Abstract
The outer membrane (OM) of Gram-negative bacteria is positioned at the frontline of the cell’s interaction with its environment and provides a barrier against influx of external toxins while still allowing import of nutrients and excretion of wastes. It is a remarkable asymmetric bilayer with a glycolipid surface-exposed leaflet and a glycerophospholipid inner leaflet. Lipid asymmetry is key to OM barrier function and several different systems actively maintain this lipid asymmetry. All OM components are synthesized in the cytosol before being secreted and assembled into a contiguous membrane on the other side of the cell wall. Work in recent years has uncovered the pathways that transport and assemble most of the OM components. However, our understanding of how phospholipids are delivered to the OM remains notably limited. Here we will review seminal works in phospholipid transfer performed some 40 years ago and place more recent insights in their context.
Keywords: Gram-negative bacteria, outer membrane, lipopolysaccharide, phospholipids, lipoproteins, β-barrel proteins
1 – Architecture of the Gram negative envelope
Bacteria are typically classified into two groups based on their characteristic Gram-staining phenotype [1]. Gram-positive organisms have a single cytoplasmic membrane surrounded by a thick peptidoglycan layer that acts as an exoskeleton and is responsible for the positive Gram-staining status. In contrast, Gram-negative bacteria have a thin peptidoglycan layer and are instead fortified by an additional, outer membrane (OM). The two, inner and outer, membranes are separated by the periplasmic space, an aqueous and highly viscous compartment that accommodates the peptidoglycan cell wall [1] (Figure 1).
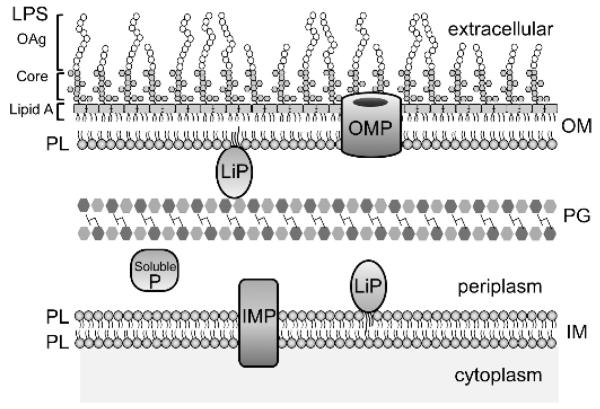
Figure 1. Architecture of the Gram-negative envelope.
The outer membrane (OM) and inner membrane (IM) are separated by an aqueous periplasm that contains the peptidoglycan (PG) cell wall. The asymmetric distribution of lipids in the OM is shown with lipopolysaccharide (LPS) in the outer leaflet and glycerophospholipid (PL) in the inner-leaflet. LPS consists of a tripartite structure of lipid A, a core oligosaccharide component, and an O-antigen (OAg) polysaccharide chain that extends into the extracellular milieu. The three major membrane proteins are shown and include integral membrane proteins (IMP), lipoproteins (LiP) and outer membrane proteins (OMP). Soluble proteins (SolubleP) also exist both in the cytoplasm and periplasm.
The double membrane envelope structure enables distinct compartmentalization of discrete sets of proteins, lipids and lipoproteins that enable these organelles to perform highly specialized functions. The OM, which will be the focus of this review, is an essential organelle that functions as a formidable and selective permeability barrier [2]. In addition to protecting the cell against harsh environments and toxic compounds (e.g. bile salts and antibiotics), the OM enables efficient uptake of nutrients and efflux of waste products and toxic compounds. The OM is a biologically distinct asymmetric lipid bilayer, with lipopolysaccharide (LPS) in the outer leaflet and glycerophospholipids (commonly referred to as phospholipids, PLs) on the inner leaflet [3,4] (Figure 1). This bilayer structure serves as a platform from which OM proteins perform their important cellular functions. Furthermore, it is the frontline from which these bacteria engage with and manipulate their environment or their host.
2 – OM composition and biogenesis
2.1 – OM proteins and lipoproteins
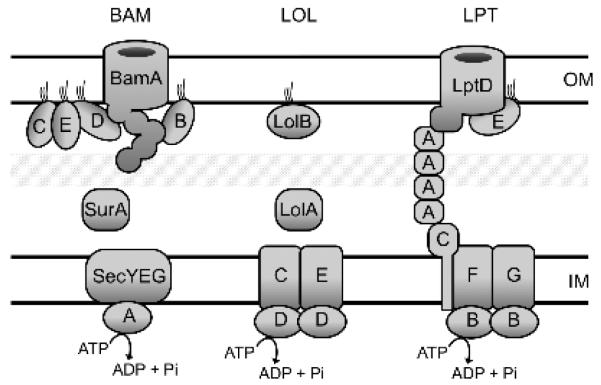
There are two major types of proteins in the OM: transmembrane β-barrel proteins (termed OMPs) and bilayer-anchored lipoproteins (Figure 1). OMPs are synthesized in the cytoplasm and targeted to the Sec translocase via an N-terminal signal sequence and then translocated into the periplasm [5,6]. Because OMPs are hydrophobic transmembrane proteins, chaperones are required to guide them across the aqueous periplasm [7]. OMPs adopt a β-barrel structure by a cylindrical folding of anti-parallel β-sheets [5,6]. In recent years, it has become clear that the five member Bam complex at the OM is responsible for OMP β-barrel folding and membrane insertion [5,6,8,9] (Figure 2).
Figure 2. OM biogenesis machinery.
Depicted are the components of three essential cellular machines required for outer membrane (OM) biogenesis. Chaperones, e.g. SurA and Skp deliver β-barrel outer membrane proteins to the β-barrel assembly machine (BAM) for assembly into the OM. While OM lipoproteins are delivered via the Lol machine. The LipoPolysaccharide Transport (LPT) Pathway transports LPS from the inner membrane (IM) to the cell surface via a hydrophobic conduit formed by the oligomerization of LptA. The molecular details of these machines are discussed in Section 2.
Lipoproteins are similarly directed to the periplasm via an N-terminal signal sequence but possess an adjoining lipobox motif at the C-terminal end of the signal sequence that enables coordinated signal sequence cleavage and triacylation of a conserved cysteine residue within the lipobox [10]. As a result, all lipoproteins newly emerging from the Sec translocon are initially tethered to the IM: first via the transmembrane signal peptide, and then as triacylated mature lipoproteins [11,12]. Some lipoproteins remain in the IM but most are designated for transport to the OM via the Lol pathway [10] (Figure 2). The identity of +2 and +3 residues adjacent to the acylated cysteine determine localization [13,14]. Recent reviews have described the current state of knowledge about the Lol pathway and lipoprotein assembly into the OM, including lipoproteins that are surface exposed [15,16].
2.2 – The OM bilayer
2.2.1 Lipopolysaccharide (LPS)
In terms of the lipid bilayer, the outer leaflet of the OM consists almost exclusively of the glycolipid LPS [3]. The tripartite structure of LPS consists of lipid A component, a core oligosaccharide component, and an O-antigen (OAg) polysaccharide chain that extends into the extracellular milieu [17] (Figure 1). The Oag component is not synthesized in the model organism E. coli K-12 [18], but it plays an important role in protecting commensal strains in their host environment and is also a crucial virulence determinant in pathogenic strains [19]. Lipid A is a diglucosamine phosphate-based lipid (Figure 3A) that is a notoriously potent activator of the innate immune system and is commonly referred to as ‘endotoxin’ [20]. LPS is anchored to the OM bilayer via the lipid A moiety [17]. The core oligosaccharide can be divided into a conserved inner core that generally consists of two units of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) and three units of L-glycero-D-manno-heptose (Hep), all linked together in discreet alpha-glycosidic linkages and subject to regulated covalent modifications with specific phosphoryl and glycosyl moieties, and a variable outer core to which the Oag polysaccharide is attached [17,20]. Phosphoryl substituents of the LPS core oligosaccharide are a key structural component thought to from strong lateral electrostatic interactions with divalent cations and contribute to OM stability [2,21-24].
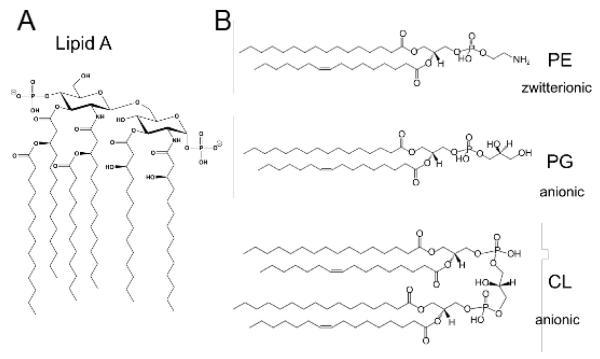
Figure 3. Major OM membrane lipids of E. coli K-12.
The structures of the most abundant membrane lipids of E. coli K-12 are depicted. The asymmetric OM bilayer consist of LPS in the outer leaflet that is anchored to the OM via a lipid A moiety (A). The inner-leaflet is comprised primarily of the zwitterionic phospholipid phosphatidylethanolamine (PE; ~70% of total lipid by weight) as well as the anionic phospholipids phosphatidylglycerol (PG; ~20%) and cardiolipin (CL; ~10%) (B). Phospholipid structures were drawn using the ‘LipidMaps’ Tools [92].
LPS is synthesized at the interface between the inner membrane and the cytoplasm. The lipid A component is synthesized via an eponymous pathway largely characterized by Chris Raetz and his colleagues [17,25] . Sugars of the core polysaccharide are sequentially attached to lipid A; the Oag component is synthesized and translocated separately [19]. Nascent LPS must be translocated across the IM bilayer. The ABC transporter MsbA has been convincingly shown to be responsible for LPS flipping activity [26-28]. Now, in the periplasmic leaflet of the IM, nascent LPS can be modified by the glycosyltransferase WaaL with Oag polysaccharide [19]. The LPS is then efficiently transported to the OM.
Recent years have seen great progress in our understanding of LPS transport and assembly into the OM, which has been recently extensively reviewed [29]. Transport is achieved by the Lpt pathway and LPS is inserted specifically into the outer leaflet of the OM (Figure 2). The current “PEZ” model of this process has emerged from the genetic and biochemical characterization of the transenvelope, seven-member LptABCDEFG complex [29]. The model suggests that sequential rounds of ATP hydrolysis move LPS through several discreet steps that inch the molecule towards the OM [29,30].
In the first step, the IM ABC transporter complex of LptBFG is thought to extract LPS from the periplasmic leaflet [31,32]. LptB is the ATPase while transmembrane proteins LptF and LptG form an inner membrane complex with each other and with LptC, a single span transmembrane protein [28,32,33]. It is not currently known whether LptFG binds LPS that has been extracted. Nonetheless, it seems clear that LptC does bind the extracted LPS and that this is an early step in transport [34].
Transit of LPS across the periplasm is facilitated by the chaperone LptA that oligomerizes to form a bridge through the periplasm [35-37]. The bridge makes direct contacts at LptC in the IM and LptD in the OM via homologous domains that mediate oligomerization [35,38]. A second round of ATP hydrolysis by LptBFG has been shown to ‘push’ LPS from LptC to LptA [30]. The current model suggests that each hydrolysis event moves the LPS molecules another step closer to the OM complex of LptD, an OMP, and LptE, a lipoprotein resident inside of LptD [39-42]. It is clear that LptD receives LPS from the transenvelope bridge and it’s possible that LPS enters the bilayer through the LptD β-barrel [41,42]. LptE must have a role during the intervening sequence of events. Indeed, biochemical evidence supports specific binding of LptE to LPS [40,43]. In fact, it seems likely that LptE binding alters the properties of the LPS molecules flowing towards the OM via the LptA bridge in a way that could facilitate insertion into the OM bilayer [43].
2.2.2 Glycerophospholipids
There are 3 major glycerophospholipids in E. coli and S. Typhimurium; the zwitterionic phospholipid phosphatidylethanolamine (PE) and the anionic phospholipids phosphatidylglycerol (PG), and cardiolipin (CL) [44] (Figure 3B). PE is the most abundant PL comprising ~70% of the total lipid (by weight), followed by PG (~20% ) and CL (~10%) [45]. The E. coli glycerophospholipids are synthesized in the cytoplasm from a phosphatidic acid (PA) precursor (1,2 diacyl-sn-glycerol-3-phosphate) via a series of enzymatic reactions defined by Eugene Kennedy [46]. PA is converted to PS with the subsequent decarboxylation of PS to PE [47]. Additionally, PA is converted to PG with subsequent synthesis of CL via the condensation of two PG molecules [48]. More recently, an alternate source for cellular CL has been identified where PE and PG are co-substrates for a new class of CL synthase [49]
The ability to separate the two membranes was an important precursor to establishing the presence of PLs in both the IM and OM [44]. These experiments showed that the major PLs PG and PE were found in both membranes with each PL distributed relatively evenly between the membranes, but with an increased ratio of PE:PG and PE:CL in the OM [44]. The cylindrical shape of PG means that it has the greatest propensity of these PL to form lamellar bilayers, whereas the conical shape of PE enables it to induce negative membrane curvature and endows it with propensity for nonlamellar transitions [45]. Similarly, although CL and PA are cylindrical like PG and can form bilayers in isolation, divalent cations Mg2+ and Ca2+ found in vivo neutralize and dehydrate the polar headgroups of these PLs thereby reducing their effective size, producing a conical shape, and giving them similar structural properties to PE [45]. Furthermore, alterations to the fatty acid content can increase or decrease the non-bilayer potential of PLs [45]. These dynamic properties of PLs help to maintain the optimal cellular function of membrane organelles and their protein constituents.
In contrast to LPS transport and assembly, very little is known about how PLs are assembled into bacterial membranes. Recent advances in PL transport have been made primarily by examining systems that act to maintain OM lipid homeostasis under conditions of stress [50,51]. These transport systems will be discussed in the following sections, however, they appear to be responsible for the migration of discrete populations of PLs rather than providing a mechanism for bulk transport of PL, which will be discussed in the final sections of this article.
3 – Maintenance of OM asymmetry
The unique asymmetric distribution of lipids in the OM is integral to its barrier function. In particular, divalent cations contribute to the barrier function by simultaneously neutralizing negatively charged phosphate moieties between adjacent LPS molecules and creating strong lateral electrostatic interactions that are responsible for excluding lipophilic and large hydrophilic molecules [2]. Disruptions to the integrity of the LPS outer leaflet of the OM can lead to compensatory flipping of PLs to the outer leaflet [3]. These mislocalized PLs create patches in the membrane bilayer that are more pervious to hydrophobic, toxic molecules [52]. As a result, OM integrity is compromised and cells become sensitive to detergents and bile salts [52]. In E. coli, several systems act to restore lipid asymmetry: the OM phospholipase PldA processively degrades surface PLs [53,54]; the OM palmitoyltransferase enzyme, PagP, converts surface-exposed PLs to sn-1-lyso-PLs during palmitoylation of lipid A and PG [55,56]; and the Mla (maintenance of lipid asymmetry) system facilitates retrograde PL trafficking of mislocalized PLs [50] (Figure 4).
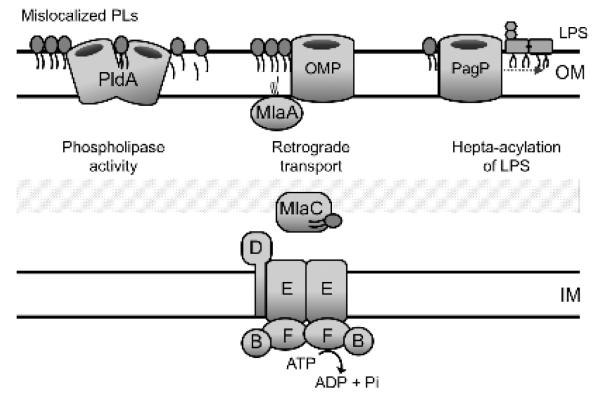
Figure 4. Maintenance of OM asymmetry.
Shown are three systems in E. coli that facilitate removal of mislocalized phospholipids from the outer leaflet. The OM phospholipase, PldA, processively degrades mislocalized PLs. The Mla system is comprised of components in each cellular compartment (MlaA,B,C,D,E,F) that collectively facilitate retrograde PL trafficking. The palmitoyltransferase, PagP, also uses mislocalized PLs as a substrate, and transfers an acyl chain to lipid A. The resultant heptaacylated LPS contributes to OM stability and the lyso-PL by-products can presumably be recycled.
PldA typically exists in an inactive monomeric form in the OM until the presence of PLs and lysophospholipids (lyso-PLs) in the outer leaflet triggers the formation of a catalytically active PldA dimer that hydrolyzes sn-1 and sn-2 acyl chains from the glycerophosphodiester backbone of both PLs and lyso-PLs [53]. PagP, similarly responds to these mislocalized PLs by palmitoylating lipid A and PG concomitantly with the production of sn-1-lyso-PLs [55] (Figure 4). The increased hydrophobicity of the resultant hepta-acylated LPS molecule is thought to reduce lipid fluidity and enhance lateral interactions between neighboring LPS molecules, thereby stabilizing the OM when divalent cations are limiting [57]. Indeed, in contrast to PldA, which requires calcium for activity, PagP levels are increased by the PhoPQ stress response under conditions of divalent cation limitation [55,58]. In addition to fortifying the OM barrier, palmitoylated lipid A can also alter the innate immune response and has proven a useful experimental tool as a probe to monitor ectopic exposure of PLs in the OM [50,59,60]. Although the fate of the lyso-PL by-products of PagP activity is unclear, systems exist to recycle these lipids [61,62], which will be discussed further in Section 4.1
The Mla pathway is a multi-protein system with components situated in each compartment of the cell envelope in order to facilitate retrograde transport of phospholipids from the outer leaflet of the OM back to the IM [50]. Null mutations in any of the identified mla genes (mlaA,B,C,D,E,F) results in OM permeability defects, with mutants displaying increased sensitivity to detergents and bile salts [50]. Notably, mla mutants cause an increased accumulation of PLs in the outer leaflet of the OM [50]. This result is important since it argues against Mla function in anterograde trafficking of PLs from the IM to the OM but does support a role for the pathway in retrograde transport. The functional overlap between the PldA phospholipase and the Mla pathway, in removal of mislocalized phospholipids, is supported by synthetic gene interactions that are observed upon loss of both these systems, as well as the ability of pldA over-expression to functionally complement inactivation of any mla gene [50].
However, there is still much to be learned about how the Mla pathway functions, in particular, how are PLs removed from the outer leaflet? How phospholipids could be flipped to the inner leaflet of the OM, which is devoid of an obvious energy source, is not yet clear. The OM lipoprotein component MlaA has been shown to interact with the OM protein porin OmpC, and deletion of this porin has a similar phenotype to mlaA null mutants under some, but not all, conditions [59]. These observations led to the proposal that the porin may facilitate transbilayer movement of mislocalized PLs [59]. Two models were proposed. Firstly, porins might promote transbilayer movement of PLs by providing a hydrophilic channel, and upon transit to the inner leaflet the PLs can then be accessed by MlaA. Alternatively, the porin may provide direct access to the outer leaflet by the MlaA lipoprotein. A recently identified dominant gain-of-function mutation in mlaA, was shown to not only inactivate the Mla system but to actively disrupt lipid homeostasis by facilitating aberrant accumulation of surface PLs [63]. How this mutant lipoprotein, termed MlaA*, functions in this regard is unclear, but it is tempting to speculate that this illustrates the potential for MlaA to facilitate bi-directional transbilayer migration of PLs in the OM. Whether this activity requires a partner protein is not yet clear.
4 – Phospholipid transport to the OM
The structural organization of the Gram negative cell envelope creates a distinct biological problem it terms of its assembly, since the OM is located at some distance from the cytoplasm, which is both the cellular compartment where all the OM constituents are synthesized and the compartmental location of the cell’s primary source of energy, ATP. As outlined in Section 2, significant progress has been made towards understanding how the protein, lipoprotein and LPS components are transported across the aqueous periplasm and assembled into the hydrophobic core of this biological membrane. A unifying feature of all of these pathways is the employment of ATP-binding cassette (ABC) transporters that can energize transport via ATP hydrolysis. Yet, a key piece of the OM biogenesis puzzle has eluded our understanding: the transport and incorporation of the PL components. Furthermore, as we will discuss, current evidence suggests the mechanism for PL transport is likely to be distinct from the other transport pathways. The following sections will review where our knowledge currently stands, highlighting the longstanding gaps and identifying key avenues that still need to be explored.
4.1 – Transbilayer movement of PLs across the inner membrane
As synthesis of PLs in E. coli occurs on the cytoplasmic side of the inner membrane, the first step on the path towards the OM is transbilayer movement of phospholipids across the IM. The typical doubling time for E. coli is approximately ~20 min [64] and this growth rate must be matched by the rate of PL synthesis and membrane assembly. Spontaneous transbilayer diffusion of phospholipids is intrinsically slow, because the polar headgroup must traverse the hydrophobic core of the membrane bilayer [65]. Studies examining the rate of PL transbilayer movement across the IM indicate that it occurs much faster than would be expected from spontaneous diffusion [66,67]. In Bacillus megaterium the rate of equilibration of newly synthesized PE with the outer leaflet of the IM had an estimated half-time of ~30 s [67]. Invariably, biological membranes must exploit factors that facilitate the required flipping efficiency and it has been proposed that membrane proteins expedite this transbilayer movement [66,67]. This idea is supported by observations that phospholipid flipping activity is protease-sensitive and that PL flipping was not observed in vesicles made solely from phospholipids [67]. However, the identity of the flippase(s) in E. coli or other organisms that are responsible for these observations has remained elusive and the mechanism of transbilayer movement of bacterial lipids remains unclear.
There is compelling genetic evidence for the role of the inner membrane protein, LplT, (Lysophospholipid transporter) in facilitating rapid transbilayer translocation of phospholipids [62]. However, LplT is unlikely to have a generalized PL flippase function. Instead, the evidence suggests that this inner membrane protein functions to reutilize lysophospholipids [62,68]. Preliminary work demonstrated that LplT functions to recycle the 2-acyl-GPE lysophospholipid by-product that is generated in the periplasm during lipoprotein acylation[62]. Specifically, LplT facilitates transbilayer movement of 2-acyl-GPE to the cytoplasm where an acyl-transferase/acyl-ACP synthetase (Aas) regenerates PE [62]. Subsequent work showed that the LplT-Aas system has a broader substrate specificity and can use lyso forms of other major bacterial membrane PLs [62]. The observation that in some bacteria LplT is fused to an acyltransferase supports the notion of this dedicated role of LplT [62]. Furthermore, LplT is not found in Gram-positive bacteria [65].
MsbA, the ABC-transport system that transports lipid A and LPS from the cytoplasmic leaflet to the periplasmic leaflet of the inner membrane [26,27] has been implicated in phospholipid flipping. In strains carrying a temperature-sensitive allele of msbA less than 10% of PLs and lipid A migrated to the OM [26]. However, it is unclear whether the decrease in PL transport is the direct effect of loss of MsbA activity or due to pleiotropic effects that likely stem from reduced transport of LPS. Although in some isolated biological membrane vesicles, PL flipping has been reported to be ATP-dependent [69], others have reported the phenomenon to be ATP-independent, refuting a potential role for IM ABC transporters in this process [70,71]
Ultimately in vitro reconstitution of PL flippase activity is lacking for MsbA and the fact that PL flipping occurs in the absence of MsbA, in Neisseria [72] (where both MsbA and LPS itself are dispensable) suggest that additional factors remain to be identified. Intriguingly, Kol et al. [73-75] have shown that PL flipping in vesicles can be promoted by a range of membrane-spanning helical peptides, which suggests the existence of a more generalized and possibly redundant mode of flipping facilitated by ∝-helical transmembrane regions of integral membrane proteins.
4.2 – Intermembrane movement of PLs
In contrast to the unidirectional transport of proteins and LPS to the OM, the reversibility of the intermembrane movement of PLs was demonstrated with the use of liposome fusion experiments in Salmonella Typhimurium [76]. These experiments exploited the ability of phospholipids from vesicles to be transferred to intact cells in the presence of calcium [77]. PS in vesicles was radiolabeled and intermembrane migration monitored through isolation of labeled-PS in both the IM and OM fractions [76]. An elegant addition to this experimental approach was the ability to detect the conversion of radiolabeled PS to PE via decarboxylation by the IM phosphatidylserine decarboxylase, Psd. These experiments were the first to demonstrate rapid equilibration of PS between the OM and IM. Subsequent pulse-chase studies enabled the native path of radiolabeled PE to be monitored [78]. Newly synthesized PE was shown to initially accumulate at the IM, followed by rapid equilibration between the membranes with a half-time for PL translocation of only ~3 minutes [78]. Phospholipid transport to the OM was found to be PMF driven and independent of protein, lipid and ATP synthesis [78].
The non-discriminatory nature of this observed PL trafficking was evident by the observed trafficking of a range of Salmonella phospholipids, even unnatural molecules such as cholesterol [76]. This promiscuity was further established in subsequent studies that examined PL transport in genetic backgrounds where various steps of the PL biosynthetic pathways are disrupted enabling significant accumulation of minor PL species, CL and PS, which were also shown to equilibrate between the IM and OM [79,80].
4.3 – Models for PL transport to the OM
Almost 40 years later and we are still searching for molecular details to account for the ground breaking studies of Osborn (1977) and Donohue-Rolfe (1980), and no proteins that clearly function in bulk PL transport to the OM have yet been identified. Various models have been proposed based on lipid trafficking pathways in eukaryotes. One such model is transport via soluble carriers (i.e. lipid transfer proteins). Indeed, this mechanism of transport has been proposed for the retrograde Mla pathway, which possesses a soluble periplasmic component that can bind lipids and is thought to act as a lipid transport protein.
In Salmonella Typhimurium, activation of the PhoPQ two-component system causes a significant increase in the amount of CL in the OM [51,56]. Dalebroux et al. (2015) have identified a protein, which they termed PbgA, that is required for this increase. Intriguingly the amino terminus of the molecule, which contains five transmembrane sequences, is essential for bacterial viability. However, the periplasmic domain is not essential, but is required for the PhoPQ-dependent increase in OM CL. The authors propose that PhoPQ activates the periplasmic domain enabling it to interact with the OM (possibly indirectly) but presumably forming a bridge that delivers CL. Dong et al. (2016) recently determined the structure of the PbgA periplasmic domain, which supports the proposed CL binding function of this domain [81]. However, the mechanism of PgbA-dependent increases in OM CL remains to be determined. The homologous E. coli protein, YejM, also has an essential amino-terminal domain and a dispensable periplasmic domain, but little else about this homolog is known [82].
However, it is clear that neither the proposed PhoPQ-dependent PbgA CL transport system nor the Mla system function to transport bulk PLs to the OM. Moreover, it seems unlikely that any transport system, or any bacterial lipid transfer protein would recognize and transport cholesterol. Similarly unlikely is transport via carrier vesicles, in a manner analogous to inter-organelle trafficking of lipids seen in eukaryotes. Presumably vesicles, typically 20 nm in diameter at their smallest size [83], would be hindered from traversing the periplasm by the meshwork of peptidoglycan cell wall in E. coli, which possesses an average pore radius of only about 2 nm [84].
Ultimately two proposed mechanisms for intermembrane transport have prevailed; (1) migration via the regulated formation of membrane hemifusions, and (2) membrane apposition and PL transfer via a hydrophobic conduit. In both these models protein factors could act to either promote zones of contact to facilitate hemifusion or membrane apposition or the formation of a hydrophobic bridge. The inception of these models can be ascribed to early electron microscopy images from Manfred Bayer [85]. Following plasmolysis of E. coli and shrinkage of the cytoplasmic membrane away from the cell wall, zones of adhesion between the inner and outer membranes were visualized at hundreds of sites, estimated to cover nearly 5% of the OM surface. These sites of contact were proposed to function as sites of export for components of the OM and spurred the seminal work by Osborn, Rolf-Donahue and Schaechter, and others. Subsequent studies challenged these observations and suggested that these ‘Bayers Junctions’ were an artifact of chemical fixation [86-89]. Bayer himself refuted these claims in a follow-up study using cryofixation [90], but the existence of these zones of adhesion remains controversial. Curiously, the Lpt system seems to conform to the idea of Bayer’s junctions: the transenvelope machine forms direct contact between the IM and OM and LPS transport continues in spheroplasted cells that lack periplasmic contents. Note however, that PL transport is inhibited by spheroplasting cells [91].
Recent support for the hemifusion model came from the study of the dominant MlaA* mutant protein [63]. As noted above, this mutant protein increases the amount of PLs in the OM rather than decreasing them. The cell responds by increasing production of LPS, and this increase in LPS destabilizes the OM. When cells starve in media with low divalent cation concentrations, OM is lost by vesiculation. Because of the underlying rigid peptidoglycan cell wall, the OM cannot shrink. To replace the lipid lost, membrane flows from the IM to the OM. This flow is quite rapid and does not appear to require energy. It is very reminiscent of the retrograde flow observed long ago by Osborn except in this case, the flow is anterograde. We suggest that the hemifusion model provides the simplest explanation for this diffusive, bidirectional flow.
5 – Future directions
The cellular machinery required to transport lipoproteins, OMPs and LPS to the OM has been identified, the proteins have been purified, their structures have been determined, and the systems have been reconstituted in vitro. However, how PLs are transported to the OM remains as mysterious now as it was 40 years ago. The problem has withstood assault by all of the modern genetic and biochemical approaches and genomic and proteomic approaches as well. So far at least high resolution microscopy, including cryo-EM, has not been helpful either. The transenvelope Lpt machine has yet to be seen, and it seems likely that hemifusions would be even less stable.
PL transport mechanisms such as the Mla system and PbgA system have been identified and further study of these systems may provide important clues that lead to the discovery of the bulk transport mechanism(s). It may well be that cells have redundant mechanisms for this critical cellular process and this will complicate standard genetic methods. Perhaps new methods will have to be invented. Alternatively, someone may design a clever genetic selection or screen.
Highlights.
The outer-membrane (OM) of Gram-negative bacteria is an asymmetric bilayer
Phospholipids form the inner-leaflet of the OM
How phospholipids reach the OM remains a mystery
Phospholipid transport is reviewed in context with other OM assembly machines
Seminal studies of phospholipid transport are revisited in light of recent insights
Acknowledgments
The authors would like to thank Russell Bishop for help with figures and Marcin Grabowicz for critical reading of the manuscript and for helpful discussions.
Funding
This work was funded by the National Institute of General Medical Sciences, grant #GM034821.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414–a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- [4].Funahara Y, Nikaido H. Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J. Bacteriol. 1980;141:1463–1465. doi: 10.1128/jb.141.3.1463-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ricci DP, Silhavy TJ. The Bam machine: a molecular cooper. Biochim. Biophys. Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rigel NW, Silhavy TJ. Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Current Opinion in Microbiology. 2012;15:189–193. doi: 10.1016/j.mib.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. Protein quality control in the bacterial periplasm. Annual Review of Microbiology. 2011;65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- [8].Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- [9].Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- [10].Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annual Review of Microbiology. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- [11].Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- [12].Fukuda A, Matsuyama S-I, Hara T, Nakayama J, Nagasawa H, Tokuda H. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 2002;277:43512–43518. doi: 10.1074/jbc.M206816200. [DOI] [PubMed] [Google Scholar]
- [13].Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- [14].Hara T, Matsuyama S-I, Tokuda H. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J. Biol. Chem. 2003;278:40408–40414. doi: 10.1074/jbc.M307836200. [DOI] [PubMed] [Google Scholar]
- [15].Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2015;370:20150030. doi: 10.1098/rstb.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilson MM, Bernstein HD. Surface-exposed lipoproteins: an emerging secretion phenomenon in Gram-negative bacteria. Trends Microbiol. 2016;24:198–208. doi: 10.1016/j.tim.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- [18].Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- [19].Kalynych S, Morona R, Cygler M. Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol. Rev. 2014;38:1048–1065. doi: 10.1111/1574-6976.12070. [DOI] [PubMed] [Google Scholar]
- [20].Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schindler M, Osborn MJ. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- [22].Yethon JA, Gunn JS, Ernst RK, Miller SI, Laroche L, Malo D, et al. Salmonella enterica serovar typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 2000;68:4485–4491. doi: 10.1128/iai.68.8.4485-4491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yethon JA, Whitfield C. Purification and characterization of WaaP from Escherichia coli, a lipopolysaccharide kinase essential for outer membrane stability. J. Biol. Chem. 2001;276:5498–5504. doi: 10.1074/jbc.M008255200. [DOI] [PubMed] [Google Scholar]
- [24].Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 1998;273:26310–26316. doi: 10.1074/jbc.273.41.26310. [DOI] [PubMed] [Google Scholar]
- [25].Raetz CRH, Guan Z, Ingram BO, Six DA, Song F, Wang X, et al. Discovery of new biosynthetic pathways: the lipid A story. J. Lipid Res. 2009;50:S103–S108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Doerrler WT, Gibbons HS, Raetz CRH. MsbA-dependent rranslocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- [27].Reyes CL, Chang G. Structure of the ABC Transporter MsbA in complex with ADP·Vanadate and lipopolysaccharide. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- [28].Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP Hydrolysis Powers Transport of Lipopolysaccharide Across the Periplasm in E. coli. Science. 2012;338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].May JM, Sherman DJ, Simpson BW, Ruiz N, Kahne D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Phil. Trans. R. Soc. B. 2015;370:20150027. doi: 10.1098/rstb.2015.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Narita S-I, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- [34].Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 2010;285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Freinkman E, Okuda S, Ruiz N, Kahne D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry. 2012;51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Merten JA, Schultz KM, Klug CS. Concentration-dependent oligomerization and oligomeric arrangement of LptA. Protein Science. 2012;21:211–218. doi: 10.1002/pro.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Santambrogio C, Sperandeo P, Villa R, Sobott F, Polissi A, Grandori R. LptA assembles into rod-like oligomers involving disorder-to-order transitions. J. Am. Soc. Mass Spectrom. 2013;24:1593–1602. doi: 10.1007/s13361-013-0687-9. [DOI] [PubMed] [Google Scholar]
- [38].Chng S-S, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chng S-S, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Freinkman E, Chng S-S, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- [43].Malojčić G, Andres D, Grabowicz M, George AH, Ruiz N, Silhavy TJ, et al. LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9467–9472. doi: 10.1073/pnas.1402746111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- [45].Dowhan W, Bogdanov M. Functional roles of lipids in membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 4th edition 2002. pp. 1–35. [Google Scholar]
- [46].Dowhan W. A retrospective: use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim. Biophys. Acta. 2013;1831:471–494. doi: 10.1016/j.bbalip.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kanfer JN, Kennedy EP. Synthesis of Phosphatidylserine by Escherichia coli. J. Biol. Chem. 1962;237:PC270–PC271. [PubMed] [Google Scholar]
- [48].Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CRH, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc M-P, et al. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe. 2015;17:441–451. doi: 10.1016/j.chom.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem. Biol. 2006;1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- [53].Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, et al. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- [54].Dekker N. Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- [55].Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- [58].Snijder HJ, Kingma RL, Kalk KH, Dekker N, Egmond MR, Dijkstra BW. Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli. J. Mol. Biol. 2001;309:477–489. doi: 10.1006/jmbi.2001.4675. [DOI] [PubMed] [Google Scholar]
- [59].Chong Z-S, Woo W-F, Chng S-S. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol. 2015;98:1133–1146. doi: 10.1111/mmi.13202. [DOI] [PubMed] [Google Scholar]
- [60].Jia W, El Zoeiby A, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- [61].Hsu L, Jackowski S, Rock CO. Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl acyl carrier protein synthetase activity. J. Biol. Chem. 1991;266:13783–13788. [PubMed] [Google Scholar]
- [62].Harvat EM, Zhang Y-M, Tran CV, Zhang Z, Frank MW, Rock CO, et al. Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J. Biol. Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- [63].Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E1565–74. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sanyal S, Menon AK. Flipping Lipids: Why an’ What’s the Reason for? ACS Chem. Biol. 2009;4:895–909. doi: 10.1021/cb900163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rothman JE, Kennedy EP. Rapid transmembrane movement of newly synthesized phospholipids during membrane assembly. Proc. Natl. Acad. Sci. U.S.A. 1977;74:1821–1825. doi: 10.1073/pnas.74.5.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hrafnsdóttir S, Nichols JW, Menon AK. Transbilayer movement of fluorescent phospholipids in Bacillus megaterium membrane vesicles. Biochemistry. 1997;36:4969–4978. doi: 10.1021/bi962513h. [DOI] [PubMed] [Google Scholar]
- [68].Lin Y, Bogdanov M, Tong S, Guan Z, Zheng L. Substrate selectivity of lysophospholipid transporter LplT involved in membrane phospholipid remodeling in Escherichia coli. J. Biol. Chem. 2016;291:2136–2149. doi: 10.1074/jbc.M115.700419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hrafnsdóttir S, Menon AK. Reconstitution and partial characterization of phospholipid flippase activity from detergent extracts of the Bacillus subtilis cell membrane. J. Bacteriol. 2000;182:4198–4206. doi: 10.1128/jb.182.15.4198-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Langley KE, Kennedy EP. Energetics of rapid transmembrane movement and of compositional asymmetry of phosphatidylethanolamine in membranes of Bacillus megaterium. Proc. Natl. Acad. Sci. U.S.A. 1979;76:6245–6249. doi: 10.1073/pnas.76.12.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Huijbregts RPH, de Kroon AIPM, de Kruijff Ben. Rapid transmembrane movement of newly synthesized phosphatidylethanolamine across the inner membrane of Escherichia coli. J. Biol. Chem. 1998;273:18936–18942. doi: 10.1074/jbc.273.30.18936. [DOI] [PubMed] [Google Scholar]
- [72].Tefsen B, Bos MP, Beckers F, Tommassen J, de Cock H. MsbA is not required for phospholipid transport in Neisseria meningitidis. J. Biol. Chem. 2005;280:35961–35966. doi: 10.1074/jbc.M509026200. [DOI] [PubMed] [Google Scholar]
- [73].Kol MA, de Kroon AI, Rijkers DT, Killian JA, de Kruijff B. Membrane-spanning peptides induce phospholipid flop: a model for phospholipid translocation across the inner membrane of E. coli. Biochemistry. 2001;40:10500–10506. doi: 10.1021/bi010627+. [DOI] [PubMed] [Google Scholar]
- [74].Kol MA, van Laak ANC, Rijkers DTS, Killian JA, de Kroon AIPM, de Kruijff B. Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition. Biochemistry. 2003;42:231–237. doi: 10.1021/bi0268403. [DOI] [PubMed] [Google Scholar]
- [75].Kol MA, van Dalen A, de Kroon AIPM, de Kruijff B. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J. Biol. Chem. 2003;278:24586–24593. doi: 10.1074/jbc.M301875200. [DOI] [PubMed] [Google Scholar]
- [76].Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J. Biol. Chem. 1977;252:7405–7412. [PubMed] [Google Scholar]
- [77].Jones NC, Osborn MJ. Interaction of Salmonella Typhimurium with phospholipid vesicles. Incorporation of exogenous lipids into intact cells. J. Biol. Chem. 1977;252:7398–7404. [PubMed] [Google Scholar]
- [78].Donohue-Rolfe AM, Schaechter M. Translocation of phospholipids from the inner to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1867–1871. doi: 10.1073/pnas.77.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Raetz CRH, Kantor GD, Nishijima M, Newman KF. Cardiolipin accumulation in the inner and outer membranes of Escherichia coli mutants defective in phosphatidylserine synthetase. J. Bacteriol. 1979;139:544–551. doi: 10.1128/jb.139.2.544-551.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Langley KE, Hawrot E, Kennedy EP. Membrane assembly: movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. J. Bacteriol. 1982;152:1033–1041. doi: 10.1128/jb.152.3.1033-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Dong H, Zhang Z, Tang X, Huang S, Li H, Peng B, et al. Structural insights into cardiolipin transfer from the inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci Rep. 2016;6:30815. doi: 10.1038/srep30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].De Lay NR, Cronan JE. Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics. 2008;178:1327–1337. doi: 10.1534/genetics.107.081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Review of Microbiology. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Demchick P, Koch AL. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 1996 doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bayer ME. Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- [86].Kellenberger E. The “Bayer bridges” confronted with results from improved electron microscopy methods. Mol. Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- [87].Hobot JA, Carlemalm E, Villiger W, Kellenberger E. Periplasmic gel: new concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. J. Bacteriol. 1984;160:143–152. doi: 10.1128/jb.160.1.143-152.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dubochet J, McDowall AW, Menge B, Schmid EN, Lickfeld KG. Electron microscopy of frozen-hydrated bacteria. J. Bacteriol. 1983;155:381–390. doi: 10.1128/jb.155.1.381-390.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Leduc M, Fréhel C, Siegel E, Van Heijenoort J. Multilayered distribution of peptidoglycan in the periplasmic space of Escherichia coli. J. Gen. Microbiol. 1989;135:1243–1254. doi: 10.1099/00221287-135-5-1243. [DOI] [PubMed] [Google Scholar]
- [90].Bayer ME. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 1991;107:268–280. doi: 10.1016/1047-8477(91)90052-x. [DOI] [PubMed] [Google Scholar]
- [91].Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- [92].Sud M, Fahy E, Subramaniam S. Template-based combinatorial enumeration of virtual compound libraries for lipids. J Cheminform. 2012;4:23. doi: 10.1186/1758-2946-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]