Abstract
Bipolar disorder (BD) is a common, complex, and heritable psychiatric disorder characterized by episodes of severe mood swings. The identification of rare, damaging genomic mutations in families with BD could inform about disease mechanisms and lead to new therapeutic interventions. To determine whether rare, damaging mutations shared identity-by-descent in families with BD could be associated with disease, exome sequencing was performed in multigenerational families of the NIMH BD Family Study followed by in silico functional prediction. Disease association and disease specificity was determined using 5 090 exomes from the Sweden-Schizophrenia (SZ) Population-Based Case-Control Exome Sequencing study. We identified 14 rare and likely deleterious mutations in 14 genes that were shared identity-by-descent among affected family members. The variants were associated with BD (p<0.05 after Bonferroni correction) and disease specificity was supported by the absence of the mutations in patients with SZ. In addition, we found rare, functional mutations in known causal genes for neuropsychiatric disorders including holoprosencephaly and epilepsy. Our results demonstrate that exome sequencing in multigenerational families with BD is effective in identifying rare genomic variants of potential clinical relevance and also disease modifiers related to coexisting medical conditions. Replication of our results and experimental validation are required before disease causation could be assumed.
Introduction
Bipolar disorder (BD) is a severe psychiatric disorder characterized by episodes of extremely elevated, expansive or irritable mood, grandiosity, flight of ideas, distractibility or agitation. These symptoms lead often to marked impairment in social and occupational functioning.1 Episodes of mania are frequently followed by severe and disabling depression. In general, BD is conceptualized as a complex disease with genetic and environmental risk factors.2 Heritability estimates range from 58% to 93% with monozygotic twin concordance of about .43.3,4 Nevertheless, the etiology of the disease remains unknown. Linkage studies and genome-wide association studies (GWAS) have suggested chromosomal and genomic regions potentially related to BD, but the identification of disease causing variants remains largely elusive.5,6 Exome-wide sequencing offers now a new opportunity to lead these investigations to a new level.
BD is a common psychiatric disorder with a population prevalence of 2–3%.7,8 However, families in which the disorder is transmitted over several generations are very rare. In the hope of finding genetic risk factors for BD with strong effect, the National Institute of Mental Health (NIMH) ascertained a number of these families in which a Mendelian mode of transmission was suggested by the pattern of disease segregation.9 However, after initial enthusiasm it was quickly realized that a single genetic risk factor with strong effect would most likely not explain the susceptibility to BD even in severely affected multigenerational families.10–12 Instead, mathematical model fitting suggested an oligogenic risk profile as the most likely cause of the disease, but indicated also substantial interfamilial heterogeneity.13 Early linkage studies were not equipped to perform well under this scenario and knowledge about the human genome was still in its infancy.
Since these early attempts had been unsuccessful in finding rare disease causing genes in BD, the search for common genomic polymorphisms as disease modifiers of BD dominated the literature. Many reviews on this subject have been published and it is beyond the scope of this paper to cover this extensive literature. Instead, it is the intend of this paper to collect and present supporting evidence for the hypothesis that rare mutations might contribute to the risk of developing BD under an oligo genic mode of inheritance.
With human genome data available and falling sequencing costs, the time seems right to revisit the original models of disease transmission in the families of the NIMH BD genetics initiative. We conducted a family-based exome sequencing study in multigenerational families of the NIMH to test the hypothesis that several rare functional mutations in gene coding regions are co-transmitted over several generations and shared identity by descent among the affected family members. We expected that the mutated genes would cluster into functional pathways suggesting potential disease path mechanisms. Large, population based samples of patients with schizophrenia and healthy controls were also available to test disease association and disease specificity.
Methods
Sample selection
The analysis presented in this article was based on publicly available data and biomaterial from families of the NIMH Bipolar Genetics Initiative.14 We selected nine affected individuals from four Caucasian families in which BD was transmitted over several generations following an apparently Mendelian mode of inheritance. In three families, we selected the two most distantly related affected family members for exome-wide sequencing. In one family, we selected three affected individuals, since the disease appeared to be transmitted through the paternal and the maternal lineage. The ethnicity of the individuals was determined based on self-report. All affected and unaffected family members, and also the independent patients had been interviewed with the Diagnostic Interview for Genetic Studies (DIGS) by trained health care professionals blinded to the clinical diagnosis. The DIGS is an extensively validated, structured clinical instrument developed by principal investigators at the NIMH for the assessment and differential diagnosis of major mood and psychotic disorders. Medical and psychiatric comorbidities were also recorded.15 Non-hierarchical Best Estimate consensus diagnoses were reached by at least three independent raters according to DSM-IV criteria.16,17 In addition, we randomly selected six unrelated individuals with BD for exome-wide sequencing, who had been evaluated under the same procedures.
Exome sequencing and bioinformatics analysis
DNA was isolated from immortalized lymphoblastoid cell lines. Genomic DNA extraction, library preparation, sequencing, and data analysis were performed using established procedures. Exome capture was carried out using the Illumina TruSeqTM Exome Enrichment Kit (Illumina Inc., San Diego, CA) and the DNA was sequenced using the HiSeq 2000 for a 100-bp paired-end run (Illumina Inc., San Diego, CA). An average of 50 million independent paired reads were generated per sample to provide a mean 10-fold coverage across the RefSeq protein-coding exons and flanking intronic sequence (±2 bp) of more than 87.5% of these bases and a mean 20-fold coverage of 78.9% of the targeted sequences (Supplementary Information). As technical controls during the sequencing process and to guard against technical artifacts, we used the DNA of 200 unrelated individuals who were sequenced in our laboratory under the same exon capture and sequencing conditions.
Variant annotation, filtering and interpretation
Single nucleotide variants (SNV) and small structural variants including insertions and deletions were annotated using Golden Helix SNP & Variation Suite (SVS) v8.1.19 Variants were filtered based on evidence for identity by descent sharing among affected family members, minor allele frequency (MAF) <= 0.01%, and predictions regarding consequence on protein function by the following in silico prediction tools: SIFT, PolyPhen 2, LRT, MutationTaster, Mutation Assessor, and FATHMM 20–26 (Figure 1). The filtered variants were then genotyped in additional affected family members. In addition, all selected variants were also genotyped in at least one unaffected family member per family. Based on these results, we selected variants that were present in the affected family members and absent in the unaffected family members. Finally, we used the exome data from the Sweden-Schizophrenia (SZ) Population-Based Case-Control Exome Sequencing dataset (dbGAP accession: phs000473.v1.p1) for a case control association analysis on the selected variants. This dataset contained exomic data of 2 545 individuals with SZ and 2 545 controls.
Figure 1. Selection algorithm for rare variants in families with bipolar disorder.
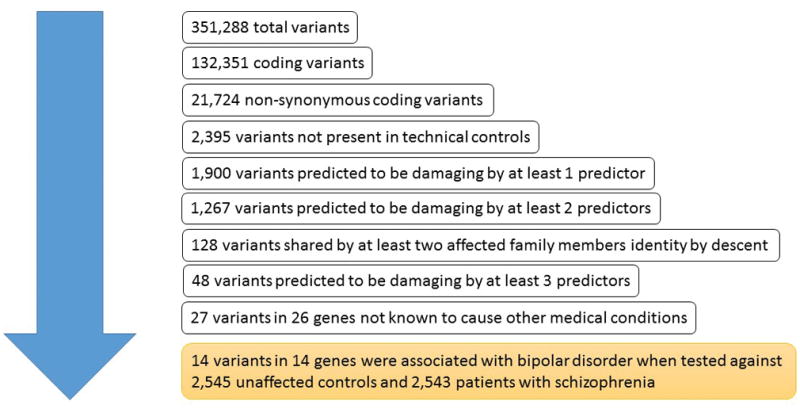
The figure delineates the algorithm that was used to select potentially disease-causing mutations in four families with bipolar disorder. SZ, schizophrenia.
Statistical analysis
To determine the statistical significance of mutation-frequency differences between cases and controls, we used the Fisher’s exact test for rare variants27 and corrected for multiple testing using the Bonferroni procedure.28 In this analysis, the family was considered to be the unit of observation since only variants shared among the affected family members were included in the analysis. Pathway analysis and gene set enrichment analysis (GSEA) of variants that were significant in the Fisher’s exact test were performed in DAVID (DAVID Bioinformatics Resource 6.7).29–32
Results
Sample characteristics
In four multigenerational families, multiple individuals were affected with a severe and complex type of BD (Table 1). The patients had been diagnosed with BD on average at 18 years of age (SD=7.7), and at the time of interview, the majority of the patients had been ill for at least 15 years. Only one fifth of the patients were male (20%). Almost all selected patients (93%) had been diagnosed with bipolar disorder type 1 (BD1) according to DSM-IV criteria, but one independent patient carried the diagnosis of bipolar disorder type 2 (BD 2). Eight patients (53%) fulfilled criteria for rapid cycling BD, a disease subtype characterized by at least four separate mood episodes over the course of one year. Ten patients (67%) had experienced symptoms of hallucinations and/or delusions and ten patients (67%) had attempted suicide at least once during the disease course. All patients had been diagnosed with one or more psychiatric comorbidities, including anxiety disorders (73%), substance use disorders (SUDs) (60%), attention-deficit hyperactivity disorder (40%), obsessive compulsive disorder (OCD) (27%), sleep disorders (27%), eating disorders (20%), and antisocial personality disorder (20%). In addition, some patients also had medical disorders that could have contributed to the phenotype variability. Among these disorders were migraine (67%), seizure disorders (33%), thyroid disorders (20%), gastrointestinal disorders (20%), metabolic disorders (13%), and cardiovascular disorders (7%). Almost half of the sample had been diagnosed with learning disability (40%).
Table 1.
Phenotype of affected individuals in four families with bipolar disorder
| N=15, % | |
|---|---|
| Age, years (SD) | 38.4 (15.6) |
| Age of onset of bipolar disorder, years (SD) | 18.2 (7.7) |
| Gender, male | 3 (20) |
| Diagnosis of bipolar disorder type 1, | 14 (93) |
| Rapid cycling | 8 (53) |
| Suicide attempts | 10 (67) |
| Psychosis | 10 (67) |
| Psychiatric comorbidity | |
| Anxiety disorder | 11 (73) |
| Attention deficit hyperactivity disorder | 6 (40) |
| Substance use disorder | 9 (60) |
| Obsessive compulsive disorder | 4 (27) |
| Antisocial personality disorder | 3 (20) |
| Medical comorbidity | |
| Seizure disorder | 5 (33) |
| Migraine | 10 (67) |
| Disorders of the endocrine system | 3 (20) |
| Disorders of the metabolic system | 2 (13) |
| Disorders of the cardiovascular system | 1 (7) |
| Disorders of the gastrointestinal system | 3 (20) |
| Learning disability | 6 (40) |
| Sleep disorder | 4 (27) |
| Eating disorder | 3 (20) |
Identification of rare, damaging, and disease-specific mutations
Whole-exome sequencing and genotyping of the 15 affected individuals identified 14 rare and likely damaging mutations that were shared identity-by-descent. The mutations were absent in the unaffected family members and also in the technical controls (Figure 1, Table 2). Seven of these mutations were novel and seven variants had been described previously in un-phenotyped population samples at very low frequency (Table 3). The variants were of high quality and predicted to be damaging for the protein structure or function by at least three functional predictors (Supplementary Information Table S1 and S2). In addition, we found one novel frameshift mutation and one known, rare deletion/insertion mutation, both with unknown functional consequences (Table 3). The mutations were private to the individual families, in which they were discovered, and none of the mutations were present in 2 545 ethnically matched controls (P≤1.6×10−3). Furthermore, none of the 2 545 exomes of patients with SZ carried the same mutations, indicating disease-specificity. Three of the mutated genes, myosin IXA (MYO9A), TBC1 domain family, member 10C (TBC1D10C), and Rho GTPase activating protein 32 (ARHGAP32) had GTPase-activating function, but in silico analysis in DAVID revealed no statistically significant clustering of the mutated genes in any known pathophysiological pathway.
Table 2.
List of mutated genes in bipolar disorder families
| Entrez # | Gene | Name | Function |
|---|---|---|---|
| 9743 | ARHGAP32 | Rho GTPase activating protein 32 | GTPase activator activity (GO:0005096), phosphatidylinositol binding (GO:0035091) |
| 60314 | C12orf10 | chromosome 12 open reading frame 10 | locomotory exploration behavior (GO:0035641) |
| 84516 | DCTN5 | dynactin 5 (p25) | Centrosome (GO:0005813) |
| 2060 | EPS15 | epidermal growth factor receptor pathway substrate 15 | calcium ion binding (GO:0005509) protein binding (GO:0005515) |
| 2568 | GABRP | Gamma-Aminobutyric Acid (GABA) A Receptor, Pi | GABA-A receptor activity (GO:0004890) |
| 115399 | LRRC56 | leucine rich repeat containing 56 | unknown |
| 4649 | MYO9A | myosin IXA | regulation of small GTPase mediated signal transduction (GO:0051056) |
| 84700 | MYO18B | myosin XVIIIB | vasculogenesis (GO:0001570) |
| 284434 | NWD1 | NACHT and WD repeat domain containing 1 | ATP binding (GO:0005524) |
| 5286 | PIK3C2A | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha | 1-phosphatidylinositol-3-kinase activity (GO:0016303) |
| 4660 | PPP1R12B | Protein Phosphatase 1, Regulatory Subunit 12B | small GTPase mediated signal transduction (GO:0007264) |
| 5829 | PXN | Paxillin | activation of MAPK activity (GO:0000187) |
| 374403 | TBC1D10C | TBC1 domain family, member 10C | regulation of Rab GTPase activity (GO:0032313) |
| 7158 | TP53BP1 | tumor protein p53 binding protein 1 | RNA polymerase II activating transcription factor binding (GO:0001102) |
| 143630 | UBQLNL | ubiquilin-like | protein binding (GO:0005515) |
| 146862 | UNC45B | Unc-45 Homolog B (C. Elegans) | chaperone-mediated protein folding (GO:0061077) |
Table 3.
Molecular characteristics of mutations in families with bipolar disorder
| Location | Chromosome | Gene | Identifier | Transcript | Exon | Coding | Protein |
|---|---|---|---|---|---|---|---|
| 11:128838929 | 11q24.3 | ARHGAP32 | novel | NM_014715 | 13 | c.5090G>T | p.Gly1697Val |
| 12:53694010 | 12q13.13 | C12orf10 | novel | NM_021640 | 2 | c.293A>G | p.Tyr98Cys |
| 16:23672532 | 16p12.2 | DCTN5 | novel | NM_001199743 | 4 | c.278T>C | p.Ile93Thr |
| 1:51826856 | 1p32.3 | EPS15 | rs148821171 | NM_001159969 | 12 | c.1589C>T | p.Ala530Val |
| 5:170238979 | 5q35.1 | GABRP | novel | NM_014211 | 10 | c.1040A>T | p.Glu347Val |
| 11:549982 | 11p15.5 | LRRC56 | novel | NM_198075 | 7 | c.407_408insT | p.Ser136fs |
| 15:72191038 | 15q23 | MYO9A | novel | NM_006901 | 25 | c.3806G>A | p.Arg1269Gln |
| 22:26224877 | 22q12.1 | MYO18B | rs373113816 | NM_032608 | 15 | c.2921G>A | p.Arg974His |
| 19:16860396 | 19p13.11 | NWD1 | rs148848880 | NM_001007525 | 6 | c.943C>T | p.Arg315Cys |
| 11:17172051 | 11p15.1 | PIK3C2A | novel | NM_002645 | 3 | c.1321T>G | p.Cys441Gly |
| 1:202398004 | 1q32.1 | PPP1R12B | rs199816573 | NM_001167857 | 6 | c.868G>A | p.Ala290Thr |
| 12:120650260 | 12q24 | PXN | novel | NM_025157 | 11 | c.1132C>T | p.Arg378Cys |
| 11:67172591 | 11q13.2 | TBC1D10C | rs201081455 | NM_198517 | 3 | c.188G>A | p.Arg63Gln |
| 15:43762077 | 15q15.3 | TP53BP1 | rs28903074 | NM_001141979 | 11 | c.1362_1367delTATCCC | p.454_456delinsPro |
| 11:5537397 | 11p15.4 | UBQLNL | rs7933557 | NM_145053 | 1 | c.275A>T | p.Asp92Val |
| 17:33504148 | 17q12 | UNC45B | rs137917897 | NM_001033576 | 16 | c.2138G>A | p.Arg713Gln |
In addition to these 15 variants, we discovered two known, rare, compound heterozygous variants in the gene solute carrier family 22 (organic cation transporter), member 1 (SLC22A1) in one severely affected individual. The first mutation (rs55918055) was inherited through the paternal lineage and the second mutation (rs34059508) was inherited through the maternal lineage. These non-synonymous coding mutations were predicted to be deleterious. We also identified mutations in known, disease-causing genes for several medical conditions that could have had disease modifying effects (Supplementary Information Tables S3 to S6). For example, a patient with seizure disorder carried a mutation in the gene prickle homolog 1 (Drosophila)] (PRICKLE1), a known gene for progressive myoclonic epilepsy 1B (EPM1B, MIM:612437). In one family, a novel mutation in the gene dispatched homolog 1 (Drosophila) (DISP1) segregated with the disease phenotype. Mutations in DISP1 are known to cause holoprosencephaly (HPE) type 2–4 (HPE2, MIM:157170; HPE3, MIM:142945; HPE4, MIM:142946; HPE5, MIM:609637), and in addition, this gene is also known as the main suspect in the Chromosome 1q41–q42 deletion syndrome (MIM:612530). Since epilepsy and holoprosencephaly could present with seizures, mood symptoms, psychosis, developmental delay, and learning disabilities, mutations in these two genes could explain some of the neuropsychiatric phenotypes that segregated in two of the families. The gene Ankyrin Repeat and Kinase Domain Containing 1 (ANKK1), which has been related to migraine and alcohol dependence,33–35 also carried a likely damaging mutation. The gene T-box 2 (TBX2) has been related to cognitive and behavioral abnormalities in the chromosome 17q23.1-q23.2 deletion syndrome (MIM:613355), and toll-like receptor 5 (TLR5) has been associated with systemic lupus erythematosus (SLEB1, MIM:601744). None of the variants could be replicated in the independent patients with BD.
Discussion
We identified rare, deleterious, and likely disease-causing mutations in gene-coding regions through unbiased, exome-wide sequencing in families with bipolar disorder (BD). Each family carried rare mutations in several genes that were shared identity-by-descent by affected family members and the variants were absent in the unaffected family members. All variants were predicted to be damaging by several in silico functional predictors. In each several rare, damaging mutations were associated with the disease. These findings are consistent with the currently favored hypothesis of oligo genic disease-causation in BD.36,37
Exome-sequencing is increasingly utilized to identify very rare and likely disease causing mutations in many neuropsychiatric disorders.38 Our focus on rare and even private mutations is consistent with current trends in genetic epidemiologic research; however, our study is one of the first to examine the exomes of BD patients from multigenerational families in an unbiased, genome-wide approach, and to evaluate the results in the context of a large number of population-based healthy controls and patients with SZ. The results of this study reveal a complex scenario of rare and private missense and loss-of-function mutations in novel candidate genes. In addition, we found mutations in known disease-causing genes for medical conditions that could have potentially had disease modifying effects, for example on intellectual ability or immune status.
Our results could be viewed in the context of previously published linkage analyses in the families of the NIMH genetics initiative. Genome-wide significant linkage signals have been reported in the chromosomal regions 16p12.2 and 17q1239, 40 (Dick et al, 2003; Chen et al., 2006). The region 16p12.2 has also been linked to the sub-phenotype psychosis and suggestive linkage has been found to the chromosomal regions 19p13 with the same phenotype40 (Chen et al., 2006). However, when considering linkage results it has to be kept in mind that linkage regions on average contain hundreds of genes, and therefore, conclusions about supporting evidence of linkage results should be viewed with great caution.
Our conclusion about a causal relationship between the described variants and BD is plausible and coherent with some pathophysiological theories. Especially GTPase-activation is a pathophysiological process that is supported by animal models and cell culture experiments.41–43 GTPases are a target of lithium, a drug frequently used to treat BD; and therefore, a role for G-proteins in disease processes of BD has long been hypothesized.44–59
The patients with BD had also been diagnosed with a number of medical and neurological disorders including seizure disorders and learning disability. Therefore, it is highly likely that some of the identified genes might in fact be related to these disorders rather than to BD itself. In fact, we were able to identify rare mutations in genes that have previously been linked to seizure disorders and holoprosencephaly. These conditions could potentially modify the disease expression and the disease course of BD. Since none of the protein-damaging mutations were present in patients with SZ, shared genetic risk factors between BD and SZ might be uncommon.
Limitations of our study are (1) the very small sample size of BD patients in this data set. This limitation could result in an underestimation or overestimation of the effect size of these rare and private mutations. Given the rare nature of the variants in the general population, replication of individual variants is highly unlikely. Another limitation of our analysis was the dependence on in silico functional predictions. Many examples indicate that these predictions might not always reflect the true biological, cell-specific consequences of a specific mutation on an individual’s genetic background. Therefore, it is recommended to test the functional consequences of the identified mutations and experimentally validate the effect in cell culture assays and in in vivo models. Despite obvious limitations, our results are consistent with previous publications in the literature. For example, several groups have identified rare functional mutations in BD families,60–62 even though statistical significance after correction for multiple testing in larger samples still needs to be established. In addition, rare structural variants have been associated with BD, but the functional consequences of these variants remain to be determined.63,64
While individual mutations and genes still require further support before generalizable conclusion can be drawn, it has become clear that BD is by far more heterogeneous than previously anticipated. Our results support a rare-variant oligo genic disease models in families with BD and stress the importance of protein-coding regions. Based on these results, we recommend to fund studies that focus on multi-generational families to identify functional mutations. Furthermore, in clinical practice, it should be recognized that in some families, BD might be transmitted with higher risk than generally anticipated in the framework of common, complex disorder, and that genetic counseling might be recommended. Exome-wide sequencing could be useful in high-risk families to identify known disease-causing mutations for neuropsychiatric disorders that might resemble BD, such as holoprosencephaly and seizure disorders.
Conclusions
The results of our study indicate that rare, deleterious mutations in gene-coding regions could be related to a BD phenotype in families, in which the disease is transmitted over several generations. Exome sequencing in multigenerational families with BD is effective in identifying rare genomic variants with potential clinical relevance. Our results further support the rare-variant oligo genic disease model of BD. The disease association of the identified mutations need to be replicated and the functional consequences of the mutations validated before the information could be used in clinical settings.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by the grant 1R01MH085744 from the National Institutes of Mental Health (NIMH), National Center for Advancing Translational Sciences Institute (CTSI) at UCLA grant UL1TR000124, as well as a NARSAD Young Investigator Award to B Kerner. AR Rao was supported by a NIH Predoctoral Training Grant (No. T32HG002536).
Footnotes
Conflict of interest:
The authors declare no conflict of interest
Author contributions: Conceived and designed the study and wrote the paper: B Kerner. Contributed to the analysis and the writing of the paper: AR Rao. Contributed analysis tools: B. Christensen, M. Yourshaw. Provided expertise: B. Christensen, S Nelson. B. Kerner, AR Rao, and Stan Nelson as thesis advisor for AR Rao had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Competing interests: The authors have declared that no competing interests exist.
Role of the Sponsors: The funders had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; nor preparation, review, or approval of the manuscript.
Supplementary information is available at Molecular Psychiatry’s website
Additional Contributions: The authors would like to acknowledge the staff at the UCLA Broad Stem Cell Research Center (BSCRC), the UCLA Clinical Genomics Center, and the Genome Sequencing (GenoSeq) Core facility at UCLA for DNA preparation, exome sequencing, and variant confirmation, for which these centers have received compensation. Data and biomaterials were collected as part of four projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative (supported by NIMH grants U01MH46282, U01MH46280, U01MH46274, R01MH59545, R01MH59534, R01MH59533, R01MH59553, R01MH60068, R01MH59548, R01MH59535, R01MH59567, R01MH059556, 1Z01MH002810-01, MH52618, MH058693, R01MH59602, R01. We are indebted to the investigators of the NIMH-Bipolar Genetics Initiative and the GAIN Initiative, as well as the families, who provided the genetic and phenotype data. Samples used for data analysis were also provided by the Swedish Cohort Collection supported by the NIMH grant R01MH077139, the Sylvan C. Herman Foundation, the Stanley Medical Research Institute, and The Swedish Research Council (grants 2009-4959 and 2011-4659). Support for the exome sequencing was provided by the NIMH Grand Opportunity grant RCMH089905, the Sylvan C. Herman Foundation, a grant from the Stanley Medical Research Institute, and multiple gifts to the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard. In addition, our study utilized data generated by the DECIPHER Consortium. A full list of centers who contributed to the generation of these data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the DECIPHER project was provided by the Wellcome Trust.
References
- 1.Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) 5. American Psychiatric Association; 2013. [DOI] [PubMed] [Google Scholar]
- 2.Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36:585–594. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song J, Bergen SE, Kuja-Halkola R, Larsson H, Landén M, Lichtenstein P. Bipolar disorder and its relation to major psychiatric disorders: a family-based study in the Swedish population. Bipolar Disord. 2015;17:184–193. doi: 10.1111/bdi.12242. [DOI] [PubMed] [Google Scholar]
- 4.Kieseppä T, Partonen T, Haukka J, Kaprio J, Lönnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161:1814–1821. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- 5.Craddock N, Sklar P. Genetics of bipolar disorder. The Lancet. 2013;381:1654–1662. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 6.Kerner B. Genetics of bipolar disorder. Appl Clin Genet. 2014;7:33–42. doi: 10.2147/TACG.S39297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kessler RC, Chiu WT, Demler O, Walters EE. Prevalence, severity, and comorbidity of twelve-month DSM-IV disorders in the National Comorbidity Survey Replication (NCS-R) Arch General Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler RC, Berglund PA, Demler O, Jin R, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication (NCS-R) Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 9.Nurnberger JI, Jr, DePaulo JR, Gershon ES, Reich T, Blehar MC, Edenberg HJ, et al. Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: a preliminary report. Am J Med Genet. 1997;74:227–237. doi: 10.1002/(sici)1096-8628(19970531)74:3<227::aid-ajmg1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 10.Crow TJ, DeLisi LE. The chromosome workshops at the 5th International Congress of Psychiatric Genetics--the weight of the evidence from genome scans. Psychiatr Genet. 1998;8:59–126. doi: 10.1097/00041444-199800820-00006. [DOI] [PubMed] [Google Scholar]
- 11.DeLisi LE, Crow TJ. Chromosome Workshops 1998: current state of psychiatric linkage. Am J Med Genet. 1999;88:215–218. Erratum in: Am J Med Genet 1999; 88: 448–449. [PubMed] [Google Scholar]
- 12.O’Rourke DH, McGuffin P, Reich T. Genetic analysis of manic-depressive illness. Am J Phys Anthropol. 1983;62:51–59. doi: 10.1002/ajpa.1330620108. [DOI] [PubMed] [Google Scholar]
- 13.Craddock N, Khodel V, Van Eerdewegh P, Reich T. Mathematical limits of multilocus models: the genetic transmission of bipolar disorder. Am J Hum Genet. 1995;57:690–702. [PMC free article] [PubMed] [Google Scholar]
- 14.NIMH Genetics - Bipolar Disorder [Internet] [cited March 3, 2015]. Available: https://www.nimhgenetics.org/available_data/bipolar_disorder/index.php.
- 15.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 16.Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM. Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry. 1982;39:879–883. doi: 10.1001/archpsyc.1982.04290080001001. [DOI] [PubMed] [Google Scholar]
- 17.Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Association; 2000. DSM-IV-TR®. [Google Scholar]
- 18.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and Statistical Genotyping of Copy-Number Variation from Whole-Exome Sequencing Depth. Am J Hum Genetics. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SNP & Variation Suite. Bozeman, MT: Golden Helix, Inc; http://www.goldenhelix.com. [Google Scholar]
- 20.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–741. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Meth. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Meth. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 24.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–211. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St Pierre J, Cadieux M, Guérault A, Quevillon M. Statistical tables to detect significance between frequencies in two small samples, with particular reference to biological assays. Rev Can Biol. 1976;35:17–23. [PubMed] [Google Scholar]
- 28.Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- 29.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 33.Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23. 1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- 34.Ridge JP, Dodd PR. Cortical NMDA receptor expression in human chronic alcoholism: influence of the TaqIA allele of ANKK1. Neurochem Res. 2009;34:1775–1782. doi: 10.1007/s11064-009-9941-8. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh J, Pradhan S, Mittal B. Identification of a novel ANKK1 and other dopaminergic (DRD2 and DBH) gene variants in migraine susceptibility. Neuromolecular Med. 2013;15:61–73. doi: 10.1007/s12017-012-8195-9. [DOI] [PubMed] [Google Scholar]
- 36.Neuman RJ, Rice JP. Two-locus models of disease. Genet Epidemiol. 1992;9:347–365. doi: 10.1002/gepi.1370090506. [DOI] [PubMed] [Google Scholar]
- 37.Gershon ES. Bipolar illness and schizophrenia as oligogenic diseases: implications for the future. Biol Psychiatry. 2000;47:240–244. doi: 10.1016/s0006-3223(99)00299-1. [DOI] [PubMed] [Google Scholar]
- 38.Binder EB. The genetic basis of mood and anxiety disorders - changing paradigms. Biol Mood Anxiety Disord. 2012;2:17. doi: 10.1186/2045-5380-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, et al. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng R, Juo SH, Loth JE, Nee J, Iossifov I, Blumenthal R, et al. Genome-wide linkage scan in a large bipolar disorder sample from the National Institute of Mental Health genetics initiative suggests putative loci for bipolar disorder, psychosis, suicide, and panic disorder. Mol Psychiatry. 2006;11:252–260. doi: 10.1038/sj.mp.4001778. [DOI] [PubMed] [Google Scholar]
- 41.Kalkman HO. Potential opposite roles of the extracellular signal-regulated kinase (ERK) pathway in autism spectrum and bipolar disorders. Neurosci Biobehav Rev. 36:2206–2213. doi: 10.1016/j.neubiorev.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK, et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry. 2014;19:1179–1185. doi: 10.1038/mp.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farhy Tselnicker I, Tsemakhovich V, Rishal I, Kahanovitch U, Dessauer CW, Dascal N. Dual regulation of G proteins and the G-protein-activated K+ channels by lithium. Proc Natl Acad Sci U S A. 2014;111:5018–5023. doi: 10.1073/pnas.1316425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan C, Toon J, Amari L, Abukhdeir AM, Hamm H, Geraldes CF, et al. Competition between lithium and magnesium ions for the G-protein transducin in the guanosine 5′-diphosphate bound conformation. J Inorg Biochem. 2004;98:691–701. doi: 10.1016/j.jinorgbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 45.Farhy Tselnicker I, Tsemakhovich V, Rishal I, Kahanovitch U, Dessauer CW, Dascal N. Dual regulation of G proteins and the G-protein-activated K+ channels by lithium. Proc Natl Acad Sci U S A. 2014;111:5018–5023. doi: 10.1073/pnas.1316425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AH. Lithium affects G-protein receptor coupling. Nature. 1988;331:388. doi: 10.1038/331388a0. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber G, Avissar S. Lithium sensitive G protein hyperfunction: a dynamic model for the pathogenesis of bipolar affective disorder. Med Hypotheses. 1991;35:237–243. doi: 10.1016/0306-9877(91)90239-u. [DOI] [PubMed] [Google Scholar]
- 48.Corena-McLeod M, Walss-Bass C, Oliveros A, Gordillo Villegas A, Ceballos C, Charlesworth CM, et al. New model of action for mood stabilizers: phosphoproteome from rat pre-frontal cortex synaptoneurosomal preparations. PLoS One. 2013;8:e52147. doi: 10.1371/journal.pone.0052147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez R. The relationship between bipolar disorder and biological rhythms. J Clin Psychiatry. 2014;75:e323–331. doi: 10.4088/JCP.13r08507. [DOI] [PubMed] [Google Scholar]
- 50.Lee HJ, Son GH, Geum D. Circadian rhythm hypotheses of mixed features, antidepressant treatment resistance, and manic switching in bipolar disorder. Psychiatry Investig. 2013;10:225–232. doi: 10.4306/pi.2013.10.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carpenter JS, Robillard R, Lee RS, Hermens DF, Naismith SL, White D, et al. The Relationship between Sleep-Wake Cycle and Cognitive Functioning in Young People with Affective Disorders. PLoS One. 2015;10:e0124710. doi: 10.1371/journal.pone.0124710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naismith SL, Lagopoulos J, Hermens DF, White D, Duffy SL, Robillard R, et al. Delayed circadian phase is linked to glutamatergic functions in young people with affective disorders: a proton magnetic resonance spectroscopy study. BMC Psychiatry. 2014;14:345. doi: 10.1186/s12888-014-0345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee HJ, Rex KM, Nievergelt CM, Kelsoe JR, Kripke DF. Delayed sleep phase syndrome is related to seasonal affective disorder. J Affect Disord. 2011;133:573–579. doi: 10.1016/j.jad.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–6411. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellivier F, Geoffroy PA, Etain B, Scott J. Sleep- and circadian rhythm-associated pathways as therapeutic targets in bipolar disorder. Expert Opin Ther Targets. 2015;19:747–763. doi: 10.1517/14728222.2015.1018822. [DOI] [PubMed] [Google Scholar]
- 56.Soreca I. Circadian rhythms and sleep in bipolar disorder: implications for pathophysiology and treatment. Curr Opin Psychiatry. 2014;27:467–471. doi: 10.1097/YCO.0000000000000108. [DOI] [PubMed] [Google Scholar]
- 57.Kõks S, Luuk H, Nelovkov A, Areda T, Vasar E. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav. 2004;3:80–89. doi: 10.1046/j.1601-183x.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 58.Kõks S, Soomets U, Plaas M, Terasmaa A, Noormets K, Tillmann V, et al. Hypothalamic gene expression profile indicates a reduction in G protein signaling in the Wfs1 mutant mice. Physiol Genomics. 2011;43:1351–1358. doi: 10.1152/physiolgenomics.00117.2011. [DOI] [PubMed] [Google Scholar]
- 59.Must A, Kõks S, Vasar E, Tasa G, Lang A, Maron E, Väli M. Common variations in 4p locus are related to male completed suicide. Neuromolecular Med. 2009;11:13–19. doi: 10.1007/s12017-008-8056-8. [DOI] [PubMed] [Google Scholar]
- 60.Song W, Li W, Noltner K, Yan J, Green E, Grozeva D, et al. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neurosci Lett. 2010;486:136–140. doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 61.Green EK, Grozeva D, Sims R, Raybould R, Forty L, Gordon-Smith K, et al. DISC1 exon 11 rare variants found more commonly in schizoaffective spectrum cases than controls. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:490–492. doi: 10.1002/ajmg.b.31187. [DOI] [PubMed] [Google Scholar]
- 62.Goes FS, Pirooznia M, Parla JS, Kramer M, Ghiban E, Mavruk S, et al. Exome Sequencing of Familial Bipolar Disorder. JAMA Psychiatry. 2016;73:590–597. doi: 10.1001/jamapsychiatry.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kember RL, Georgi B, Bailey-Wilson JE, Stambolian D, Paul SM, Bućan M. Copy number variants encompassing Mendelian disease genes in a large multigenerational family segregating bipolar disorder. BMC Genet. 2015;16:27. doi: 10.1186/s12863-015-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta D, Iwamoto K, Ueda J, Bundo M, Adati N, Kojima T, et al. Comprehensive survey of CNVs influencing gene expression in the human brain and its implications for pathophysiology. Neurosci Res. 2014;79:22–33. doi: 10.1016/j.neures.2013.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


