show haplotypes with imputed SNPs and monomorphic sites with missing data (white). Columns represent SNPs with color code relative to the threespine stickleback reference genome, a freshwater stickleback female [6]. Gene positions are indicated with boxes above the figure. The haplotype matrix and EHH values can be found in S1 Data.
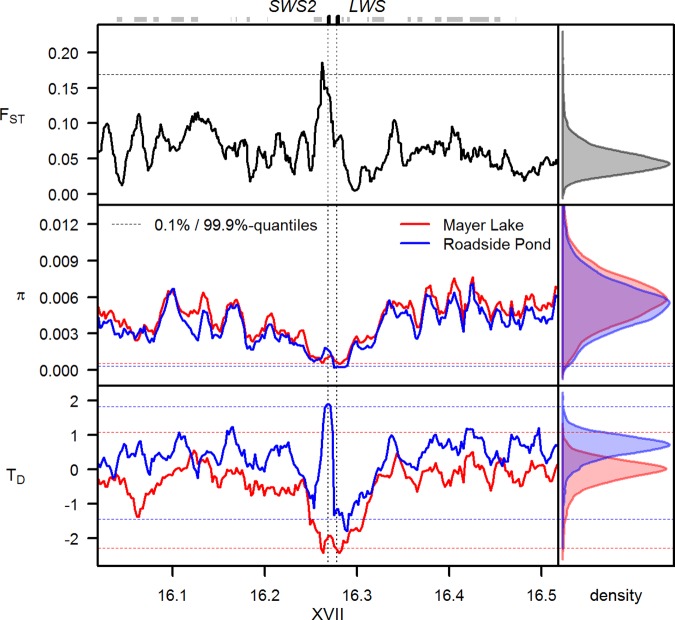
Patterns of nucleotide diversity, differentiation, site frequency spectra, and allele-frequency change across the genome from the selection experiment data support the presence of a selective sweep signature in the region containing the two opsins SWS2 and LWS and two tightly linked genes, HCFC1A and ENSGACG00000022160 (Figs 3 and 4, S1 Fig). In the blackwater source population Mayer Lake, nucleotide diversity is significantly reduced compared to the genome-wide expectation, and Tajima’s D is strongly negative, as expected under a selective sweep (Fig 3). The transplant into a clearwater habitat, however, has led to an increase in frequency of the alternate haplotype (Fig 2) and therefore to significant differentiation in this region between the two populations, as measured by FST, ranking this region among the highest 0.1% differentiated regions in the genome (Fig 3). While nucleotide diversity and Tajima’s D on LWS are still reduced in the source population Mayer Lake from the initial selective sweep, the rising frequency of the alternate haplotype has led to a positive Tajima’s D centered on SWS2. These high Tajima’s D values are among the top 0.1% outliers even against the positively shifted genome-wide distribution of Tajima’s D, which was caused by the bottleneck experienced during the population transplant [48]. This positive Tajima’s D is a consequence of both reduced diversity due to the first sweep shared with Mayer Lake (cf. Fig 3) and a rapid increase of the alternate haplotype in the transplant population to a similar frequency as the haplotype favored in Mayer Lake (Fig 2). A transient phase of a “reverse selective sweep” for the alternate haplotype, associated with the shift in light regime in the selection experiment, or the combined effect of a past selective sweep and a bottleneck may have caused this pattern. The fact that this genomic region is among the top 0.1% FST outlier windows and that linked low-frequency alleles associated with SWS2 and LWS are among the top 1% allele-frequency changes in the genome (Fig 4) support selection for the alternate haplotype in the transplant population. Note that linkage disequilibrium is not increased beyond the region containing the two opsins SWS2 and LWS and the two linked genes, perhaps due to high recombination in this chromosomal segment, making it unlikely that selection on genes further up- or downstream was involved (S1 Fig., [51, 52]).
Fig 3. Signature of selective sweep in blackwater-adapted stickleback and “reversed sweep” after transplant to clearwater habitat.
Significantly reduced nucleotide diversity (middle left) and Tajima’s D (TD, bottom left) in the blackwater source population Mayer Lake compared to genome-wide expectations (right) indicate a selective sweep in this population, consistent with the signature seen across the adaptive radiation (cf. Figs 1 and 2). After transplant to a clearwater habitat, however, the alternate haplotype has increased in frequency (Fig 2), leading to one of the strongest differentiation signals in the genome (FST, top left panel) and a significantly positive Tajima’s D. Such a pattern is consistent with a transient phase of a reversed selective sweep. Dashed horizontal lines indicate the 0.1%- and 99.9%-quantiles for each statistic based on their genome-wide distribution in regions with similar recombination rates (right panels); boxes above the figure indicate the position of genes, with black boxes and vertical dashed lines highlighting the position of the two cone opsins SWS2 and LWS. Sliding-window FST, nucleotide diversity and Tajima’s D values and genome-wide distributions of each statistic can be found in S1 Data.
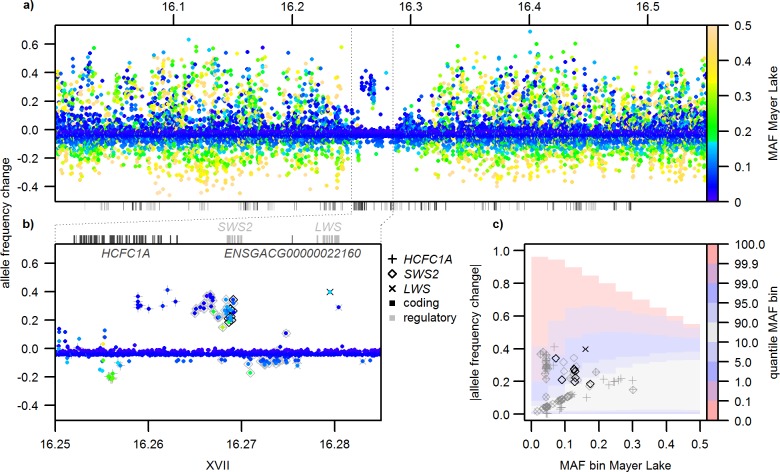
Fig 4. Allele-frequency change around SWS2 and LWS in the selection experiment.
Distribution of single SNP allele-frequency change in the selection experiment (Roadside Pond minus Mayer Lake) around the selective sweep region on chromosome XVII (a,b). Note the unexpectedly strong increase of low frequency alleles linked to blue-shifting SWS2 coding variation after the transplant to a clear water pond. Color codes in (a) and (b) show “starting” allele frequencies, i.e., minor allele frequencies (MAFs) in Mayer Lake, color codes in (c) indicates the genome-wide distribution of allele-frequency change for Mayer Lake MAF bins of 0.05 width. Symbol shapes and colors in (b) and (c) indicate associations with different genes and predicted effects. Black and grey boxes below (a) and above (b) are exons of genes surrounding the sweep region. Depicted allele frequencies and allele-frequency change quantiles can be found in S1 Data.
Selection on coding variation favoring red-shifted SWS2
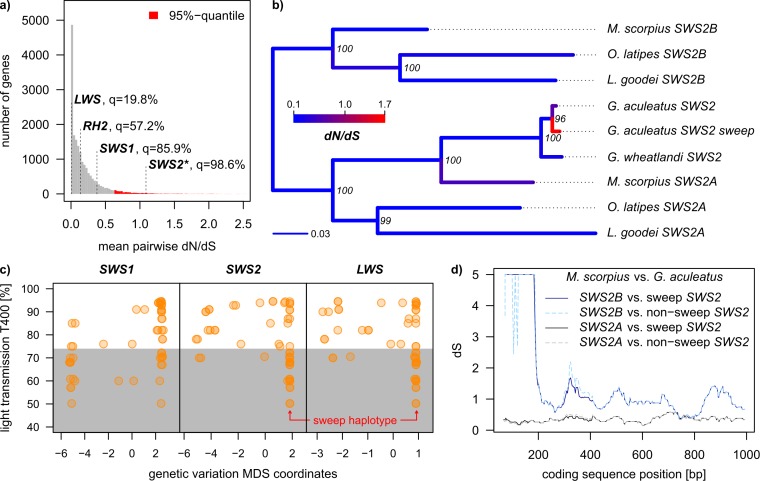
We identified segregating amino acid polymorphisms in all four cone opsins and genes linked to the selective sweep around SWS2 and LWS in both the adaptive radiation and selection experiment datasets (Fig 5). The blue-sensitive SWS2 opsin contains the highest number of amino acid polymorphisms, with seven alternative amino acid residues occurring at high frequency and in nearly perfect linkage (Fig 5), in contrast to only four synonymous substitutions. The sweep haplotype identified in the adaptive radiation dataset carries the same alleles as the reference genome for all seven amino acid polymorphism in SWS2, leading to a nearly perfect association of the SWS2 polymorphisms with the sweep haplotype (χ2 tests, all SWS2 Bonferroni-corrected p < 0.05, Fig 2), while no association is found between the LWS polymorphism and the sweep haplotype. Segregating amino acid polymorphisms in SWS2 are both numerous and occur at high frequency in the adaptive radiation SNP dataset, leading to a high mean pairwise ratio of non-synonymous to synonymous substitutions (dN/dS) estimate of 1.08 between the 56 haplotypes in this dataset, an exceptional value when compared to other functional protein-coding genes in the threespine stickleback genome (Fig 6A).
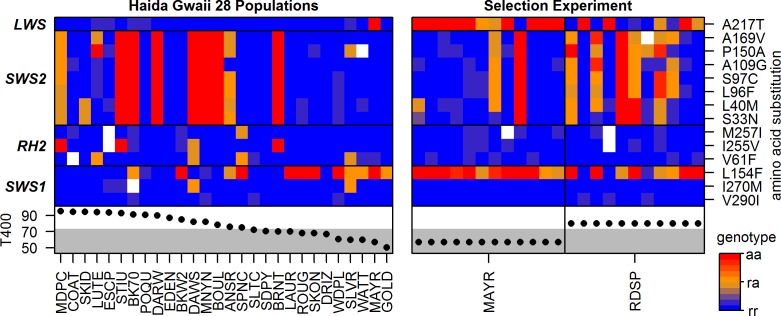
Fig 5. Cone opsin amino acid polymorphisms across the Haida Gwaii stickleback radiation and selection experiment.
Note the high frequency and nearly perfect linkage of SWS2 amino acid polymorphisms. Columns represent single individuals per population, rows represent amino acid polymorphisms. Color codes show the genotype probabilities and amino acid alleles relative to the threespine stickleback reference genome (S97C: reference = S, alternate = C; rr: homozygous for reference amino acid, aa: homozygous for alternate amino acid, ra: heterozygous genotype). Amino acid positions are relative to the bovine rhodopsin protein. Light transmission ratios at 400 nm (T400) for the different water bodies are given in percent in the lower panels, with the grey area representing blackwater lakes [16]. Genotype probabilities can be found in S1 Data, T400 values in Table 1.
Fig 6. Accelerated protein evolution, evolutionary origin of threespine stickleback SWS2, and association with blackwater habitation.
(a) SWS2 shows an unexpected high frequency of amino acid polymorphisms given the total number of mutations in SWS2. The distribution of mean pairwise relative protein sequence divergence (mean ratio of non-synonymous to synonymous substitutions [dN/dS]) is based on 17,846 functional protein coding genes in the stickleback genome. Each value is based on pairwise comparisons between all 56 haplotypes in the “adaptive radiation dataset” containing one individual per population from the Haida Gwaii adaptive radiation, two mainland freshwater, and a marine site (see materials and methods). (b) Rooted phylogenetic tree of SWS2 opsins in stickleback and related taxa, which have retained two ancient SWS2 paralogs. Both red-shifted (“sweep”) and blue-shifted threespine stickleback haplotypes are derived from the ancestral SWS2A paralog, and the haplotype under selection has rapidly accumulated amino acid changes (dN/dS = 1.64). Branches are color coded with maximum-likelihood dN/dS estimates, and node labels show Bayesian branch credibility. (c) Genetic variation at SWS2, LWS, and SWS1, each summarized in single multidimensional scaling (MDS) coordinates, is associated with light transmission at 400nm (T400), but only variation at the sweep haplotype carrying coding and noncoding SWS2 and noncoding LWS variation is associated with the colonization of blackwater habitats. The grey area indicates blackwater lakes, following [16]. (d) Gene conversion did not contribute to molecular convergence between stickleback haplotypes and SWS2 paralogs in other fish: synonymous divergence (dS) between the two threespine stickleback haplotypes is larger to the SWS2B paralog than to the SWS2A paralog, except for a region around bp 700–800, a region without intraspecific amino acid variation. Mean pairwise dN/dS values for all genes shown in (a), MDS values for the three opsins SWS1, SWS2, and LWS in (c), and dS values in (d) can be found in S1 Data.
Thanks to earlier mutagenesis and protein structure studies, we can predict functional consequences for four SWS2, one LWS, and two RH2 amino acid polymorphisms. Three SWS2 polymorphisms are at opsin key sites 96, 97, and 109 and cause shifts in the peak light absorption of SWS2 [53] and the rod opsin RH1 [54]. Notably, the sweep haplotype reference alleles at SWS2-specific key sites, S97 and A109, lead to a “red-shift” in the absorption spectrum, i.e., a peak absorbance at a longer wavelength, while the alternate haplotype alleles at sites C97 and G109, lead to a “blue-shift,” a shorter wavelength absorption maximum at SWS2 [53]. Furthermore, these two key sites and three other polymorphic amino acids in SWS2 (site 40) and RH2 (sites 179 and 203) face the “retinal binding pocket” of the opsin proteins, in which a functional effect is likely [55]. In SWS1 and LWS, no polymorphism falls into a key or retinal binding pocket site, but the single LWS substitution (A217T) replaces a hydrophobic with a hydrophilic residue and may thus have functional consequences. While this polymorphism is not associated with the sweep haplotype in the adaptive radiation dataset (Fig 5), it has increased in frequency linked with the blue-shifted SWS2 haplotype in the selection experiment alongside noncoding SNPs around HCFC1A (Figs 4 and 5).
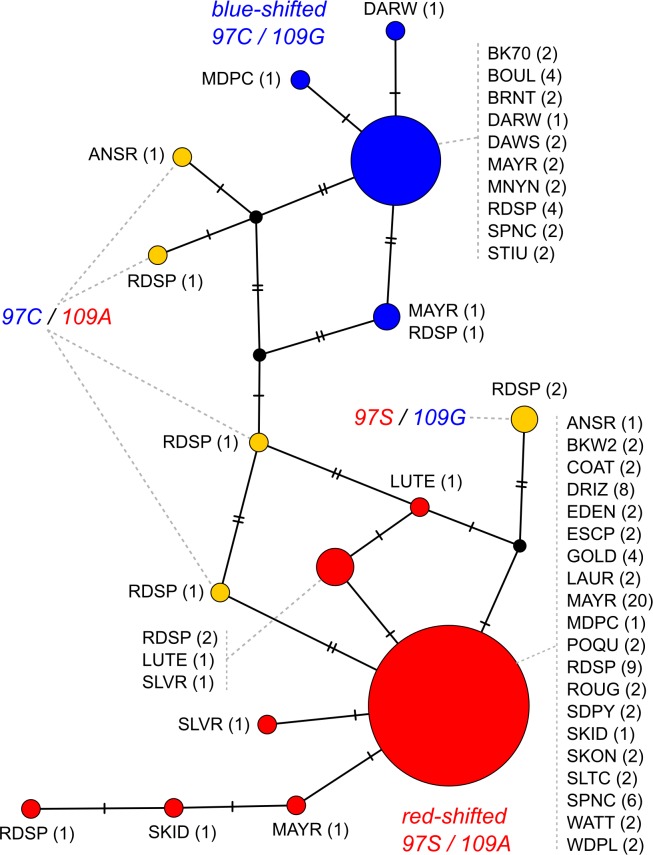
Population genomic patterns and functional predictions suggest that the amino acid polymorphisms in SWS2 are the most likely target of the selective sweep among Haida Gwaii stickleback and again in the selection experiment, while noncoding variation in both opsins and the two linked genes, the latter lacking coding variation, may have hitchhiked on the selected haplotype. Notably, the same haplotype with the same red-shifted SWS2 key sites is found across the Haida Gwaii adaptive radiation: nearly all populations show identical SWS2 coding sequence haplotypes, either the same blue-shifted or red-shifted haplotype (Fig 7). Selection thus favored the same red-shifted SWS2 cone opsin sequence across multiple Haida Gwaii populations and the alternate and widespread blue-shifted SWS2 haplotype in the selection experiment (Figs 2, 5 and 7).
Fig 7. Median-joining network of SWS2 coding sequence haplotypes.
Virtually all populations across the Haida Gwaii adaptive radiation recruited the same red- or blue-shifted SWS2 allele. Population codes are shown in Table 1, numbers in brackets indicate the number of haplotypes among all 58 sequenced individuals, and the color code indicates haplotypes predicted to be red- or blue-shifted or intermediate, based on the two SWS2 opsin key sites at position 97 and 109.
Blackwater colonization associated with coding SWS2 and noncoding SWS2/LWS variation
We asked whether the selective sweep for a red-shifted SWS2 cone opsin was associated with the colonization of blackwater. For this, we tested for an association between genetic variation at the four cone opsins and three predictors and covariates of visual environment: light transmission at 400 nm (T400), lake area, and lake depth. Light transmission at 400 nm is a predictor of light intensity and spectrum, with lower transmission indicating a red-shifted light spectrum in Haida Gwaii lakes [18]. Lake area is a strong predictor of between lake differences in predation regime and might capture the interaction of visual environment and predation landscape influencing selection on color vision [16]. The third covariate, lake depth, is a predictor for variation in light spectra found within a single lake, with more divergent light spectra and thus potential for disruptive selection on color vision in deeper lakes [16, 18]. Genetic variation at cone opsins SWS2, SWS1, and LWS was significantly associated with T400 but not with lake depth or area (Table 2, Fig 6C). When we tested more specifically for an association with blackwater (T400 ≤ 74%), we found only genetic variation at SWS2 and LWS—sweep haplotype variation—to be significantly associated with blackwater (Table 2, Fig 6C). Correlation with blackwater was strongest for four coding and seven noncoding SNPs in SWS2, one noncoding SNP also being an upstream regulatory SNP for LWS (single SNP χ2, p < 0.01). In addition, 28 noncoding SNPs around the UV-sensitive SWS1 opsin showed such a strong correlation, likely due to a high frequency of certain haplotypes in clearwater habitat (Fig 6C). In conclusion, the selective sweep on a single haplotype carrying a red-shifted SWS2 cone opsin coding sequence and linked noncoding variation in SWS2 and LWS may be associated with successful repeated colonization of blackwater habitat across the Haida Gwaii adaptive radiation.
Table 2. General linear model results for genotype–environment association tests.
| T400 | Depth | Area | |||||||
| Gene | β | t4,50 | p | β | t4,50 | p | β | t4,50 | p |
| SWS1 | 0.16 | 4.09 | <0.01 | −0.02 | −0.44 | 0.66 | 0.15 | 0.43 | 0.67 |
| RH2 | −0.02 | −0.41 | 0.68 | 0.05 | 0.81 | 0.42 | −1.26 | −2.75 | 0.01 |
| SWS2 | -0.10 | −3.36 | <0.01 | 0.07 | 1.72 | 0.09 | 0.16 | 0.60 | 0.55 |
| LWS | −0.06 | −3.39 | <0.01 | 0.04 | 1.97 | 0.06 | 0.00 | 0.02 | 0.99 |
| Clear- versus Blackwater | Depth | Area | |||||||
| Gene | β | t4,50 | p | β | t4,50 | p | β | t4,50 | p |
| SWS1 | 2.44 | 2.46 | 0.02 | 0.03 | 0.52 | 0.60 | −0.08 | −0.22 | 0.83 |
| RH2 | −0.81 | −0.68 | 0.50 | 0.06 | 0.91 | 0.37 | −1.26 | −2.82 | 0.01 |
| SWS2 | −2.91 | −4.37 | <0.01 | 0.06 | 1.79 | 0.08 | 0.23 | 0.92 | 0.36 |
| LWS | −1.33 | −3.50 | <0.01 | 0.03 | 1.70 | 0.10 | 0.05 | 0.38 | 0.71 |
The upper half of the table shows test results with light spectrum as a continuous variable, the lower half shows test results with light spectrum as a categorical variable: clearwater versus blackwater (T400 ≤ 74%). Bold numbers indicate significant tests after Bonferroni-correction for multiple testing.
The selection experiment supports a role of the sweep haplotype and associated coding variation in SWS2 during adaptation to different light regimes: The blackwater-associated SWS2 haplotype with the red-shifted SWS2 allele occurs at high frequency in the blackwater population, Mayer Lake, where a selective sweep signature is persistent (Figs 2–5). In contrast, the frequency of the alternate haplotype has rapidly increased after 13 generations in the clearwater habitat, with blue-shifting SWS2 key site substitutions rising from 13% to 40% frequency, with significant population differentiation, and with positive Tajima’s D centered on SWS2 (Figs 4 and 5). Shifts in allele frequency at SWS2 key site substitutions and linked regulatory sites ranks them among the top 5% and 1%, respectively, for allele-frequency change across the genome in the selection experiment (Fig 4), making the blue-shifted SWS2 haplotype a genome-wide outlier and thus a likely target of reversed selection due to habitat shift. Under a pure selection model, an allele-frequency shift of 27% over approximately 13 generations would correspond to an evolutionary change of 1.12 haldanes and to a selection coefficient of 0.28 (see materials and methods). The experimental transfer of a blackwater population to a clearwater habitat thus lead to evolution in the expected direction, given the habitat association across the adaptive radiation: a change from red- to blue-shifted SWS2 allele after the transfer into clearwater habitat.
Evolutionary origin of the red-shifted SWS2 haplotype in stickleback
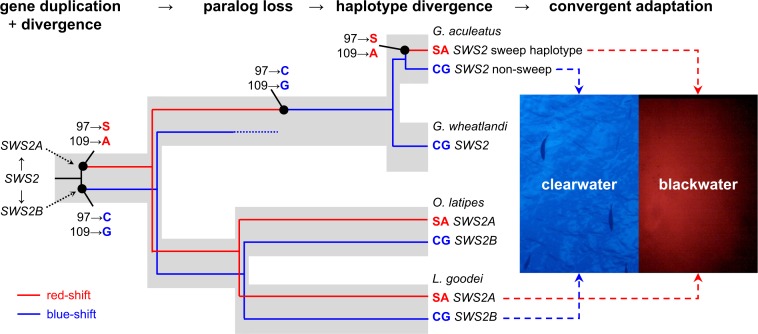
The blue light–sensitive SWS2 gene has undergone two duplication and divergence cycles [47], of which the first, in the ancestor of spiny-rayed fish, has led to a blue-shifted SWS2B paralog and a red-shifted SWS2A paralog [22, 26, 53, 56–58]. The single SWS2 gene copy of threespine stickleback is derived from an SWS2A paralog, while the other paralogs have been lost in the stickleback lineage [22, 47]. Strikingly, the two SWS2 key site amino acid polymorphisms found in our study are also key sites that have led to red- and blue-shifts in the SWS2A and SWS2B paralogs, respectively (Fig 6B, [47, 53]): at these two key sites, the sweep haplotype in threespine stickleback shows the same amino acids as the ancestral red-shifted SWS2A paralog, and the alternate haplotype shows the same amino acids as the blue-shifted SWS2B paralog lost in the stickleback lineage (Fig 8). Also, at three of the remaining five SWS2 substitutions segregating among threespine stickleback (sites 33, 150 and 169), the amino acids on the sweep haplotype are the same as in SWS2A paralogs of medaka (Oryzias latipes) and bluefin killifish (Lucania goodei), while some amino acids on the nonsweep haplotype are identical to their SWS2B paralogs.
Fig 8. Convergent evolution of the blue-sensitive SWS2 opsin at the molecular, functional, and ecological level.
The duplication of SWS2 in the ancestor of most spiny-rayed fish 198 million years ago was followed by a red-shift in SWS2A and a blue-shift in SWS2B [22, 47], paralogs that are divergently expressed among bluefin killifish (L. goodei) living in blackwater and clearwater habitats [24]. Two key amino acid polymorphisms of the ancient paralogs causing shifts in their absorption spectra have reevolved within threespine stickleback and are now divergently selected between blackwater and clearwater habitats in Haida Gwaii—convergent evolution at the molecular, functional, and ecological level. Clearwater spectra (left photo) are blue-shifted with increasing depth, typical of marine habitats and oligotrophic clearwater lakes on Haida Gwaii [18]. Tannin-stained blackwater (right photo) absorbs almost all up- and sidewelling light, making it a nearly nocturnal habitat, except for red-shifted downwelling light visible in a small cone above the observer. Unedited photos taken with a GoPro Hero 4 (GoPro Inc.) pointed towards the zenith at approximately 20 m depth in clearwater (Palau) and at 4 m depth in blackwater (Drizzle Lake).
We tested whether the similarity of the red- and blue-shifted stickleback SWS2 haplotypes with each of the ancestral paralogs was due to shared ancestry, gene conversion, or convergent evolution. First, we reconstructed the evolutionary origin of the two threespine stickleback SWS2 haplotypes to ask whether both threespine stickleback haplotypes are derived from an SWS2A ancestor. For this, we embedded the two most common SWS2 threespine stickleback haplotypes (nsweep = 74, nnon-sweep = 23 of 116 haplotypes from all 58 sequenced individuals, Fig 7) into a phylogeny with an orthologous SWS2 sequence from blackspotted stickleback (Gasterosteus wheatlandi) and both SWS2A and SWS2B paralogs from shorthorn sculpin (Myoxocephalus scorpius), medaka, and bluefin killifish. We chose these taxa because their SWS2 paralogs have not been excessively affected by gene conversion [47]. The phylogeny confirmed that both threespine stickleback SWS2 haplotypes and blackspotted stickleback SWS2 were derived from an SWS2A paralog, in line with findings of earlier studies on the origin of the stickleback SWS2 [22, 47], ruling out shared ancestry as an explanation for the similarity between SWS2 paralogs and stickleback haplotypes (Fig 6B).
Second, we asked whether gene conversion between SWS2A and SWS2B paralogs in the lineage leading to threespine stickleback may have contributed to the similarity between SWS2 paralogs and the threespine stickleback SWS2 haplotypes. For this, we computed divergence at synonymous sites (dS) between the threespine stickleback haplotypes and both paralogs of the shorthorn sculpin in sliding windows across the gene (Fig 6D). The divergence distribution shows that the SWS2B paralog is more divergent from both stickleback haplotypes than the SWS2A paralog almost throughout the whole gene, inconsistent with ancestral gene conversion. A single region of reduced synonymous divergence in the last third of the protein suggests ancient gene conversion but does not overlap with the amino acid polymorphisms among Haida Gwaii stickleback. This gene conversion signal has been ascribed to the shorthorn sculpin lineage and not the threespine stickleback lineage in a larger phylogenetic analysis [47]. Ancestral gene conversion thus cannot explain key site similarity between segregating stickleback haplotypes and divergent ancestral paralogs. Instead, new, recurrent mutations in threespine stickleback must have led to convergent amino acid changes with the ancestral paralogs.
We tested whether the SWS2 sweep haplotype in threespine stickleback has accumulated amino acid–changing mutations at an extraordinarily rapid rate (“positive selection”). Branch-specific dN/dS estimates in the SWS2 phylogeny show an elevated dN/dS ratio of 1.64 on the branch leading to the selective sweep haplotype (Fig 6B), indicative of accelerated amino acid substitution and positive selection. A branch-site model test for positive selection on protein sequence, however, could not distinguish between positive selection on this branch and a null model without positive selection (ΔLRT = 0.65, p = 0.13). As the terminal threespine stickleback branches contain only a few substitutions (N * dN = 5.2 and S * dS = 1.0 on the sweep haplotype branch and N * dN = 1.9 and S * dS = 1.0 on the nonsweep haplotype branch), the low divergence of the two haplotypes has likely limited our power to distinguish the two models using the branch-site test, which is most powerful for divergent sequences from interspecific comparisons [59, 60].
Discussion
Our study reveals that threespine stickleback have adapted wavelength sensitivity through selection at the SWS2 locus. A single red-shifted SWS2 allele has been favored across the adaptive radiation and blackwater lakes, most of which were colonized in replicate from marine ancestors and are almost exclusively inhabited by individuals with this red-shifted SWS2 allele. The evolution of a red-shifted SWS2 opsin thus likely facilitated the colonization of blackwater lakes and subsequent establishment in this extreme habitat. Tannin-stained blackwater is characterized by a red-shifted light spectrum with reduced transmission of short wavelengths [18]. A blue-sensitive opsin spectrally tuned to a longer wavelength will thus increase an individuals’ ability to detect any residual short wavelength light in blackwater. Such an adaptation mechanism is supported by genomic signatures of selection, by functional effect predictions and by genotype-environment associations across the adaptive radiation and in the selection experiment. Also, previously observed phenotypic differences [18] support this mechanism: cones expressing SWS2 in blackwater stickleback from some of these and other populations had an absorption spectrum red-shifted by ~10 nm [18], a stronger shift than is explainable by alternate chromophore use. The combined results from our study and Flamarique et al. [18] thus show that threespine stickleback adapted visual perception to blackwater habitats by spectral tuning of SWS2 key sites, causing a higher sensitivity to the remaining short wavelength light, and increased expression of LWS in double cones, maximizing the sensitivity to background light.
Remarkably, the molecular mechanism underlying this recent adaptation in threespine stickleback recapitulates the duplication and divergence of SWS2 around 198 million years ago in the spiny-rayed fish ancestor [22, 53]. Amino acid replacements at SWS2 key sites are identical and thus convergent between the threespine stickleback SWS2 alleles and the SWS2A and SWS2B paralogs, respectively [18, 47, 53]. Such convergent spectral tuning at key sites of cone opsins has been found previously but exclusively at larger evolutionary timescales, such as between damselfish species [40], between butterflies and vertebrates [41], butterflies and bees [42], humans and poeciliid fish [31], or across the animal kingdom [43]. Convergent spectral tuning between such vastly different time scales—on one side, a microevolutionary, intraspecific level and on the other side, a 198 million-y-old duplication-divergence process—has not yet been shown to our knowledge.
Not only the mechanism of spectral tuning at SWS2 is convergent, but also the environmental context: two other fish species inhabiting both tannin-stained blackwater and clearwater habitats, bluefin killifish and black bream Acanthopagrus butcheri, show expression divergence between the red-shifted SWS2A and blue-shifted SWS2B paralogs [24, 26, 37, 61]. Bluefin killifish populations occur either in blackwater or clearwater habitats [24, 37], and black bream live in clearwater as juveniles and migrate to blackwater habitats where they spend their adult life [26, 61]. In both species, the red-shifted SWS2A cone abundance is higher in blackwater habitats and the blue-shifted SWS2B cone abundance is higher in clearwater habitats, which is due to reduced SWS2B expression and an increased SWS2A expression relative to SWS2B, respectively [24, 61]. While the two key amino acids at sites 97 and 109 each are convergent between bluefin killifish paralogs and stickleback alleles living in either blackwater or clearwater (Fig 8, [37, 47]), black bream has substituted these with other amino acids but still shows the same function for the two paralogs (red- and blue-shift) [26] and thus ecological, phenotypic, and functional convergence.
Adaptation to blackwater environment via SWS2 spectral tuning and therein improved visual capacities can have a multitude of consequences for survival and reproduction. The light environment in blackwater lakes is limited to downwelling, red-shifted light and thus visual detection of predators and prey is much reduced, leading to short action and reaction distances. Any improved detection of prey or predators, for example, via increased sensitivity to color contrast at residual short wavelengths, would be favored by natural selection. Bluefin killifish from blackwater environments indeed showed increased color contrast attention towards blue objects in blackwater [62]. Increased color contrast attention might also be favored by sexual selection: the “blue morph” in bluefin killifish is more abundant in blackwater, and blue males are preferred by individuals raised in stained water [63, 64]. Similarly, stickleback inhabiting blackwater systems have lost red nuptial throat color and instead show black throats, contrasting with the background and with blue eyes, and these two traits are under sexual selection by choosy females [17, 19]. Spectral tuning of blue-sensitive SWS2 may thus be under both natural and sexual selection in threespine stickleback and other blackwater-adapted fish species.
Our selection experiment confirmed that the segregating SWS2 alleles are favored by selection in different light environments: after only 13 generations in a clearwater habitat, the red-shifted SWS2 allele associated with the blackwater sweep haplotype decreased in frequency while the alternate blue-shifted allele swept to high frequency, indicating a reverse, ongoing sweep in clearwater habitats. The direction of change is in line with both functional predictions and genotype–environment association across the adaptive radiation. The evolutionary change of 1.12 haldanes estimated from this allele-frequency shift is much larger than the change in feeding morphology or predator defense morphology traits, which show a mean change of 0.22 haldanes over 12 generations [48]. This could arise from inherent differences between genotype- and phenotype-based estimates, such as the increased variation due to a complex genetic basis and environmental effects in phenotypic estimates [65]. Change in allele frequency at SWS2 by 27% is comparable to the strongest relative changes in trait means: gill raker length was reduced by 43%, lateral plate three height by 18%, and lateral plate two frequency and dorsal spine length by 16% [48]. Selection on color vision thus led to similarly rapid or slightly faster evolutionary change compared to other, feeding and predator defense–related traits; and genetic and phenotypic change was in the direction predicted from independently evolved populations across the adaptive radiation, recapitulating the same habitat contrast.
Repeated use of the same red-shifted SWS2 haplotype during replicated adaptation to blackwater lakes in Haida Gwaii suggest that these adaptive mutations have been present as standing genetic variation in the marine population prior to colonization. Indeed, the marine individual in our dataset is heterozygous for all SWS2 amino acid polymorphisms, confirming the presence of both red-shifted sweep and blue-shifted nonsweep haplotypes in a marine population (Fig 2). Also, freshwater populations outside Haida Gwaii, such as one individual from mainland British Columbia in our study (Table 1, Figs 5 and 7) and the reference genome, a female freshwater stickleback from Alaska, carry the same red-shifted SWS2 haplotype, while another mainland individual carries the blue-shifted SWS2 haplotype. Maintenance of two spectrally tuned opsin alleles as standing genetic variation might be a “microevolutionary rescue” solution to the loss of multiple functionally divergent SWS2 paralogs in the lineage leading to threespine stickleback, which could explain convergent evolution with the ancestral SWS2 paralogs. To maintain such divergent SWS2 alleles, more complex selection scenarios than selection in blackwater might be necessary, including disruptive or fluctuating selection or selection for a red-shifted allele in other freshwater habitats than blackwater lakes. Visual spectra in freshwater habitats rapidly change with depth, dissolved organic particles, and other biophysical properties, making more complex scenarios likely. Further study of cone opsin variation in additional freshwater and marine populations, taking ecological knowledge of visual environments into account, may provide better insight into the maintenance of variation and repeatability of adaptation to light spectra.
By combining population genomic data, functional genomic analysis and a selection experiment, we have uncovered the genetic mechanisms underlying repeated adaptation of color vision to divergent visual environments in threespine stickleback. This mechanism of adaptation is convergent at the molecular, functional, and ecological level with other fish species that have used 198 million-y-old paralogs to adapt to similar blackwater environments. Convergent evolution at the same gene happened thus at vastly different timescales, involving two mechanisms: repeated de novo mutation, leading to convergent amino acid changes, and the reuse of standing genetic variation for repeated adaptation in an adaptive radiation. Our study thus supports the emerging view that mechanisms underlying adaptive evolution are often highly repeatable and likely predictable [4] and that evolutionary tinkering with the same, constrained toolset can lead to convergent adaptation, both within species and between distantly related groups.
Materials and methods
Ethics statement
Stickleback collection followed guidelines for scientific fish collection in British Columbia, Canada, under Ministry of Environment permits SM09-51584 and SM10-62059. Collections in Naikoon Provincial Park and Drizzle Lake Ecological Reserve were carried out under park use permits: 103171, 103172, 104795, and 104796.
Sampling, sequencing, and variant calling
Among the more than 100 stickleback populations previously studied from the Haida Gwaii archipelago [16], a subset of 25 populations was chosen to comprise the full range of biophysical attributes of the freshwater habitats on Haida Gwaii, including water spectra, lake area, bathymetry, and predation regime. Stickleback from these 25 freshwater sites, two freshwater sites in coastal British Columbia [66], and one marine site were collected between 1993 and 2012, using minnow traps or recovery from salmon stomachs (marine sample, Table 1, for coordinates, see [16, 66]; coordinates BKW2: 53.375089°N, −130.177378°W). We selected one to four individuals per population for whole-genome resequencing. From the selection experiment [48], we chose 12 fish from the source population Mayer Lake and 11 from the population introduced into Roadside Pond (equivalent to “Mayer Pond” in [48]), the latter sampled in 2012, 19 y or approximately 13 generations after the release of 100 Mayer Lake fish, assuming a generation time of 1.5 y being intermediate between Mayer Lake (2 y generation time) and Roadside Pond (1 y generation time). In total, 58 individuals, 56 females and two males, were resequenced to 6x depth using paired-end Illumina reads as described in [6] at the Broad Institute.
We aligned reads to the Broad S1 reference [6] using BWA v0.5.9 [67] with parameters -q 5 -l 32 -k 2 -o 1 and recalibrated base qualities using the GATK v1.4 tools CountCovariates and TableRecalibration [68], with read group, quality score, cycle, and dinucleotide covariates in the recalibration model. This resulted in 2,992,040,331 aligned and recalibrated reads. Variants were called using GATK’s UnifiedGenotyper for each chromosome separately and all 58 individuals combined, with default parameters for SNP and indel calling, respectively. We removed variants with quality normalized by depth ≥ 2, read position rank sum test value ≥ −20, and allele-specific strand bias ≤ 200, using GATK’s VariantFiltration from the dataset. We recalibrated variants using the GATK’s VariantRecalibrator and ApplyRecalibration with a VQSR-LOD cutoff of 98.5%. Filtered and recalibrated variants were lifted over to an improved ordering of scaffolds in the reference stickleback genome [51] using Picard v2.2.1 (http://broadinstitute.github.io/picard). Also, we realigned base quality recalibrated reads to this improved reference using samtools v1.3 [69] and BWA v0.7.12 with the same alignment parameters as above, in order to enable read-backed phasing and read-based genotype likelihood–based analyses (see below), resulting in 2,935,821,595 aligned reads, which have been deposited on the NCBI short read archive under accession SRP100209.
We obtained a set of high-quality SNPs by removing all variants failing variant recalibration, variants with quality < 45 and with a mean depth > 9.51 (= average mean depth plus 1.5 times the interquartile range of the mean depth distribution), variants with less than four reads of each allele, variants with more than two alleles, and indels, using bcftools v1.3.1 [69]. This dataset was partitioned by chromosome, and males (individuals in populations Banks 70, Laurel) were removed from the sex chromosome XIX partition. SNPs were further split into an “adaptive radiation” and “selection experiment” SNPs partition. The “adaptive radiation” SNP partition contained one randomly picked individual from each of the 28 populations except the transplant population Roadside Pond (Table 1) in order to perform further analyses with equal sample size for all natural populations. The “selection experiment” SNP partition contained all 12 and 11 individuals from Mayer Lake and Roadside Pond, respectively. In both adaptive radiation and selection experiment SNP datasets, genotypes with less than four reads and sites with more than 50% missing genotypes were removed using vcftools v0.1.15 [70], resulting in 15.3% and 16.2% missing genotypes, respectively. Both the adaptive radiation and selection experiment SNP datasets were phased and missing genotypes imputed with the read-backed phasing algorithm implemented in SHAPEIT v2.r790 [71]. Phase-informative reads covered 9.3% of all heterozygote genotypes and 32.7% of all graph segments.
Population genomic analyses
We scanned the genomic regions containing the four cone opsins for signatures of selective sweeps by using variation across the whole genome to identify outlier regions. We computed two haplotype-based statistics, integrated haplotype score iHS and H12 [49, 50], for the phased adaptive radiation SNPs. These statistics have been developed to detect signatures of incomplete hard and soft selective sweeps, based on extended haplotype homozygosity around an allele under selection compared to its alternate allele (iHS, [49]) or based on the haplotype frequency spectrum expected under a selective sweep (H12, [50]). Applied to the adaptive radiation dataset, these statistics will capture selective sweeps shared by multiple members of the adaptive radiation. iHS for each SNP in the genome was computed in selscan v1.1.0b [72] with default parameters and standardized in 5% allele frequency bins. In addition, we calculated the percentage of absolute iHS values > 2 in nonoverlapping 10 kb windows with more than 10 iHS estimates [49]. H12 was computed in bins spanning 81 SNPs using scripts published alongside the definition of H12 [50]. We used an outlier approach to identify significant iHS and H12 regions. We identified the top 0.1% genome-wide outliers for SNP-iHS, window-iHS, and H12 in recombination rate bins (<0.5, 0.5–2, 2–3.5, 3.5–5, >5 cM/Mb) because of the sensitivity of these statistics to variation in recombination rate. Local recombination rates were estimated from the “FTC x LITC”-cross recombination map published in [51] with a cubic splines smoothing approach described in [73]. For a significant outlier region indicating a selective sweep centered on opsins SWS2 and LWS, we used the top H12 estimates to identify the haplotype under selection or “sweep haplotype.” We visualized haplotype structure around a selective sweep in both adaptive radiation and selection experiment datasets using the extended haplotype homozygosity (EHH) statistic [74] calculated in selscan with default parameters.
We further traced the evolution of the selective sweep region around SWS2 and LWS in the selection experiment. For this, we computed population differentiation (FST) between Mayer Lake and Roadside Pond as well as nucleotide diversity (π) and Tajima’s D (TD) in each population across the genome and linkage disequilibrium (r2) in the selective sweep region for the unphased selection experiment SNPs. We first estimated the folded two-dimensional site-frequency spectrum (2D-SFS) from genotype likelihoods at all sites, from aligned reads with mapping-quality ≥ 17 and bases with quality ≥ 17 using angsd v0.911 [75, 76]. Using this 2D-SFS, we computed π and TD in 10 kb nonoverlapping as well as 10 kb wide, 2 kb step sliding windows across the genome for each population using angsd [76]. Then we estimated population allele frequencies with angsd and used them with the 2D-SFS to compute FST in 10 kb nonoverlapping as well as 10 kb wide, 2 kb step sliding windows across the genome using realSFS from the angsd software package [76, 77]. As for iHS and H12, we identified the top 0.1% outliers among nonoverlapping windows, based on the genome-wide distribution of π and TD in recombination rate bins. We computed linkage disequilibrium (r2) across the selective sweep region for SNPs with minor allele frequency (MAF) ≥ 5% and maximum 20% missing data in both populations using vcftools.
We identified synonymous- and nonsynonymous variation in the coding sequence of the four cone opsins SWS1, SWS2, RH2, and LWS in both the adaptive radiation and selection experiment SNPs. For the adaptive radiation SNPs, we tested whether coding variation at cone opsins was associated with the sweep haplotype identified above by using both alleles at each nonsynonymous SNP and chi-square tests with Bonferroni-corrected p-values. We also estimated the mean ratio of pairwise sequence divergence at synonymous and nonsynonymous sites (mean pairwise dN/dS) for all pairs of haplotypes in the adaptive radiation dataset using PAML v4.8 [78] and following the approach by Yang and Nielsen [79]. This statistic measures the relative frequency of segregating amino acid polymorphism to silent mutations [80]. We assessed whether any of the four cone opsins showed an unusually high frequency of amino acid changes by computing the distribution of mean pairwise dN/dS for all functional amino acid–coding genes on assembled chromosomes in our dataset (n = 17,846 genes).
Genotype–environment association
We tested whether genetic variation at the four cone opsins was associated with variation in light spectrum, using three environmental proxies of light spectrum, percent light transmission at 400 nm (T400), lake depth in meters, and log-transformed lake area in square meters. We assigned SNPs to up- and downstream regulatory regions, introns, exons, and 3′/5′-untranslated regions of each cone opsin gene using SnpEff v4.2 [81] and combined all SNP alleles per gene into a single multidimensional scaling (MDS) coordinate in R v3.3.1 [82]. We used the MDS coordinate as a response variable in a general linear model with three predictor variables: T400, lake depth, and log-transformed lake area. To test more specifically for an association with blackwater environment, we repeated the general linear model analysis with a categorical light transmission variable “clearwater” for lakes with T400 > 74% and “blackwater” with T400 ≤ 74%, following [16]. Significance of effects was determined after Bonferroni-adjustment for multiple testing. We qualitatively assessed which SNPs are most strongly correlated with blackwater habitat from single-SNP chi-square tests for each gene-associated SNP.
For the selection experiment populations Mayer Lake and Roadside Pond, we estimated allele frequencies based on genotype likelihoods with angsd v0.911 [75] using raw-aligned reads of mapping quality > 17 for sites with quality > 17 and the GATK genotype likelihood model [68]. We computed allele-frequency changes for all variable sites in the genome and the empirical quantiles for absolute allele-frequency changes at SNPs surrounding SWS2 in Mayer Lake–based MAF bins of width 0.05. We also computed evolutionary change in haldanes at SWS2 key sites following equation 1 in [65], with raw allele frequency mean, standard deviations, and a generation time of 12.7 as input. Furthermore, we estimated the expected selection coefficient under a pure selection model, following equation 3.2 in [83], assuming incomplete dominance h = 0.5 and using a per generation allele-frequency change by dividing the observed allele-frequency change by 12.7 generations.
Evolutionary history of SWS2
We reconstructed the evolutionary history of the cone opsin associated with a selective sweep, SWS2, using a Bayesian phylogenetic approach implemented in MrBayes v3.2.6 [84] and the same evolutionary model and run parameters as in [47]. For phylogenetic reconstruction, we used the two most common threespine stickleback SWS2 haplotypes, one associated and the other not associated with the sweep haplotype; an SWS2 sequence of blackspotted stickleback ([85], SRA-accession DRR013347); and both SWS2A and SWS2B paralogs from shorthorn sculpin ([47], genbank accession KM978046.1), medaka ([56], genbank accessions AB223056.1, AB223057.1), and bluefin killifish ([53], genbank accessions AY296737.2, AY296736.1). To get the blackspotted stickleback SWS2 sequence, we aligned whole-genome sequence data with a subset of the threespine stickleback reference sequence spanning the SWS2 coding sequence and 1 kb sequence up- and downstream of the start and stop codon, respectively, using bowtie2 v2.2.3 [86] with parameters -N 1 and -L 20, called genotypes using the GATK v3.5 tool UnifiedGenotyper [68] with default parameters and genotype likelihood model “BOTH,” converted the variants to FASTA format using the GATK-tool FastaAlternateReferenceMaker, and reintroduced missing sites using bedtools v2.25.0 [87]. We then aligned threespine stickleback and blackspotted stickleback SWS2 with the SWS2A and SWS2B paralogs of the other species manually in BioEdit v7.2.5 [88] and clipped the alignment to codons present in all sequences. We created the same alignment of codons for all 116 phased threespine stickleback haplotypes in our dataset to compute a neighbor-joining network using standard parameters in PopART v1.7 (http://popart.otago.ac.nz).
We tested whether selection on the threespine stickleback SWS2 haplotype lead to a rapid accumulation of amino acid–changing mutations compared to synonymous mutations (“positive selection”) on this haplotype. We used the phylogeny from above, estimated branch-specific dN, dS, and dN/dS and performed a branch-site test for positive selection with a subset of the phylogeny containing only stickleback SWS2 and other species’ SWS2A paralogs [60, 89], using PAML v4.8 [78] and the threespine stickleback’s sweep-associated haplotype as the foreground branch. Following [47], we also tested for gene conversion between SWS2A and SWS2B paralogs in the threespine stickleback ancestor by calculating dS between threespine stickleback SWS2 haplotypes and shorthorn sculpin SWS2A and SWS2B paralogs in 30 bp sliding windows with 1 bp step size in DnaSP v.5.10.01 [90]. After excluding gene conversion, we assessed whether amino acid substitutions at opsin key sites in the stickleback SWS2 haplotypes paralleled the divergence of ancestral SWS2A and SWS2B paralogs. We also added a partial coding sequence of black bream SWS2A and SWS2B paralogs ([26], genbank accessions DQ354580.1, DQ354581.1) to assess potential molecular and functional convergence with two other species inhabiting both clear- and blackwater (black bream, bluefin killifish).
Supporting information
Both axes give positions along chromosome XVII and boxes on top and right of the figure indicate the position of genes.
(TIFF)
Acknowledgments
We thank Bruce Deagle, Craig B. Lowe, Shannon D. Brady, Jason Turner, Kerstin Lindblad-Toh, and the Broad Genomics Platforms for help with sequences and samples.
Abbreviations
- dN/dS
ratio of non-synonymous to synonymous substitutions
- EHH
extended haplotype homozygosity
- MAF
minor allele frequency
Data Availability
Aligned sequences have been deposited on the NCBI short read archive under accession SRP100209.
Funding Statement
National Research Council Canada http://www.nrc-cnrc.gc.ca (grant number NRC2354). This research was largely supported by an NSERC operating grant to TER. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. National Institute of Health https://www.nih.gov/ (grant number 3P50HG002568-09S1 ARRA).Received by DMK. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. National Institute of Health https://www.nih.gov/ (grant number 3P50HG002568). Received by DMK. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23(1):26–32. 10.1016/j.tree.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 2.Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- 3.Storz JF. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 2016;17(4):239–50. 10.1038/nrg.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323(5915):746–51. 10.1126/science.1158997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadeau NJ, Pardo-Diaz C, Whibley A, Supple MA, Saenko SV, Wallbank RW, et al. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature. 2016;534(7605):106–10. 10.1038/nature17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313(5783):101–4. 10.1126/science.1126121 [DOI] [PubMed] [Google Scholar]
- 8.Rosenblum EB, Hoekstra HE, Nachman MW. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 2004;58(8):1794–808. [DOI] [PubMed] [Google Scholar]
- 9.Nachman MW, Hoekstra HE, D'Agostino SL. The genetic basis of adaptive melanism in pocket mice. Proc Natl Acad Sci U S A. 2003;100(9):5268–73. 10.1073/pnas.0431157100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38(1):107–11. 10.1038/ng1700 [DOI] [PubMed] [Google Scholar]
- 11.Chan YF, Villarreal G, Marks M, Shapiro M, Jones F, Petrov D, et al. From trait to base pairs: Parallel evolution of pelvic reduction in three-spined sticklebacks occurs by repeated deletion of a tissue-specific pelvic enhancer at Pitx1. Mech Develop. 2009;126:S14–S5. [Google Scholar]
- 12.Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67(5):1235–50. 10.1111/evo.12081 [DOI] [PubMed] [Google Scholar]
- 13.Deagle BE, Jones FC, Absher DM, Kingsley DM, Reimchen TE. Phylogeography and adaptation genetics of stickleback from the Haida Gwaii archipelago revealed using genome-wide single nucleotide polymorphism genotyping. Mol Ecol. 2013;22(7):1917–32. 10.1111/mec.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moodie GEE, Reimchen TE. Glacial refugia, endemism, and stickleback populations of Queen Charlotte Islands, British-Columbia . Can Field Nat. 1976;90(4):471–4. [Google Scholar]
- 15.Oreilly P, Reimchen TE, Beech R, Strobeck C. Mitochondrial-DNA in Gasterosteus and pleistocene glacial refugium on the Queen-Charlotte-Islands, British-Columbia. Evolution. 1993;47(2):678–84. [DOI] [PubMed] [Google Scholar]
- 16.Reimchen TE, Bergstrom C, Nosil P. Natural selection and the adaptive radiation of Haida Gwaii stickleback. Evol Ecol Res. 2013;15(3):241–69. [Google Scholar]
- 17.Reimchen TE. Loss of nuptial color in threespine sticklebacks (Gasterosteus aculeatus). Evolution. 1989;43(2):450–60. [DOI] [PubMed] [Google Scholar]
- 18.Flamarique IN, Cheng CL, Bergstrom C, Reimchen TE. Pronounced heritable variation and limited phenotypic plasticity in visual pigments and opsin expression of threespine stickleback photoreceptors. J Exp Biol. 2013;216(Pt 4):656–67. 10.1242/jeb.078840 [DOI] [PubMed] [Google Scholar]
- 19.Flamarique IN, Bergstrom C, Cheng CL, Reimchen TE. Role of the iridescent eye in stickleback female mate choice. J Exp Biol. 2013;216(15):2806–12. [DOI] [PubMed] [Google Scholar]
- 20.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364(1531):2881–95. 10.1098/rstb.2009.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowmaker JK. Evolution of vertebrate visual pigments. Vision Res. 2008;48(20):2022–41. 10.1016/j.visres.2008.03.025 [DOI] [PubMed] [Google Scholar]
- 22.Rennison DJ, Owens GL, Taylor JS. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 2012;62(3):986–1008. 10.1016/j.ympev.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 23.Cummings ME, Partridge JC. Visual pigments and optical habitats of surfperch (Embiotocidae) in the California kelp forest. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2001;187(11):875–89. [DOI] [PubMed] [Google Scholar]
- 24.Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J. Population variation in opsin expression in the bluefin killifish, Lucania goodei: a real-time PCR study. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2004;190(2):147–54. 10.1007/s00359-003-0478-z [DOI] [PubMed] [Google Scholar]
- 25.Hofmann CM, O'Quin KE, Smith AR, Carleton KL. Plasticity of opsin gene expression in cichlids from Lake Malawi. Mol Ecol. 2010;19(10):2064–74. 10.1111/j.1365-294X.2010.04621.x [DOI] [PubMed] [Google Scholar]
- 26.Shand J, Davies WL, Thomas N, Balmer L, Cowing JA, Pointer M, et al. The influence of ontogeny and light environment on the expression of visual pigment opsins in the retina of the black bream, Acanthopagrus butcheri. J Exp Biol. 2008;211(Pt 9):1495–503. 10.1242/jeb.012047 [DOI] [PubMed] [Google Scholar]
- 27.Temple SE, Veldhoen KM, Phelan JT, Veldhoen NJ, Hawryshyn CW. Ontogenetic changes in photoreceptor opsin gene expression in coho salmon (Oncorhynchus kisutch, Walbaum). J Exp Biol. 2008;211(Pt 24):3879–88. 10.1242/jeb.020289 [DOI] [PubMed] [Google Scholar]
- 28.Cheng CL, Novales Flamarique I. Opsin expression: new mechanism for modulating colour vision. Nature. 2004;428(6980):279. [DOI] [PubMed] [Google Scholar]
- 29.Cheng CL, Flamarique IN. Chromatic organization of cone photoreceptors in the retina of rainbow trout: single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J Exp Biol. 2007;210(Pt 23):4123–35. 10.1242/jeb.009217 [DOI] [PubMed] [Google Scholar]
- 30.Jacobs GH. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision-A significant trend in the evolution of mammalian vision. Visual Neurosci. 2013;30(1–2):39–53. [DOI] [PubMed] [Google Scholar]
- 31.Ward MN, Churcher AM, Dick KJ, Laver CR, Owens GL, Polack MD, et al. The molecular basis of color vision in colorful fish: four long wave-sensitive (LWS) opsins in guppies (Poecilia reticulata) are defined by amino acid substitutions at key functional sites. BMC Evol Biol. 2008;8:210 10.1186/1471-2148-8-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M, Tripathi N, Henz SR, Lindholm AK, Weigel D, Breden F, et al. Opsin gene duplication and diversification in the guppy, a model for sexual selection. Proc Biol Sci. 2007;274(1606):33–42. 10.1098/rspb.2006.3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann CM, Carleton KL. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr Comp Biol. 2009;49(6):630–43. 10.1093/icb/icp079 [DOI] [PubMed] [Google Scholar]
- 34.Wang FY, Yan HY, Chen JS, Wang TY, Wang D. Adaptation of visual spectra and opsin genes in seabreams. Vision Res. 2009;49(14):1860–8. 10.1016/j.visres.2009.04.023 [DOI] [PubMed] [Google Scholar]
- 35.Larmuseau MH, Raeymaekers JA, Ruddick KG, Van Houdt JK, Volckaert FA. To see in different seas: spatial variation in the rhodopsin gene of the sand goby (Pomatoschistus minutus). Mol Ecol. 2009;18(20):4227–39. 10.1111/j.1365-294X.2009.04331.x [DOI] [PubMed] [Google Scholar]
- 36.Rennison DJ, Owens GL, Heckman N, Schluter D, Veen T. Rapid adaptive evolution of colour vision in the threespine stickleback radiation. Proc Biol Sci. 2016;283(1830):20160242 10.1098/rspb.2016.0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J. Genetic and environmental variation in the visual properties of bluefin killifish, Lucania goodei. J Evol Biol. 2005;18(3):516–23. 10.1111/j.1420-9101.2005.00886.x [DOI] [PubMed] [Google Scholar]
- 38.Weadick CJ, Loew ER, Rodd FH, Chang BS. Visual pigment molecular evolution in the Trinidadian pike cichlid (Crenicichla frenata): a less colorful world for neotropical cichlids? Mol Biol Evol. 2012;29(10):3045–60. 10.1093/molbev/mss115 [DOI] [PubMed] [Google Scholar]
- 39.Phillips GA, Carleton KL, Marshall NJ. Multiple genetic mechanisms contribute to visual sensitivity variation in the Labridae. Mol Biol Evol. 2016;33(1):201–15. 10.1093/molbev/msv213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann CM, Marshall NJ, Abdilleh K, Patel Z, Siebeck UE, Carleton KL. Opsin evolution in damselfish: convergence, reversal, and parallel evolution across tuning sites. J Mol Evol. 2012;75(3–4):79–91. 10.1007/s00239-012-9525-0 [DOI] [PubMed] [Google Scholar]
- 41.Briscoe AD. Functional diversification of lepidopteran opsins following gene duplication. Mol Biol Evol. 2001;18(12):2270–9. [DOI] [PubMed] [Google Scholar]
- 42.Briscoe AD. Homology modeling suggests a functional role for parallel amino acid substitutions between bee and butterfly red- and green-sensitive opsins. Mol Biol Evol. 2002;19(6):983–6. [DOI] [PubMed] [Google Scholar]
- 43.Chang BS, Crandall KA, Carulli JP, Hartl DL. Opsin phylogeny and evolution: a model for blue shifts in wavelength regulation. Mol Phylogenet Evol. 1995;4(1):31–43. 10.1006/mpev.1995.1004 [DOI] [PubMed] [Google Scholar]
- 44.Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–28. 10.1146/annurev-biophys-101209-104901 [DOI] [PubMed] [Google Scholar]
- 45.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19(4):385–419. [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama S, Radlwimmer FB. The "five-sites" rule and the evolution of red and green color vision in mammals. Mol Biol Evol. 1998;15(5):560–7. [DOI] [PubMed] [Google Scholar]
- 47.Cortesi F, Musilova Z, Stieb SM, Hart NS, Siebeck UE, Malmstrom M, et al. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc Natl Acad Sci U S A. 2015;112(5):1493–8. 10.1073/pnas.1417803112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leaver SD, Reimchen TE. Abrupt changes in defence and trophic morphology of the giant threespine stickleback (Gasterosteus sp.) following colonization of a vacant habitat. Biol J Linn Soc. 2012;107(3):494–509. [Google Scholar]
- 49.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 2015;11(2):e1005004 10.1371/journal.pgen.1005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, Miller CT. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3. 2015;5(7):1463–72. 10.1534/g3.115.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roesti M. Recombination in the threespine stickleback genome—patterns and consequences (vol 22, pg 3014, 2013). Mol Ecol. 2013;22(20):5270-. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama S, Takenaka N, Blow N. A novel spectral tuning in the short wavelength-sensitive (SWS1 and SWS2) pigments of bluefin killifish (Lucania goodei). Gene. 2007;396(1):196–202. 10.1016/j.gene.2007.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 2008;105(36):13480–5. 10.1073/pnas.0802426105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carleton KL, Spady TC, Cote RH. Rod and cone opsin families differ in spectral tuning domains but not signal transducing domains as judged by saturated evolutionary trace analysis. J Mol Evol. 2005;61(1):75–89. 10.1007/s00239-004-0289-z [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in medaka (Oryzias latipes). Gene. 2006;371(2):268–78. 10.1016/j.gene.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 57.Owens GL, Windsor DJ, Mui J, Taylor JS. A fish eye out of water: ten visual opsins in the four-eyed fish, Anableps anableps. PLoS ONE. 2009;4(6):e5970 10.1371/journal.pone.0005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parry JW, Carleton KL, Spady T, Carboo A, Hunt DM, Bowmaker JK. Mix and match color vision: tuning spectral sensitivity by differential opsin gene expression in Lake Malawi cichlids. Curr Biol. 2005;15(19):1734–9. 10.1016/j.cub.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 59.Gharib WH, Robinson-Rechavi M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 2013;30(7):1675–86. 10.1093/molbev/mst062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang ZH, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19(6):908–17. [DOI] [PubMed] [Google Scholar]
- 61.Shand J, Hart NS, Thomas N, Partridge JC. Developmental changes in the cone visual pigments of black bream Acanthopagrus butcheri. J Exp Biol. 2002;205(Pt 23):3661–7. [DOI] [PubMed] [Google Scholar]
- 62.Johnson AM, Stanis S, Fuller RC. Diurnal lighting patterns and habitat alter opsin expression and colour preferences in a killifish. Proc Biol Sci. 2013;280(1763):20130796 10.1098/rspb.2013.0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuller RC. Lighting environment predicts the relative abundance of male colour morphs in bluefin killifish (Lucania goodei) populations. Proc Biol Sci. 2002;269(1499):1457–65. 10.1098/rspb.2002.2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuller RC, Noa LA. Female mating preferences, lighting environment, and a test of the sensory bias hypothesis in the bluefin killifish. Anim Behav. 2010;80(1):23–35. [Google Scholar]
- 65.Hendry AP, Kinnison MT. Perspective: The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53(6):1637–53. [DOI] [PubMed] [Google Scholar]
- 66.Reimchen TE, Nosil P. Replicated ecological landscapes and the evolution of morphological diversity among Gasterosteus populations from an archipelago on the west coast of Canada. Can J Zool. 2006;84(5):643–54. [Google Scholar]
- 67.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93(4):687–96. 10.1016/j.ajhg.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szpiech ZA, Hernandez RD. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 2014;31(10):2824–7. 10.1093/molbev/msu211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, et al. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 2016;12(2):e1005887 10.1371/journal.pgen.1005887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–7. 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- 75.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15(1):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS ONE. 2012;7(7):e37558 10.1371/journal.pone.0037558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sanchez E, Albrechtsen A, et al. Quantifying population genetic differentiation from next-generation sequencing data. Genetics. 2013;195(3):979–92. 10.1534/genetics.113.154740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 79.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17(1):32–43. [DOI] [PubMed] [Google Scholar]
- 80.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4(12):e1000304 10.1371/journal.pgen.1000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. http://www.R-project.org/. [Google Scholar]
- 83.Gillespie JH. Population genetics: a concise guide. 2nd ed. Baltimore, Md: Johns Hopkins University Press; 2004. [Google Scholar]
- 84.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida K, Makino T, Yamaguchi K, Shigenobu S, Hasebe M, Kawata M, et al. Sex chromosome turnover contributes to genomic divergence between incipient stickleback species. PLoS Genet. 2014;10(3):e1004223 10.1371/journal.pgen.1004223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quinlan AR. BEDTools: The Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11 2 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf). 1999;41:95–8. [Google Scholar]
- 89.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–18. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 90.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Both axes give positions along chromosome XVII and boxes on top and right of the figure indicate the position of genes.
(TIFF)
Data Availability Statement
Aligned sequences have been deposited on the NCBI short read archive under accession SRP100209.