Summary
Phytopathogens have developed elaborate mechanisms to attenuate the defense response of their host plants, including convergent evolution of complex pathways for production of the gibberellin (GA) phytohormones, which were actually first isolated from the rice fungal pathogen Gibberella fujikuroi. The rice bacterial pathogen Xanthomonas oryzae pv. oryzicola (Xoc) has been demonstrated to contain a biosynthetic operon with cyclases capable of producing the universal GA precursor ent-kaurene. Genetic (knock-out) studies indicate that the derived diterpenoid serves as a virulence factor for this rice leaf streak pathogen, serving to reduce the jasmonic acid (JA) mediated defense response.
Here the function of the remaining genes in the Xoc operon are elucidated and the distribution of the operon in X. oryzae investigated in over 100 isolates.
The Xoc operon leads to production of the bioactive GA4, an additional step beyond production of the penultimate precursor GA9 mediated by the homologous operons recently characterized from rhizobia. Moreover, this GA biosynthetic operon was found to be widespread in Xoc (>90%), but absent in the other major oryzae pathovar.
These results indicate selective pressure for production of GA4 in the distinct lifestyle of Xoc, and the importance of GA to both fungal and bacterial pathogens of rice.
Keywords: biogeographical distribution, cytochromes P450, gibberellin biosynthesis, plant–microbe interactions, virulence factor
Introduction
Pathogenic microbes are under intense selection to manipulate their hosts, particularly to attenuate the host defense response. An example of the complexity that can arise from this selective pressure appears to be the independent evolution of biosynthetic pathways leading to the production of plant hormones by phytopathogens. Indeed, the gibberellins (GAs), production of which requires over ten transformations from the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP), were first isolated from the rice fungal pathogen Gibberella fujikuroi (anamorph Fusarium fujikuroi) (Hedden & Sponsel, 2015). The production of GAs by G. fujikuroi leads to the characteristic excessive growth phenotype of the resulting bakanae or foolish seedling disease, and has recently been demonstrated to act as virulence factor for this phytopathogen (Wiemann et al., 2013).
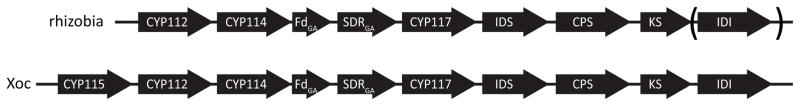
It has long been recognized that certain bacteria also produce GAs (Bottini et al., 2004). Such biosynthetic capacity has been speculatively ascribed to a cytochrome P450 (CYP) rich operon widely distributed in rhizobia (Fig. 1) (Tully et al., 1998; Keister et al., 1999), which has been demonstrated by characterization of the encoded enzymes. The three associated synthases/cyclases lead to production of ent-kaurene, one is a GGPP synthase (GGPS), the next is an ent-copalyl diphosphate synthase (CPS), while the last is an ent-kaurene synthase (KS) (Morrone et al., 2009; Hershey et al., 2014). The three CYPs (CYP112, CYP114, and CYP117), along with the ferredoxin (FdGA) and short-chain alcohol dehydrogenase (SDRGA), have recently been shown to transform ent-kaurene to GA9 (although this is not thought to exhibit hormonal activity – i.e., be bioactive) in several of the rhizobia in which the operon is found (Nett et al., 2016; Tatsukami & Ueda, 2016).
Fig. 1.
Schematic representation of the gibberellin (GA) biosynthetic operon. Abbreviations for the genes in the arrows are: CYP, cytochrome P450; Fd, ferredoxin; SDR, short-chain alcohol dehydrogenase/reductase; IDS, isoprenyl diphosphate synthase; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; and isopentenyl diphosphate isomerase (IDI), which is not found in all copies of the operon in rhizobia; the arrow indicates the direction of transcription.
In addition to appearing in a sub-set of symbiotic rhizobia from the Rhizobiales order in the Alphaproteobacteria class (Hershey et al., 2014), a similar operon also can be found in a few phytopathogens from the separate Gammaproteobacteria class, indicating that this has been more widely distributed by horizontal gene transfer. It has been suggested that the rice bacterial leaf streak pathogen Xanthomonas oryzae pv. oryzicola (Xoc), which contains a copy of this operon, might produce GA(s) as a virulence factor. However, only the capacity to produce ent-kaurene from GGPP, by functional characterization of the associated CPS and KS, has been reported to-date. Intriguingly, knocking out the CPS, KS, or CYP112 was sufficient to attenuate the virulence of Xoc, allowing rice to mount a more robust jasmonic acid (JA) mediated defense response (Lu et al., 2015). Nevertheless, the fact that this operon was only found in the single oryzicola pathovar that had been sequenced at that time, and not in any of the several genome sequences available for other X. oryzae pathovars, left the broader importance of this operon and resulting ent-kaurene derived diterpenoid in these rice bacterial pathogens, and even Xoc more specifically, in question.
Here biochemical characterization is reported demonstrating that the enzymes encoded by this operon in Xoc can function to produce the bioactive GA4. This represents further transformation of the GA9 produced by the previously characterized rhizobial operon, with the relevant activity of 3β-hydroxylation carried out by the additional CYP (CYP115) found in the Xoc operon (Fig. 1). Moreover, although seemingly restricted to the oryzicola pathovar in X. oryzae, investigation of the biogeographical distribution of the GA biosynthetic operon with over 100 isolates of Xoc found widespread conservation that would be consistent with an important role for production of GA4 specifically in this rice bacterial leaf streak pathogen.
Materials and Methods
Unless otherwise specified, chemicals were purchased from Sigma and Fisher Scientific. GA12-aldehyde, GA12, GA9 and GA4 were purchased from OlChemIm Ltd., while ent-7α-hydroxykaurenoic acid, GA15 and GA24 were kindly provided by Dr Peter Hedden (Rothamsted Research).
Genes were cloned by amplification from genomic DNA of Xoc strain BLS256 using Q5 Hot Start High-Fidelity DNA polymerase (NEB) according to the product manual, and using 5 μl of the high GC-content enhancer and gene specific primers (Supporting Information Table S1). The forward primer included a CACC sequence before the start codon to allow cloning via directional TOPO isomerization into pET100 (Invitrogen) according to the instructions supplied by the manufacturer. Clones were sequenced to verify the correct DNA sequence. CYP114 was cloned either alone or in tandem with the neighboring ferredoxin, using the forward primer for CYP114 and the reverse primer for FdGA.
For recombinant in vivo feeding studies the pET100 based expression constructs for CYP117, CYP112, CYP114, CYP114+FdGA, CYP115 and SDRGA were transformed into Escherichia coli expression strain BL21-Star (Invitrogen). Three individual colonies were inoculated into 10 ml TB broth (12 g l−1 casein, 24 g l−1 yeast extract, 0.4% glycerol), including 50 μg ml−1 carbenicillin, and grown at 18°C for 2 d with constant shaking at 200 rpm. From these starter cultures, 2 ml were transferred into 25 ml fresh TB broth, including 50 μg ml−1 carbenicillin. Cultures expressing CYPs were induced at an OD of 0.8, while that for SDRGA was induced at an OD 0.6, with 1 mM IPTG. Upon induction, 5 ml 1 M phosphate buffer pH 7.5, 1 mM aminolevulenic acid, 1 mM riboflavin, 0.1 mM FeCl3 and 10 μM of specific substrates dissolved in a 1 : 1 mixture of DMSO and methanol (v : v) were added. Cultures were then grown at 18°C with constant shaking at 200 rpm for 3 d.
To analyze recombinant enzymatic activity, the fed cultures were acidified to pH 3.0 with 5 N HCl to neutralize the expected carboxylated products, then extracted three times with 50 ml hexanes (CYP117) or 50 ml ethyl acetate (CYP112, CYP114, CYP114+FdGA, CYP115 and SDRGA). The organic phases were combined in a 250 ml round bottom flask and evaporated in a rotary evaporator to dryness. The flask contents were washed three times with 2 ml hexanes or ethyl acetate, with the exception of CYP117, reduced to c. 0.5 ml volume and purified using 1 ml silica gel in a glass Pasteur pipette, with elution via a stepwise gradient starting at 100% hexane and ending at 100% ethyl acetate, with 25% increments between solvent combinations. Fractions were methylated using diazomethane, and for reactions with CYP115, silylated with BSTFA+TMCS (99:1). Derivatized samples were dried under a stream of nitrogen, dissolved in n-hexane and analyzed by GC-MS, carried out with a 3900 Saturn GC (Varian), equipped with an HP-5MS column and coupled to a Saturn 2100T ion trap mass spectrometer detector (Varian). The injector of the GC was kept at a temperature of 250°C, the temperature gradient of the column started at 50°C, held for 3 min, which was then raised by 15°C min−1 to 300°C, again held for 3 min.
Bioinformatic searches for copies of the GA biosynthetic operon in X. oryzae were carried by BLAST searches of this species at NCBI and JGI using the characteristic cps, ks and cyp112 genes as query sequences. Insertion sequences elements in the GA operon vicinity were first found using ISfinder (Siguier et al., 2006), and then more completely elucidated by manual curation.
For screening, previously described West African (Wonni et al., 2014) and Asian (Raymundo et al., 1999) X. oryzae strains were obtained from a collection maintained at Colorado State University (Fort Collins, USA). The presence of the GA biosynthetic operon was directly investigated by PCR amplification using four overlapping fragments that together cover the entire operon (Fig. S1). Primers were designed to anneal within the genes. Fragments were amplified using Phusion High-Fidelity DNA polymerase (Thermo Scientific), according to the manufactures’ instructions, in reactions optimized for GC-rich templates by the addition of 3% DMSO. Amplified fragments were detected via gel electrophoresis, using 0.8% TAE gels and GelRed stain (Biotium Inc.). In most cases all four fragments could be detected, although for six isolates only three fragments (and in one case only one), were observed. Given the extensive overlap and difficulty in amplification (presumably due to the high GC content), it was assumed that detection of any fragment indicated the presence of the operon. Nucleotide polymorphisms in the primer target sequences also may result in failure of amplification.
Results
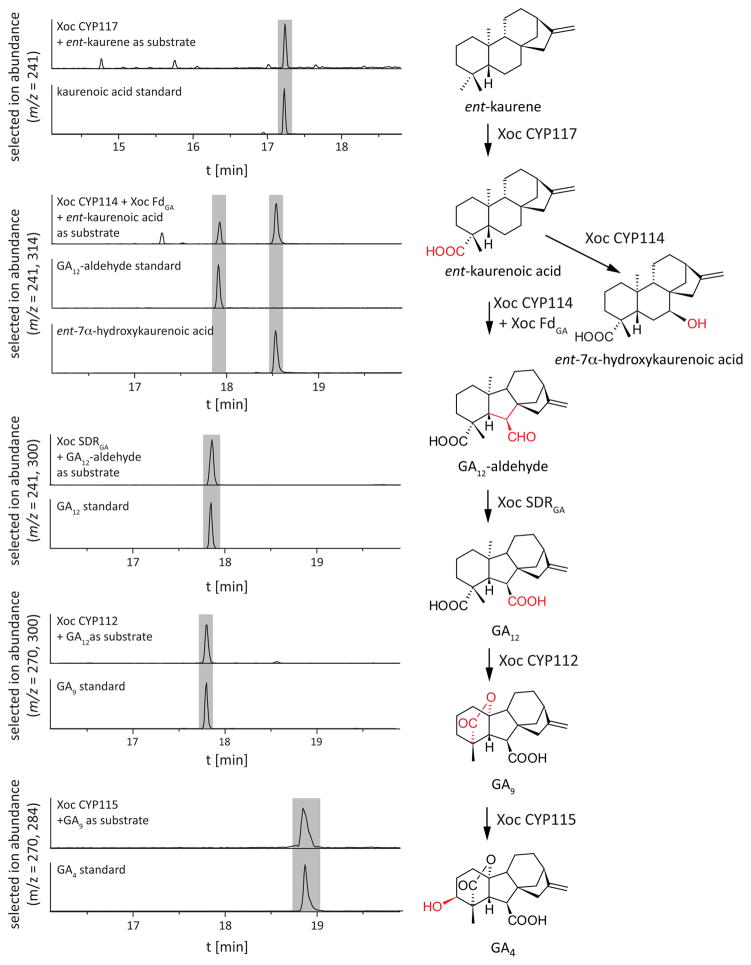
The CPS and KS from the Xoc operon together produce ent-kaurene from GGPP (Lu et al., 2015). Here we report functional characterization of the remaining oxidative enzymes from the operon (i.e., CYP112, CYP114, CYP115, CYP117, SDRGA and FdGA; see Fig. 1). For this purpose, each of the CYPs and the SDRGA were individually expressed in E. coli. CYP114 was also co-expressed with FdGA, which has been shown to be required for full activity of this CYP, while the other CYPs presumably are efficiently reduced by endogenous Fd (Nett et al., 2016). The resulting recombinant strains were fed each biosynthetic intermediate/precursor, in parallel cultures that were then extracted for analysis by gas-chromatography mass-spectrometry (GC-MS) to determine what (if any) enzymatic transformations had been catalyzed.
The results of these studies demonstrated that the enzymes in common between the Xoc operon and those from rhizobia (Fig. 1) catalyze the same reactions, such that the pathway from ent-kaurene to GA9 elucidated by these studies is analogous to that recently reported for the rhizobial species Bradyrhizobium japonicum and Sinorhizobium fredii (Nett et al., 2016). Briefly, CYP117 catalyzed the three oxidative transformations required to transform ent-kaurene to ent-kaurenoic acid (Fig. 2). This was further converted to GA12-aldehyde by CYP114, which includes ring contraction from the 6-6-6-5 kaurane backbone to the 6-5-6-5 ring structure characteristic of the gibberellanes (Figs 2, S2). We predict that this transformation proceeds via initial conversion of ent-kaurenoic acid to ent-7α-hydroxykaurenoic acid, with the subsequent oxidative ring contraction reaction dependent on co-expression of FdGA, because cultures expressing CYP114 alone only produced ent-7α-hydroxykaurenoic acid (Fig. S3). SDRGA efficiently catalyzes the further oxidation of GA12-aldehyde to GA12 (Figs 2, S2). CYP112 then catalyzes the conversion of GA12 to GA9, which also requires three reactions, first hydroxylation to the C-20 alcohol derivative GA15, then oxidation to the aldehyde GA24, before catalysis of the coupled loss of C-20 with formation of a γ-lactone ring (Figs 2, S2).
Fig. 2.
GC-MS of XocCYP117, CYP114+FdGA, SDRGA, CYP112 and CYP115 from Escherichia coli extracts. (Left panel) GC-MS chromatograms of hexane extracts from E. coli cultures expressing the indicated enzymes and chromatograms of standard compounds as a reference. (Right panel) Reactions catalyzed by the respective enzymes.
Notably, the operon in Xoc contains an additional CYP, CYP115, which is not generally present in rhizobia (Lu et al., 2015). CYP115 proved to catalyze 3β-hydroxylation of GA9 to form GA4 (Figs 2, S2). Thus, beyond the penultimate precursor GA9 observed with rhizobia (Mendez et al., 2014), the operon in Xoc can not only be confidently assigned to GA biosynthesis, but also to further lead to production of the bioactive GA4.
Bioinformatic searches of the sequence information currently available for X. oryzae indicated that the GA biosynthetic operon is present in Xoc, but not those for the closely related and other major pathovar X. oryzae pv. oryzae (Xoo), whose genomes have been fully sequenced (Lee et al., 2005; Ochiai et al., 2005; Salzberg et al., 2008; Booher et al., 2015; Quibod et al., 2016). In Xoc, examination of the available genome sequences (Bogdanove et al., 2011; Wilkins et al., 2015) revealed that the GA biosynthetic operon is flanked by multiple insertion sequence (IS) elements. Although there is some difference in the exact composition of these, all Xoc isolates have at least one IS element on each side of the operon (Fig. S4).
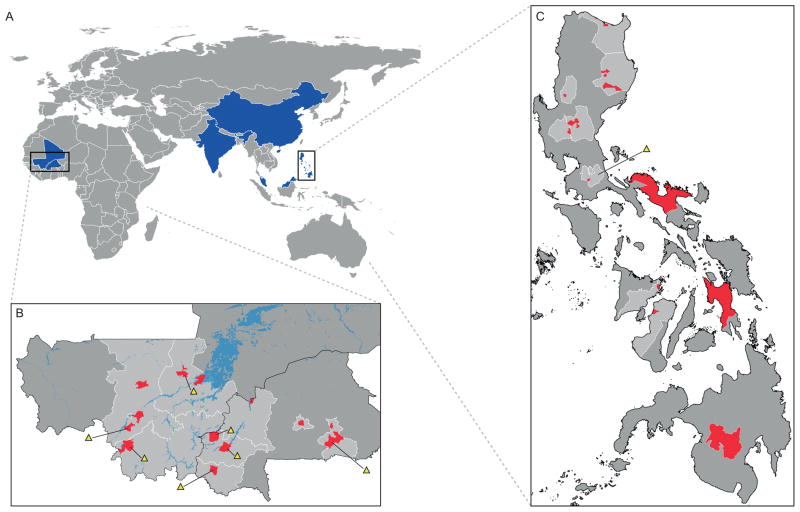
The presence of mobile elements flanking the GA biosynthetic operon denotes potential mobility, and raises questions about the ephemerality of GA4 production in Xoc. Thus, it seemed worth investigating how widespread the GA biosynthetic operon is within Xoc. For this purpose, 116 isolates from different regions where the resulting bacterial leaf streak disease is of agronomic concern, both West Africa and Asia, were screened by PCR for the presence of the GA biosynthetic operon. Included among these are ten isolates whose complete genome sequences are currently available (Wilkins et al., 2015). The isolates from West Africa were collected from rice and wild grasses showing disease symptoms between 2003 and 2011 in Mali and Burkina Faso (35 and 32 isolates, respectively), while those from Asia were collected from rice only, with 41 isolates from the Philippines, collected between 1978 and 1990, along with three from Malaysia, four from China and one from India (Tables 1, S2).
Table 1.
Total number of Xanthomonas oryzae pv. oryzicola isolates screened for the presence of the gibberellin (GA) biosynthetic operon, by country
| Country | Number of isolates
|
||
|---|---|---|---|
| total | GA + | GA − | |
| Mali | 35 | 31 | 4 |
| Burkina Faso | 32 | 27 | 5 |
| Philippines | 41 | 40 | 1 |
| Malaysia | 3 | 3 | 0 |
| China | 4 | 4 | 0 |
| India | 1 | 1 | 0 |
|
| |||
| Total | 116 | 106 | 10 |
Interestingly, the GA biosynthetic operon is highly prevalent in Xoc, as it is present in 106 of 116 isolates screened (91.3%), being absent in only 10 isolates (Table S2). On a regional level, the operon is absent only in a single isolate from the Philippines, and is absent somewhat more frequently in isolates from West Africa (9 of 67; 13.4%) (Fig. 3). While the isolates that do not contain the GA biosynthetic operon were all found on Oryza sativa rather than other species from the Oryza genus, this may not be significant, as only a limited number of isolates from these wild grasses were tested. To further test the generality of the observation that the GA biosynthetic operon is not present in the 13 complete genomes available for Xoo, five Xoo isolates from Asia and an isolate of X. oryzae (no pathovar designation) from the United States were tested, and found to not contain the operon (Table S2).
Fig. 3.
Origin of Xanthomonas oryzae pv. oryzicola isolates screened for the presence of the gibberellin (GA) biosynthetic operon. (a) Countries where Xanthomonas oryzae pv. oryzicola were isolated. Blue areas indicate Mali and Burkina Faso (West Africa) as well as India, Malaysia, China and the Philippines (Asia). Details on the collection area of West African (b) and Philippines (c) isolates. Red areas indicate provinces or towns (when available); yellow triangles indicate regions where isolates were found which do not have the GA biosynthetic operon.
Discussion
The biochemical results reported here demonstrate that the GA biosynthetic operon in Xoc leads to the production of bioactive GA4 (Fig. 2). This is consistent with the previous demonstration that the operon acts to produce a virulence factor that reduces the JA-mediated defense response (Lu et al., 2015), as an antagonistic relationship between GA and JA has already been established (Robert-Seilaniantz et al., 2007; Bari & Jones, 2009; Wasternack & Hause, 2013). The importance of such production of GA4 for Xoc is indicated by the very high prevalence of the GA biosynthetic operon in this pathovar over both a broad geographical range and different periods of time also shown here (Fig. 3; Table S2).
While the GA biosynthetic operon is somewhat less prevalent in the West African isolates tested here, it is still widely found in this region (>85%). Interestingly, the virulence factor AvrRxo1, which (along with its chaperone Arc1) is similarly flanked by several IS elements (Zhao et al., 2004; Liu et al., 2014), also is more prevalent in Asian than African isolates of X. oryzae (Triplett et al., 2016). Nevertheless, particularly given the lack of a clear pattern in more specific location or date of collection of these isolates, any underlying rationale for the slightly decreased prevalence of the GA biosynthetic operon in this region remains uncertain.
More critically, it also is unclear why the GA biosynthetic operon seems to be tightly restricted to Xoc and is not found in Xoo. This is especially puzzling given that exogenous application of bioactive GA3 has been reported to increase the susceptibility of rice to Xoo (Qin et al., 2013). Nevertheless, the selective distribution of the operon in X. oryzae presumably reflects a differential ability of GA4 to act as a virulence factor in the distinct lifestyles of these pathovars. Notably, Xoc nominally enters rice leaves through stomata and propagates throughout the mesophyll parenchyma causing leaf streak disease, while Xoo enters through hydathodes and propagates in the xylem causing leaf blight (Nino-Liu et al., 2006). While both Xoc and Xoo also can enter through wounds, given the association of the JA response with wounding, it can be hypothesized that such damaged tissue may provide a more important route for the entry of Xoc than Xoo.
Beyond Xoc and rhizobia, the GA biosynthetic operon has only been reported to be present in various pathovars of Xanthomonas translucens (Fig. S5), where it includes CYP115 (Lu et al., 2015). These phytopathogens infect either forage grasses or small grain cereal crops such as wheat, barley and rye, and cause leaf streak disease. Thus, their tissue specificity and mode of infection resemble that of Xoc (Stromberg et al., 1999; Wichmann et al., 2013; Pesce et al., 2015). Assuming that production of GA4 also serves to increase virulence for these phytopathogens, it can be similarly hypothesized that damaged tissue may be an important route of entry for X. translucens as well.
Intriguingly, while not otherwise particularly closely related (Naushad et al., 2015), Xoc and X. translucens both infect plants from a sub-family of the Poaceae/grass family, specifically the BEP clade (GPWGII, 2012). Accordingly, it can be further speculated that the selective presence of the GA biosynthetic operon in these phytopathogens is a function of the high water-repellency of their host plant leaves (Neinhuis & Barthlott, 1997), due to their vertical/upright nature and hydrophobic surface (Koch et al., 2008), which limits bacterial colonization and access to stomata (Beattie, 2011). Consistent with an important role for wounding in the entry of these pathogens, outbreaks of bacterial leaf streak disease have been correlated with the aftereffects of typhoons in rice (Nino-Liu et al., 2006), and with mowing in forage grass pastures (Pesce et al., 2015).
Lastly, it is notable that the final 3β-hydroxylation reaction catalyzed by the additional CYP115 found in the Xoc GA biosynthetic operon differentiates this from previously characterized operons (Fig. 1), which are all from rhizobia that seem to produce only the penultimate precursor GA9 (Mendez et al., 2014; Nett et al., 2016; Tatsukami & Ueda, 2016). The presence of a short fragment of CYP115 in rhizobia (Tully et al., 1998; R. S. Nett et al., unpublished), suggests that this gene was lost due to some negative effect on the symbiotic interaction of these rhizobia with their host plants. Indeed, the GA biosynthetic operon is not found in all rhizobia (Hershey et al., 2014), and knock-out mutants are still able to nodulate their respective host plants (Tully & Keister, 1993; Nett et al., 2016; Tatsukami & Ueda, 2016). The presence of Cyp115 and, therefore, the ability to produce bioactive GA4, may reflect the difference between the relationships of these bacteria with their host plants. As bioactive GA suppresses the JA defense response (Robert-Seilaniantz et al., 2007), rhizobia presumably benefit from maintaining the health of their host legume and it can be hypothesized that these symbionts may leave the final step in production of bioactive GA to the plant in order to not compromise the ability of the host to respond to infection by pathogenic microbes. Regardless of such speculation, the results reported here highlight the use of bioactive GAs as virulence factors by both fungal and bacterial pathogens of rice (Wiemann et al., 2013; Lu et al., 2015), which indicates the importance of the balance between GA and JA in the defense response of this important cereal crop plant to these disparate microbial disease agents, and is consistent with previous evidence that these hormonal interactions differ between rice and other plants such as Arabidopsis thaliana (De Bruyne et al., 2014).
Supplementary Material
Acknowledgments
The authors thank Dr Gwyn Beattie (Iowa State University) for insightful discussion of the lifestyles of Xoc and X. translucens with relationship to host plant architecture, and Dr Peter Hedden (Rothamsted Research) for authentic standards. This work was supported by grants from the NIH (GM109773 to R.J.P.), USDA (NIFA-AFRI grant 2014-67013-21720 to R.J.P.), and from FAPESP (2008/52074-0 to M-A.V.S.), a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DFG; NA 1261/1-1 to R.N.), and FAPESP Doctorate fellowship 2012/ 24954-0 (to P.C.G.T.).
Footnotes
Author contributions
RN cloned the genes, performed the heterologous expression, analyzed data and wrote the manuscript, RSN aided in data analysis and writing the manuscript, RJP aided in data analysis and writing the manuscript, PT performed DNA amplifications, analyzed data and wrote the manuscript, JEL and VV provided the Xoc and Xoo DNA collection, MAVS aided in data analysis and writing the manuscript.
Additional Supporting Information may be found online in the Supporting Information tab for this article:
Fig. S1 Strategy used for PCR screening of Xoc isolates.
Fig. S2 Mass spectra of peaks from GC-chromatograms.
Fig. S3 Activity of recombinant XocCyp117, Cyp114+/-FdGA and Cyp112.
Fig. S4 Insertional sequence (IS) elements flanking the GA biosynthetic operon in Xoc.
Fig. S5 Phylogeny of the GA biosynthetic operon.
Table S1 Primers used in this study
Table S2 Isolates of X. oryzae pv. oryzicola screened for GA biosynthetic operon
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Beattie GA. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol. 2011;49:533–555. doi: 10.1146/annurev-phyto-073009-114436. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, Patil PB, Van Sluys MA, Ryan RP, Meyer DF, Han SW, et al. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J Bacteriol. 2011;193(19):5450–5464. doi: 10.1128/JB.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher NJ, Carpenter SC, Sebra RP, Wang L, Salzberg SL, Leach JE, Bogdanove AJ. Single molecule real-time sequencing of Xanthomonas oryzae genomes reveals a dynamic structure and complex TAL (transcription activator-like) effector gene relationships. Microb Genom. 2015;1(4) doi: 10.1099/mgen.000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini R, Cassan F, Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol. 2004;65(5):497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- De Bruyne L, Höfte M, De Vleesschauwer D. Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Molecular Plant. 2014;7(6):943–959. doi: 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193(2):304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- Hedden P, Sponsel V. A century of gibberellin research. J Plant Growth Regul. 2015;34(4):740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey DM, Lu X, Zi J, Peters RJ. Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J Bact. 2014;196:100–106. doi: 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keister DL, Tully RE, Van Berkum P. A cytochrome P450 gene cluster in the Rhizobiaceae. J Gen Appl Microbiol. 1999;45(6):301–303. doi: 10.2323/jgam.45.301. [DOI] [PubMed] [Google Scholar]
- Koch K, Bhushan B, Barthlott W. Diversity of structure, morphology and wetting of plant surfaces. Soft Matter. 2008;4:1943–1963. [Google Scholar]
- Lee BM, Park YJ, Park DS, Kang HW, Kim JG, Song ES, Park IC, Yoon UH, Hahn JH, Koo BS, et al. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Research. 2005;33(2):577–586. doi: 10.1093/nar/gki206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chang Q, Feng W, Zhang B, Wu T, Li N, Yao F, Ding X, Chu Z. Domain dissection of AvrRxo1 for suppressor, avirulence and cytotoxicity functions. PLoS ONE. 2014;9(12):e113875. doi: 10.1371/journal.pone.0113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Hershey DM, Wang L, Bogdanove AJ, Peters RJ. An ent-kaurene derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. New Phytol. 2015;406:295–302. doi: 10.1111/nph.13187. [DOI] [PubMed] [Google Scholar]
- Mendez C, Baginsky C, Hedden P, Gong F, Caru M, Rojas MC. Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry. 2014;98:101–109. doi: 10.1016/j.phytochem.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583(2):475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- Naushad S, Adeolu M, Wong S, Sohail M, Schellhorn HE, Gupta RS. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek. 2015;107(2):467–485. doi: 10.1007/s10482-014-0344-8. [DOI] [PubMed] [Google Scholar]
- Neinhuis C, Barthlott W. Characterization and distribution of water-repellent, self-cleaning plant surfaces. Ann Bot. 1997;79:667–677. [Google Scholar]
- Nett RS, Montanares M, Marcassa A, Lu X, Nagel R, Charles TC, Hedden P, Rojas MC, Peters RJ. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2232. Author, if possible, please update the doi with the volume and page range. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 2006;7(5):303–324. doi: 10.1111/j.1364-3703.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- Ochiai H, Inoue V, Takeya M, Sasaki A, Kaku H. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jarq-Japan Agricultural Research Quarterly. 2005;39(4):275–287. [Google Scholar]
- Pesce C, Bolot S, Cunnac S, Portier P, Fischer-Le Saux M, Jacques M-A, Gagnevin L, Arlat M, Noël LD, Carrère S, et al. High-quality draft genome sequence of the Xanthomonas translucens pv. cerealis pathotype strain CFBP 2541. Genome Announcements. 2015;3(1):e01574–01514. doi: 10.1128/genomeA.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Liu JH, Zhao WS, Chen XJ, Guo ZJ, Peng YL. Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol Plant Microbe Interact. 2013;26(2):227–239. doi: 10.1094/MPMI-05-12-0138-R. [DOI] [PubMed] [Google Scholar]
- Quibod IL, Perez-Quintero A, Booher NJ, Dossa GS, Grande G, Szurek B, Vera Cruz C, Bogdanove AJ, Oliva R. Effector diversification contributes to Xanthomonas oryzae pv. oryzae phenotypic adaptation in a semi-isolated environment. Sci Rep. 2016;6:34137. doi: 10.1038/srep34137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymundo AK, Briones AM, Ardales EY, Perez MT, Fernandez LC, Leach JE, Mew TW, Ynalvez MA, McLaren CG, Nelson RJ. Analysis of DNA polymorphism and virulence in Philippine strains of Xanthomonas oryzae pv. oryzicola. Plant Dis. 1999;83:434–440. doi: 10.1094/PDIS.1999.83.5.434. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10(4):372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. doi: 10.1186/1471-2164-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Research. 2006;34(Database issue):D32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromberg KD, Kinkel LL, Leonard KJ. Relationship between phyllosphere population sizes of Xanthomonas translucens pv. translucens and bacterial leaf streak severity on wheat seedlings. Phytopathology. 1999;89(2):131–135. doi: 10.1094/PHYTO.1999.89.2.131. [DOI] [PubMed] [Google Scholar]
- Tatsukami Y, Ueda M. Rhizobial gibberellin negatively regulates host nodule number. Scientific Reports. 2016;6:27998. doi: 10.1038/srep27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett LR, Shidore T, Long J, Miao J, Wu S, Han Q, Zhou C, Ishihara H, Li J, Zhao B, et al. AvrRxo1 is a bifunctional type III secreted effector and toxin-antitoxin system component with homologs in diverse environmental contexts. PLoS ONE. 2016;11(7):e0158856. doi: 10.1371/journal.pone.0158856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully RE, Keister DL. Cloning and mutagenesis of a cytochrome P-450 locus from Bradyrhizobium japonicum that is expressed anaerobically and symbiotically. Appl Environ Microbiol. 1993;59:4136–4142. doi: 10.1128/aem.59.12.4136-4142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully RE, van Berkum P, Lovins KW, Keister DL. Identifcation and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum. Biochim Biophys Acta. 1998;1398:243–255. doi: 10.1016/s0167-4781(98)00069-4. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany. 2013;111(6):1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann F, Vorholter F-J, Hersemann L, Widmer F, Blom J, Niehaus K, Reinhard S, Conradin C, Kolliker R. The noncanonical type III secretion system of Xanthomonas translucens pv. graminis is essential for forage grass infection. Mol Plant Pathol. 2013;14(6):576–588. doi: 10.1111/mpp.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Sieber CM, von Bargen KW, Studt L, Niehaus EM, Espino JJ, Huss K, Michielse CB, Albermann S, Wagner D, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9(6):e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KE, Booher NJ, Wang L, Bogdanove AJ. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front Plant Sci. 2015;6:536. doi: 10.3389/fpls.2015.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonni I, Cottyn B, Detemmerman L, Dao S, Ouedraogo L, Sarra S, Tekete C, Poussier S, Corral R, Triplett L, et al. Analysis of Xanthomonas oryzae pv. oryzicola population in Mali and Burkina Faso reveals a high level of genetic and pathogenic diversity. Phytopathology. 2014;104:520–531. doi: 10.1094/PHYTO-07-13-0213-R. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ardales EY, Raymundo A, Bai J, Trick HN, Leach JE, Hulbert SH. The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol Plant Microbe Interact. 2004;17(7):771–779. doi: 10.1094/MPMI.2004.17.7.771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.