Abstract
Structural birth defects are a leading cause of mortality and morbidity in children world-wide, affecting as much as 6% of all live births. Among these conditions, neural tube defects (NTDs), including spina bifida and anencephaly, arise from a combination of complex gene and environment interactions that are as yet poorly understood within human populations. Rapid advances in massively parallel DNA sequencing and bioinformatics allow for analyses of the entire genome beyond the 2% of the genomic sequence covering protein coding regions. Efforts to collect and analyze these large datasets hold promise for illuminating gene network variations and eventually epigenetic events that increase individual risk for failure to close the neural tube. In this review, we discuss current challenges for DNA genome sequence analysis of NTD affected populations, and compare experience in the field with other complex genetic disorders for which large datasets are accumulating. The ultimate goal of this research is to find strategies for optimizing conditions that promote healthy birth outcomes for individual couples.
Keywords: Whole genome sequencing (WGS), Whole exome sequencing (WES), complex genetic disorders, variant analysis, biogeography, intergenic (non-coding) sequence analysis
Introduction
Birth defects are a global problem affecting ~6% of all births (Christensen et al., 2016). In the United States, birth defects are the leading cause of pediatric hospitalizations (Yoon et al., 1997), medical expenditures (Waitzman et al., 1994), and death in the first year of life (Statistics., 2015). Further, they continue to rank as a top cause of death for children aged 1–4 years (#2 cause of death), 5–14 years (#3) and 15–24 years (#6) (Statistics., 2015). Birth defects are, therefore, one of the most important childhood healthcare issues.
Neural tube defects (NTDs), primarily failed caudal neural tube closure (spina bifida) or an open cranium with missing cortex (anencephaly), are among the most severe structural anomalies. Among live births, NTDs have a prevalence in the US of 1 in 3,000 and a world wide prevalence ranging from 1 in 1,000 (in Europe and the Middle East) to 3 in 1,000 (in northern China as of 2014 with folate supplementation campaigns, down from 10 per 1,000 for years 2000–2004) (Salih et al., 2014; Khoshnood et al., 2015; Liu et al., 2016). Despite their public health significance, little is known about the etiology of NTDs in humans. While folic acid fortification of the food supply has been associated with reductions in the prevalence of NTDs, these developmental defects are far from being eradicated. For example, in the US, the prevalence of NTDs declined by only 19% following mandatory folic acid fortification (Honein et al., 2001), and worldwide it has been estimated that 75% of the folic acid preventable NTDs are not prevented due to inadequate or absent fortification (Youngblood et al., 2013).
This modest population-based response to folate dietary fortification and supplementation was unexpected, since the earlier clinical epidemiological studies indicated an up to 70% reduction in NTD recurrence with periconceptional supplementation with folic acid (MRC, 1991), or indeed similar reduction in first occurrence with empirical maternal folate supplementation (Czeizel and Dudas, 1992). The possible interpretations of this experience are far ranging. Some NTDs are likely due to folate deficiency in the maternal diet, but this has been difficult to document as some studies find no difference in serum folate levels in mothers with NTD affected compared to those with healthy pregnancies (Molloy et al., 1985) and the protective effect of folic acid supplementation has been seen regardless of whether maternal blood folate levels fit the clinical definition of folate deficiency (Wald et al., 1996). Alternative explanations for this seeming underperfomance of folic acid fortification include genetic variation that (i) impacts the efficiency of folate utilization/transport/metabolism, so that current levels of folate supplementation are inadequate (ii) the underlying geneic risk involves folic acid-independent pathways (iii) individual variation in folate metabolic pathway function may render exposure to excess folate to have unintended consequences for embryo viability (Stover and Garza, 2002; Gray et al., 2010; Marean et al., 2011). All of these possibilities argue for a need for a comprehensive understanding of genetic and physiological variables with which to individualize recommendations for folic acid and other micronutrient supplementation.
Further, despite considerable evidence of a genetic contribution to NTDs in humans, there are no clinically actionable NTD-related genes, while the few known non-genetic NTD risk factors (including lack of folic acid supplementation and low dietary intake of folate) account for less than a third of all NTDs (Agopian et al., 2012). This gap in our understanding of the causes of NTDs in humans presents a critical barrier to the development of new strategies for preventing these serious conditions. With the development of more effective next generation DNA sequencing capabilities, it is possible now to interrogate complex disorders such as NTDs and gain a better understanding of the genomic architecture that underlies susceptibility to this group of congenital malformations.
In this review, we discuss recent insights into technical and analytical requirements for illumination of the complex genetics contributing to human NTDs. This is a timely consideration as advances in whole exome and whole genome sequencing (WES and WGS, respectively) have enabled the generation of genome data from small amounts of input DNA and at a cost enabling testing of substantial cohorts. Common complex genetic diseases such as schizophrenia, bipolar disorder, and autism spectrum disorder (ASD) are leading the way with analyses of large cohorts. However, recent efforts in applying genome-wide sequencing and analysis approaches to NTDs are beginning to expand the repertoire of developmental disorders for which the genetic underpinnings in human patients may soon be revealed.
Anticipated complexity of genetic contributions to NTDs
Several lines of evidence support the view that NTDs are of complex genetic causality. Among these are twin concordance data showing 6.8% concordant NTDs among same sex twins (many assumed to be monozygotic), and 3.7% when considering all twins (Windham and Sever, 1982). A couple with one NTD affected pregnancy have a 1-in-20 risk of recurrence, while after two affected pregnancies the recurrence risk is 1-in-10, and this risk does not increase with further affected progeny. Additionally, a number of case-control studies have identified alleles that are associated with increased risk of NTDs in humans, depending on the population. Examples include the thermolabile MTHFR C677T variant (Shields et al., 1999) and several deleterious variants in the planar cell polarity (PCP) pathway (Kibar et al., 2007; Kibar et al., 2011; Robinson et al., 2012; Lei et al., 2013; Lei et al., 2014) or the WNT signaling cascade (Lei et al., 2015). Moreover, recent WES data from 43 severe NTD cases suggest that de novo mutations are significant contributors to spina bifida (Lemay et al., 2015).
That numerous gene defects may contribute to NTD is supported by work in animal models that collectively indicate over 400 genes in the mouse that are associated with failed neural tube closure (Harris and Juriloff, 2010) (http://www.informatics.jax.org/searchtool/Search.do?query=neural+tube+defects&submit=Quick+Search). Moreover, numerous studies have shown that genetic background in mouse models has a major impact on the penetrance of NTDs in individuals bearing an NTD associated mutation. Indeed, modifier loci have been mapped in several mouse mutant lines (Juriloff et al., 2001; Korstanje et al., 2008). Thus, genetically determined predisposition is likely to be oligo- or poly-genic with relatively small effect size genome variants combining to determine individual risk. Few population based genetic studies have been reported in the NTD literature and most involve examination of individual or relatively few candidate genes selected on the basis of mouse model data and evaluated across small cohorts. In contrast, population studies of two other common developmental disorders, autism spectrum disorder (ASD) and schizophrenia, have involved thousands of subjects. Those two behavioral disorders provide some insight into what we may expect to find using genomic approaches to study NTDs.
With a current US prevalence of approximately 1 in 3,000 live births, NTDs are among the most common of the serious structural birth defects, second only to congenital heart anomalies (Statistics., 2015). However, they are not as common as ASD with prevalence as high as 1 in 68 children or schizophrenia, which affects 0.5–1% of the adult population (Saha et al., 2005; Christensen et al., 2016). Genome wide association studies (GWAS) and exome data across populations indicate that schizophrenia and ASD are complex disorders in which genetic variants underlie susceptibility to the condition, including chromosome-wide deletions/duplications, small mutations and insertions/deletions (indels), and de novo mutations. A GWAS of SNP array data encompassing 13,000 individuals suffering from schizophrenia, followed by replication of SNPs at 168 locations in 7,413 cases and 19,726 controls revealed 22 loci with genome wide significance, 13 of which had not been found in previous studies (Ripke et al., 2013). Studies of trios indicate that while most genetic risk of schizophrenia comes from inherited alleles, a small proportion of cases are associated with de novo mutations including copy number variants (CNVs) disproportionally involving synaptic proteins (Fromer et al., 2014). These and numerous additional genome-wide studies of schizophrenia indicate a polygenic model in which multiple common and rare variants contribute to a threshold above which the disorder is manifested. Indeed, a meta-analysis of data encompassing around 50,000 healthy and schizophrenic subjects has strongly supported such a threshold model (Cross-Disorder Group of the Psychiatric Genomics et al., 2013)
A somewhat different picture is emerging for ASD (reviewed in (Mullins et al., 2016)). Several GWA studies based on array CGH and whole exome sequencing (WES) have led to estimates that ASD risk alleles involve between 400–1,200 genes (De Rubeis and Buxbaum, 2015; Geschwind and State, 2015). In contrast to schizophrenia, a seemingly large proportion of ASD (as much as 22%) appears to be accounted for by de novo mutations (Iossifov et al., 2014). In both behavioral disorders, the compilation of risk alleles converge in a relatively discrete collection of functional pathways, with components of synaptic function and regulators of the epigenetic landscape prominent between both disorders (Cross-Disorder Group of the Psychiatric Genomics et al., 2013; Mullins et al., 2016). These behavioral disorders studies suggest that genetic factors conferring NTD risk will similarly be found clustered within functional molecular pathways that are important for successful neurulation.
GWAS data for schizophrenia, autism spectrum and other common medical disorders have shown that the original assumptions were incorrect that common disorders would be caused by common variants having large effect sizes. Data outcomes demonstrated that with few exceptions, the disease effect size of common variants is much smaller than previously assumed. Notable exceptions include the APOEε4 allele found in 14% of the population that confers a >4 fold risk of developing Alzheimer's Disease (Ringman and Coppola, 2013) or the CFH allele that increases the risk of macular degeneration in the aged by 2–5 times (Fritsche et al., 2014). No such common alleles with large effect sizes have emerged in genetic studies of ASD, schizophrenia or candidate gene investigations of NTDs. Thus, it appears that much of the disease burden in NTD will derive from rare variants, likely to occur as a mix of inherited and de novo mutations. This has borne out in the results of a recent WES investigation of severe NTDs in 43 sporadic cases and their parents (Lemay et al., 2015).
In addition to SNPs and indels, complete sequence analyses should include assessments of copy number variants (CNVs), a mutation class that has been associated with ASD as well as NTDs (Bassuk et al., 2013; Chen et al., 2013). While CNVs are still often detected by high density microarrays, methods for detection of CNVs using massively parallel, next-generation sequencing (NGS) data can be used to identify CNVs with reasonable sensitivity, especially for deletion CNVs (Krumm et al., 2012; Tan et al., 2014; Wang et al., 2014; Miyatake et al., 2015; Sudmant et al., 2015).
Factors to account for in genome wide studies
Assessment of data quality and choice of sequence alignment method
Critical to any massively parallel sequencing project is the meticulous assessment of data quality that allows for confidence in the variants called. Robust sequence data and analysis are dependent on even and comprehensive coverage of the exome/genome, with an average of 40× coverage common for WES and 30× for WGS with an expected 98% representation of the reference genome (83% at a depth of at least 20×) (Taylor et al., 2015). However, subsequent work has demonstrated that these coverage guidelines for WGS data are only germane to SNVs and very small indels, since larger indels (>5bp) require at least 60× coverage (Fang et al., 2014). Moreover, validation experiments have shown the WGS data can detect indels with greater accuracy than WES approaches (84% vs. 57% validation), primarily due to the greater evenness of coverage and simpler experimental protocols for sample processing and library construction.
In a recent analysis of the success of WGS across 156 cases encompassing a broad spectrum of illnesses, the greatest accuracy was achieved when jointly calling variants across samples (especially important for trios), filtering against both internal and external databases, and using several annotation tools (Taylor et al., 2015). Currently the most widely used sequence alignment tools are GATK (Genome Analysis Toolkit) (DePristo et al., 2011) and CASAVA (Illumina's Consensus Assessment of Sequence and Variation program), and the variant-calling methods that employ local sequence assembly approaches (like GATK) are the most accurate (Fang et al., 2014). Variants are readily annotated with the ANNOVAR package (Wang et al., 2010) by comparing new sequences against publically available reference datasets, including Ensembl (Cunningham et al., 2015), refseq, dbSNP, and functional enhancers annotated in the VISTA browser (Visel et al., 2007).
Finally, work in both DNA and RNA sequencing has shown the need to avoid batch effects, or if unavoidable, to employ means to remove them. This includes tracking metadata and annotation for the types of sequencers that are used (SEQC Consortium, 2014, Li, Tighe, et al., 2014), the algorithms for processing the data (Mason, Porter, Smith, 2014), and also the means of removing false positives for potential batch effects (Li et al, 2014). There are synthetic controls that be now be used for RNA-sequencing, such as the External RNA Control Consortium (ERCCs) spike-ins (Munro et al, 2014) and also the clinical-standard DNA sample called the Genome in a Bottle (GIAB) sample from the National Institute of Standards and Technology (NIST), which serve as a set of positive and negative controls for genome sequencing and processing (Zook et al., 2016). Methods and controls for epigenetics that can quantify and track changes such as epigenetic clonality (Li et al, 2016) are not yet developed but are under active development at NIST and the FDA.
Biogeography, Ancestry and familial relatedness
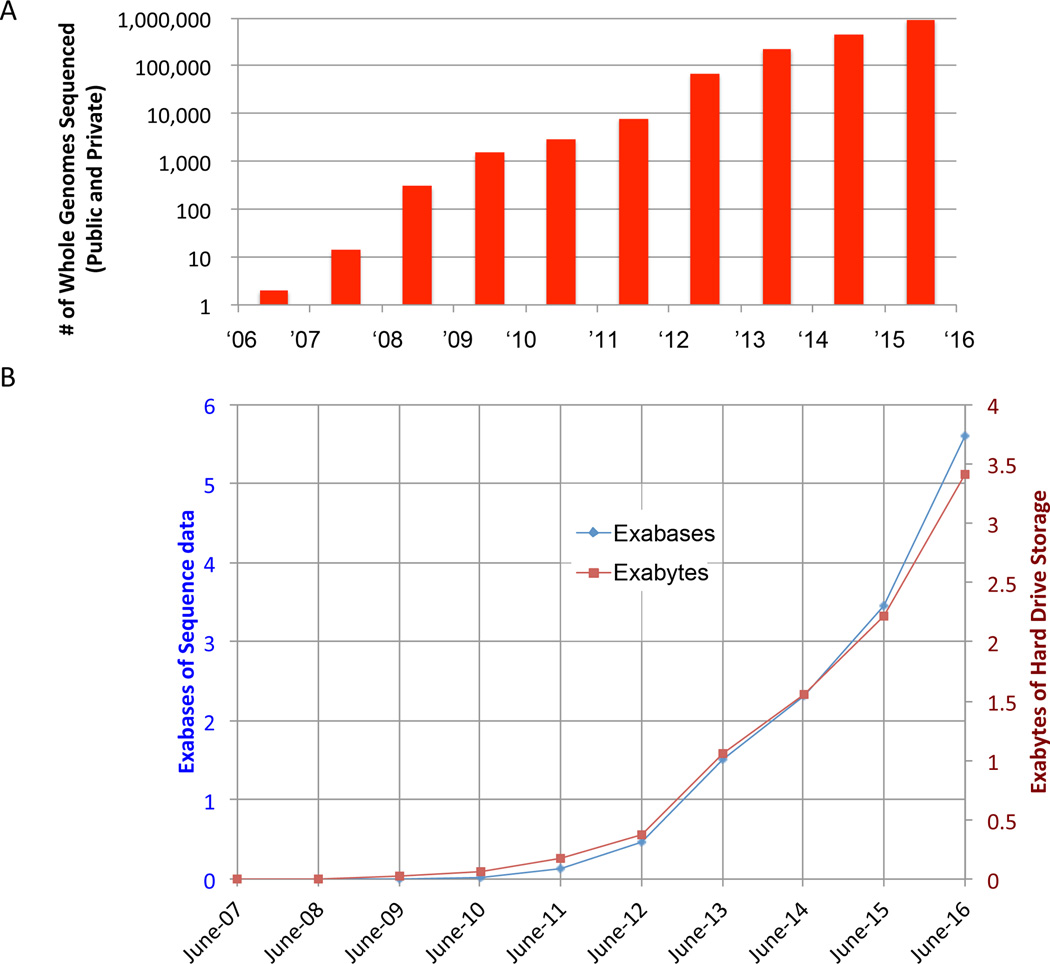
There is no doubt that ethnic background is a significant factor in the assessment of case-control comparisons as allele frequencies can vary significantly according to population admixture (Kidd et al., 2012; Tennessen et al., 2012). Nevertheless, there is reason to believe that while admixture will impact the power of association, the heterogeneity in the growing databases (HapMap, 1000 Genomes Project, ExAC, etc.) will ensure that the effects will likely be small with low expectation of introducing false positive results (Clark et al., 2005). Indeed, the available population databases and the available human exomes or genomes have grown by orders of magnitude over the past 5 years and the size of datasets are accelerating to a point that population admixture effects are of diminishing impact as sources of false positives in the search for potentially pathological rare variants (Figure 1A,B). Moreover, at the current size of ~100GB per compressed human genome and the exponential or super-exponential increase in the rate of DNA/RNA sequence generation, within 10 years we will likely reach a yottabyte (a trillion terabytes) of sequence data (Stephens et al, 2015). This is a particularly important point when considering analytical approaches to genome sequence data derived from patient populations that will, of necessity, be small compared to common diseases like diabetes.
Figure 1. Rapid Growth of Genomic and Sequence Data.
(A) The total number of whole genome sequences for humans has grown at an exponential or super-exponential rate in the past ten years, with an estimated nearly one million genomes to be completed by the end of 2016. (B) The size of sequence data in exabytes (right axis, red) and the total number of bases (left axis, blue) are also increasing at an exponential or super-exponential pace, indicating that a yottabyte of sequence data should be generated by the late 2020s. Exabyte (EB)=1×1018 bytes = a million terabytes; Yottabyte (YB)=1×1024 bytes = a trillion terabytes
Ideally, it would be advantageous to filter experimental datasets for population admixture before pooling to assess for rare variants. In some non-familial population treatments, individuals may be removed from comparison if their relatedness to the data pool examined (using PLINK (Purcell et al., 2007) or BEAGLE (Browning and Browning, 2010) is too high (π_hat>0.9) or too low (π_hat<0.2) compared to individuals in the collection (Stevens et al., 2011; Ruderfer et al., 2014). However, this will reduce the statistical power of the comparisons, and is only practical in variant searches in complex genetic disorders for which large numbers of cases are available (Ruderfer et al., 2014; Ruderfer et al., 2015). For smaller cohorts, admixture is not routinely assessed, but rather rare and private variants are the focus of study (Lemay et al., 2015; Turner et al., 2016). This becomes an increasingly valid approach as the publically available control databases enlarge (Figure 1). Currently, datasets in frequent use include the 1000 Genomes Project (containing over 2,500 individuals) (Abecasis et al., 2012), the NHBLI Exome Project (containing over 6,500 exomes) (Fu et al., 2013), and the Exome Aggregation Consortium (ExAC, containing over 60,500 exomes, (Consortium). Just released in mid-2016 is the Simons Simplex Collection (SSC) with over 6,000 whole genomes.
When a particular variant arises in multiple cases and not in the public databases, it becomes necessary to evaluate the relatedness of the affected individuals. There are several approaches to such an assessment. A first analysis is to use BEAGLE (v3.3.2) to calculate identical by decent (IBD) segments in the region surrounding the variant in question, compared between pairs of cases. PRIMUS (v1.8.0) can then be used to calculate the relatedness degree (first, second, third etc.) of the samples. In addition haplotypes in the region surrounding the locus in question can be imported using GATK to recall the genomes locally and globally at sites in the 1000 genomes database. After the local variants are converted to the PLINK format (v1.90b3w), Haploviewer (v4.1) can be used to calculate and graphically visualize haplotype blocks.
Once first-degree relationships are excluded, it becomes particularly interesting to use population admixture to infer the geographic origins of individuals with rare variants found in common. One such effective tool is the Geographical Population Structure (GPS) algorithm (Elhaik et al., 2014). This examines some 114,000 SNPs that overlap the Genochip SNPs (Elhaik et al., 2013) to convert the genetic distances between a test individual and nine putative reference populations. These genetic distances can then be used to infer ancestry (Das et al., 2016) and likely country of origin for a person. Using this approach, it is possible to pinpoint the recent geographical origin of individuals to a region or even community within a country; indeed these methods have even been used to re-construct the U.S. Census demographic data from DNA left on public surfaces across New York City (Afshinnekoo et al., 2015). While not conclusive proof, that individuals bearing a particular variant share geographic population structure leaves open the possibility of a founder effect and replication studies of samples collected from that region may yield convincing validation.
Rare variant enrichment as indicator of risk
A combination of open source tools may be used to identify variants present in cases and absent in controls or publically available sequence of healthy individuals. These include BEDtools, GATK, and vcftools (Quinlan and Hall, 2010; Danecek et al., 2011; DePristo et al., 2011). Disease-associated variants are expected to impair the normal function of its encoded gene and may be either likely deleterious (nonsense or frameshift SNPs) or missense variants that are predicted to be damaging when assessed by a number of computational tools (e.g., SIFT (Kumar et al., 2009), PolyPhen-2 (Adzhubei et al., 2010), CADD (combined annotation dependent depletion (Kircher et al., 2014)). These likely deleterious mutations can then be assessed for several features--for example, whether their transcribed or encoded proteins are expressed during embryogenesis and are known to be expressed in the neural tube or surrounding mesenchyme. Another indication of an NTD association may be whether the variant is rare, occurring only in cases and not internal controls or available databases, or found with a mean allele frequency of less than 0.01 (MAF<1%). It is important to ensure that correction is made for multiple comparisons to estimate the false discovery rate or FDR correction such as the Benjamini-Hochberg method (Benjamini and Hochberg, 1995).
Coding variants and assessment of mutation burden
Beyond simple enrichment estimates, it is possible to estimate the burden of coding mutations in cases with approaches such as SKAT (sequence kernel association test) (Ionita-Laza et al., 2013). This approach can assess the cumulative effect of both common and rare variants in a particular gene coding sequence. It has the attractive characteristic of permitting combining SNP array and exome data and can also be applied to deep re-sequencing data.
Another approach to assessment of deleterious variants is the estimation of genic tolerance. The premise underlying Residual Variation Intolerance (RVIS) (Gussow et al., 2016) is that genes known to contain few common variations in healthy individuals are relatively intolerant of deleterious changes. For example, genes known to be associated with monogenic inherited disorders are significantly less likely to contain sequence variations (i.e. are more highly conserved across a population) than genes that are not currently connected to any known disease (Petrovski et al., 2013). Scores are compared across the entire coding region. Moreover, by paring down to specific exons, it is possible to identify pathogenic mutations within subregions of genes (Gussow et al., 2016).
Functional pathway enrichment of coding variants as indicator of risk
The challenge continues to identify candidate genes for association to NTDs in human cases using genome-wide sequencing in relatively modest sized cohorts, in which only hundreds of cases may be available. An interesting strategy is to assess molecular pathways for enrichment of likely deleterious mutations. This can be approached using Gene Ontology (GO) relationships and Ingenuity Pathway Analysis (IPA) (O'Roak et al., 2012). Once candidates within these pathways are identified, it will then be possible to launch direct sequencing using new high throughput methods.
Intergenic features and new methods needed to assess individual risk
The advent of relatively low-cost, rapid methods for DNA sequencing of whole exomes using small amounts of input DNA have revolutionized the analysis of genome variation. However, WES data represent only approximately 2% of the human genome. Intergenic regions encompassing the other 98% must ultimately be interrogated in WGS data to provide sensitive assessment of NTD risk conferred by variation in the entire genome. This is more challenging for a number of reasons, including the faster evolutionary sequence divergence within non-coding sequences, the relatively high degree of repetitive sequences with little or no known function, the broad cell type and temporal specificity of regulatory region action that complicates expression mapping of functional genomic domains, epigenetic modifications in non-coding regions, and the importance of 3D relationships of cis- and trans- genomic DNA interactions that can modulate the transcriptome. Two approaches to investigation of intergenic DNA regions are the interrogation of sequence variations within ENCODE-defined enhancer regions, and the use of expression quantitative trait loci (eQTLs) to assess functionally relevant rare sequence variants.
Rare non-coding variants, enhancer peak annotation
The Encyclopedia of DNA elements (ENCODE) consortium continues to amass a comprehensive map of enhancer and other elements in the human genome (Rosenbloom et al., 2012; Won et al., 2013) or mouse genome (Stamatoyannopoulos et al., 2012), along with work from the BluePrint Project in the European Union (http://www.blueprint-epigenome.eu/). Numerous techniques are used, among them chromatin IP-sequencing (ChIP-seq), enhancer traps for identification of elements that regulate restricted anatomical localization of gene expression, and methods for determining stretches of DNA that are open for transcription (e.g. Assay for Transposase Accessible Chromatin (ATAC)-seq). Many of these enhancer elements are accessible through the VISTA genome browser (Visel et al., 2007; Dubchak et al., 2014). When dealing with modest case cohorts, a variant analysis can be used that first searches for rare variants (MAF<1%) and then for statistically significant enrichment of variants in cases compared to controls. Larger case collections (500 or more) may be amenable to linkage disequilibrium assessment of variant association with NTDs. One can then determine the expression pattern of the genes most proximal to the enriched element to suggest whether genome variation in that element may impact expression of genes known to be utilized in relevant embryonic structures expected to impact neurulation. As for candidate variants in coding regions, it will ultimately be necessary to demonstrate that mutation in that element indeed contributes to failed neural tube closure.
Utility of eQTLs in assessing impact of intergenic variation
The effect of genome variation on gene expression can be assessed using eQTLs using an outbred (e.g. human) population for which one has both WGS and RNAseq data from each individual in that population (reviewed in (Albert and Kruglyak, 2015). Comparison between genotype and expression levels is used to find association between variants and gene expression at the transcriptional level. Statistical treatment tests whether a given variant that occurs in a particular subset of individuals has a reproducible association with a particular gene expression level that differs from another subgroup, and can identify which RNA(s) are affected. This is amassed for every DNA variant across the genome to produce an eQTL scan. This massive effort has thus far generated human eQTL datasets for a number of organ systems including circulating blood cells and brain tissue regions, though many age groups and larger cohorts are needed. However, SNPs and indels associated with a disease under study can be examined for enrichment or linkage disequilibrium in the case vs. control cohort and for location of those variants within regions that have been connected by eQTLs to regulation of particular genes. The target genes regulated by that region may be a great distance from the non-coding element and may be either cis- or trans-acting. This approach has been successfully used in several disease studies, including for Parkinson's Disease (Zou et al., 2012; Ramasamy et al., 2014).
Concluding remarks
Technological advances have only recently enabled the generation of WGS data from the typically small input amounts of DNA available from samples collected from infants. Resources of large and growing DNA databases offer critical tools for case-control studies, while many groups are now training efforts on the collection of patient-parent trios for the assessment of de novo mutation vs. inherited risk alleles. We can look forward to important genome-wide studies of NTD risk that will of necessity begin with relatively modest sample numbers but that will be used for meta-analyses by consortia of investigators willing to pool their datasets. Replication studies will no doubt follow as high throughput resequencing methodologies are perfected. Further development of computational tools is needed for the assessment of complex gene-gene interactions and eventually gene-environment interactions modulating the epigenetic landscape. This will require not only expansion of the ENCODE resources but also eQTL maps at various ages in appropriate tissues in both human and animal models, especially mouse. Studies of highly prevalent complex genetic disorders are leading the way for analytical approaches. However, strategies for the evaluation of complex genetic disorders that affect somewhat more modest sized populations must be agreed upon. It seems likely that the most fruitful approaches will rely on genome wide analyses in hundreds of cases to find candidates in an unsupervised manner followed by direct sequencing on a large scale, involving thousands of subjects.
Acknowledgments
Funding Sources: National Institutes of Health (P01HD067244, R01NS076465, HD081216 and HD083809)
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshinnekoo E, et al. Geospatial Resolution of Human and Bacterial Diversity with City-Scale Metagenomics. Cell Syst. 2015;1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agopian AJ, Lupo PJ, Tinker SC, Canfield MA, Mitchell LE National Birth Defects Prevention S. Working towards a risk prediction model for neural tube defects. Birth Defects Res A Clin Mol Teratol. 2012;94:141–146. doi: 10.1002/bdra.22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nature reviews Genetics. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Muthuswamy LB, Boland R, Smith TL, Hulstrand AM, Northrup H, Hakeman M, Dierdorff JM, Yung CK, Long A, Brouillette RB, Au KS, Gurnett C, Houston DW, Cornell RA, Manak JR. Copy number variation analysis implicates the cell polarity gene glypican 5 as a human spina bifida candidate gene. Hum Mol Genet. 2013;22:1097–1111. doi: 10.1093/hmg/dds515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Browning SR, Browning BL. High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet. 2010;86:526–539. doi: 10.1016/j.ajhg.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Gao Y, Zhao H, Sheng X, Zou J, Lip V, Xie H, Guo J, Shao H, Bao Y, Shen J, Niu B, Gusella JF, Wu BL, Zhang T. Detection of copy number variants reveals association of cilia genes with neural tube defects. PLoS One. 2013;8:e54492. doi: 10.1371/journal.pone.0054492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Baio J, Braun KV, Bilder D, Charles J, Constantino JN, Daniels J, Durkin MS, Fitzgerald RT, Kurzius-Spencer M, Lee LC, Pettygrove S, Robinson C, Schulz E, Wells C, Wingate MS, Zahorodny W, Yeargin-Allsopp M. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EA Exome Aggregation Consortium (ExAC) http://exacbroadinstituteorg v 0.3. [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R Genomes Project Analysis G. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Wexler P, Pirooznia M, Elhaik E. Localizing Ashkenazic Jews to primeval villages in the ancient Iranian lands of Ashkenaz. Genome Biol Evol. 2016 doi: 10.1093/gbe/evw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Buxbaum JD. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet. 2015;24:R24–R31. doi: 10.1093/hmg/ddv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Balasubramanian S, Wang S, Cem M, Sulakhe D, Poliakov A, Bornigen D, Xie B, Taylor A, Ma J, Paciorkowski AR, Mirzaa GM, Dave P, Agam G, Xu J, Al-Gazali L, Mason CE, Ross ME, Maltsev N, Gilliam TC. An integrative computational approach for prioritization of genomic variants. PLoS One. 2014;9:e114903. doi: 10.1371/journal.pone.0114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Greenspan E, Staats S, Krahn T, Tyler-Smith C, Xue Y, Tofanelli S, Francalacci P, Cucca F, Pagani L, Jin L, Li H, Schurr TG, Greenspan B, Spencer Wells R Genographic C. The GenoChip: a new tool for genetic anthropology. Genome Biol Evol. 2013;5:1021–1031. doi: 10.1093/gbe/evt066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Tatarinova T, Chebotarev D, Piras IS, Maria Calo C, De Montis A, Atzori M, Marini M, Tofanelli S, Francalacci P, Pagani L, Tyler-Smith C, Xue Y, Cucca F, Schurr TG, Gaieski JB, Melendez C, Vilar MG, Owings AC, Gomez R, Fujita R, Santos FR, Comas D, Balanovsky O, Balanovska E, Zalloua P, Soodyall H, Pitchappan R, Ganeshprasad A, Hammer M, Matisoo-Smith L, Wells RS Genographic C. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun. 2014;5:3513. doi: 10.1038/ncomms4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Wu Y, Narzisi G, O'Rawe JA, Barron LT, Rosenbaum J, Ronemus M, Iossifov I, Schatz MC, Lyon GJ. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6:89. doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O'Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project NES. Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet neurology. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JD, Nakouzi G, Slowinska-Castaldo B, Dazard JE, Sunil Rao J, Nadeau JH, Elizabeth Ross M. Functional interactions between the LRP6 WNT co-receptor and folate supplementation. Hum Mol Genet. 2010;19:4560–4572. doi: 10.1093/hmg/ddq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 2016;17:9. doi: 10.1186/s13059-016-0869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res A Clin Mol Teratol. 2010;88:653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Honein MA, Paulozzi LJ, Mathews TJ, Erickson JD, Wong LY. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. Jama. 2001;285:2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff DM, Gunn TM, Harris MJ, Mah DG, Wu MK, Dewell SL. Multifactorial genetics of exencephaly in SELH/Bc mice. Teratology. 2001;64:189–200. doi: 10.1002/tera.1064. [DOI] [PubMed] [Google Scholar]
- Khoshnood B, Loane M, de Walle H, Arriola L, Addor MC, Barisic I, Beres J, Bianchi F, Dias C, Draper E, Garne E, Gatt M, Haeusler M, Klungsoyr K, Latos-Bielenska A, Lynch C, McDonnell B, Nelen V, Neville AJ, O'Mahony MT, Queisser-Luft A, Rankin J, Rissmann A, Ritvanen A, Rounding C, Sipek A, Tucker D, Verellen-Dumoulin C, Wellesley D, Dolk H. Long term trends in prevalence of neural tube defects in Europe: population based study. BMJ. 2015;351:h5949. doi: 10.1136/bmj.h5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibar Z, Salem S, Bosoi CM, Pauwels E, De Marco P, Merello E, Bassuk AG, Capra V, Gros P. Contribution of VANGL2 mutations to isolated neural tube defects. Clin Genet. 2011;80:76–82. doi: 10.1111/j.1399-0004.2010.01515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, Kirillova I, De Marco P, Merello E, Hayes JM, Wallingford JB, Drapeau P, Capra V, Gros P. Mutations in VANGL1 associated with neural-tube defects. N Engl J Med. 2007;356:1432–1437. doi: 10.1056/NEJMoa060651. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, Degenhardt JD, Brisbin A, Sheth V, Chen R, McLaughlin SF, Peckham HE, Omberg L, Bormann Chung CA, Stanley S, Pearlstein K, Levandowsky E, Acevedo-Acevedo S, Auton A, Keinan A, Acuna-Alonzo V, Barquera-Lozano R, Canizales-Quinteros S, Eng C, Burchard EG, Russell A, Reynolds A, Clark AG, Reese MG, Lincoln SE, Butte AJ, De La Vega FM, Bustamante CD. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korstanje R, Desai J, Lazar G, King B, Rollins J, Spurr M, Joseph J, Kadambi S, Li Y, Cherry A, Matteson PG, Paigen B, Millonig JH. Quantitative trait loci affecting phenotypic variation in the vacuolated lens mouse mutant, a multigenic mouse model of neural tube defects. Physiological genomics. 2008;35:296–304. doi: 10.1152/physiolgenomics.90260.2008. [DOI] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O'Roak BJ, Malig M, Coe BP, Project NES. Quinlan AR, Nickerson DA, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Yang W, Ross ME, Shaw GM, Finnell RH. Identification of novel CELSR1 mutations in spina bifida. PLoS One. 2014;9:e92207. doi: 10.1371/journal.pone.0092207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Duhon C, Yang W, Ross ME, Shaw GM, Finnell RH. Mutations in planar cell polarity gene SCRIB are associated with spina bifida. PLoS One. 2013;8:e69262. doi: 10.1371/journal.pone.0069262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Fathe K, McCartney D, Zhu H, Yang W, Ross ME, Shaw GM, Finnell RH. Rare LRP6 Variants Identified in Spina Bifida Patients. Hum Mutat. 2015;36:342–349. doi: 10.1002/humu.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay P, Guyot MC, Tremblay E, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, De Marco P, Merello E, Massicotte C, Desilets V, Michaud JL, Rouleau GA, Capra V, Kibar Z. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J Med Genet. 2015;52:493–497. doi: 10.1136/jmedgenet-2015-103027. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang L, Li Z, Jin L, Zhang Y, Ye R, Liu J, Ren A. Prevalence and trend of neural tube defects in five counties in Shanxi province of Northern China, 2000 to 2014. Birth Defects Res A Clin Mol Teratol. 2016;106:267–274. doi: 10.1002/bdra.23486. [DOI] [PubMed] [Google Scholar]
- Marean A, Graf A, Zhang Y, Niswander L. Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Hum Mol Genet. 2011;20:3678–3683. doi: 10.1093/hmg/ddr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake S, Koshimizu E, Fujita A, Fukai R, Imagawa E, Ohba C, Kuki I, Nukui M, Araki A, Makita Y, Ogata T, Nakashima M, Tsurusaki Y, Miyake N, Saitsu H, Matsumoto N. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an 'exome-first' approach. J Hum Genet. 2015;60:175–182. doi: 10.1038/jhg.2014.124. [DOI] [PubMed] [Google Scholar]
- Molloy AM, Kirke P, Hillary I, Weir DG, Scott JM. Maternal serum folate and vitamin B12 concentrations in pregnancies associated with neural tube defects. Arch Dis Child. 1985;60:660–665. doi: 10.1136/adc.60.7.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRC V. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- Mullins C, Fishell G, Tsien RW. Unifying Views of Autism Spectrum Disorders: A Consideration of Autoregulatory Feedback Loops. Neuron. 2016;89:1131–1156. doi: 10.1016/j.neuron.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, Consortium UKBE, North American Brain Expression C. Coin L, de Silva R, Cookson MR, Singleton AB, Hardy J, Ryten M, Weale ME. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringman JM, Coppola G. New genes and new insights from old genes: update on Alzheimer disease. Continuum (Minneap Minn) 2013;19:358–371. doi: 10.1212/01.CON.0000429179.21977.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene ND, Copp AJ, Stanier P. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat. 2012;33:440–447. doi: 10.1002/humu.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Diekhans M, Fujita PA, Goldman M, Gravell RC, Harte RA, Hinrichs AS, Kirkup VM, Kuhn RM, Learned K, Maddren M, Meyer LR, Pohl A, Rhead B, Wong MC, Zweig AS, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Lim ET, Genovese G, Moran JL, Hultman CM, Sullivan PF, McCarroll SA, Holmans P, Sklar P, Purcell SM. No evidence for rare recessive and compound heterozygous disruptive variants in schizophrenia. Eur J Hum Genet. 2015;23:555–557. doi: 10.1038/ejhg.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of Psychiatric Genomics C, Bipolar Disorder Working Group of Psychiatric Genomics C, Cross-Disorder Working Group of Psychiatric Genomics C. Gejman PV, O'Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landen M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Molecular psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih MA, Murshid WR, Seidahmed MZ. Epidemiology, prenatal management, and prevention of neural tube defects. Saudi Med J. 2014;35(Suppl 1):S15–S28. [PMC free article] [PubMed] [Google Scholar]
- Shields DC, Kirke PN, Mills JL, Ramsbottom D, Molloy AM, Burke H, Weir DG, Scott JM, Whitehead AS. The "Thermolabile" Variant of Methylenetetrahydrofolate Reductase and Neural Tube Defects: An Evaluation of Genetic Risk and the Relative Importance of the Genotypes of the Embryo and the Mother. Am J Hum Genet. 1999;64:1045–1055. doi: 10.1086/302310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics. NCfH. Brief. Hyattsville, MD: USDHHS, National Center for Health Statistics; 2015. Health, United States, 2015. 2016 http://www.cdc.gov/nchs/hus/contents2015.htm-016. [Google Scholar]
- Stevens EL, Heckenberg G, Roberson ED, Baugher JD, Downey TJ, Pevsner J. Inference of relationships in population data using identity-by-descent and identity-by-state. PLoS Genet. 2011;7:e1002287. doi: 10.1371/journal.pgen.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover PJ, Garza C. Bringing individuality to public health recommendations. J Nutr. 2002;132:2476S–2480S. doi: 10.1093/jn/132.8.2476S. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, Jiang Q, Allen AS, Zhu M. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35:899–907. doi: 10.1002/humu.22537. [DOI] [PubMed] [Google Scholar]
- Taylor JC, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015 doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Hormozdiari F, Duyzend MH, McClymont SA, Hook PW, Iossifov I, Raja A, Baker C, Hoekzema K, Stessman HA, Zody MC, Nelson BJ, Huddleston J, Sandstrom R, Smith JD, Hanna D, Swanson JM, Faustman EM, Bamshad MJ, Stamatoyannopoulos J, Nickerson DA, McCallion AS, Darnell R, Eichler EE. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am J Hum Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitzman NJ, Romano PS, Scheffler RM. Estimates of the economic costs of birth defects. Inquiry. 1994;31:188–205. [PubMed] [Google Scholar]
- Wald NJ, Hackshaw AD, Stone R, Sourial NA. Blood folic acid and vitamin B12 in relation to neural tube defects. British journal of obstetrics and gynaecology. 1996;103:319–324. doi: 10.1111/j.1471-0528.1996.tb09735.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Evans JM, Bhagwate AV, Prodduturi N, Sarangi V, Middha M, Sicotte H, Vedell PT, Hart SN, Oliver GR, Kocher JP, Maurer MJ, Novak AJ, Slager SL, Cerhan JR, Asmann YW. PatternCNV: a versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics. 2014;30:2678–2680. doi: 10.1093/bioinformatics/btu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windham GC, Sever LE. Neural tube defects among twin births. Am J Hum Genet. 1982;34:988–998. [PMC free article] [PubMed] [Google Scholar]
- Won KJ, Zhang X, Wang T, Ding B, Raha D, Snyder M, Ren B, Wang W. Comparative annotation of functional regions in the human genome using epigenomic data. Nucleic Acids Res. 2013;41:4423–4432. doi: 10.1093/nar/gkt143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med. 1997;151:1096–1103. doi: 10.1001/archpedi.1997.02170480026004. [DOI] [PubMed] [Google Scholar]
- Youngblood ME, Williamson R, Bell KN, Johnson Q, Kancherla V, Oakley GP., Jr 2012 Update on global prevention of folic acid-preventable spina bifida and anencephaly. Birth Defects Res A Clin Mol Teratol. 2013;97:658–663. doi: 10.1002/bdra.23166. [DOI] [PubMed] [Google Scholar]
- Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, Malphrus KG, Palusak R, Lincoln S, Bisceglio G, Georgescu C, Kouri N, Kolbert CP, Jen J, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Alzheimer's Disease Genetics C. Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]