Summary
Land plant phytochromes perceive red and far-red light to control growth and development, using the linear tetrapyrrole (bilin) chromophore phytochromobilin (PΦB). Phytochromes from streptophyte algae, sister species to land plants, instead use phycocyanobilin (PCB). PCB and PΦB are synthesized by different ferredoxin-dependent bilin reductases (FDBRs): PΦB is synthesized by HY2, whereas PCB is synthesized by PcyA. The pathway for PCB biosynthesis in streptophyte algae is unknown. We used phylogenetic analysis and heterologous reconstitution of bilin biosynthesis to investigate bilin biosynthesis in streptophyte algae.
Phylogenetic results suggest that PcyA is present in chlorophytes and prasinophytes but absent in streptophytes. A system reconstituting bilin biosynthesis in Escherichia coli was modified to utilize HY2 from the streptophyte alga Klebsormidium flaccidum (KflaHY2). The resulting bilin was incorporated into model cyanobacterial photoreceptors and into phytochrome from the early-diverging streptophyte alga Mesostigma viride (MvirPHY1).
All photoreceptors tested incorporate PCB rather than PΦB, indicating that KflaHY2 is sufficient for PCB synthesis without any other algal protein. MvirPHY1 exhibits a red–far-red photocycle similar to those seen in other streptophyte algal phytochromes.
These results demonstrate that streptophyte algae use HY2 to synthesize PCB, consistent with the hypothesis that PΦB synthesis arose late in HY2 evolution.
Keywords: aquatic photosynthesis, biliverdin IXα, Mesotaenium caldariorum, photoreceptor, spectral tuning, terrestrial colonization, tetrapyrrole biosynthesis
Introduction
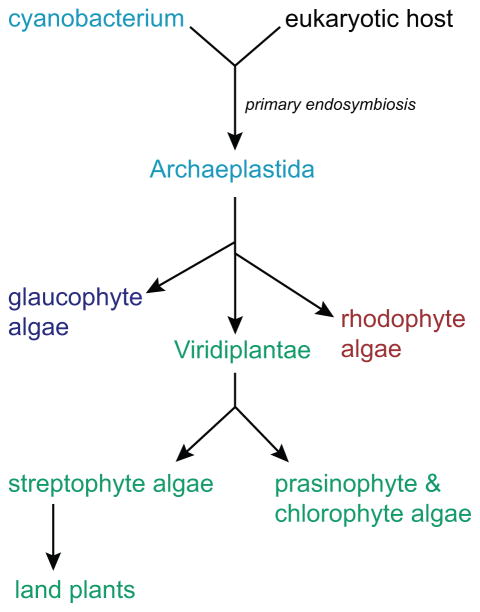
The colonization of terrestrial environments by multicellular land plants permitted the rise of terrestrial ecosystems and human societies. Recent research suggests that many of the adaptations thought to have permitted algae to evolve into plants were already present in streptophyte algae (Hori et al., 2014), the algal lineage that gave rise to land plants (Wickett et al., 2014). Streptophyte algae diverged from the prasinophyte/chlorophyte algal clade within the Viridiplantae (Leliaert et al., 2012), one of three extant lineages of the eukaryotic supergroup Archaeplastida (Fig. 1). The Archaeplastida also includes glaucophyte and rhodophyte algae, all of which may descend from a single primary endosymbiosis of a free-living cyanobacterium (Price et al., 2012; Burki et al., 2016). The transition from water to land was also accompanied by a change in light environment, because algae would typically be exposed to light enriched for blue and green wavelengths due to attenuation of ultraviolet, red, and far-red light by water (Morel, 1988; Braun & Smirnov, 1993). As sensors of red (615–675 nm) and far-red (675–740 nm) light, phytochromes are especially well suited to measuring the depletion of photosynthetically active light by neighboring plants (Lee & Graham, 1986) and to regulating shade avoidance responses that improve the competitive fitness of land plants (Rockwell et al., 2006; Franklin & Quail, 2010; Chen & Chory, 2011; Casal, 2013). The importance of this photoreceptor family in photosynthetic eukaryote evolution is underscored by recent evidence supporting the hypothesis that phytochrome was present in the common ancestor of all three extant Archaeplastida lineages (Duanmu et al., 2014; Li et al., 2015). However, the biological functions of phytochromes have not been elucidated in eukaryotic algae.
Fig. 1.
Extant lineages of Archaeplastida. A simplified view of the evolution of extant Archaeplastida lineages is shown, based on recent analyses (Price et al., 2012; Duanmu et al., 2014).
Algal phytochromes exhibit greater spectral diversity than plant phytochromes. Phytochromes from prasinophyte algae can respond to yellow or orange light, whereas glaucophyte algal phytochromes can respond to blue or red light (Rockwell et al., 2014a). The rough correlation between the spectral properties of phytochromes from different algae and their light environments (Forest, 2014; Rockwell et al., 2014a) implicates the ambient light environment as an important driving force in evolving photoreceptor spectral properties. Consistent with this hypothesis, the light absorbing properties of streptophyte algae and land plant phytochromes also differ. Streptophyte algal phytochromes exhibit red–far-red photocycles like land plant phytochromes, but both photostates of algal phytochromes are blue-shifted relative to those of land plants (Kidd & Lagarias, 1990; Jorissen et al., 2002).
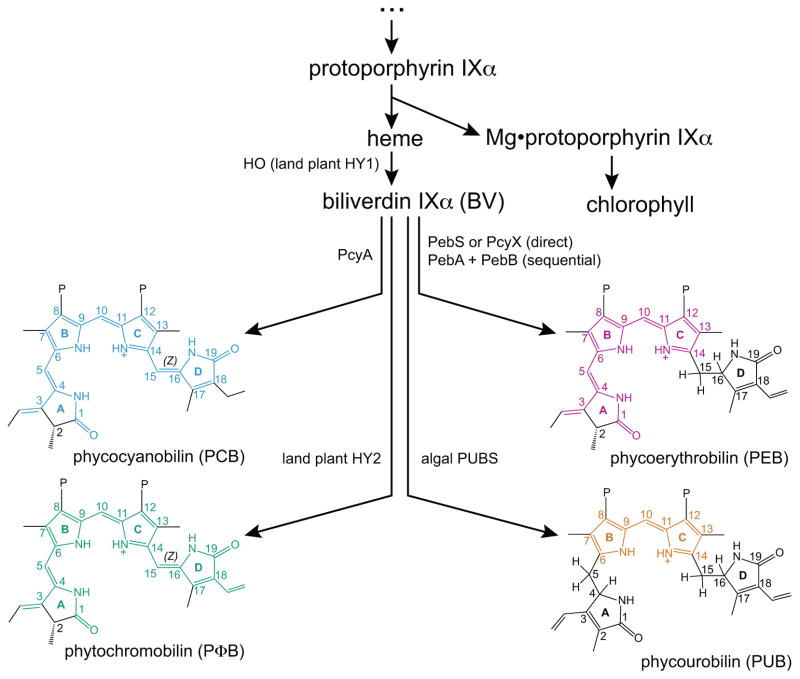
This spectral shift may arise due to differences in chromophore content: phytochrome from the derived streptophyte alga Mesotaenium caldariorum incorporates phycocyanobilin (PCB, structure shown in Fig. 2), whereas phytochromes from land plants instead incorporate phytochromobilin (PΦB, Fig. 2: Lagarias & Rapoport, 1980; Wu et al., 1997; Zeidler et al., 1998). PCB and PΦB are both synthesized from heme in two steps (Fig. 2). In the first step, heme is oxidatively metabolised to biliverdin IXα (BV) by heme oxygenases such as land plant HY1 (Davis et al., 1999; Muramoto et al., 1999; Frankenberg & Lagarias, 2003; Duanmu et al., 2013). BV is subsequently reduced by ferredoxin-dependent bilin reductases (FDBRs) to yield phytobilins (Frankenberg et al., 2001; Kohchi et al., 2001; Dammeyer & Frankenberg-Dinkel, 2008). Seven different FDBRs are known, varying in substrate specificity and regiospecificity of reduction (Fig. 2). Two of these FDBRs, PebS and PcyX, have only been found in cyanophages to date (Dammeyer et al., 2008; Ledermann et al., 2016). In cyanobacteria and chlorophyte algae, PCB is known to be synthesized from BV by PcyA (Frankenberg & Lagarias, 2003; Duanmu et al., 2013). In land plants, PΦB is synthesized from BV by HY2 (Frankenberg et al., 2001; Kohchi et al., 2001; Sawers et al., 2004). The presence of PCB rather than PΦB in M. caldariorum (Wu et al., 1997) thus implicated the presence of PcyA rather than HY2 in streptophyte algae.
Fig. 2.
Bilin biosynthesis by ferredoxin-dependent bilin reductases (FDBRs). Heme and chlorophyll are synthesized by a common trunk pathway. After initial breakdown of heme by heme oxygenase (encoded by the HY1 gene in land plants), different FDBRs produce different bilins. The chromophoric conjugated π systems of different bilins are colored, approximately matching their apparent visual colors. Algal and plant proteins encoded in the nucleus are often referred to in capital letters (e.g. PEBB), with exceptions (Petersen et al., 2006), whereas cyanobacterial enzymes are designated like other bacterial proteins (e.g. PebB). Enzymes such as PcyA would thus be formally designated as PcyA in cyanobacteria but PCYA in algae. For simplicity, we use bacterial nomenclature for such proteins throughout. Bilins are shown in the 3E configuration. P, propionate.
Surprisingly, recent transcriptomic and genomic studies have shown that streptophyte algae lack identifiable PcyA orthologs. BLAST (Altschul et al., 1997) searches of expressed sequence tag (EST) data have only detected HY2-related transcripts in early-diverging streptophyte algae, implicating the loss of PcyA early in streptophyte evolution (Timme et al., 2012; Rockwell et al., 2014b). Consistent with this hypothesis, the genome of the streptophyte alga Klebsormidium flaccidum contains HY2 but lacks PcyA (Hori et al., 2014). The biosynthetic origin of PCB in M. caldariorum, and by extension in other streptophyte algae, therefore remains an open question. In the present work, we address this question by reconstituting PCB biosynthesis from streptophyte algae via heterologous expression in an in vivo system. Our results demonstrate that algal HY2 enzymes can synthesize PCB, implicating more recent evolution of PΦB product specificity within the streptophyte HY2 lineage that may have coincided with the appearance of land plants.
Materials and Methods
Bioinformatics
Selected cyanobacterial and genomic land plant FDBR sequences were extracted from an existing FDBR alignment available as part of the distribution package for homolmapper software (Rockwell & Lagarias, 2007). FDBR sequences were identified using BLAST searches of genomes of Klebsormidium flaccidum, Cyanidioschyzon merolae, and Galdieria sulphuraria and of publicly available transcriptomes from the large-scale MMETSP (Keeling et al., 2014) and OneKP (Matasci et al., 2014) projects (accessible at http://marinemicroeukaryotes.org/ and https://sites.google.com/a/ualberta.ca/onekp/, respectively). The Cyanophora paradoxa PcyA sequence was assembled using both the draft genome (http://cyanophora.rutgers.edu/cyanophora/home.php) and BLAST searches of the Genbank EST database for this organism. Neither source was complete, but comparison of the two allowed construction of a complete gene model (Supporting Information Fig. S1a). The Genbank EST database was also used to identify FDBRs in the streptophyte algae Penium margaritaceum and Chaetosphaeridium globosum (Timme et al., 2012). FDBRs with complete or almost complete catalytic core regions were manually added to the pre-existing alignment, using published FDBR structures (Hagiwara et al., 2006; Tu et al., 2007; Busch et al., 2011) as a guide. This procedure removed N- and C-terminal extensions found in some algal sequences, since such extensions are known not to affect function in PcyA from the chlorophyte alga Chlamydomonas reinhardtii (Duanmu et al., 2013). Large insertions in PUBS from Mantoniella antarctica SL-175 (Fig. S1b) were omitted from the final alignment. Red chlorophyll catabolite reductase (RCCR) sequences from selected land plant genomes and transcriptomes were manually added to the FDBR alignment as an outgroup, using the crystal structure for Arabidopsis thaliana RCCR (Sugishima et al., 2010) as a reference to yield the alignment in Notes S1. Using an in-house script, sites with ≥5% gaps were removed and structural information was added to yield an alignment of 97 sequences and 211 amino acid sites in PHYLIP format (Notes S2). Accession information for FDBR sequences in this study is presented in Table S1. This alignment was used with PhyML-structure (Le & Gascuel, 2010) to infer a maximum likelihood phylogeny using an EX-EHO substitution model in partition mode with four rate substitution categories and with empirically estimated gamma shape parameter and proportion of invariant sites (command line settings: -m EX_EHO -M PART -a e -c 4 -v e -o tlr). Support was estimated using the approximate likelihood ratio test (aLRT) as implemented in PhyML (Guindon et al., 2009). Notes S1 and S2 are supplied as a pair of flat unix text files in a compressed archive generated with the standard unix utilities gzip and tar (MD5 checksum 887fa0320bc0d89a32817e6f21e359a1).
Plasmid construction
To reconstitute algal bilin biosynthesis, plasmid pKT271 (Mukougawa et al., 2006) was chosen as a starting point. BLAST searches against the K. flaccidum genome (Hori et al., 2014) had identified a complete HY2 gene, permitting commercial synthesis of a synthetic gene (Genscript) lacking the predicted chloroplast transit peptide but encoding the catalytic core region (amino acids 108–403 of Genbank accession GAQ84116: Fig. S2) and having flanking BamHI and SalI sites. A unique BamHI site was introduced 5′ to the cyanobacterial PcyA gene in the parent plasmid pKT271 (Mukougawa et al., 2006), and the synthetic KflaHY2 gene was cloned using BamHI and SalI to yield plasmid pBam-KflaHY2. A plastid-directed heme oxygenase (KflaHY1) was similarly identified in the K. flaccidum genome, and a synthetic gene (Genscript) encoding the catalytic core of KflaHY1 (amino acids 92–315 of Genbank accession GAQ79033: Fig. S3) was cloned into unique NdeI and XhoI sites in pBam-KflaHY2 to yield plasmid pBam-full_Kfla, allowing complete reconstitution of the K. flaccidum bilin biosynthesis pathway in Escherichia coli.
We used coexpressed biliprotein photoreceptors as acceptors for the bilin synthesized by KflaHY2 in E. coli cells. NpR5113g23 is a fragment of tandem photoreceptor containing two cyanobacteriochrome (CBCR) GAF domains, each of which have known properties (Rockwell et al., 2012a,c). NpR5113g23 (amino acids 206–561 of the Npun_R5113 locus of Nostoc punctiforme) was amplified from N. punctiforme genomic DNA using the polymerase chain reaction and was cloned into the published plasmid pBAD-Cph1-CBD (Rockwell et al., 2009), for E. coli expression of cyanobacterial phytochrome Cph1, using unique NcoI and SmaI sites to replace the Cph1 open reading frame. NpR5113g23 sequences were then amplified from the resulting plasmid and cloned into the published plasmid pET28-RcaE (Hirose et al., 2013), for E. coli expression of CBCR RcaE, using unique NcoI and BamHI restriction sites to replace the RcaE open reading frame. NpR5113g2 and Cph1 were similarly subcloned into pET28-RcaE using the published intein-chitin-binding-domain (intein-CBD) expression plasmids (Rockwell et al., 2009, 2012c) as PCR templates. MvirPHY1 (amino acids 3-608 of the M. viride phytochrome gene, Genbank accession AKN34540; Fig. S4) was procured as a synthetic gene (Genscript) and cloned into pBAD-Cph1-CBD and pET28-RcaE using the same sites and appropriate primers. All constructs were verified by oligonucleotide sequencing.
Photoreceptor expression, purification, and characterization
Characterization of NpR5113g2 with PCB chromophore and of NpR5113g3 with phycoviolobilin (PVB) chromophore has been previously described (Rockwell et al., 2012a,c, 2014c). To obtain NpR5113g2 with PΦB chromophore, we followed the previous procedure but used pPL-PΦB (Fischer et al., 2005) to drive bilin biosynthesis. The NpR5113g2 construct contains a C-terminal intein-CBD tag for affinity purification. The same affinity purification scheme was used to obtain a reference standard of MvirPHY1 containing PCB. For all intein-CBD fusion proteins, expression in E. coli strain LMG194 and induction of PCB or PΦB biosynthesis were as described (Gambetta & Lagarias, 2001; Fischer et al., 2005). Intein-CBD fusion proteins were purified on chitin resin (NEB) as previously described, with final dialysis into TKKG buffer (25 mM TES-KOH pH 7.8, 100 mM KCl, 10% (v/v) glycerol) after step elution using dithiothreitol (Rockwell et al., 2009).
His-tagged NpR5113g23, Cph1, and MvirPHY1 were expressed in E. coli strains BL21[DE3] or C41 (Miroux & Walker, 1996) using either cyanobacterial PcyA (Mukougawa et al., 2006) or KflaHY2 to reconstitute the last step of bilin biosynthesis in K. flaccidum. His-tagged NpR5113g2 was expressed in E. coli strain C41 using K. flaccidum HY1 and HY2 to reconstitute the entire pathway of K. flaccidum bilin biosynthesis. His-tagged proteins were purified on purified on His-bind Ni2+-NTA resin (Novagen) using an imidazole gradient and dialyzed into 20 mM sodium phosphate (pH 7.5), 50 mM NaCl, 10% (v/v) glycerol and 1 mM EDTA (Rockwell et al., 2012a; Hirose et al., 2013). Purified proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using standard procedures and apparatus (Bio-Rad) followed by semi-dry transfer to PVDF membranes, staining with amido black for visualizing total protein, and zinc blotting (Berkelman & Lagarias, 1986). Absorption spectra were acquired on a Cary 50 spectrophotometer. Photoconversion was triggered in the absorption cuvette using a xenon source equipped with band-pass interference filters (400 ± 35 nm, 420 ± 5 nm, 500 ± 20 nm, 550 ± 35 nm, 600 ± 20 nm, 650 ± 20 nm) or with a 695 nm long-pass filter. For denaturation assays, a 100 μl aliquot of protein was added to 1 ml of 7 M guanidinium chloride/1% HCl (v/v). Denatured samples were illuminated using the xenon lamp equipped with a 320 nm long-pass filter.
Results
Streptophyte algae contain HY2
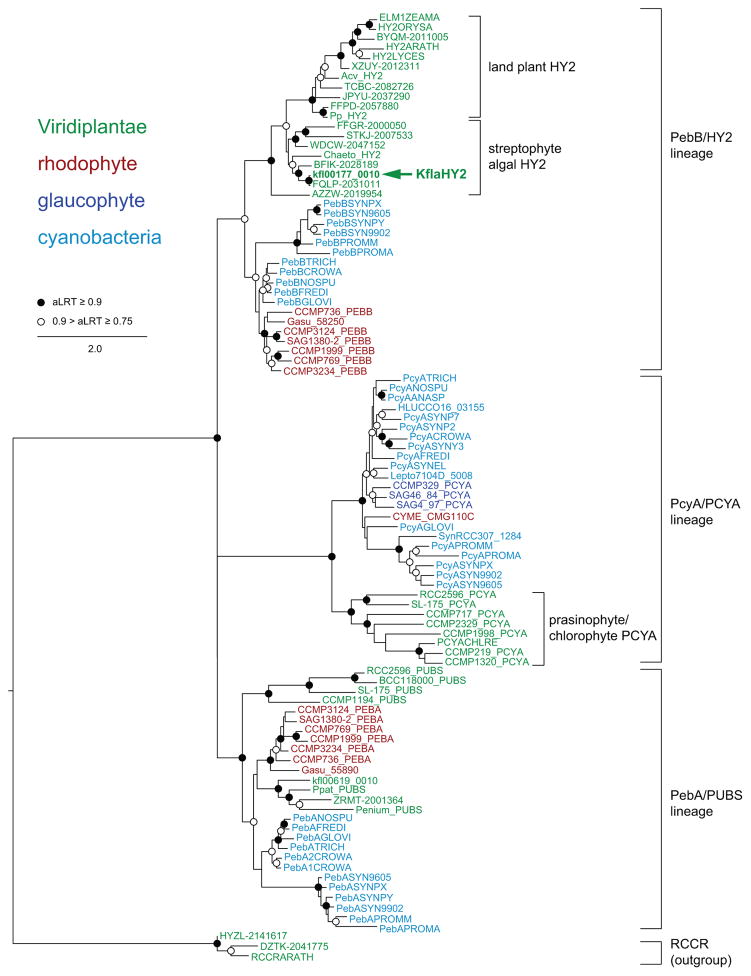
The previous assessment that streptophytes lack a PcyA gene (Rockwell et al., 2014b) was based on preliminary BLAST searches of then-available EST data (Timme et al., 2012). To extend this analysis, we used publicly available transcriptomic data from the OneKP project (Matasci et al., 2014) to identify additional FDBR sequences from a range of algae. Rather than attempting a complete analysis of FDBR evolution, we sought to identify the FDBRs present in streptophyte algae to understand the biosynthesis of PCB in such algae. We therefore focused on identifying sequences in early-diverging streptophyte genera such as Chlorokybus, Mesostigma, Spirotaenia, Entransia, and Klebsormidium (Wickett et al., 2014). Complete or almost complete FDBR sequences were identified in OneKP transcriptomes from Chlorokybus atmophyticus, Entransia fimbriata, and Klebsormidium subtile as well as in the genome of K. flaccidum (Hori et al., 2014). FDBR sequences were also found in Mesostigma viride and Spirotaenia minuta transcriptomes, but these sequences were incomplete and hence were not used in phylogenetic analysis. FDBR sequences from prasinophyte, glaucophyte, chlorophyte, and rhodophyte algae had been previously identified in transcriptomes sequenced under the auspices of the MMETSP project (Keeling et al., 2014; Rockwell et al., 2014b). Complete or almost complete eukaryotic FDBR sequences were aligned to cyanobacterial FDBR sequences, yielding an alignment containing FDBRs from all Archaeplastida lineages and from representative cyanobacteria. RCCR sequences were added to the alignment as an outgroup. The resulting alignment (Notes S1, S2) was combined with structural information to perform a structurally informed maximum-likelihood phylogeny in PhyML-structure (Le & Gascuel, 2010). As shown in Fig. 3, the results agree with previous analyses of FDBR evolution (Frankenberg et al., 2001; Rockwell et al., 2014b), recovering three FDBR lineages: PcyA, PebA/PUBS, and PebB/HY2.
Fig. 3.
Streptophyte algae contain HY2 and lack PcyA. A maximum-likelihood tree was inferred for ferredoxin-dependent bilin reductases (FDBRs) from cyanobacteria and Archaeplastida using PhyML-structure (Le & Gascuel, 2010) as described in the Materials and Methods section. Support was estimated using the approximate likelihood ratio test (aLRT) as implemented in PhyML-Structure (Guindon et al., 2009). Accession information for all sequences is in Supporting Information Table S1, and the alignment is shown in Notes S1 and S2. The arrow indicates KflaHY2 (annotated as kfl00177_0010 on the Klebsormidium flaccidum genome).
The PcyA lineage comprises cyanobacterial and algal PcyA sequences (see Fig. 2 legend for nomenclature). All members of this lineage characterized to date carry out the 4-electron reduction of BV to PCB (Fig. 2), including cyanobacterial, phage, and chlorophyte examples (Frankenberg & Lagarias, 2003; Dammeyer et al., 2008; Duanmu et al., 2013), but noting the caveat that cyanobacterial PcyX sequences were not included in our phylogeny and are thought to belong to this lineage (Ledermann et al., 2016). Eukaryotic PcyA genes were found in glaucophyte, prasinophyte, and chlorophyte algae (Fig. 3; Table S1). A PcyA gene was also found in the genome of the atypical rhodophyte Cyanidioschyzon merolae (Fig. 3). No streptophyte members of the PcyA lineage were identified.
The PebA/PUBS lineage comprises PebA and PUBS enzymes. PebA enzymes carry out the 2-electron reduction of BV to 15,16-dihydrobiliverdin (15,16-DHBV), targeting the 15,16-double bond (Fig. 2). PUBS enzymes carry out the same reaction but also reduce the 4,5-double bond (Chen et al., 2012), resulting in the 4-electron reduction of BV to PUB (Fig. 2). Cyanobacteria and rhodophytes other than C. merolae contain PebA sequences. Members of the Viridiplantae instead contain PUBS sequences (Fig. 3). This includes streptophyte algae, mosses such as Physcomitrella patens, in which PUBS was first described (Chen et al., 2012), and prasinophytes such as Prasinococcus capsulatus, Mantoniella antarctica, and Ostreococcus spp. (Table S1). PUBS was found in prasinophyte and streptophyte algae, but not in flowering plants or derived chlorophyte algae such as Chlamydomonas. No evidence for PebA or PUBS was found in glaucophyte algae. Prasinophyte and streptophyte PUBS sequences did not form a monophyletic clade in this analysis. However, it is known that members of both PUBS clades act as PUBS and not PebB enzymes, because in vitro PUBS activity has been demonstrated for purified enzymes from P. patens, Selaginella moellendorffii, and Ostreococcus lucimarinus after heterologous expression in E. coli (Chen et al., 2012). These studies show that members of the PebA/PUBS lineage are present in rhodophytes and early diverging Viridiplantae but suggest that these enzymes are absent in glaucophytes.
The third FDBR lineage found in cyanobacteria and Archaeplastida comprises PebB and HY2 enzymes. Both enzymes catalyze reduction of the C3-endovinyl substituent, but they use different substrates (Frankenberg & Lagarias, 2003; Dammeyer & Frankenberg-Dinkel, 2006): PebB acts on 15,16-DHBV to produce PEB, whereas land plant HY2 converts BV to PΦB (Fig. 2). PebB and HY2 are sister lineages, with the monophyly of each robustly supported (Fig. 3). PebB was found in cyanobacteria and in rhodophytes other than C. merolae. HY2 sequences were found in land plants, including early-diverging land plants such as Nothoceros vincentianus (a hornwort), P. patens and Ceratodon purpureus (mosses), and Marchantia polymorpha (a liverwort) (Wickett et al., 2014). HY2 was also present in streptophyte algae (Fig. 3). HY2 from Chlorokybus atmophyticus is sister to all other streptophyte HY2 sequences (Fig. 3, accession AZZW-2019954). Although this topology is not fully supported, it is consistent with organismal relationships of streptophyte algae and land plants (Wickett et al., 2014). Only HY2 was found in Mesotaenium endlicherianum, suggesting that PCB of the closely related M. caldariorum is not synthesized by PcyA. Transcriptomic data thus provide good support for the presence of HY2 and the absence of PcyA in streptophyte algae. Moreover, only HY2 and PUBS sequences were found in the only available streptophyte algal genome, that of Klebsormidium flaccidum (Hori et al., 2014). These results suggest that PCYA was lost from ancestors of the streptophyte algal lineage, whereas HY2 arose within this lineage.
Reconstitution of Klebsormidium bilin biosynthesis
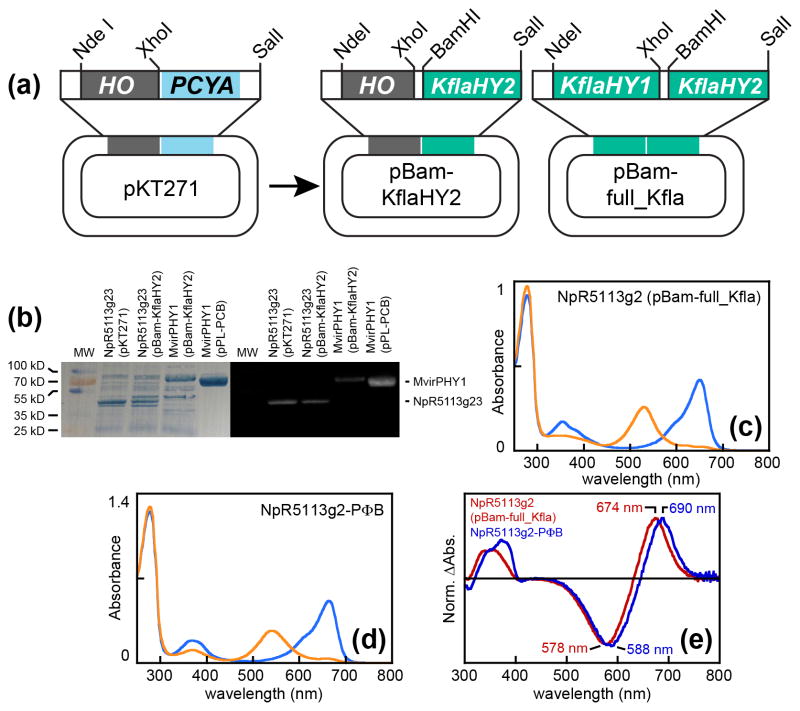
The confirmation that streptophyte algae contain HY2 and not PcyA is inconsistent with the known synthesis of PCB in M. caldariorum (Wu et al., 1997). It remains possible that M. caldariorum contains an atypical PcyA gene absent in other streptophyte algae. However, HY2 itself could be an alternative source of PCB in M. caldariorum were algal HY2 proteins to exhibit different regiospecificity than the HY2 proteins of land plants. HY2 and PcyA both reduce the A-ring endo-vinyl group of BV, with PcyA carrying out the additional reduction of the D-ring exo-vinyl group (Fig. 2). It thus seemed plausible that algal HY2 enzymes might also carry out the second reduction to yield PCB. Similar changes in FDBR regiospecificity can be seen in the PebA/PUBS lineage (Chen et al., 2012) and in the recently described cyanophage enzyme PcyX (Ledermann et al., 2016), which is related to PcyA but converts BV to phycoerythrobilin (PEB) instead of PCB (Fig. 2). To elucidate the reaction product of an algal HY2 enzyme, we replaced the cyanobacterial PcyA gene in a published system for PCB biosynthesis in E. coli (Mukougawa et al., 2006) with the core catalytic region of the HY2 gene from Klebsormidium flaccidum (KflaHY2, Fig. 4a). Due to possible incompatibility between the cyanobacterial heme oxygenase and KflaHY2, we also constructed a plasmid in which both cyanobacterial enzymes were replaced with HY1 and HY2 enzymes from K. flaccidum (Fig. 4a). Reconstitution in E. coli permits incorporation of the resulting bilin into diverse phytochromes and CBCRs, many of which have been characterized with both PCB and PΦB (Yeh et al., 1997; Rockwell et al., 2011, 2012a–c). Characterization of the ‘acceptor’ photoreceptor thus permits identification of the KflaHY2 reaction products, as has been done for the HY2 ortholog ELM1 in Zea mays (Sawers et al., 2004). We first purified holoprotein photoreceptors using KflaHY2 and cyanobacterial heme oxygenase for chromophore biosynthesis (Fig. 4b). These results demonstrate that KflaHY2 is able to support synthesis of bilin chromophores in E. coli and also enable identification of the KflaHY2 reaction product by characterizing these photoreceptors.
Fig. 4.
Reconstitution of algal bilin biosynthesis in Escherichia coli. (a) Construction of bilin biosynthesis plasmids used in this study is shown. (b) Purified biliprotein photoreceptors are shown after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (left) and zinc blotting (right) to assess bilin content (Berkelman & Lagarias, 1986). MW, molecular weight standards. (c) Absorption spectra are shown for NpR5113g2 in the 15Z red-absorbing dark state (blue) and 15E green-absorbing photoproduct state (orange) after expression with bilin biosynthesis driven by KflaHY1 and KflaHY2. (d) Absorption spectra are shown for NpR5113g2 in the 15Z red-absorbing dark state (blue) and 15E green-absorbing photoproduct state (orange) after coexpression with biosynthetic machinery for phytochromobilin (PΦB) (Fischer et al., 2005). (e) Normalized photochemical difference spectra are shown for denatured NpR5113g2 incorporating bilin produced using Klebsormidium flaccidum bilin biosynthetic machinery (red trace) or PΦB biosynthetic machinery (dark blue trace), with peak wavelengths indicated.
Klebsormidium HY2 synthesizes PCB
We compared recombinant photoreceptors incorporating bilin produced by KflaHY2 with those incorporating bilin produced using published systems for production of PCB or PΦB in E. coli (Gambetta & Lagarias, 2001; Fischer et al., 2005; Mukougawa et al., 2006). NpR5113g2, a previously characterized red/green CBCR (Rockwell et al., 2012c, 2014c), was examined after expression in the presence of both KflaHY1 and KflaHY2. The resulting protein preparation exhibited lower chromophorylation than published preparations using cyanobacterial bilin biosynthesis enzymes, but the red/green photocycle was readily observed (Fig. 4c; Table 1). We also used the previously published expression construct (Rockwell et al., 2012c) in combination with a published system for reconstitution of PΦB synthesis (Fischer et al., 2005) to obtain a reference sample incorporating PΦB (Fig. 4d; Table 1). Peak wavelengths for NpR5113g2 incorporating the KflaHY2 reaction product were 650 nm for the 15Z red-absorbing dark state and 530 nm for the 15E green-absorbing photoproduct (Table 1). These wavelengths were similar to those previously reported for the PCB adduct (650 nm and 528 nm: Table 1) and were distinct from those observed for the PΦB adduct (664 nm and 542 nm).
Table 1.
In vitro spectral properties of biliprotein photosensors in this study1
| Photoreceptor | Affinity Tag | FDBR | SAR | 15Z λmax | 15E λmax |
|---|---|---|---|---|---|
| NpR5113g22 | intein-CBD | PcyA | 1.2 | 650 | 528 |
| NpR5113g2 | intein-CBD | AtHY2 | 0.4 | 664 | 542 |
| NpR5113g2 | poly-His | KflaHY2 | 0.5 | 650 | 530 |
| NpR5113g33 | poly-His | PcyA | 0.3 | 422 | 496 |
| NpR5113g23 | poly-His | PcyA | 0.4 | 650, 422 | 528, 496 |
| NpR5113g23 | poly-His | KflaHY2 | 0.2 | 650, 424 | 528, 496 |
| Cph1 | poly-His | PcyA | 0.2 | 660 | 704 |
| Cph1 | poly-His | KflaHY2 | 0.1 | 666 | 714 |
| MvirPHY1 | intein-CBD | PcyA | 0.9 | 646 | 718 |
| MvirPHY1 | poly-His | KflaHY2 | 0.1 | 646 | 716 |
For NpR5113g23, values for domain 2 are reported first and values for domain 3 are derived from the photochemical difference spectrum after subtraction of signals from domain 2 (Fig. 5). Specific absorbance ratio (SAR) was calculated as the ratio of the peak absorbance of the longest-wavelength chromophore band for the 15Z photostate to the peak absorbance of the aromatic amino acid band at 280 nm. NpR5113g2 was expressed in the presence of KflaHY2 and KflaHY1, whereas other KflaHY2 expressions used KflaHY2 and cyanobacterial heme oxygenase. 2Values from Rockwell et al. (2012c). 3Values from Rockwell et al. (2012a).
We next characterized NpR5113g23, a larger fragment comprising 2 CBCR GAF domains of the Npun_R5113 photoreceptor from N. punctiforme that includes both NpR5113g2 and the adjacent blue/teal CBCR NpR5113g3 (Rockwell et al., 2012a). NpR5113g3 autocatalytically isomerizes PCB to PVB, a bilin with a saturated C5 methine bridge and a shorter conjugated system. NpR5113g3 is hence quite sensitive to the presence of the additional 18-vinyl moiety of PΦB (Rockwell et al., 2012a,b). NpR5113g23 holoprotein was readily obtained after expression with KflaHY2 as the only algal protein present (Fig. 4b), albeit with lower chromophore incorporation than was obtained with the parent system using PcyA from Synechocystis sp. PCC 6803 (Table 1). This result may reflect poor coupling of the cyanobacterial heme oxygenase with the streptophyte FDBR, of the streptophyte FDBR with E. coli ferredoxin, or of the streptophyte FDBR with heterologous photoreceptors. Fortunately, this system readily yielded sufficient protein for spectroscopic characterization.
Both domains of NpR5113g23 incorporated chromophore (Fig. 5a). Illumination with red light (650 ± 20 nm) yielded specific photoconversion of the red/green domain, equivalent to isolated NpR5113g2 with PCB. The resulting photochemical difference spectrum was indistinguishable from that observed for NpR5113g2 incorporating PCB and distinct from that observed for NpR5113g2 incorporating PΦB (Fig. 5b). The blue/teal photocycle of NpR5113g3 could not be independently observed due to spectral overlap: the teal-absorbing state of NpR5113g3 overlaps the green-absorbing state of NpR5113g2, and the blue-absorbing state of NpR5113g3 overlaps the S0–S2 (Soret) transition of the red-absorbing state of NpR5113g2. However, subtraction of the contaminating red/green component was readily achieved by scaling the difference spectrum produced by red light relative to changes in the red-absorbing band of NpR5113g2 (Fig. 5c). The resulting difference spectrum for NpR5113g3 was again consistent with that obtained for the control sample incorporating PCB precursor with subsequent autocatalytic formation of PVB (Fig. 5d).
Fig. 5.
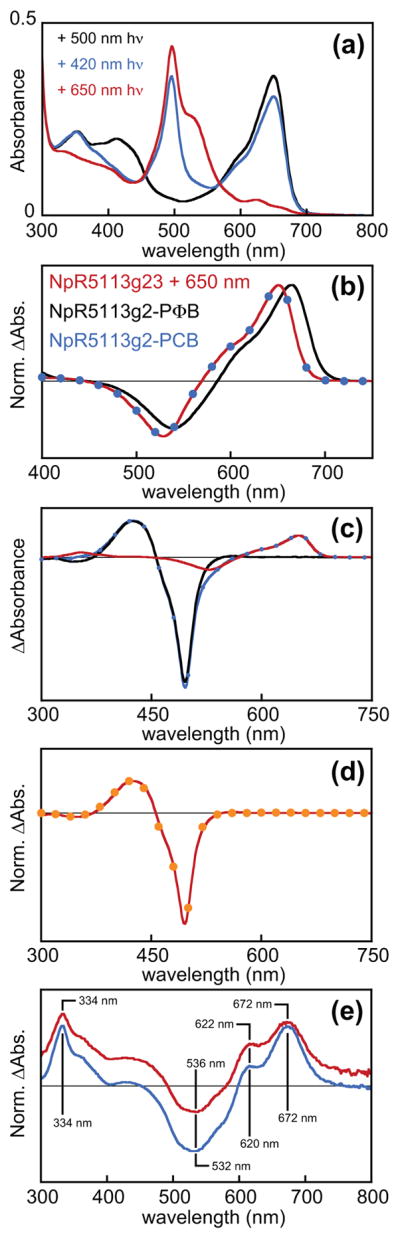
Characterization of NpR5113g23. (a) NpR5113g23 was expressed in Escherichia coli using KflaHY2 to drive the last step in bilin biosynthesis. Absorption spectra are shown for purified protein after illumination with teal light (500 ± 20 nm, black), blue light (420 ± 5 nm, blue) and red light (650 ± 20 nm, red). (b) The normalized photochemical difference spectrum observed for NpR5113g23 after illumination with red light (650 ± 20 nm, red) is compared with that for NpR5113g2 containing phycocyanobilin (PCB) (blue circles) or phytochromobilin (PΦB) (black) chromophore. (c) Subtraction of the scaled NpR5113g23 difference spectrum under red light (b, red) from the difference spectrum triggered by blue light (420 ± 5 nm, blue) is shown, with the resulting spectrum in black. (d) After subtraction of the NpR5113g2 signal (c), the resulting photochemical difference spectrum (red) is compared with that of isolated NpR5113g3 (orange circles). (e) Normalized photochemical difference spectra are shown for denatured NpR5113g23 expressed using cyanobacterial PcyA (blue) or KflaHY2 (red) for bilin biosynthesis. Peak wavelengths are indicated, and the spectrum of the KflaHY2 sample was shifted on the y-axis for clarity.
The presence of PCB in the red/green domain NpR5113g2 and of PVB in NpR5113g3 was confirmed using a well-established acid denaturation assay (Zhao et al., 1995; Zhao & Scheer, 1995; Yoshihara et al., 2006; Shang et al., 2010; Ishizuka et al., 2011; Rockwell et al., 2012a; Duanmu et al., 2013; Song et al., 2014; Narikawa et al., 2015a,b). Denaturation under these conditions does not prevent photoisomerization from the 15E configuration of the bilin chromophore to the 15Z configuration but does remove the spectral tuning effects of the native protein on the chromophore. It is thus possible to photoconvert 15E populations after denaturation as a means of assigning chromophore configuration and bilin content for a given photoreceptor (Ishizuka et al., 2011; Rockwell et al., 2011, 2012a–c, 2015a; Narikawa et al., 2015a,b). For NpR5113g23, both domains can be converted to the 15E photostate by illumination with violet light followed by red light. Denaturation of the resulting population and subsequent illumination with white light will regenerate 15Z bilin. The resulting photochemical difference spectrum for the denatured sample contains signals from both PCB and PVB. We therefore compared photochemical difference spectra for denatured NpR5113g23 containing chromophore synthesized either by KflaHY2 or by cyanobacterial PcyA. The two difference spectra exhibited similar peak and trough wavelengths (Fig. 5e), confirming that KflaHY2 synthesizes PCB that is then isomerized to PVB by the NpR5113g3 domain but not by the NpR5113g2 domain. Further support for this conclusion came from denaturation of isolated NpR5113g2, with the preparation incorporating bilin produced using KflaHY1 and KflaHY2 clearly containing PCB and not PΦB (Fig. 4e).
We next characterized two phytochromes after recombinant expression in the presence of KflaHY2 and cyanobacterial heme oxygenase. The first was the cyanobacterial phytochrome Cph1 from Synechocystis sp. PCC 6803, a well-characterized model protein for understanding phytochrome structure and function (Hughes et al., 1997; Yeh et al., 1997; Borucki et al., 2003; Fischer & Lagarias, 2004; Fischer et al., 2005; Strauss et al., 2005; Hahn et al., 2006; Rohmer et al., 2006; Essen et al., 2008; Rockwell et al., 2009; Song et al., 2011a,b; Kim et al., 2012, 2013, 2014a, b). Cph1 exhibits considerable heterogeneity. For example, peak wavelengths for the red-absorbing 15Z Pr dark state with PCB range from 654 nm to 663 nm, whereas peak wavelengths for Pr incorporating PΦB range from 668 nm to 675 nm (Yeh et al., 1997; Gambetta & Lagarias, 2001; Fischer et al., 2005; Rockwell et al., 2009). Upon incorporation of bilin produced by KflaHY2, we observed an intermediate peak wavelength of 666 nm for the Cph1 Pr state (Table 1). Photoconversion to Pfr was readily observed, even though chromophorylation was reduced relative to a control preparation coexpressed with PcyA driving PCB biosynthesis (Fig. 6a; Table 1). The photochemical difference spectra for the two samples were also not equivalent. For the KflaHY2 sample, the Pfr peak intensity was notably lower relative to that of Pr (Fig. 6b). This was not the case for the PcyA sample, for which the two peak intensities were approximately equal.
Fig. 6.

Characterization of phytochromes incorporating phycocyanobilin (PCB) produced by KflaHY2. (a) Absorption spectra are shown for Cph1 in the Pr (blue) and Pfr (orange) photostates. KflaHY2 was used to drive bilin biosynthesis, and protein was expressed as a poly-His construct. (b) Normalized difference spectra are shown for Cph1 expressed using cyanobacterial PcyA (blue) or KflaHY2 (red) for bilin biosynthesis. (c) Absorption spectra are shown for MvirPHY1 in the Pr (blue) and Pfr (orange) photostates. Cyanobacterial PcyA was used to drive PCB biosynthesis, and protein was expressed as an intein-chitin-binding-domain (intein-CBD) fusion protein. (d) Absorption spectra are shown for MvirPHY1 in the Pr (blue) and Pfr (orange) photostates. KflaHY2 was used to drive bilin biosynthesis, and protein was expressed as a poly-His construct. (e) Normalized difference spectra are shown for MvirPHY1 expressed using cyanobacterial PcyA (blue) or KflaHY2 (red) for bilin biosynthesis.
The second phytochrome tested with KflaHY2 was MvirPHY1 from M. viride, a member of the earliest diverging streptophyte algal lineage (Wickett et al., 2014). Control protein incorporating PCB exhibited ready photoconversion between Pr and Pfr states having peak wavelengths of 646 and 718 nm, respectively (Fig. 6c; Table 1). Incorporation of bilin produced by KflaHY2 resulted in a photocycle with poor chromophorylation but having almost identical peak wavelengths (646 nm and 716 nm, respectively: Fig. 6d; Table 1). Comparison of the difference spectra for the two samples demonstrated that the KflaHY2 sample again exhibited a lower Pfr peak intensity (Fig. 6e).
We therefore analyzed both phytochromes using the denaturation assay. In the case of MvirPHY1, the photochemical difference spectra for the two samples were identical after denaturation (Fig. 7a). The MvirPHY1 sample produced using KflaHY2 was also indistinguishable from that for NpR6012g4 (Fig. 7b), a red/green CBCR known to undergo 15,16-photoisomerization of a PCB chromophore (Rockwell et al., 2015b,c). The denatured difference spectrum for Cph1 incorporating PCB produced using PcyA was similarly equivalent to that of NpR6012g4 (Fig. 7c), whereas that for Cph1 produced using KflaHY2 was slightly red-shifted (Fig. 7d). However, this red shift fell within the known error of PCB peak wavelengths for this assay, and the observed peak wavelengths were clearly distinct from PΦB peak wavelengths (Fig. 7e). We therefore conclude that KflaHY2 carries out the reduction of BV to PCB and not PΦB in K. flaccidum.
Fig. 7.
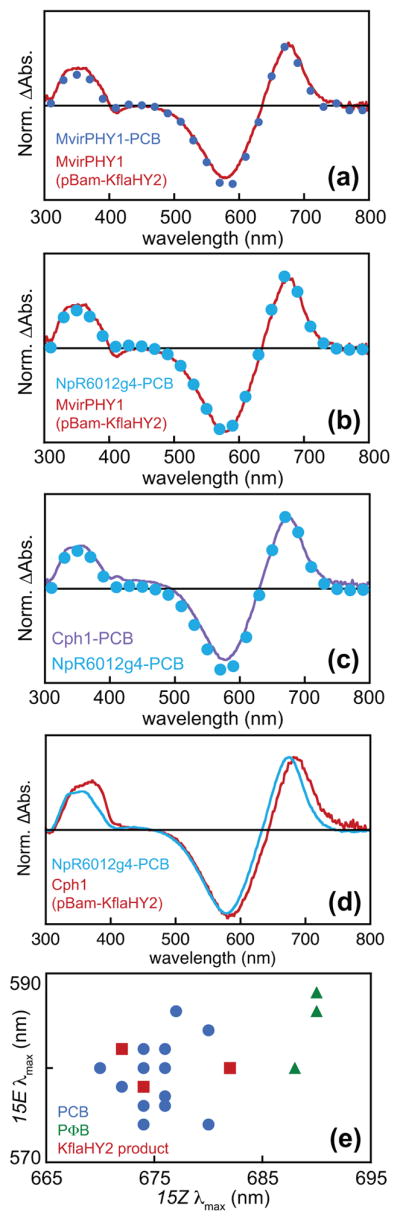
Characterization of denatured phytochromes. (a) Normalized difference spectra are shown for denatured MvirPHY1 expressed using cyanobacterial PcyA (blue circles) or KflaHY2 (red trace) for bilin biosynthesis. (b) Normalized difference spectra are shown for denatured MvirPHY1 expressed using KflaHY2 (red) and for NpR6012g4 incorporating phycocyanobilin (PCB) (teal circles). (c) Normalized difference spectra are shown for denatured Cph1 (violet) and for NpR6012g4 incorporating PCB (teal circles). (d) Normalized difference spectra are shown for denatured Cph1 expressed using KflaHY2 (red trace) and for NpR6012g4 incorporating PCB (teal trace). (e) Peak wavelengths from the photochemical difference spectra are plotted against each other (15Z, x-axis; 15E, y-axis) for denatured samples incorporating PCB (blue circles), phytochromobilin (PΦB) (green triangles), and PCB produced by KflaHY2 (red squares).
Discussion
Our studies reveal that synthesis of PCB in streptophyte algae is mediated by an FDBR belonging to the HY2 lineage rather than the PcyA lineage found in prasinophytes and chlorophytes. These results demonstrate that FDBR reaction products cannot be easily predicted by amino acid sequence and/or by phylogenetic analysis. The distinct product specificity of PebB and HY2 is easy to explain: cyanobacterial PebB does not recognize BV as substrate (Frankenberg & Lagarias, 2003), and a similar preference in rhodophyte PebB would explain the known pathway of bilin biosynthesis in rhodophytes (Beale, 1993). However, our work shows that an early-diverging streptophyte algal HY2 enzyme can catalyze the four-electron reduction of BV to PCB. This result strikingly contrasts with the two-electron reductions catalyzed by PebB (15,16-dihydrobiliverdin to PEB) and by land plant HY2 (BV to PΦB). Together with the presence of PCB in M. caldariorum (Wu et al., 1997) and of HY2 in M. endlicherianum (Table S1), our studies implicate HY2 as the source of PCB in the streptophyte algal lineage.
A complete picture of FDBR evolution is beyond the scope of the present investigation, but our studies suggest that the product regiospecificity of HY2 changed from PCB to PΦB within the streptophyte lineage. The use of PCB vs PΦB in liverworts and hornworts, two of the earliest-diverging land plant lineages, remains unknown; however, the moss Ceratodon purpureus uses PΦB for its phytochromes (Zeidler et al., 1998). Similarly, HY2 reaction products have not been established for most streptophyte algae. Thus, resolution of the origin of land plant HY2 function within the streptophyte lineage will require additional studies. The functions of FDBRs and bilins in diverse algal lineages also remain an interesting and unresolved issue. It was recently shown that the bilin biosynthesis pathway is essential for phototrophic growth in C. reinhardtii, an organism lacking phytochromes and other known bilin-binding proteins (Duanmu et al., 2013). The only FDBR present in C. reinhardtii is PcyA, and it is known to synthesize PCB (Duanmu et al., 2013). If PCB is ancestrally essential for photosynthesis in green algae, the loss of PcyA in streptophytes would only have been tolerated if was functionally redundant with the ancestral HY2 enzyme such that there was no loss of PCB biosynthesis.
We observed slight but significant differences between phytochromes incorporating PCB synthesized by PcyA or PCB synthesized by KflaHY2. The peak wavelengths for Pr and Pfr differ for the two PCB populations in the cyanobacterial model phytochrome Cph1. We also observe a systematic difference in both Cph1 and MvirPHY1: with PCB produced by PcyA, the peak Pr and Pfr intensities in the phytochrome difference spectrum are approximately equal, whereas the Pfr peak absorption is lower than that of Pr with PCB produced by KflaHY2 (Fig. 6). Thus, the Pfr extinction coefficient is lower relative to that of Pr for PCB produced by KflaHY2 than for PCB produced by PcyA. These differences are not retained upon denaturation, because the peak wavelengths for denatured samples incorporating bilin produced by KflaHY2 are all consistent with PCB (Fig. 7e).
We have not elucidated the basis for this difference in the current study, but it may arise due to differences in the PCB being produced by the two FDBRs. Synechocystis PcyA produces a mixture of 3Z and 3E PCB (Frankenberg et al., 2001), whereas the orthologous enzyme from the eukaryotic chlorophyte alga Chlamydomonas reinhardtii produces 3E PCB in excess (Duanmu et al., 2013). Crystal structures of Cph1 and phytochrome B from Arabidopsis thaliana both reveal the expected R stereochemistry for the covalent linkage to chromophore (Lagarias & Rapoport, 1980; Essen et al., 2008; Burgie et al., 2014), consistent with incorporation of 3E bilin precursors. Were KflaHY2 to produce the 3Z isomer of PCB in excess, then the resulting phytochrome holoprotein would contain the opposite S stereochemistry at the site of covalent attachment and would be expected to exhibit slightly different protein–chromophore interactions. Differences between 3Z and 3E PCB precursors have not been studied in phytochromes. However, in vitro assembly of oat phytochrome A with 3Z and 3E PΦB revealed slight differences between the two isomers (Terry et al., 1995): notably, the Pr and Pfr peak intensities for 3E chromophore were approximately equal, whereas the Pfr peak intensity is notably lower than that of Pr for 3Z chromophore. Therefore, the slight differences observed with Pfr extinction coefficients in our current work may also arise from differences in the isomers of PCB produced by different FDBRs.
Our studies also present the first spectroscopic characterization of a phytochrome from an early-diverging streptophyte alga, MvirPHY1 from M. viride. Mesostigma viride retains many features not associated with streptophytes, such as flagella and scales (Buchmann & Becker, 2009). Nevertheless, MvirPHY1 exhibits a red–far-red photocycle very similar to that of M. caldariorum phytochrome, yet distinct from the orange–far-red and yellow–far-red photocycles of phytochromes from early-diverging prasinophyte phytochromes (Kidd & Lagarias, 1990; Wu et al., 1997; Rockwell et al., 2014a). The more variable prasinophyte photocycles may reflect the ability of prasinophytes to live in deep water habitats that are depleted in red and far-red light (Forest, 2014; Rockwell et al., 2014a). The apparent lack of spectral diversity in streptophyte phytochromes may reflect a greater competition for light with land plants at the interface between aquatic and terrestrial environments or reflect the advantage of the red–far-red cycle in environments with significant levels of far-red light, such as the extremely shallow aquatic environments and moist soils where early-diverging streptophyte algae are found today (Hoshaw & Hilton, 1966; Rogers et al., 1980; Wickett et al., 2014). Experimental characterization of phytochromes and FDBRs from additional algae and early-diverging land plants, and of phytochrome gene expression and function in different light environments, are needed to test this hypothesis.
Supplementary Material
Fig. S1 Gene models for Notes S1.
Fig. S2 Synthetic gene sequence for the catalytic core of KflaHY2.
Fig. S3 Synthetic gene sequence for the catalytic core of KflaHY1.
Fig. S4 Synthetic gene sequence for the photosensory core module of MvirPHY1.
Table S1 Accession information for FDBR and RCCR sequences
Notes S1 Flat text alignment of FDBR sequences.
Notes S2 Formatted input file for FDBR phylogeny.
Acknowledgments
This work was supported by NIH grant R01 GM068552 (to J.C.L.) and NSF DEB-1407158 (to F-W.L.). Cloning of NpR5113g23 was supported by a grant from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, United States Department of Energy (DOE DE-FG02-09ER16117 to J.C.L.). The authors thank Prof. Debashish Bhattacharya (Rutgers University) for helpful discussions and Elsie Campbell and Prof. Jack Meeks (UC Davis) for the generous gift of N. punctiforme genomic DNA.
Footnotes
Author contributions
N.C.R. and J.C.L. designed the research and S.M. helped guide phylogenetic design. N.C.R. and S.S.M. performed the research. F-W.L. and S.M. provided gene models and sequences. N.C.R. wrote the manuscript. All authors commented on the manuscript.
Additional Supporting Information may be found online in the Supporting Information tab for this article:
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale SI. Biosynthesis of phycobilins. Chemical Reviews. 1993;93(2):785–802. [Google Scholar]
- Berkelman TR, Lagarias JC. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Analytical Biochemistry. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- Borucki B, Otto H, Rottwinkel G, Hughes J, Heyn MP, Lamparter T. Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochemistry. 2003;42(46):13684–13697. doi: 10.1021/bi035511n. [DOI] [PubMed] [Google Scholar]
- Braun CL, Smirnov SN. Why is water blue? Journal of Chemical Education. 1993;70:612–614. [Google Scholar]
- Buchmann K, Becker B. The system of contractile vacuoles in the green alga Mesostigma viride (Streptophyta) Protist. 2009;160(3):427–443. doi: 10.1016/j.protis.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proceedings of the National Academy of Sciences, USA. 2014;111(28):10179–10184. doi: 10.1073/pnas.1403096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proceedings of the Royal Society B-Biological Sciences. 2016;283(1823):20152802. doi: 10.1098/rspb.2015.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch AW, Reijerse EJ, Lubitz W, Frankenberg-Dinkel N, Hofmann E. Structural and mechanistic insight into the ferredoxin-mediated two-electron reduction of bilins. Biochemical Journal. 2011;439(2):257–264. doi: 10.1042/BJ20110814. [DOI] [PubMed] [Google Scholar]
- Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annual Review of Plant Biology. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends in Cell Biology. 2011;21(11):664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Su YS, Tu SL. Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proceedings of the National Academy of Sciences, USA. 2012;109(21):8310–8315. doi: 10.1073/pnas.1201744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammeyer T, Bagby SC, Sullivan MB, Chisholm SW, Frankenberg-Dinkel N. Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Current Biology. 2008;18(6):442–448. doi: 10.1016/j.cub.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Dammeyer T, Frankenberg-Dinkel N. Insights into phycoerythrobilin biosynthesis point toward metabolic channeling. Journal of Biological Chemistry. 2006;281(37):27081–27089. doi: 10.1074/jbc.M605154200. [DOI] [PubMed] [Google Scholar]
- Dammeyer T, Frankenberg-Dinkel N. Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms. Photochemical and Photobiological Sciences. 2008;7(10):1121–1130. doi: 10.1039/b807209b. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci, USA. 1999;96(11):6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Bachy C, Sudek S, Wong CH, Jimenez V, Rockwell NC, Martin SS, Ngan CY, Reistetter EN, van Baren MJ, et al. Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Sciences, USA. 2014;111:15827–15832. doi: 10.1073/pnas.1416751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Casero D, Dent RM, Gallaher S, Yang W, Rockwell NC, Martin SS, Pellegrini M, Niyogi KK, Merchant SS, et al. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proceedings of the National Academy of Sciences, USA. 2013;110(9):3621–3626. doi: 10.1073/pnas.1222375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proceedings of the National Academy of Sciences, USA. 2008;105(38):14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Lagarias JC. Harnessing phytochrome’s glowing potential. Proceedings of the National Academy of Sciences, USA. 2004;101(50):17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry. 2005;44(46):15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest KT. Vivid watercolor paintbox for eukaryotic algae. Proceedings of the National Academy of Sciences, USA. 2014;111(15):5448–5449. doi: 10.1073/pnas.1403530111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. The Plant Cell. 2001;13(4):965–978. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg NF, Lagarias JC. Biosynthesis and biological function of bilins. In: Kadish KM, Smith KM, Guilard R, editors. The porphyrin handbook. Chlorophylls and bilins: biosynthesis structure and degradation. New York, NY, USA: Academic Press; 2003. pp. 211–235. [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany. 2010;61(1):11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proceedings of the National Academy of Sciences, USA. 2001;98(19):10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Hagiwara Y, Sugishima M, Takahashi Y, Fukuyama K. Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXα, a key enzyme in the biosynthesis of phycocyanobilin. Proc Natl Acad Sci, USA. 2006;103(1):27–32. doi: 10.1073/pnas.0507266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Strauss HM, Landgraf FT, Gimenez HF, Lochnit G, Schmieder P, Hughes J. Probing protein-chromophore interactions in Cph1 phytochrome by mutagenesis. Febs J. 2006;273(7):1415–1429. doi: 10.1111/j.1742-4658.2006.05164.x. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Rockwell NC, Nishiyama K, Narikawa R, Ukaji Y, Inomata K, Lagarias JC, Ikeuchi M. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proceedings of the National Academy of Sciences, USA. 2013;110(13):4974–4979. doi: 10.1073/pnas.1302909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshaw RW, Hilton RL. Observations on the sexual cycle of the saccoderm desmid Spirotaenia condensata. Journal of the Arizona Academy of Science. 1966;4:88–92. [Google Scholar]
- Hughes J, Lamparter T, Mittmann F, Hartmann E, Gartner W, Wilde A, Borner T. A prokaryotic phytochrome. Nature. 1997;386(6626):663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Kamiya A, Suzuki H, Narikawa R, Noguchi T, Kohchi T, Inomata K, Ikeuchi M. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry. 2011;50(6):953–961. doi: 10.1021/bi101626t. [DOI] [PubMed] [Google Scholar]
- Jorissen H, Braslavsky SE, Wagner G, Gärtner W. Heterologous expression and characterization of recombinant phytochrome from the green alga Mougeotia scalaris. Photochemistry and Photobiology. 2002;76(4):457–461. doi: 10.1562/0031-8655(2002)076<0457:heacor>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biology. 2014;12(6):e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd DG, Lagarias JC. Phytochrome from the green alga Mesotaenium caldariorum – purification and preliminary characterization. Journal of Biological Chemistry. 1990;265(12):7029–7035. [PubMed] [Google Scholar]
- Kim PW, Pan J, Rockwell NC, Chang CW, Taylor KC, Lagarias JC, Larsen DS. Ultrafast E to Z photoisomerization dynamics of the Cph1 phytochrome. Chemical Physics Letters. 2012;549:86–92. doi: 10.1016/j.cplett.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PW, Rockwell NC, Freer LH, Chang CW, Martin SS, Lagarias JC, Larsen DS. Unraveling the primary isomerization dynamics in cyanobacterial phytochrome Cph1 with multi-pulse manipulations. Journal of Physical Chemistry Letters. 2013;4(16):2605–2609. doi: 10.1021/jz401443q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PW, Rockwell NC, Martin SS, Lagarias JC, Larsen DS. Dynamic inhomogeneity in the photodynamics of cyanobacterial phytochrome Cph1. Biochemistry. 2014a;53(17):2818–2826. doi: 10.1021/bi500108s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PW, Rockwell NC, Martin SS, Lagarias JC, Larsen DS. Heterogeneous photodynamics of the Pfr state in the cyanobacterial phytochrome Cph1. Biochemistry. 2014b;53(28):4601–4611. doi: 10.1021/bi5005359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. The Plant Cell. 2001;13(2):425–436. doi: 10.1105/tpc.13.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarias JC, Rapoport H. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. Journal of the American Chemical Society. 1980;102:4821–4828. [Google Scholar]
- Le SQ, Gascuel O. Accounting for solvent accessibility and secondary structure in protein phylogenetics is clearly beneficial. Systematic Biology. 2010;59(3):277–287. doi: 10.1093/sysbio/syq002. [DOI] [PubMed] [Google Scholar]
- Ledermann B, Béjà O, Frankenberg-Dinkel N. New biosynthetic pathway for pink pigments from uncultured oceanic viruses. Environ Microbiol. 2016 doi: 10.1111/1462-2920.13290. [DOI] [PubMed] [Google Scholar]
- Lee DW, Graham R. Leaf optical properties of rainforest sun and extreme shade plants. American Journal of Botany. 1986;73(8):1100–1108. [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and molecular evolution of the green algae. Critical Reviews in Plant Sciences. 2012;31(1):1–46. [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nature Communications. 2015;6:7852. doi: 10.1038/ncomms8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. Journal of Molecular Biology. 1996;260(3):289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Morel A. Optical modeling of the upper ocean in relation to its biogenous matter content (Case I waters) Journal of Geophysical Research. 1988;93(C9):10749–10768. [Google Scholar]
- Mukougawa K, Kanamoto H, Kobayashi T, Yokota A, Kohchi T. Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Letters. 2006;580(5):1333–1338. doi: 10.1016/j.febslet.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. The Plant Cell. 1999;11(3):335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R, Fushimi K, Ni Ni W, Ikeuchi M. Red-shifted red/green-type cyanobacteriochrome AM1_1870g3 from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Biochemical & Biophysical Research Communications. 2015a;461(2):390–395. doi: 10.1016/j.bbrc.2015.04.045. [DOI] [PubMed] [Google Scholar]
- Narikawa R, Nakajima T, Aono Y, Fushimi K, Enomoto G, Ni Ni W, Itoh S, Sato M, Ikeuchi M. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Scientific Reports. 2015b;5:7950. doi: 10.1038/srep07950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Teich R, Becker B, Cerff R, Brinkmann H. The GapA/B gene duplication marks the origin of Streptophyta (charophytes and land plants) Mol Biol Evol. 2006;23(6):1109–1118. doi: 10.1093/molbev/msj123. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335(6070):843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D, Worden AZ, Lagarias JC. Eukaryotic algal phytochromes span the visible spectrum. Proceedings of the National Academy of Sciences, USA. 2014a;111(10):3871–3876. doi: 10.1073/pnas.1401871111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. Flexible mapping of homology onto structure with homolmapper. BMC Bioinformatics. 2007;8:123. doi: 10.1186/1471-2105-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC, Bhattacharya D. Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Frontiers in Ecology and Evolution. 2014b;2:66. doi: 10.3389/fevo.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proceedings of the National Academy of Sciences, USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012a;51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry. 2014c;53(19):3118–3130. doi: 10.1021/bi500037a. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012b;51(17):3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Red/green cyanobacteriochromes: sensors of color and power. Biochemistry. 2012c;51(48):9667–9677. doi: 10.1021/bi3013565. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochemical & Photobiological Sciences. 2015a;14(5):929–941. doi: 10.1039/c4pp00486h. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lim S, Lagarias JC, Ames JB. Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: a hydrophobic pocket for the C15-E,anti chromophore in the photoproduct. Biochemistry. 2015b;54(24):3772–3783. doi: 10.1021/acs.biochem.5b00438. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lim S, Lagarias JC, Ames JB. Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: a protonated bilin ring system in both photostates. Biochemistry. 2015c;54(16):2581–2600. doi: 10.1021/bi501548t. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Shang L, Martin SS, Lagarias JC. Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proceedings of the National Academy of Sciences, USA. 2009;106(15):6123–6127. doi: 10.1073/pnas.0902370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CE, Mattox KR, Stewart KD. The zoospore of Chlorokybus atmophyticus, a charophyte with sarcinoid growth habit. American Journal of Botany. 1980;67:774–783. [Google Scholar]
- Rohmer T, Strauss H, Hughes J, de Groot H, Gartner W, Schmieder P, Matysik J. 15N MAS NMR studies of cph1 phytochrome: chromophore dynamics and intramolecular signal transduction. J Phys Chem B. 2006;110(41):20580–20585. doi: 10.1021/jp062454+. [DOI] [PubMed] [Google Scholar]
- Sawers RJ, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR, Kohchi T, Terry MJ, Brutnell TP. The Elm1 (ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiology. 2004;136(1):2771–2781. doi: 10.1104/pp.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Rockwell NC, Martin SS, Lagarias JC. Biliverdin amides reveal roles for propionate side chains in bilin reductase recognition and in holophytochrome assembly and photoconversion. Biochemistry. 2010;49(29):6070–6082. doi: 10.1021/bi100756x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Psakis G, Kopycki J, Lang C, Matysik J, Hughes J. The D-ring, not the A-ring, rotates in Synechococcus OS-B′ phytochrome. Journal of Biological Chemistry. 2014;289(5):2552–2562. doi: 10.1074/jbc.M113.520031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Psakis G, Lang C, Mailliet J, Gartner W, Hughes J, Matysik J. Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proceedings of the National Academy of Sciences, USA. 2011a;108(10):3842–3847. doi: 10.1073/pnas.1013377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Psakis G, Lang C, Mailliet J, Zaanen J, Gartner W, Hughes J, Matysik J. On the collective nature of phytochrome photoactivation. Biochemistry. 2011b;50(51):10987–10989. doi: 10.1021/bi201504a. [DOI] [PubMed] [Google Scholar]
- Strauss HM, Hughes J, Schmieder P. Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry. 2005;44(23):8244–8250. doi: 10.1021/bi050457r. [DOI] [PubMed] [Google Scholar]
- Sugishima M, Okamoto Y, Noguchi M, Kohchi T, Tamiaki H, Fukuyama K. Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction. J Mol Biol. 2010;402(5):879–891. doi: 10.1016/j.jmb.2010.08.021. [DOI] [PubMed] [Google Scholar]
- Terry MJ, McDowell MT, Lagarias JC. (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. Journal of Biological Chemistry. 1995;270(19):11111–11118. doi: 10.1074/jbc.270.19.11111. [DOI] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE. 2012;7(1):e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Rockwell NC, Lagarias JC, Fisher AJ. Insight into the radical mechanism of phycocyanobilin-ferredoxin oxidoreductase (PcyA) revealed by X-ray crystallography and biochemical measurements. Biochemistry. 2007;46(6):1484–1494. doi: 10.1021/bi062038f. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA. 2014;111(45):E4859–4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, McDowell MT, Lagarias JC. Phycocyanobilin is the natural precursor of the phytochrome chromophore in the green alga Mesotaenium caldariorum. Journal of Biological Chemistry. 1997;272(41):25700–25705. doi: 10.1074/jbc.272.41.25700. [DOI] [PubMed] [Google Scholar]
- Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- Yoshihara S, Shimada T, Matsuoka D, Zikihara K, Kohchi T, Tokutomi S. Reconstitution of blue-green reversible photoconversion of a cyanobacterial photoreceptor, PixJ1, in phycocyanobilin-producing Escherichia coli. Biochemistry. 2006;45(11):3775–3784. doi: 10.1021/bi051983l. [DOI] [PubMed] [Google Scholar]
- Zeidler M, Lamparter T, Hughes J, Hartmann E, Remberg A, Braslavsky S, Schaffner K, Gartner W. Recombinant phytochrome of the moss Ceratodon purpureus: heterologous expression and kinetic analysis of Pr→Pfr conversion. Photochem Photobiol. 1998;68(6):857–863. doi: 10.1111/j.1751-1097.1998.tb05296.x. [DOI] [PubMed] [Google Scholar]
- Zhao KH, Haessner R, Cmiel E, Scheer H. Type I reversible photochemistry of phycoerythrocyanin involves Z/E-isomerization of alpha-84 phycoviolobilin chromophore. Biochimica Et Biophysica Acta-Bioenergetics. 1995;1228(2–3):235–243. [Google Scholar]
- Zhao KH, Scheer H. Type I and type II reversible photochemistry of phycoerythrocyanin alpha-subunit from Mastigocladus laminosus both involve Z, E isomerization of phycoviolobilin chromophore and are controlled by sulfhydryls in apoprotein. Biochimica Et Biophysica Acta-Bioenergetics. 1995;1228(2–3):244–253. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Gene models for Notes S1.
Fig. S2 Synthetic gene sequence for the catalytic core of KflaHY2.
Fig. S3 Synthetic gene sequence for the catalytic core of KflaHY1.
Fig. S4 Synthetic gene sequence for the photosensory core module of MvirPHY1.
Table S1 Accession information for FDBR and RCCR sequences
Notes S1 Flat text alignment of FDBR sequences.
Notes S2 Formatted input file for FDBR phylogeny.