Abstract
Background
A variant in steroidogenic factor-1 (SF-1, encoded by the gene NR5A1), p.Arg92Trp, has recently been reported in multiple families with 46,XX ovotesticular or testicular DSD. This amino-acid change impacts the DNA-binding domain and perturbs gonadal differentiation pathways.
Methods
Whole-exome sequencing was performed on a 46,XX subject with ovotesticular DSD.
Results
Exome results identified a heterozygous NR5A1 variant, p.Arg92Gln, in the 46,XX ovotesticular DSD proband. This arginine-to-glutamine change has been previously reported in the homozygous state in a 46,XY patient with gonadal and adrenal dysgenesis, though 46,XY and 46,XX heterozygous carriers of this variant have not been previously reported to have any clinical phenotype.
Conclusions
The NR5A1 p.Arg92Gln variant, which has thus far only been seen in a family with 46,XY DSD, most likely contributes to the ovotesticular DSD in this case. In light of the recent reports of unrelated 46,XX subjects with testicular or ovotesticular DSD with the NR5A1 variant, p.Arg92Trp, it appears that other mutations in the DNA binding domain have the potential to impact the factors determining testicular and ovarian differentiation. This case demonstrates the variability of phenotypes with the same genotype and broadens our understanding of the role of SF-1 in gonadal differentiation.
Key terms: SF-1, disorder of sex development, ovotestes, exome sequencing
Introduction
Steroidogenic factor-1 (SF-1), encoded by the NR5A1 gene, was identified in the early 1990’s as a nuclear receptor with an essential role in adrenal and gonadal development [1,2]. Homozygous pathogenic NR5A1 variants cause complete gonadal dysgenesis and adrenal insufficiency[3]. Heterozygous NR5A1 variants can also cause gonadal dysfunction in 46,XY individuals and have been shown to be associated with a wide spectrum of phenotypes ranging from infertility without genital anomalies to significant undervirilization with hypospadias, microphallus, and/or undescended testes to overt genital ambiguity[4]. In 46,XX individuals, many of the same heterozygous NR5A1 variants have also been associated with gonadal dysfunction, presenting as primary ovarian insufficiency[5].
Recently, Bashamboo et al. presented four families with a specific variant in NR5A1, p.Arg92Trp, with ovotesticular or testicular disorders of sex development (DSD) in 46,XX individuals[6]. This was followed by three additional cases with the same variant reported by Baetens et al. [7]. Individuals with ovotesticular DSD have both testicular and ovarian tissue and are most often seen in the setting of a 46,XX karyotype. The amino acid altered by this variant is located in the accessory DNA-binding domain and is highly conserved. While the role of NR5A1 in early gonadal development had been well established, these reports demonstrated a previously undescribed role for NR5A1 in gonadal fate determination, with the missense variant p.Arg92Trp leading to disruption of ovarian-specific developmental pathways, namely, the Wnt/β-catenin pathway, which normally suppresses the expression of testis genes[6,7]. To date, only this single NR5A1 variant has been described in patients with 46,XX ovotesticular or testicular DSD.
Methods and Subjects
Clinical History
This study was approved by the Boston Children’s Hospital Institutional Review Board. Written informed consent was obtained for all participants.
The patient is a 46,XX individual of European ancestry (England, Scotland, Ireland, France) born with ambiguous genitalia and raised as a girl. She had not had any prior surgeries. She was initially seen at our institution at age 3 and was noted to have significant clitoromegaly (2.5 cm length, 1.1 cm width) without palpable gonads. She had an anogenital ratio of 0.5 and rugated labia majora. Her evaluation for congenital adrenal hyperplasia (CAH) revealed normal baseline and stimulated adrenal hormones and precursors. Ultrasound and MRI imaging identified a uterus measuring 2.0 × 0.7 × 0.4 cm and possible abdominal gonads, with oval-shaped gonadal structures located at the junction of the pelvis and inguinal regions, measuring 0.8 × 0.7 × 0.6 cm on the left and 1.0 × 0.7 × 0.6 cm on the right. The structure on the left contained at least two cysts, potentially consistent with ovarian follicles.
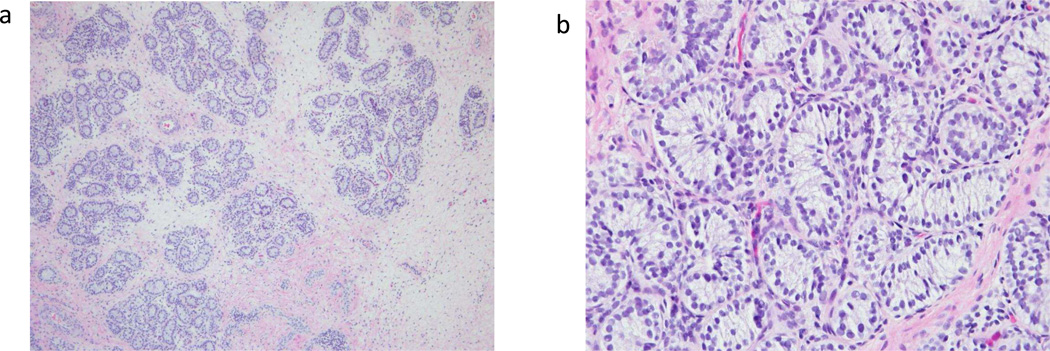
To assess for the presence of testicular tissue, additional evaluation included an AMH measurement as well as an hCG stimulation test. AMH was 6.47 ng/ml (reference range 0.256–6.34 ng/ml, conversion factor to pmol/L: 7.1429) and baseline serum total testosterone was 2 ng/dL (conversion factor to nmol/L: 0.0347). After 3 days of stimulation with intramuscular hCG 1500 units daily, testosterone rose to 39 ng/dL. Based on this evidence of testosterone-producing tissue, the subject underwent laparoscopy, which identified bilateral gonadal structures that appeared to be ovotestes. The testicular-appearing material was resected from both gonads, and pathology showed dysgenetic testicular tissue with absent germ cells (Figure 1). Cytogenetics on the testicular tissue confirmed a 46,XX karyotype.
Figure 1.
a) Left gonad at intermediate magnification - testicular tissue demonstrating seminiferous tubules
b) Right gonad at high magnification – seminiferous tubules without visible germ cells
Genetic Analyses
Whole-exome sequencing of blood- and testis-derived genomic DNA was performed at the Broad Institute (Cambridge, Massachusetts, USA) on the proband and her parents, as previously described[8]. For hybrid selection, we used the custom Illumina Content Exome capture kit (Illumina, San Diego, California, USA). Sequencing reads were aligned to the hg19 reference genome[9]. We applied the Genome Analysis Toolkit (GATK) for base quality score recalibration and indel (insertion-deletion) realignment[10]. Variant quality score recalibration was simultaneously performed for SNP and indel discovery according to GATK Best Practices recommendations [11,12]. We used SnpEff (http://snpeff.source forge.net/) for functional annotation. We filtered for variants that were found in less than 1% of the reference population based on allele frequencies from the Exome Aggregation Consortium (http://exac.broadinstitute.org/)[13]. Based on the quality standards at the Broad Institute, at least 80% of the exome had 20X coverage. Results were reviewed for variants in known DSD-related genes [14]. Sanger sequencing was performed to confirm the variant of interest.
Results
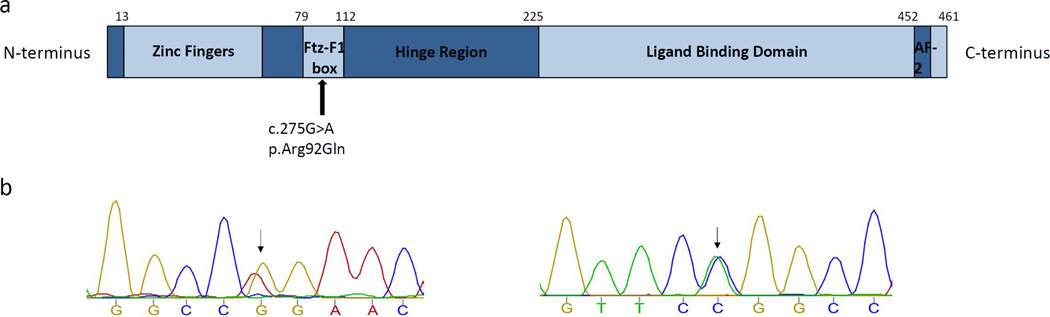
Exome sequencing on the 46,XX subject with bilateral ovotestes identified an extremely rare missense variant in NR5A1, a gene recently implicated in ovotesticular DSD. The variant, c.275G>A, p.Arg92Gln (Figure 2), was inherited from the proband’s father [5,15], a phenotypically normal adult male who has fathered two children. The variant was not present in the ExAC database but had previously been reported in an unrelated family with a homozygous 46,XY proband with gonadal dysgenesis and adrenal insufficiency and unaffected 46,XX heterozygotes (mother and sister of affected proband) [16]. In silico prediction of the missense variant using Polyphen-2 yielded a score of 0.932, considered “probably damaging” by the software[17]. We compared whole-exome sequencing results from both blood- and testis-derived DNA from the proband to identify somatic mutations that may be contributing to the phenotype; this identified no differences to suggest somatic mutations.
Figure 2.
a) SF-1 protein structure, with the location of the heterozygous mutation in the 46,XX ovotesticular DSD subject indicated. = Activation function domain 2; Ftz- F1 = fushi tarazu factor 1 box. (adapted from Wong, J Mol Endocrinol, 1996 and Camats, JCEM, 2012) b) Chromatograms obtained by Sanger sequencing indicating a G>A transition in exon 4 of NR5A1 (Chr 9:127262964) the subject. On the left is the forward read, and on the right is the reverse read.
*Figure not to scale.
Discussion
We have identified a paternally inherited, heterozygous variant in NR5A1, p.Arg92Gln, in a patient with 46,XX ovotesticular DSD. The p.Arg92Gln change is at the same position as variants in recently reported 46,XX ovotesticular and testicular DSD cases but involves a different amino-acid change[6,7]. Based on other reported families, this change is not always pathogenic in the heterozygous state [16,18], nor does it impact synergy of SF-1 with β-catenin based on in vitro studies[6]. Early studies looking at DNA binding in the p.Arg92Gln variant showed an impact on DNA-binding specificity when present with other reported pathogenic variants such as p.Gly35Glu[19]. Other studies have confirmed partial loss of function of the p.Arg92Gln variant with respect to activation of SF-1 target gene promoters[20]. A recent report looking at Turkish families with primary adrenal insufficiency identified another family carrying the same p.Arg92Gln variant with unaffected heterozygous parents and a homozygous 46,XX phenotypically female daughter with adrenal insufficiency [18]. This report does not describe the gonadal function of the mother or the affected daughter.
The fact that the p.Arg92Gln variant is absent in large databases but has now been seen in three different families with abnormalities in gonadal and/or adrenal function provides evidence that multiple missense mutations at this amino-acid residue can impact the function of SF-1. Reviewing this heterozygous variant in the context of the American College of Medical Genetics and Genomics (ACMG) criteria[21], we find that it is “likely pathogenic” for the 46,XX ovotesticular DSD phenotype given that it is absent from control databases, alters an amino-acid residue previously associated with the same phenotype, and has been seen as clearly pathogenic when homozygous in a patient with adrenal and gonadal insufficiency. Additional evidence includes in silico prediction, with a Polyphen-2 score in the “probably damaging” range, as well as a disease phenotype consistent with a variant in this gene.
The recent report of four families with 46,XX testicular or ovotesticular DSD by Bashamboo et. al. is the first to describe a missense mutation in NR5A1 leading to these 46,XX ovotesticular or testicular phenotypes[6]. This was quickly followed by Baetens et. al. with three additional unrelated (ovo)testicular DSD subjects with the identical p.Arg92Trp variant[7]. The reports provide evidence of the p.Arg92Trp variant leading to decreased synergy of SF-1 with β-catenin. The model proposed for p.Arg92Trp in the setting of 46,XX DSD involves decreased expression of NR0B1 and other anti-testis genes, secondary to the decreased synergy with β-catenin. This subsequently leads to increased expression of SOX9 and other pro-testis genes, resulting in testicular or ovotesticular DSD[6]. Notably, some of these additional families include unaffected 46,XX carriers, demonstrating incomplete penetrance of the p.Arg92Trp variant.
Although we have not directly tested the molecular function of the p.Arg92Gln variant, we presume that the p.Arg92Gln variant, previously reported in 46,XX carriers without identified phenotypes, can sometimes lead to similar perturbations in signaling as the p.Arg92Trp variant and can thereby lead to ovotesticular DSD in some individuals. It is possible that other genetic variants, whether in NR5A1 or other genes, or nongenetic factors could explain this incomplete penetrance, although we did not find any other NR5A1 variants in our subject. There is also strong evidence of variable penetrance of other NR5A1 variants, particularly in 46,XY DSD cases[22], and it is likely that additional examples demonstrating variable penetrance will be observed with the p.Arg92Gln variant as more individuals carrying this variant are identified.
Our work provides further support for the unfolding story showing NR5A1 as a cause of 46,XX ovotesticular DSD and suggests that the p.Arg92Gln variant may do so separately from effects on Wnt signaling [6]. Instead, the altered amino-acid apparently influences another aspect of SF-1 action leading to the ovotesticular DSD phenotype through a mechanism that has yet to be identified. Further study of the functional effects of this variant may reveal how NR5A1 influences gonadal fate specification. In particular, functional defects common to both p.Arg92Gln and p.Arg92Trp (as opposed to being associated with only one of the variants) are more likely to be relevant to the phenotype.
Our case, along with other recently published reports, highlights the contribution of NR5A1 to multiple steps in the development of ovarian or testicular tissue from the undifferentiated gonad, with a previously unappreciated role in gonadal fate specification in addition to the well-established involvement of NR5A1 in gonadal development. These recent ovotesticular DSD cases have been crucial to providing insights for future research.
Established Facts.
NR5A1 has an essential role in adrenal and gonadal development, and individuals with homozygous mutations in NR5A1 have adrenal and gonadal dysgenesis.
Heterozygous pathogenic NR5A1 variants in 46,XY individuals have been shown to be associated with varying degrees of undervirilization, ranging from infertility to significant undervirilization.
Heterozygous pathogenic variants in 46,XX individuals have been associated with premature ovarian insufficiency.
Recent reports have associated testicular and ovotesticular DSD in 46,XX individuals with a specific NR5A1 variant, p.Arg92Trp.
Another report identified homozygosity of a very rare variant at the same amino acid, p.Arg92Gln, in a patient with gonadal dysgenesis and adrenal insufficiency.
Novel Insights.
Heterozygosity of the deleterious NR5A1 variant p.Arg92Gln appears to be associated with ovotesticular DSD in a 46,XX subject
Acknowledgments
We are grateful to the subject and her family for their time and contributions in advancing the field. We thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. We also thank Dr. Alexandra Kovach for her help with the pathology images.
Grants or fellowships supporting the writing of the paper: This work was supported by National Institutes of Health (NIH) Grant 5T32DK007699-32 (J.S.), the March of Dimes Foundation, as well as the Manton Center for Orphan Disease Research. This work was also conducted with support from Harvard Catalyst. The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
Footnotes
Disclosure summary: The authors have no relevant conflicts of interest.
References
- 1.Morohashi KI, Honda SI, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 2.Rice DA, Mouw AR, Bogerd AM, Parker KL. A shared promoter element regulates the expression of three steroidogenic enzymes. Mol Endocrinol. 1991 Oct;5:1552–1561. doi: 10.1210/mend-5-10-1552. [DOI] [PubMed] [Google Scholar]
- 3.Suntharalingham JP, Buonocore F, Duncan AJ, Achermann JC. DAX-1 (NR0B1) and steroidogenic factor-1 (SF-1, NR5A1) in human disease. Best Pract Res Clin Endocrinol Metab. 2015;29:607–619. doi: 10.1016/j.beem.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashamboo A, Ferraz-De-Souza B, Loureno D, Lin L, Sebire NJ, Montjean D, et al. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet. 2010;87:505–512. doi: 10.1016/j.ajhg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camats N, Pandey AV, Fernández-Cancio M, Andaluz P, Janner M, Torán N, et al. Ten novel mutations in the NR5A1 gene cause disordered sex development in 46,XY and ovarian insufficiency in 46,XX individuals. J Clin Endocrinol Metab. 2012 Jul;97:E1294–E1306. doi: 10.1210/jc.2011-3169. [DOI] [PubMed] [Google Scholar]
- 6.Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Seneviratne SN, et al. A recurrent p.Arg92Trp variant in steroidogenic factor-1 (NR5A1) can act as a molecular switch in human sex development. Hum Mol Genet. 2016 Jul 4; doi: 10.1093/hmg/ddw186. 0:ddw186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baetens D, Stoop H, Peelman F, Todeschini A-L, Rosseel T, Coppieters F, et al. NR5A1 is a novel disease gene for 46,XX testicular and ovotesticular disorders of sex development. Genet Med. 2016 doi: 10.1038/gim.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swartz JM, Ciarlo R, Guo MH, Abrha A, Diamond DA, Chan Y-M, et al. Two Unrelated Undervirilized 46,XY Males with Inherited NR5A1 Variants Identified by Whole-Exome Sequencing. Horm Res Paediatr. 2016 doi: 10.1159/000448754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 Oct 30; doi: 10.1038/nature19057. Available from: http://biorxiv.org/content/early/2015/10/30/030338.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baxter RM, Arboleda Va, Lee H, Barseghyan H, Adam MP, Fechner PY, et al. Exome Sequencing for the Diagnosis of 46,XY Disorders of Sex Development. J Clin Endocrinol Metab. 2015 Nov 10;100:E333–E344. doi: 10.1210/jc.2014-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong M, Ramayya MS, Chrousos GP, Driggers PH, Parker KL. Cloning and sequence analysis of the human gene encoding steroidogenic factor 1. J Mol Endocrinol. 1996;17:139–147. doi: 10.1677/jme.0.0170139. [DOI] [PubMed] [Google Scholar]
- 16.Achermann JC, Ozisik G, Ito M, Orun UA, Harmanci K, Gurakan B, et al. Gonadal determination and adrenal development are regulated by the orphan nuclear receptor steroidogenic factor-1, in a dose-dependent manner. J Clin Endocrinol Metab. 2002 Apr;87:1829–1833. doi: 10.1210/jcem.87.4.8376. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guran T, Buonocore F, Saka N, Ozbek MN, Aycan Z, Bereket A, et al. Rare causes of primary adrenal insufficiency: Genetic and clinical characterization of a large nationwide cohort. J Clin Endocrinol Metab. 2016;101:284–292. doi: 10.1210/jc.2015-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M, Achermann JC, Jameson JL. A naturally occurring steroidogenic factor-1 mutation exhibits differential binding and activation of target genes. J Biol Chem. 2000;275:31708–31714. doi: 10.1074/jbc.M002892200. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Philibert P, Ferraz-de-Souza B, Kelberman D, Homfray T, Albanese A, et al. Heterozygous missense mutations in steroidogenic factor 1 (SF1/Ad4BP, NR5A1) are associated with 46,XY disorders of sex development with normal adrenal function. J Clin Endocrinol Metab. 2007;92:991–999. doi: 10.1210/jc.2006-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warman DM, Costanzo M, Marino R, Berensztein E, Galeano J, Ramirez PC, et al. Three new SF-1 (NR5A1) gene mutations in two unrelated families with multiple affected members: Within-family variability in 46,XY subjects and low ovarian reserve in fertile 46,XX subjects. Horm Res Paediatr. 2011;75:70–77. doi: 10.1159/000320029. [DOI] [PubMed] [Google Scholar]