Abstract
Two genes, md1 and md2, coding for multidrug resistance ATP-binding cassette transporters were identified in Mycoplasma hominis PG21. Expression of these two genes, quantified by quantitative competitive reverse transcription-PCR, was significantly increased in ethidium bromide-resistant strains of M. hominis compared to that in M. hominis PG21.
Mycoplasma hominis is a cause of human urogenital tract infections for the treatment of which fluoroquinolones represent an efficient antimicrobial class. Three mechanisms of bacterial resistance to fluoroquinolones have been described, target-related mechanisms, by either alteration or protection, and active efflux (7). Active efflux of fluoroquinolones is mediated by endogenous multidrug resistance (MDR) efflux pumps, increased expression of which can develop an MDR phenotype (15). In M. hominis, resistance by target alteration has already been described in vivo (2) and in vitro (1). Furthermore, we recently reported an active efflux system, possibly an ATP-binding cassette (ABC)-type efflux pump, in ethidium bromide (EtBr)-selected strains of M. hominis showing an MDR phenotype with increased MICs of ciprofloxacin and EtBr (16).
Few bacterial ABC MDR efflux systems have been characterized; all are homologous to the known human P glycoprotein LmrA in Lactococcus lactis (19), MsbA in Escherichia coli (3), HorA in Lactobacillus brevis (17), VcaM in Vibrio cholerae (8), and more recently the heterodimeric ABC transporter EfrAB in Enterococcus faecalis (10). Mycoplasmal genome sequencing revealed the presence of two adjacent ABC-type genes identified as putative MDR genes, mg014 and mg015 in Mycoplasma genitalium and pmd1 and msbA in Mycoplasma pneumoniae (13, 18). To determine the genetic support of the ciprofloxacin and EtBr active efflux identified in M. hominis, we searched for homologous genes in the M. hominis genome.
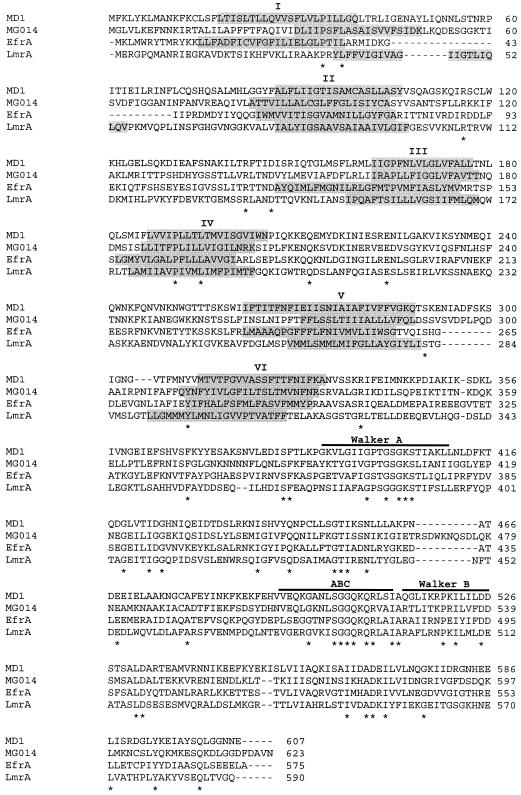
A consensus primer, MD2-1 (5′-GGTCCTACAGGAACTGGAAAA-3′), located in the Walker A motif of the ATP-binding domain and a degenerated primer, MD2R (5′-ATAATYTCHTCWTYWGTDGCAT-3′), located downstream of this motif just before the ABC signature were deduced from the alignment of MDR-like genes of M. genitalium (5) and M. pneumoniae (6). PCR amplification of the genomic DNA from the M. hominis PG21 reference strain with primers MD2-1 and MD2R led to a 224-bp DNA fragment showing homology with the mg014 gene of M. genitalium. Two recombinant plasmids selected by colony hybridization with the radiolabeled 224-bp DNA fragment were obtained from two HindIII and XbaI genomic libraries of M. hominis PG21. Inserts of these two recombinant plasmids were sequenced by primer walking. The 10,616-bp DNA sequence obtained was shown to contain eight putative open reading frames, two of which, E and F, were assigned as MDR-like genes in M. hominis and named md1 and md2, respectively. Analysis of the upstream region of gene md1 revealed a putative promoter and a consensus Shine-Dalgarno sequence located upstream of the ATG start codon. The TAG stop codon of the md1 gene is preceded by the ATG start codon of md2, and the two genes overlap by 8 nucleotides. A short stem-loop structure, followed by a run of T residues corresponding to a rho-independent transcription terminator frequently found in mollicutes (11), was found only downstream of the md2 gene stop codon. The predicted MD1 and MD2 proteins contained 607 and 625 amino acids, respectively, corresponding to calculated molecular masses of 68 and 70 kDa. Kyte-Doolittle hydropathy plots detected one hydrophilic carboxyl-terminal domain and one hydrophobic amino-terminal domain in both proteins. The hydrophobic domain contained six potential transmembrane segments (TMS) as described for LmrA in L. lactis (19), HorA in L. brevis (17), and other bacterial ABC MDR pumps (3, 8, 10). An ATP-binding domain was found in the carboxyl-terminal domain of both proteins, including the characteristic Walker A and B motifs and the ABC signature sequence (18). MD1 and MD2 showed 27.3% sequence identity and 68.3% similarity. ClustalW comparison of the MD1 and MD2 proteins with the other ABC-type MDR proteins identified in bacteria and the two halves of the human P glycoprotein is summarized Table 1. Proteins MD1 and MD2 showed the best levels of identity and similarity with E. faecalis MDR proteins EfrA and EfrB, respectively. It should be noted that the efrA and efrB genes seem to be organized in an operon like md1 and md2, with the two genes overlapping and being followed by a transcription terminator-like sequence (10). The sequence alignments of the MD1, MG014, EfrA, and LmrA proteins are shown Fig. 1. TMS prediction with the TMpred program indicated that the six TMSs of the MD1 protein were at positions similar to those of the other ABC MDR pumps (18, 19). The Walker A and B motifs and the ABC signature sequence were conserved in all four proteins (Fig. 1).
TABLE 1.
Percentages of identity and similarity between the MD1 and MD2 proteins of M. hominis and other ABC-type MDR transporters
| Transporter | Organism | Identity similarity (%)
|
Reference | |
|---|---|---|---|---|
| MD1 | MD2 | |||
| MD1 | M. hominis | 100, 100 | 27.3, 68.3 | This study |
| MD2 | M. hominis | 27.3, 68.3 | 100, 100 | This study |
| EfrA | E. faecalis | 29, 71.2 | 27.8, 67.8 | 12 |
| EfrB | E. faecalis | 24.2, 66.8 | 34.4, 72.8 | 12 |
| HorA | L. brevis | 13.9, 57.2 | 28, 67 | 17 |
| LmrA | L. lactis | 13.7, 56.9 | 26.6, 69.1 | 19 |
| MsbA | E. coli | 23.6, 65.9 | 29.1, 66.5 | 9 |
| VceM | V. cholerae | 21.8, 61.3 | 22.4, 62 | 8 |
| MDR1Na | Homo sapiens | 25.2, 68.3 | 26.9, 66.1 | 4 |
| MDR1Ca | Homo sapiens | 23.8, 65.5 | 25.2, 67 | 4 |
MDR1C and MDR1N correspond to the carboxy-terminal and amino-terminal halves of the human P glycoprotein, respectively.
FIG. 1.
ClustalW alignment of the deduced amino acid sequences of MD1 from M. hominis (GenBank accession no. AY169817), MG014 from M. genitalium (5), EfrA from E. faecalis (12), and LmrA from L. lactis (19). Asterisks indicate identical residues. Shading indicates the putative transmembrane α-helices predicted by TMpred from the INFOBIOGEN website. The ABC signature sequence and Walker A and B motifs are indicated by horizontal lines.
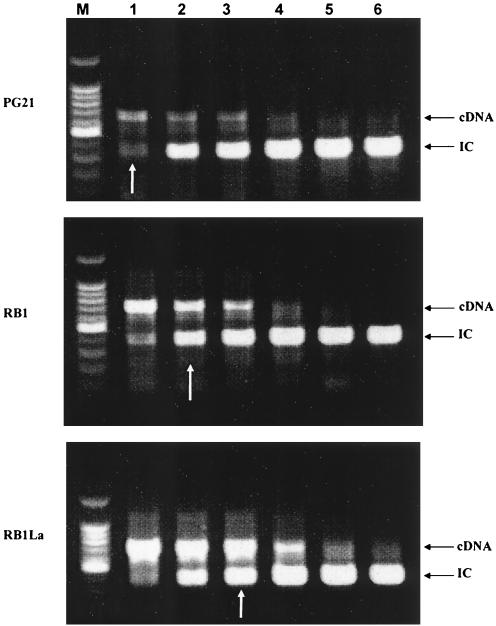
Expression of md1 and md2 in M. hominis PG21 and the MDR phenotype strains RB1 and RB1La selected on EtBr (16) was studied and quantified by quantitative competitive reverse transcription (RT)-PCR (14) (Fig. 2). RNAs were isolated from mycoplasma cultures in the exponential growth phase with the High Pure RNA isolated kit (Roche Diagnostics GmbH) and quantified by spectrophotometry. mRNAs were reverse transcribed into cDNA with the Enhanced Avian RT-PCR kit (Sigma). Internal competitor (IC) DNAs were generated by PCR amplification from M. hominis PG21 with primers a20MD1 (5′-TGTCAAAGCCATCAGAGTGCG-3′) and b20c20MD1 (5′-AATGAAGAAGCAACAGCTCCTTTGCTTGTTGTGGTTCCTC-3′) for the md1 IC and primers a20MD2 (5′-TAGTGCTTTAACATCGCTTGG-3′) and b20c20MD2 (5′-AGGTCCTACAATAGCAAACACCAACGCAAATGCTCCGCCAAC-3′) for the md2 IC. Each IC was added at concentrations of 0.03 to 2 pg to a constant amount of cDNA. The PCR amplification was performed on the IC DNA-cDNA mixture with primers a20MD1 and c20MD1 (5′-AATGAAGAAGCAACAGCTCC-3′) for md1 and primers a20MD2 and c20MD2 (5′-AGGTCCTACAATAGCAAACAC-3′) for md2. After agarose gel electrophoresis, EtBr-stained PCR products were visually quantified with a UV lamp by comparing the relative amounts of the two products, which are distinct in size. For md1 expression, strain PG21 expressed 0.03 pg of mRNA whereas the RB1 and RB1La strains expressed 0.07 and 0.2 pg of mRNA, respectively, i.e., two- and sevenfold more than the PG21 strain (Fig. 2). In the same way, expression of md2 was 3- and 10-fold increased for RB1 and RB1La, respectively, compared to that of PG21 (data not shown). These results indicated constitutive expression of both genes in control strain PG21 and overexpression in the MDR phenotype strains. The most EtBr-resistant strain, RB1La, which had the lowest CIP and EtBr uptake levels (16), also had the highest level of md1 and md2 expression, strengthening the association of these genes with the MDR phenotype observed in M. hominis.
FIG. 2.
Quantitative competitive RT-PCR of the md1 gene in M. hominis strains PG21, RB1, and RB1La. White arrows indicate the cDNA quantity detected for each strain compared to the internal control (IC) range. Lanes: M, 100-bp molecular mass marker; 1, 0.03 pg; 2, 0.06 pg; 3, 0.2 pg; 4, 0.5 pg; 5, 1 pg; 6, 2 pg.
To explain this overexpression, we looked for mutations in the putative md1 promoter region of the RB1 and RB1La strains. However, no point mutation was detected in this region. In the same way, no mutation was found within the md1 and md2 sequences or in the 5-kbp region upstream of md1. No gene homologous to transcriptional regulator families regulating the expression of bacterial MDR pumps has been found in the mycoplasmal genomes completely sequenced. The mechanism of regulation of the expression of genes md1 and md2 remains to be determined. Genetic inactivation of these two genes in EtBr-resistant strains would prove their involvement in the MDR efflux of M. hominis. However, gene disruption through homologous recombination has not been successfully applied to M. hominis. Therefore, direct evidence of the role of the MDR genes identified in this study in M. hominis would benefit from the development of genetic tools for this microorganism.
Nucleotide sequence accession number.
The nucleotide sequence data reported for M. hominis have been submitted to the GenBank database and assigned accession no. AY169817.
REFERENCES
- 1.Bébéar, C. M., H. Renaudin, A. Charron, J. M. Bové, C. Bébéar, and J. Renaudin. 1998. Alterations in topoisomerase IV and DNA gyrase in quinolone-resistant mutants of Mycoplasma hominis obtained in vitro. Antimicrob. Agents Chemother. 42:2304-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bébéar, C. M., J. Renaudin, A. Charron, H. Renaudin, B. de Barbeyrac, T. Schaeverbeke, and C. Bébéar. 1999. Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob. Agents Chemother. 43:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. J., J. E. Chin, K. Ueda, D. P. Clark, I. Pastan, M. M. Gottesman, and I. B. Roninson. 1986. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47:381-389. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 6.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huda, N., E. W. Lee, J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 47:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karow, M., and C. Georgopoulos. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7:69-79. [DOI] [PubMed] [Google Scholar]
- 10.Lee, E. W., M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3733-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muto, A., and C. Ushida. 2002. Transcription and translation, p. 323-345. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 12.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 14.Piddock, L. J. V., M. M. Johnson, S. Simjee, and L. Pumbwe. 2002. Expression of efflux pump gene pmrA in fluoroquinolone-resistant and -susceptible clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raherison, S., P. Gonzalez, H. Renaudin, A. Charron, C. Bébéar, and C. M. Bébéar. 2002. Evidence of active efflux in resistance to ciprofloxacin and to ethidium bromide by Mycoplasma hominis. Antimicrob. Agents Chemother. 46:672-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto, K., A. Margolles, H. W. van Veen, and W. N. Konings. 2001. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J. Bacteriol. 183:5371-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Veen, H. W., and W. N. Konings. 1998. The ABC family of multidrug transporters in microorganisms. Biochim. Biophys. Acta 1365:31-36. [DOI] [PubMed] [Google Scholar]
- 19.van Veen, H. W., K. Venema, H. Bolhuis, I. Oussenko, J. Kok, B. Poolman, A. J. Driessen, and W. N. Konings. 1996. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. USA 93:10668-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]