Abstract
High-dose clindamycin (CLDM) and benzylpenicillin (PCG) are the recommended chemotherapeutic remedies for toxic shock-like syndrome caused by group A streptococci. One reason for this is that it has been shown that CLDM suppresses the expression of some exoproteins, e.g., SpeB, SpeA, and streptolysin O (Slo). We analyzed the effects of antibiotics on the production of whole exoproteins by two-dimensional gel electrophoresis. Unexpectedly, we found that the levels of several exoproteins, Slo, NAD+-glycohydrolase (Nga), M protein, and Sic, were increased by CLDM treatment, although we also confirmed previous findings that the levels of various exoproteins, including SpeB, were decreased. The increases in exoprotein levels were also detected by using other protein synthesis inhibitor antibiotics: erythromycin, kanamycin, tetracycline, chloramphenicol, and linezolid. Peptidoglycan synthesis inhibitors (such as PCG, cefazolin, and imipenem), DNA replication inhibitors (such as gatifloxacin), and an RNA polymerase inhibitor (rifampin) did not have significant effects on exoprotein production. The combination of CLDM and PCG had no advantageous effects with regard to exoprotein production compared to the effect achieved with CLDM alone. We also analyzed the transcriptional levels of slo and nga by reverse transcription-PCR and found that this change was also detected at the transcriptional level. Furthermore, the phenomenon was seen not only in strains of the M1 serotype but also in strains of the other M serotypes. Our study suggests that the clinical effectiveness of CLDM might be due to the inhibition of the production of a limited number of exoproteins.
Streptococcus pyogenes is a gram-positive bacterium that infects the upper respiratory tract, including the tonsils and pharynx, and is responsible for postinfection diseases such as rheumatic fever and glomerulonephritis. In addition, S. pyogenes causes streptococcal toxic shock-like syndrome (TSLS) (5, 15, 17, 19, 23).
Management of TSLS requires aggressive antibiotic treatment, supportive therapies, and surgical procedures. S. pyogenes continues to be susceptible to penicillin and other β-lactam antibiotics, but clinical failures with penicillin have been reported (21). The present consensus regarding the use of antibiotic treatment for TSLS includes the use of a high dose of clindamycin (CLDM) together with penicillin. CLDM is a lincosamine derivative that acts by binding to the 50S subunit of the bacterial ribosome and inhibiting protein synthesis. It has been shown that the synthesis of several streptococcal exoproteins, including virulence factors, is inhibited by subinhibitory concentrations of CLDM (6, 16, 18, 20, 21). However, those studies examined only a limited number of exoproteins, and no comprehensive studies have analyzed the effects of antibiotics on the production of all exoproteins at the same time. Two-dimensional gel electrophoresis (2-DE) is a powerful method for the detection of proteins not only qualitatively but also quantitatively. We applied this technique to the analysis of exoprotein production by S. pyogenes cultured with the maximum concentrations of antibiotics, including CLDM and benzylpenicillin (PCG), that did not suppress bacterial growth, as determined by measurement of the absorbance.
MATERIALS AND METHODS
Bacteria.
The S. pyogenes M1 serotype strain 1529, M3 serotype strain 1268, M4 serotype strain 1266, M5 serotype strain 1547, and M12 serotype strain GG01 used in this study were all clinical isolates from hospitalized patients with S. pyogenes infections in Japan. The bacteria were cultured in brain heart infusion broth (BHI; Eiken Chemical Co., Tokyo, Japan) containing 0.3% yeast extract (Difco Laboratories, Detroit, Mich.) for 7 to 8 h for early-stationary-phase analysis (optical density [OD], approximately 1.0) and 18 h for late-stationary-phase analysis (OD, approximately 1.0) at 37°C without agitation. The culture volumes were 3 ml for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and 25 ml for 2-DE analysis.
Antibiotics.
The following antibiotics were used in this study: clindamycin hydrochloride (Sigma Chemical Co., St. Louis, Mo.), linezolid (Pharmacia & Upjohn Co.), PCG potassium (Meiji Seika Co., Tokyo, Japan), kanamycin monosulfate (Meiji Seika Co.), cefazolin sodium hydrate (Fujisawa Pharmaceutical Co., Osaka, Japan), imipenem (Banyu Pharmaceutical Co., Tokyo, Japan), erythromycin (Shionogi Pharmaceutical Co., Osaka, Japan), gatifloxacin hydrate (Kyorin Pharmaceutical Co., Osaka, Japan), tetracycline (Sigma Chemical Co.), chloramphenicol (Sigma Chemical Co.), and rifampin (Sigma Chemical Co.). All antibiotics were added to the medium at the same time when the bacterial culture was started.
Determination of antibiotic concentrations for exoprotein analysis.
Bacteria were cultured in BHI that contained various concentrations of antibiotics. The maximum antibiotic concentration that did not suppress bacterial growth, determined from the absorbance of the culture at 660 nm measured with a colorimeter (Asahi Science Co., Tokyo, Japan), was determined; and this concentration was used for the study of exoprotein production.
One-dimensional SDS-PAGE analysis of exoproteins and cellular proteins.
S. pyogenes was cultured in 3 ml of BHI. After 18 h of culture, with or without antibiotic at the different concentrations, when the bacteria were in the late stationary phase of growth, 1 ml of the 3-ml culture was centrifuged and the supernatant was precipitated with trichloroacetic acid (final concentration, 10%). After an acetone wash, the precipitate was dissolved in 50 μl of SDS-PAGE buffer. In the case of the bacterial cellular protein, the centrifuged bacterial cells from 1 ml of culture were directly dissolved in 50 μl of SDS-PAGE buffer. Twenty microliters each of the 50-μl supernatant and the cellular protein samples were electrophoresed. The gels were stained with Coomassie brilliant blue.
2-DE analysis.
S. pyogenes was cultured in 25 ml of BHI. The length of culture was approximately 7 to 8 h for analysis of early-stationary-phase cells and 18 h for analysis of late-stationary-phase cells. Throughout the culture, the bacteria had been exposed to one or two kinds of antibiotics; the control, however, was not exposed to antibiotics. Exoproteins from the culture supernatant were prepared as described previously (10, 11). Briefly, all sample pellets derived from 25 ml of the bacterial culture supernatant (approximately 500 to 1,500 μg) were dissolved in 300 μl of immobilized pH gradient dehydration solution (Amersham Biosciences Co., Piscataway, N.J.), which consisted of 7.8 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, 0.6% dithiothreitol, and 0.5% immobilized pH gradient buffer. Aliquots (250 μl) of the samples were loaded onto 13-cm Immobiline DryStrip gels (pH 3 to 10; Amersham Biosciences Co.). The first-dimensional electrophoresis conditions and second-dimensional SDS-PAGE separation were the same as those described previously (10, 11). The experiments were repeated at least three times to confirm their reproducibilities.
Peptide mass mapping (PMM) analysis.
Protein spots were excised from the gels and placed into tubes. The gel pieces were washed twice with a solution of 100 mM ammonium bicarbonate and destained with a solution of 100 mM ammonium bicarbonate and 30% acetonitrile. The destained gel pieces were dried by vacuum centrifugation. For carbamidmethyl modification, the dried gel pieces were rehydrated in a solution of 100 mM dithiothreitol and 100 mM ammonium bicarbonate for 30 min at 60°C. After removal of the solution, the gel pieces were then incubated in a solution of 100 mM iodoacetamide and 100 mM ammonium bicarbonate for 30 min at 37°C. The gel pieces were dried again by vacuum centrifugation. The dried gel pieces were rehydrated in 20 μl of trypsin digest solution (20 μg/ml of mass spectrometry grade Trypsin Gold [Promega Co., Madison, Wis.] in 100 mM ammonium bicarbonate). After removal of the excess trypsin solution, the gel pieces were incubated in 20 μl of 100 mM ammonium bicarbonate overnight at 37°C. Each of the peptide mixtures was extracted from gel pieces with an acetonitrile solution containing 0.1% (vol/vol) trifluoroacetic acid. Extracted solutions were dried by vacuum centrifugation and stored at 4°C before mass analysis.
Nanoelectrospray tandem mass analysis was performed with an LCQ Advantage nanospray ionization ion trap mass spectrometer (Thermo Finnigan Co., San Jose, Calif.) combined with a MAGIC2002 high-pressure liquid chromatography (HPLC) system (Michrom BioResources Inc., Auburn, Calif.). The peptide mixture sample was injected onto the MAGIC2002 HPLC system, which was equipped with a C18 column (length, 50 mm; 5 μm; particle size, 200 Å; Michrom BioResources Inc.). Ionization was performed with a capillary voltage of 2,500 V. Precursor ion scanning was carried out with a mass-to-charge ratio (m/z) of 300 to 2,000 prior to tandem mass spectrometry (MS/MS) analysis. Multiple MS/MS spectrum data were collected with the program TurboSEQUEST (Thermo Finnigan Co.); merged as SEQUEST format files; and submitted to the program Mascot Search, available on the Matrix Science website (http://www.matrixscience.com/), for the MS/MS ion search. Identification of the proteins in the spots conformed to the description on the Matrix Science website.
Assessment of mRNA levels by semiquantitative RT-PCR.
For reverse transcription-PCR (RT-PCR), bacterial cells were cultured in 5 ml of BHI for 4 to 5 h and harvested when the OD was approximately 0.8 (late log phase). As in the protein analysis, the bacteria were exposed to antibiotics throughout the cultivation. Total RNA was prepared from 0.8 ml of the 5 ml of cultured bacterial cells with an RNA-protected Bacteria Reagent (Qiagen, Hilden, Germany), followed by treatment with RNase-free DNase (Qiagen). A total of 100 ng of total RNA was reverse transcribed in the presence of slo-, nga-, and gryA-specific primers with the SuperScript first-strand synthesis system, according to the instructions of the manufacturer (Invitrogen Co., Carlsbad, Calif.) in a 10-μl reaction volume. The primers used for amplification of the slo gene were Slo-EcoRI (5′-GGAATTCCTACTTATAAGTAATCGAAC-3′) and Slo-BamHI (5′-CGGATCCGAATCGAACAAACAAAACAC-3′). The primers used for amplification of the nga gene were Nga-BamHI (5′-CGGATCCGTTAGTGGCAAAGAAAATAA-3′) and Nga-XhoI (5′-CCTCGAGTTACTTGGTATCTTGCATTT-3′). The primers used for amplification of the gyrA gene were 5′-CGACTTGTCTGAACGCCAAA-3′ and 5′-TTATCACGTTCCAAACCAGTCAA-3′. A total of 0.4 μl of the 10-μl reverse transcriptase reaction mixture was used for the next PCR (reaction volume, 10 μl). Amplification and detection of specific products were performed with the following cycle profile: 25 cycles for slo, nga, and gyrA annealing at 55°C (nga and gryA) or 50°C (slo) and extension at 72°C for 90 s (slo and nga) or 20 s (gyrA). The amount of contaminating chromosomal DNA in each sample was determined in control reactions without reverse transcriptase. The quantity of cDNA used in the experiments for each gene was normalized to the quantity of gyrA cDNA in each sample. Triplicate assays were performed with RNA from at least two independent cultures.
RESULTS
Determination of antibiotic concentrations for 2-DE study.
We cultured S. pyogenes 1529 with several concentrations of antibiotics; and the maximum dose of antibiotic which did not inhibit the growth of the bacteria, as judged by measurement of the absorbance at 660 nm, was used for further study. The concentration of PCG and CLDM used both alone and together was 7.0 × 10−3 μg/ml. Neither synergistic nor additive effects were detected. The concentrations of antibiotics used for the study are listed in Table 1.
TABLE 1.
Antibiotic concentrations used for 2-DE
| Organism and antibiotic | Concn (μg/ml) |
|---|---|
| S. pyogenes 1529 (type M1) with antibiotics alone | |
| PCG | 1.75 × 10−3 |
| CLDM | 7.0 × 10−3 |
| CEZ | 3.2 × 10−2 |
| IPM | 9.8 × 10−4 |
| KM | 3.0 |
| TC | 3.0 × 10−2 |
| CP | 1.0 |
| EM | 1.5 × 10−2 |
| LZ | 2.5 × 10−1 |
| Gatifloxacin | 3.2 × 10−2 |
| Rifampin | 1.5 × 10−2 |
| S. pyogenes 1529 (type M1) with antibiotics in combination | |
| PCG | 7.0 × 10−3 |
| CLDM | 7.0 × 10−3 |
| S. pyogenes strains of other M types and CLDM | |
| Type M3 strain 1268 | 7.5 × 10−3 |
| Type M4 strain 1266 | 3.9 × 10−2 |
| Type M5 strain 1547 | 7.8 × 10−3 |
| Type M12 strain GG01 | 7.8 × 10−3 |
SDS-PAGE analysis of exoprotein production.
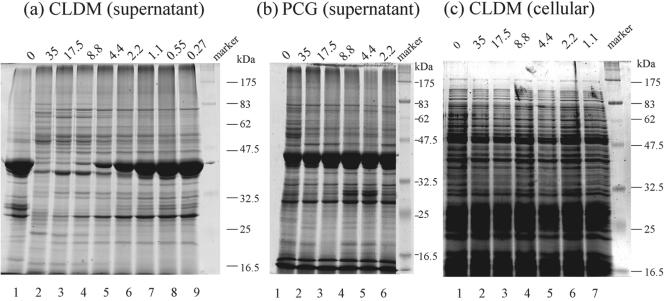
First, exoproteins from the culture supernatant and cellular proteins in the late stationary phase were analyzed by SDS-PAGE (Fig. 1). As shown in Fig. 1a, a dose-dependent decrease in the amount of a 40-kDa protein was detected with CLDM treatment. A CLDM dose of less than 1.1 × 10−4 μg/ml had almost no effect (compare lane 1 and lanes 7 to 9). On the other hand, PCG had no significant effect on the 40-kDa protein at any dose (Fig. 1b). Although we detected variations in the intensities of some protein bands other than the 40-kDa band, the results were not reproducible. No significant changes in the amounts of cellular (cytoplasmic and membrane-bound) proteins were detected by SDS-PAGE analysis with CLDM treatment (Fig. 1c).
FIG. 1.
Effects of CLDM and PCG on exoprotein production, as determined by SDS-PAGE analysis. S. pyogenes 1529 was incubated with CLDM (a and c) or PCG (b). (a and b) Results obtained with supernatant proteins from overnight cultures; (c) results obtained with cellular (cytoplasmic and membrane-bound) proteins from S. pyogenes 1529. The concentrations of antibiotics (10−4 μg/ml) used are given above each lane.
Effects of CLDM and PCG on exoprotein production analyzed by 2-DE.
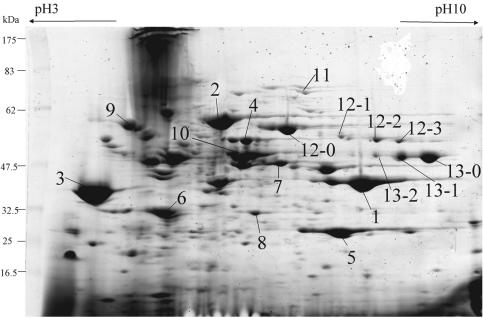
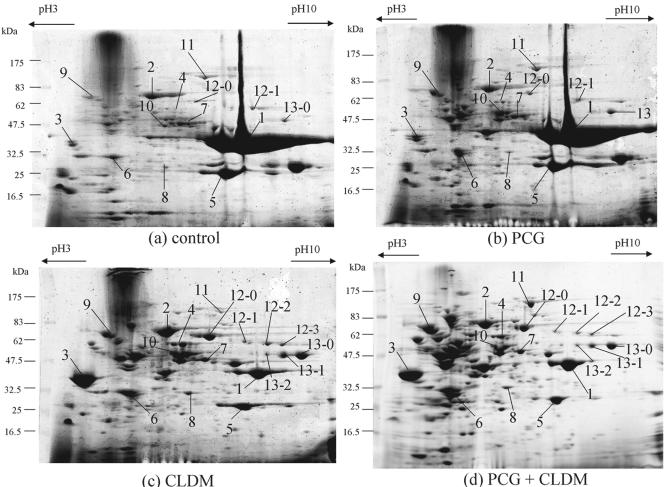
Exoproteins were then analyzed by 2-DE, because SDS-PAGE could not distinguish each exoprotein separately in detail. Antibiotics were added from the beginning of culture, and the bacteria were harvested after 18 h, when the bacteria were in the late stationary phase of growth. After separation by 2-DE, several protein spots were identified by PMM analysis. The profiles of the exoproteins and their corresponding names are shown in Fig. 2 and Table 2. In the late stationary phase, the exoprotein profile achieved by treatment with the maximum PCG dose (Fig. 3b) that did not suppress bacterial growth was almost the same as that of a control culture with no antibiotics, except for increases in the amounts of heat shock protein 70 (Fig. 3b, spot 9) and many acidic proteins, most of which were identified as intracellular proteins by PMM analysis (data not shown). It is possible that these proteins were released from the cells because of the breakdown of peptidoglycan by the inhibitory effect of PCG. CLDM treatment remarkably decreased the levels of SpeB (Fig. 3c, spot 1) and slightly decreased the levels of SpeF (Fig. 3c, spot 5); however, Sic levels were increased significantly (Fig. 3c, spot 3). The levels of streptolysin O (Slo) (Fig. 3c, spot 12), NAD+-glycohydrolase (Nga) (Fig. 3c, spot 13), M1 protein (Fig. 3c, spot 10), α-amylase (Fig. 3c, spot 2), Isp2 (Fig. 3c, spot 4), streptokinase A (Fig. 3c, spot 7), CAMP factor (Fig. 3c, spot 8), and heat shock protein 70 (Fig. 3c, spot 9) also increased. Several new protein spots (Fig. 3, spots 12-0, 12-1, 12-2, 12-3, 13-0, 13-1, and 13-2) appeared after treatment with CLDM and were also identified as Slo and Nga by PMM analysis. These spots seemed to be portions of the mature proteins probably digested by proteases or incompletely synthesized because of terminated translation caused by CLDM. Very similar results were obtained for the other M1 serotype strain, strain SF370, which was used for the genome project (7) (data not shown). Treatment with the combination of CLDM and PCG produced a result similar to that achieved with CLDM treatment alone; a difference was increases in the amounts of intercellular acidic proteins (Fig. 3d).
FIG. 2.
2-DE analysis of exoproteins from S. pyogenes 1529 identified by PMM after 2-DE separation. Bacteria were cultivated without antibiotics until the late stationary phase. The protein spots identified by PMM analysis were numbered. These numbers are same as those in Fig. 3 to 6 and Table 2. Each group of four protein spots numbered 12 and three protein spots numbered 13 was derived from one protein. Leftward arrow, acidic side; rightward arrow, basic side.
TABLE 2.
Identities and densities of protein spots after treatment with various antibioticsa
| Spot no. | Identical or homologous protein | Peptidoglycan synthesis inhibitor
|
Protein synthesis inhibitor
|
Other drugs
|
Combination of PCG and CLDM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCG | CEZ | IPM | KM | TC | CP | EM | CLDM | LZ | RFP | GFLX | |||
| 1 | SpeB (streptococcal pyrogenic exotoxin B) | − | − | − | DDD | DDD | D | D | DDD | DDD | − | − | DDD |
| 2 | α-Amylase | − | v | D | D | − | v | − | II | v | I | − | v |
| 3 | Sic (streptococcal inhibitor of complement) | − | − | − | III | III | III | III | III | III | − | − | III |
| 4 | Isp2 (immunogenic secreted protein2) | − | − | − | I | I | I | I | III | III | − | − | III |
| 5 | SpeF (streptococcal pyrogenic exotoxin F) | − | − | − | D | D | D | − | D | D | − | − | D |
| 6 | Mf3 (deoxyribonuclease) | − | − | I | − | I | − | I | III | − | v | − | II |
| 7 | Ska (streptokinase A) | − | − | − | DD | I | − | − | II | − | D | I | II |
| 8 | CAMP factor | − | − | − | − | D | − | I | I | I | I | − | I |
| 9 | Hsp70 (heat shock protein 70) | II | I | III | − | − | − | I | II | − | − | I | II |
| 10 | M1 protein | − | − | − | − | v | I | I | II | I | D | − | I |
| 11 | EndoS (secreted endoglycosidase) | − | v | I | DD | − | − | I | D | − | I | I | I |
| 12-0, -1, -2, -3 | Slo (streptolysion O) | − | − | − | I | III | II | III | III | III | I | − | III |
| 13-0, -1, -2 | Nga (NAD+-glycohydrolase) | I | v | − | − | II | − | II | III | II | I | II | III |
Abbreviations and symbols: I, proteins whose levels increased two to four times compared with those under the control condition; II, proteins whose levels increased four to eight times; III, proteins whose levels increased more than eight times; D, proteins whose levels decreased one-half to one-fourth compared with those under the control condition; DD, proteins whose levels decreased one-fourth to one-eighth; DDD, proteins whose levels decreased less than one-eighth; −, no significant change; v, variable changes; RFP, rifampin; GFLX, gatifloxacin.
FIG. 3.
2-DE analysis of the effects of PCG, CLDM, and the combination of PCG and CLDM on the production of exoproteins from bacteria harvested in the late stationary phase. S. pyogenes 1529 was cultured for 18 h with no drugs (a), PCG (b), CLDM (c), and both PCG and CLDM (d). The bacteria were exposed to antibiotics throughout the cultivation. The numbers of the protein spots are the same as those described in the legend to Fig. 2 and Table 2.
We then analyzed the effects of antibiotics on the exoproteins in bacteria in the early stationary phase by 2-DE (Fig. 4). In this experiment the antibiotics were also added from the beginning of culture, and the bacteria were harvested when the growth curve reached early stationary phase (OD, approximately 1.0). In principle, the amount of exoprotein produced should be less in this phase compared to the amount produced in the late stationary phase. With CLDM treatment, the decreases in SpeB (Fig. 4, spot 1) and SpeF (Fig. 4, spot 5) levels were not clearly detected. It is possible that production of SpeB and SpeF occurs mainly in the late stationary phase and that the difference caused by the inhibitory effect of CLDM was not observed then. However, CLDM already had an influence on the production of some exoproteins in the early stationary phase, so that the levels of Slo (Fig. 4b, spot 12) and Sic (Fig. 4b, spot 3) were observed to increase.
FIG. 4.
2-DE analysis of the effect of CLDM on the production of exoproteins from bacteria harvested in the early stationary phase. S. pyogenes 1529 was cultured until the early stationary phase with no drugs (a) or CLDM (b) and harvested. Antibiotics were added at the beginning of the culture, and the bacteria were exposed to antibiotics throughout the culture. The numbers of the protein spots are the same as those described in the legend to Fig. 2 and Table 2.
Effects of other antibiotics on exoprotein production.
Next we carried out similar experiments with other antibiotics to analyze whether the effect of increased levels of exoprotein production was CLDM specific. First, the effects of peptidoglycan synthesis inhibitors belonging to the same category as PCG were analyzed. We used cefazolin (CEZ), which is classified as a narrow-spectrum cephem and a carbapenem antibiotic, imipenem (IPM), which is used to treat serious infectious diseases. The effects of these antibiotics did not differ from that of PCG (data not shown).
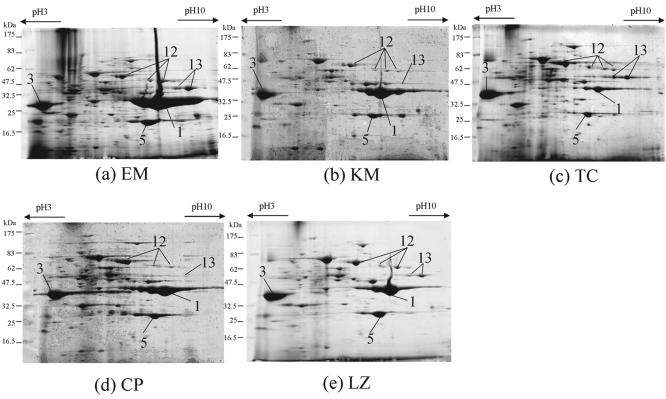
We then studied the effects of protein synthesis inhibitors belonging to the same category as CLDM (Fig. 5), as follows: erythromycin (EM) (Fig. 5a), classified as a macrolide that is very similar to CLDM; kanamycin (KM) (Fig. 5b), classified as an aminoglycoside; tetracycline (TC) (Fig. 5c); chloramphenicol (CP) (Fig. 5d); and linezolid (LZ) (Fig. 5e). Each antibiotic inhibited the translation of peptides from mRNA, but the target molecules differed. Reductions in the levels of SpeB (Fig. 5, spot 1) and SpeF (Fig. 5, spot 5), which were also detected with CLDM, were found with all antibiotics except EM (Fig. 5a); and the level of Sic (Fig. 5, spot 3) was increased by all protein synthesis inhibitors. The level of Slo (Fig. 5, spot 12) was increased by treatment with all drugs except KM (Fig. 5b), and the level of Nga (Fig. 5, spot 13) was increased by EM (Fig. 5a) and TC (Fig. 5c).
FIG. 5.
2-DE analysis of the effect of protein synthesis inhibitors on the production of exoproteins. S. pyogenes 1529 was cultured for 18 h. Bacteria were exposed to EM (a), KM (b), TC (c), CP (d), and LZ (e) throughout the cultivation. The numbers of the protein spots are the same as those described in the legend to Fig. 2 and Table 2.
We also studied the effects of rifampin, which is an RNA polymerase inhibitor, and gatifloxacin, which is classified as a fluoroquinolone. Both of these drugs are bacterial DNA replication inhibitors. These drugs had almost no effect on exoprotein production, similar to the effect of PCG and the control without antibiotics (data not shown).
A summary of the effects of the antibiotics on the production of exoproteins is shown in Table 2. Protein synthesis inhibitors had more of an influence on the production of exoproteins than the other classes of antibiotics did, although the effects on the increases or decreases on the levels of production of exoproteins were variable between each protein synthesis inhibitor.
Analysis of other M-type S. pyogenes strains that cause TSLS.
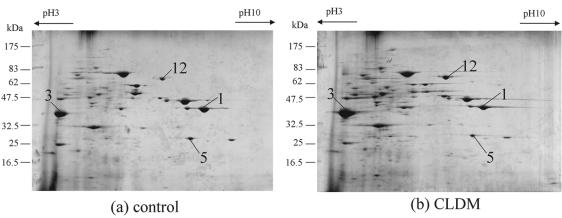
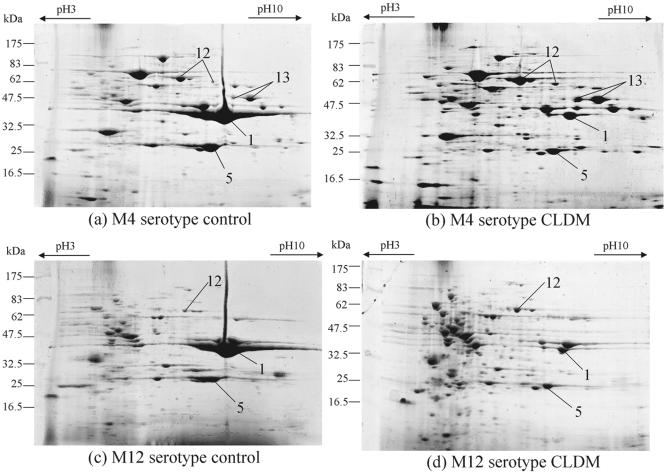
We also analyzed the effect of CLDM on the other M-type S. pyogenes strains that cause TSLS to determine whether the effect of CLDM on exoprotein production was specific to the M1 serotype (Fig. 6). Serotypes M3, M4, M5, and M12 of S. pyogenes were used for the study. We did not detect significant changes in exoprotein levels with the M3 or M5 strains (data not shown). Clear decreases in SpeB (Fig. 6, spot 1) and SpeF (Fig. 6, spot 5) levels and increases in Slo (Fig. 6, spot 12) and Nga (Fig. 6, spot 13) levels were detected for M4 and M12 strains, as was shown for serotype M1 strain 1529 (Fig. 3c, spots 12 and 13), suggesting that the effect of CLDM is not M-type specific.
FIG. 6.
2-DE analysis of the exoproteins of the clinically isolated S. pyogenes strains causing TSLS. (a) and (b) M4 serotype S. pyogenes strain 1266 without and with CLDM, respectively; (c) and (d) M12 serotype S. pyogenes strain GG01 without and with CLDM, respectively. The bacteria were cultivated for 18 h until the late stationary phase of growth. CLDM was added at the beginning of the culture. The numbers for the protein spots are the same as those described in the legend to Fig. 2 and Table 2.
Assessment of exoprotein transcription levels with CLDM treatment.
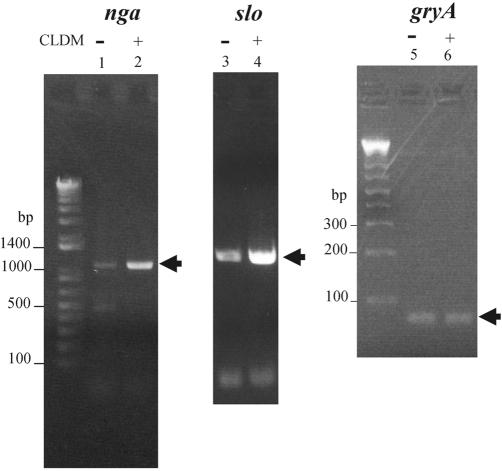
We used semiquantitative RT-PCR analysis of transcription with CLDM treatment to determine whether the levels of transcription of slo and nga were increased. To rule out the possibility that reverse-transcribed cDNA contained genomic DNA, we also used a negative control that was not reverse transcribed (data not shown). Judging from the data that CLDM already had an effect on exoprotein levels in the early stationary phase, we analyzed the RNA levels in late log phase. The bacteria were cultivated for 4 to 5 h with or without CLDM until the OD reached approximately 0.8, and total RNA was extracted. In agreement with the results of 2-DE protein analysis, slo and nga mRNA levels increased and the internal control gyrA mRNA level was unchanged (Fig. 7). The same results, i.e., increases in slo and nga mRNA levels, were obtained by Northern blotting analysis (data not shown). These results suggest that CLDM also affected the levels of transcription of several exoprotein genes.
FIG. 7.
RT-PCR analysis of the nga (lanes 1 and 2), slo (lanes 3 and 4), and gyrA (lanes 5 and 6) genes. Total RNA was isolated from late-log-phase cultures of S. pyogenes 1529. Lanes 1, 3, and 5, controls (no drugs); lanes 2, 4, and 6, with CLDM. gyrA was used as an internal control.
DISCUSSION
The number of clinical cases of TSLS caused by S. pyogenes and the other streptococci, including group C and G streptococci, has been increasing in Japan (12). The progression of this disease is very rapid, and the prognosis is still very poor, despite intensive medical treatment. The main therapeutic strategy is antibiotic treatment. Many kinds of antibiotics are available, but the recommended regimen is high-dose penicillin and high-dose CLDM together. The reason for using penicillin is its very strong bactericidal activity, and there have been no reports of the appearance of penicillin-resistant S. pyogenes to date. The reason for the use of CLDM is that it transfers to tissues well, and it has been reported that CLDM suppresses the expression of exotoxins in S. pyogenes (6, 16, 20-22). Although the suppressive effect of CLDM has also been reported in S. aureus (13), the number of proteins analyzed was small. As far as we know, a comprehensive study to determine the effect of antibiotics on the expression of whole exoproteins in S. pyogenes has never been carried out. We introduced the 2-DE method for this analysis. Our results support those of previous studies that showed that the levels of some exoproteins were decreased with CLDM treatment (6, 16, 20-22). Unexpectedly, we also found that the levels of some exoproteins were unchanged or even increased with CLDM treatment. These results were obtained not only with CLDM treatment but also with treatment with the other classes of protein synthesis inhibitors. The tendencies for change and the degrees of changes in the production of the exoproteins were different (Table 2). Because all protein synthesis inhibitors directly suppress the 70S ribosome and inhibition mechanisms are different according to the type of drug, it is possible that the mechanism for each drug is affected by the tendency for exoprotein production and the degree of exoprotein production. In the experiments with the protein synthesis inhibitors CP (Fig. 6d), EM (Fig. 6a), and CLDM (Fig. 3c), CLDM reduced the levels of production of SpeB and affected more kinds of exoproteins than CP and EM did. They all have common reaction points, inhibit peptidyltransferase, and interfere with the elongation reaction on the 50S ribosome (9, 24). However, it has been reported that the reaction mechanisms are different (24). CLDM, which extends to the peptidyltransferase center, causes the dissociation of peptidyl-tRNAs containing two, three, or four amino acid residues (24), whereas EM, which does not reach the peptidyltransferase center, induces the dissociation of peptidyl-tRNAs containing six, seven, or eight amino acid residues (24). Our study suggests that these slightly different mechanisms of CLDM and EM are related to their different effects on exoprotein production. The complexes composed of the 70S ribosome, mRNA, and drugs are important factors that affect the selective production of exoproteins. Moreover, it might be possible that other regulation pathways control translation and/or transcription by this complex.
We also analyzed the levels of transcription of slo and nga and found that the same changes were detected at the transcriptional level (Fig. 7). In principle, protein synthesis inhibitors interact directly with the ribosome and stop peptide translation from mRNA. Therefore, it is puzzling that these drugs work at the transcriptional level. With regard to S. aureus, it was suggested (13) that CLDM blocks the translation of several regulation factors, and as a result, the levels of transcription of exoproteins increase or decrease. Increases in exoprotein levels may have been observed because the synthesis of suppressive regulators is inhibited, and as a result, this may remove the suppressive effects and allow the effect of decreased translation to be overcome by the direct effects of antibiotic treatment. The other possibility is that the proteins whose levels are increased are necessary for the bacteria to survive in stressful surroundings, such as in the presence of antibiotics, and that the mechanism for increases in protein levels occurs at the transcriptional level. These points require further analysis.
As far as the different effects of CLDM on M serotype strains are concerned, increases in Nga (spot 13) and Slo (spot 12) levels were detected in serotype M1 strains (Fig. 3) and serotype M4 and M12 strains (Fig. 6), but not in serotype M3 or M5 strains. The M1, M4, and M12 strains that we used produced fairly large amounts of SpeB (spot 1) without CLDM, but the M3 and M5 strains that we used did not (data not shown). One possibility is that SpeB, which works as a serine protease, degrades Nga and Slo; and the decrease in SpeB levels in the presence of CLDM causes the increases in Nga and Slo levels. Since M3 and M5 do not produce much SpeB compared with the amounts produced by M1, M4, and M12, this change does not occur. However, we believe that this is not the case, because increases in Nga and Slo levels were also detected in the presence of both protease inhibitors and CLDM (data not shown). In addition, we tested an rgg-knockout strain which does not produce much SpeB (4) and found that Nga and Slo levels still increased with CLDM treatment (data not shown). However, we cannot rule out the possibility of the involvement of other proteases, so this mechanism requires further study.
As mentioned above, the combination of high-dose CLDM and PCG is recommended as chemotherapy for TSLS. Previously, it was reported that this combination has no antagonistic effects but that the combination does not have a bactericidal advantage over either PCG or CLDM alone (22). In our study, the concentration used for each drug in the combination was almost the same as or higher than that of each drug alone (Table 1). Although we detected slight changes in the abundance of proteins by SDS-PAGE analysis, some of the bands were found to originate from the intracellular proteins by 2-DE analysis (data not shown) and the results were reproducible. Thus, we could not detect any advantage in using the combination to suppress exoproteins compared with the ability of CLDM alone to do so. The advantage of these drugs is probably PCG's very strong bactericidal effect and CLDM's easy penetration of tissue.
Our study revealed that protein synthesis inhibitors increased the levels of production of some exoproteins. In particular, the increases in Sic (spot 3), Slo (spot 12), and Nga (spot 13) levels were remarkable. Sic (streptococcal inhibitor of complement) interferes with complement function (1), inactivates some antibacterial peptides involved in bacterial clearance (8), and plays an important role in bacterial adherence and internalization (14). It is possible that the increase in Sic levels caused by protein synthesis inhibitors delays bacterial clearance and enhances the ability of a drug to invade bacteria. Slo and Nga appear to be functionally linked. Slo exerts it cytolytic function by forming large homopolymeric pores in membranes (2). Nga, which is transferred by the pore made by Slo, contributes to bacterial pathogenesis by modulating host cell signaling pathways to inhibit internalization, augment Slo-mediated cytotoxicity, and induce target cell apoptosis (3). It is also possible that the increase in Sic levels facilitates the invasion of target cells by Nga and, furthermore, that the increase in Nga levels enhances pathogenicity. Since sensitivity to virulence is different from one person to another, CLDM might have an adverse effect on the treatment of TSLS, as judged only from exoprotein production. Furthermore, this concern is important, because there is a possibility that the drug concentration in some tissues was lower than the MIC. Further studies are needed to assess the choice of antibiotics for the treatment of TSLS.
Acknowledgments
This work was supported by the fund for comprehensive research on aging and health (13kou-01) from the Ministry of Health, Labour and Welfare, Japan.
REFERENCES
- 1.Akesson, P., A. G. Sjoholm, and L. Bjorck. 1996. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 271:1081-1088. [DOI] [PubMed] [Google Scholar]
- 2.Billington, S. J., B. H. Jost, and J. G. Songer. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182:197-205. [DOI] [PubMed] [Google Scholar]
- 3.Bricker, A. L., C. Cywes, C. D. Ashbaugh, and M. R. Wessels. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol. Microbiol. 44:257-269. [DOI] [PubMed] [Google Scholar]
- 4.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cone, L. A., D. R. Woodard, P. M. Schlievert, and G. S. Tomory. 1987. Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes. N. Engl. J. Med. 317:146-149. [DOI] [PubMed] [Google Scholar]
- 6.Coyle, E. A., R. Cha, and M. J. Rybak. 2003. Influences of linezolid, penicillin, and clindamycin, alone and in combination, on streptococcal pyrogenic exotoxin a release. Antimicrob. Agents Chemother. 47:1752-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti, J. J., B. A. Roe, S. W. Clifton, S. P. Lin, X. Wang, M. Zhan, A. Reece, A. N. Suvorov, and W. M. McShan. 1997. The Streptococcus pyogenes genome sequencing project. A progress report. Adv. Exp. Med. Biol. 418:961-963. [DOI] [PubMed] [Google Scholar]
- 8.Frick, I. M., P. Akesson, M. Rasmussen, A. Schmidtchen, and L. Bjorck. 2003. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J. Biol. Chem. 278:16561-16566. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa, T., K. Torii, S. Hashikawa, Y. Iinuma, and M. Ohta. 2002. Cloning and characterization of the deoxyribonuclease sdα gene from Streptococcus pyogenes. Curr. Microbiol. 45:13-17. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa, T., K. Torii, S. Hashikawa, Y. Iinuma, and M. Ohta. 2002. Cloning and characterization of two novel DNases from Streptococcus pyogenes. Arch. Microbiol. 177:451-456. [DOI] [PubMed] [Google Scholar]
- 12.Hashikawa, S., Y. Iinuma, M. Furushita, T. Ohkura, T. Nada, K. Torii, T. Hasegawa, and M. Ohta. 2004. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J. Clin. Microbiol. 42:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoe, N. P., R. M. Ireland, F. R. DeLeo, B. B. Gowen, D. W. Dorward, J. M. Voyich, M. Liu, E. H. Burns, Jr., D. M. Culnan, A. Bretscher, and J. M. Musser. 2002. Insight into the molecular basis of pathogen abundance: group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc. Natl. Acad. Sci. USA 99:7646-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoge, C. W., B. Schwartz, D. F. Talkington, R. F. Breiman, E. M. MacNeill, and S. J. Englender. 1993. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA 269:384-389. [PubMed] [Google Scholar]
- 16.Mascini, E. M., M. Jansze, L. M. Schouls, J. Verhoef, and H. Van Dijk. 2001. Penicillin and clindamycin differentially inhibit the production of pyrogenic exotoxins A and B by group A streptococci. Int. J. Antimicrob. Agents 18:395-398. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz, B., R. R. Facklam, and R. F. Breiman. 1990. Changing epidemiology of group A streptococcal infection in the USA. Lancet 336:1167-1171. [DOI] [PubMed] [Google Scholar]
- 18.Sriskandan, S., A. McKee, L. Hall, and J. Cohen. 1997. Comparative effects of clindamycin and ampicillin on superantigenic activity of Streptococcus pyogenes. J. Antimicrob. Chemother. 40:275-277. [DOI] [PubMed] [Google Scholar]
- 19.Stevens, D. L. 1994. Invasive group A streptococcal infections: the past, present and future. Pediatr. Infect. Dis. J. 13:561-566. [DOI] [PubMed] [Google Scholar]
- 20.Stevens, D. L., A. E. Bryant, and S. P. Hackett. 1995. Antibiotic effects on bacterial viability, toxin production, and host response. Clin. Infect. Dis. 20(Suppl. 2):S154-S157. [DOI] [PubMed] [Google Scholar]
- 21.Stevens, D. L., A. E. Gibbons, R. Bergstrom, and V. Winn. 1988. The eagle effect revisited: efficacy of clindamycin, erythromycin, and penicillin in the treatment of streptococcal myositis. J. Infect. Dis. 158:23-28. [DOI] [PubMed] [Google Scholar]
- 22.Stevens, D. L., K. J. Madaras-Kelly, and D. M. Richards. 1998. In vitro antimicrobial effects of various combinations of penicillin and clindamycin against four strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:1266-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens, D. L., M. H. Tanner, J. Winship, R. Swarts, K. M. Ries, P. M. Schlievert, and E. Kaplan. 1989. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N. Engl. J. Med. 321:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Tenson, T., M. Lovmar, and M. Ehrenberg. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005-1014. [DOI] [PubMed] [Google Scholar]