Abstract
A study designed to gain baseline information on strains of Escherichia coli displaying resistance to cefoxitin in Canada is described. A total of 29,323 E. coli isolates were screened at 12 participating hospital sites as part of an extended-spectrum beta-lactamase surveillance initiative. A total of 411 clinically significant, nonrepeat isolates displaying reduced susceptibilities to the NCCLS-recommended beta-lactams were submitted to a central laboratory over a 1-year period ending on 30 September 2000. Two hundred thirty-two isolates were identified as resistant to cefoxitin. All cefoxitin-resistant strains were subtyped by pulsed-field gel electrophoresis, and of these, 182 strains revealed a unique fingerprint and 1 strain was untypeable. PCR and sequence analysis of the ampC promoter region revealed 51 different promoter or attenuator variants and 14 wild-type promoters. Three promoter regions were interrupted by insertion elements, two contained IS10 elements, and one contained an IS911 variant. PCR and sequence analysis for the detection of acquired AmpC resistance (by the acquisition of ACT-1/MIR-1, CMY-2, or FOX) revealed that 25 strains contained CMY-2, including 7 of the strains found to have wild-type promoters. The considerable genetic variability in both the strain fingerprint and the promoter region suggests that AmpC-type resistance may emerge spontaneously by mutation of sensitive strains rather than by the spread of strains or plasmids in the hospital setting.
Escherichia coli contains a chromosomal ampC gene which has a weak promoter as well as a transcriptional attenuator (13, 14). Strains carrying the wild-type gene produce a low basal amount of AmpC and are susceptible to ampicillin. Analysis of cefoxitin-resistant strains has revealed that the ampC promoter and attenuator regions often carry mutations leading to the overproduction of the enzyme (3, 4, 5, 7, 10, 14, 20, 21, 22, 29). These AmpC-overproducing strains are not only resistant to ampicillin but also usually have reduced susceptibilities to expanded-spectrum cephalosporins. Genetically, the most common mutations are those that create a strong promoter that more closely resembles the E. coli consensus promoter (12) and mutations that destabilize the attenuator.
Plasmid-mediated ampC genes were first reported in 1988 (25, 31). The plasmid-mediated ampC genes are derived from inducible chromosomal genes that have become mobilized (11, 24, 25). The first genetically characterized plasmid-mediated AmpC was MIR-1, which was mobilized from an Enterobacter isolate (23). These plasmid-mediated genes are of special interest because their mobility allows them to emerge in one genus or species and spread to different organisms. The prevalence of plasmid-mediated AmpC-type resistance at the national level in most countries is unknown because studies have not examined the strains at the molecular level of detail required to elucidate the different mechanisms involved. A recent report from the United States, however, showed that among 752 Klebsiella spp. and E. coli strains from 70 sites in 25 states, 7 to 8.5% of the Klebsiella spp. and 4% of the E. coli strains contained plasmid-mediated AmpC-type enzymes (2). In this report, we present the first detailed characterization of cefoxitin-resistant strains of E. coli acquired from nosocomial sources at the national level in Canada. We used sequence analysis to determine the frequency of promoter or attenuator mutations and PCR to look for plasmid-mediated ampC genes. Molecular epidemiological methods were also used to determine the extent of clonal spread of cefoxitin-resistant strains.
MATERIALS AND METHODS
Surveillance network.
Established in 1995, the Canadian Nosocomial Infection Surveillance Program (CNISP) is a collaborative effort involving members of the Canadian Hospital Epidemiology Committee (a subcommittee of the Canadian Infectious Diseases Society), the National Microbiology Laboratory (NML), and the Centre for Infectious Disease Prevention and Control, Health Canada. At present, CNISP comprises 35 hospital sites. The data presented in this 1-year study represent results obtained from 12 participating CNISP tertiary-care hospitals which volunteered their sites for study. To maintain site confidentiality, data were tabulated into two geographical regions: the west, which included British Columbia (two sites), Alberta (one site), and Saskatchewan (one site), and the east, which included Ontario (five sites), Quebec (two sites), and Nova Scotia (one site).
Study design.
This was a prospective laboratory-based surveillance study (conducted from 1 October 1999 to 30 September 2000) in which all nonrepeat strains of E. coli determined to be clinically significant by the participating hospital laboratories were tested with the center's normal panel of beta-lactam antibiotics for extended-spectrum beta-lactamase (ESBL) production by use of the screening criteria described by NCCLS (17, 18, 19). Strains identified as possible ESBL producers were submitted to NML for further characterization. Denominator data, consisting of the total number of E. coli strains, were collected for the surveillance period to determine the rates of incidence of cefoxitin-resistant strains. Patient-specific information collected from the patients' charts included the service that the patient was on when the specimen was identified, date of birth, gender, date of specimen collection, and type of specimen.
Bacterial strains.
All isolates were identified at the participating sites by the routine methods performed at each laboratory. Strains meeting the study criteria were submitted to NML, where, upon receipt, they were stored at −70°C in Microbank vials (Pro-Lab Diagnostics, Richmond Hill, Ontario, Canada). The identities of all isolates submitted were confirmed with Vitek GNI cards (bioMérieux, Hazelwood, Mo.). The control strains used in this study included Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 27853, and E. coli ATCC 25922.
Antimicrobial susceptibility testing.
Potential ESBL-producing isolates were confirmed to be cefoxitin resistant (MICs ≥ 32 mg/liter) by the broth microdilution method, as described by NCCLS (17). The MICs of cephalosporins and aztreonam, with and without clavulanic acid, as well as the MICs of meropenem for all cefoxitin-resistant strains detected in this study were determined by broth microdilution (17). In addition, potential ESBLs were confirmed by the disk diffusion method as described by the NCCLS (18, 19) with disks containing ceftazidime (CAZ), ceftazidime-clavulanic acid, cefotaxime, and cefotaxime-clavulanic acid (Mast Diagnostics, Merseyside, United Kingdom). Susceptibilities to other classes of antimicrobials were determined with Vitek GNS-121 panels (bioMérieux).
Molecular subtyping by PFGE.
Cefoxitin-resistant strains were subtyped by pulsed-field gel electrophoresis (PFGE) by the standardized protocol for E. coli (O157:H7) (30), and the fingerprints were analyzed with a BioNumerics software program (version 2.5; Applied Maths, Saint Martens-Latem, Belgium). Gels were normalized by using the molecular weight standard strain Salmonella enterica serovar Braenderup Universal Marker (kindly provided by B. Swaminathan, Centers for Disease Control and Prevention, Atlanta, Ga.). A 1.0% tolerance with 1.5% optimization was used during cluster analysis by the unweighted pair group method, and DNA relatedness was calculated on the basis of the Dice coefficient.
Molecular characterization of Ambler class C resistance determinants.
In order to minimize the number of strains to be characterized, it was assumed that all strains that were identified at the same site and that had indistinguishable DNA fingerprints were identical.
PCR for amplification of the ampC promoter region of the E. coli strains was performed with the primer pairs and conditions described previously (4). Amplicons were purified with a Microcon PCR kit (Millipore) and were sequenced at the DNA Core Facility at NML. The resulting ampC promoter sequences were compared to the corresponding E. coli K-12 sequence (GenBank database accession number U00096) by using the DNASTAR MegAlign program (version 4.05).
To identify acquired AmpC-type resistance, individual PCRs were carried out with the cycling conditions described above and the following primers at 0.5 μM each: CIT-A (5′-ATGCAGGAGCAGGCTATTC-3′) and CIT-B (5′-TGGAGCGTTTTCTCCTGAAC-3′) for CMY-2-related genes, FOX-A (5′-TGTGGACGGCATTATCCAG-3′ ) and FOX-B (5′-AAAGCGCGTAACCGGATTG-3′) for FOX-related genes, and ENT-A (5′-AGTAAAACCTTCACCTTCACCG-3′) and ENT-B (5′-ATGCGCCTCTTCCGCTTTC-3′) for ACT-1/MIR-1-related genes.
RESULTS
Epidemiologic characteristics of study strains.
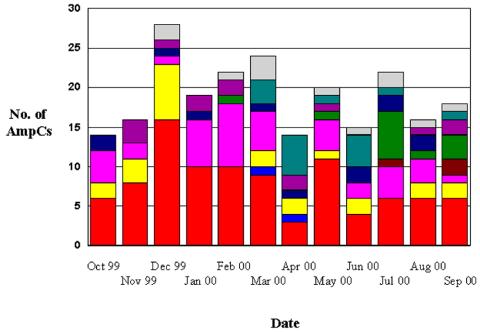
A total of 29,323 E. coli strains from the 12 participating sites were screened for potential ESBL producers. A total of 411 (1.4%) E. coli isolates met the NCCLS screening criteria as potential ESBL producers. These strains were submitted to NML for antimicrobial susceptibility testing and advanced characterization. Results for ESBL-producing strains have been published previously (16). This collection was further screened for the AmpC phenotype. A strain was considered an AmpC producer if it displayed resistance to cefoxitin by broth microdilution. A total of 232 strains were found to be resistant to cefoxitin, yielding an incident rate of 0.8/100 (range, 0 to 5.1%). With the exception of the first month of the study, a steady number of AmpC-producing strains was identified over the 1-year period of the study (Fig. 1). One hundred twenty-two (53%) of the AmpC-producing isolates were from the west. One hundred twenty-eight females (55%) were infected with E. coli isolates with AmpC-type resistance. The age distribution of the 104 females that reported a date of birth was as follows: 0 to 21 years, n = 9; 22 to 65 years, n = 34; and over 65 years, n = 61. The ages of 97 males were reported; and the distribution was as follows: 0 to 21 years, n = 13; 22 to 65 years, n = 31; and over 65 years, n = 53.
FIG. 1.
Number of cefoxitin-resistant strains isolated by month and by site over the length of the study. Colors represent the total number of strains at a specific site.
The site of isolation of the specimen was reported for 209 isolates (Table 1), with the highest number of strains isolated from the urinary tract (n = 162). Information regarding ward location was collected for 201 isolates: 58 from acute medical care units, 36 from outpatient units, 31 from emergency departments, 28 from surgical wards, 9 from intensive care units, 5 from oncology units, 4 from pediatric wards, 2 from medical or surgical wards, and 28 from other units.
TABLE 1.
Site of isolation of cefoxitin-resistant strains
| Site of isolation | No. of E. coli isolates froma:
|
|
|---|---|---|
| Males | Females | |
| Blood (n = 18) | 13 | 5 |
| Wound (n = 12) | 11 | 1 |
| Abscess (n = 1) | 1 | 0 |
| Urine or urinary tract (n = 162) | 61 | 101 |
| Upper respiratory tract (n = 7) | 5 | 2 |
| Lower respiratory tract (n = 3) | 2 | 1 |
| Other (n = 6) | 4 | 2 |
| Total | 97 | 112 |
A total of 209 cefoxitin-resistant E. coli isolates recovered from 232 reported sites of isolation.
Antimicrobial susceptibilities of study strains.
Of the 232 strains resistant to cefoxitin (AmpC producers), the cefoxitin MICs were 32 mg/liter for 45% (n = 104), 64 mg/liter for 25% (n = 59), and >64 mg/liter for the remaining 30% (n = 69) of the strains. A comparison of the antimicrobial susceptibilities of the AmpC- and the non-AmpC-producing isolates is shown in Table 2. The rates of resistance to the fluoroquinolones and nitrofurantoin were significantly higher (P < 0.001) among the cefoxitin-resistant strains than among the cefoxitin-sensitive E. coli isolates. Interestingly, the rates of resistance to the three aminoglycosides tested remained relatively unchanged between cefoxitin-resistant and -sensitive E. coli strains (Table 2).
TABLE 2.
Antimicrobial resistance patterns of study strainsa
| Antimicrobial agent | % (no.) of resistant strainsb
|
|
|---|---|---|
| AmpC producers (n = 232) | Non-AmpC producers (n = 183) | |
| Amoxicillin-clavulanate | 66 (152) | 43 (79) |
| Ampicillin | 96 (223) | 82 (150) |
| Cefazolin | 55 (127) | 36 (65) |
| Ceftazidime | 1.3 (3) | 1.1 (2) |
| Ceftriaxone | 2.1 (5) | 11 (21) |
| Piperacillin-tazobactam | 4.7 (11) | 2.2 (4) |
| Cefotetan | 0 (0) | 0 (0) |
| Imipenem | 0 (0) | 0 (0) |
| Ciprofloxacin | 45 (105) | 19 (35) |
| Levofloxacin | 44 (102) | 19 (34) |
| Nitrofurantoin | 7.3 (17) | 1.6 (3) |
| Gentamicin | 27 (63) | 25 (45) |
| Amikacin | 1.3 (3) | 2.2 (4) |
| Tobramycin | 26 (60) | 18 (33) |
| Trimethoprim-sulfamethoxazole | 51 (118) | 33 (60) |
As determined with Vitek GNS 121 cards.
Cefoxitin MICs were ≥32 μg/ml for AmpC producers and <32 μg/ml for non-AmpC producers, as determined by broth microdilution.
Molecular characterization of study strains.
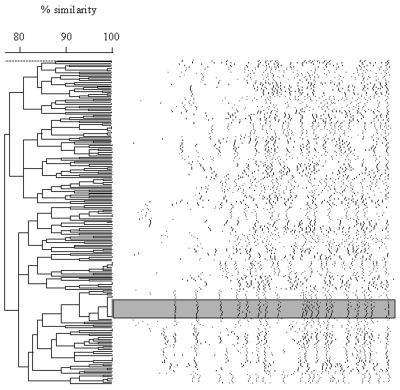
All cefoxitin-resistant strains were fingerprinted by PFGE. A dendrogram showing the different molecular subtypes is shown in Fig. 2. One hundred eighty-two unique subtypes were identified among the 232 E. coli cefoxitin-resistant isolates. One isolate was untypeable. On the basis of macrorestriction analysis, the intrahospital dissemination of E. coli strains was suggested to have occurred at 5 of the 12 sites. With the exception of one site, the outbreaks at these sites involved 2 to 4 isolates; the one site that was the exception recorded an outbreak involving 17 isolates (Fig. 2). There were two examples of interhospital dissemination: the first one involved a clone identified once at two sites (western and eastern Canada), and the second involved four strains identified at three sites in western Canada.
FIG. 2.
Dendrogram depicting DNA macrorestriction patterns of cefoxitin-resistant E. coli strains generated with XbaI. The shaded region highlights the largest outbreak cluster described in the text.
Sequence analysis of the ampC promoter region.
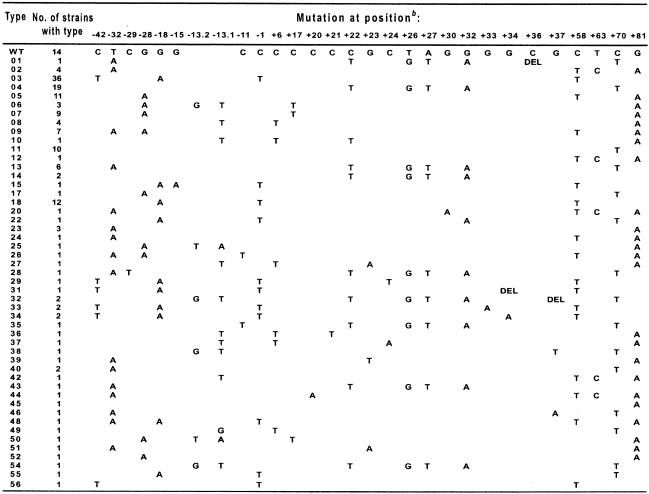
The ampC gene promoter regions of all of the 183 unique strains, as determined by macrorestriction analysis, were subjected to sequence analysis. A 191-bp fragment containing the ampC promoter region and the first 61 bp of the coding region was amplified from all of the cefoxitin-resistant strains sequenced and compared to the same region from E. coli K-12 (GenBank database accession number U00096). Among the 183 regions sequenced, 166 were found to contain at least one or multiple nucleotide changes or an insertion or deletion compared to the E. coli K-12 sequence, and the sequences of 14 strains were found to be identical to the K-12 sequence (Table 3). The remaining three regions were found to contain insertion sequence (IS) elements in the promoter region (see below). Among the 166 regions with changes, there were a total of 49 unique sequences, each of which was given a specific type number (Table 3). Sixty-eight percent (113 of 166) of the strains contained 1 of 9 types, with 32 types found only in a single strain. Nucleotide changes and/or insertion or deletions could be found throughout the region at 28 positions, including 3 positions (positions +63, +70, and +81) in the ampC-coding region. Some positions appeared to be more subject to mutation, with changes most often found at positions +58 (85 strains), −1 (59 strains), +81 (58 strains), −18 (58 strains), +70 (49 strains), and −42 (43 strains).
TABLE 3.
Mutations at given positions in ampC promoter region for specific promoter typesa
Position numbering as defined by Jaurin et al. (13). Type numbers were assigned arbitrarily. WT, wild-type promoter from E. coli K-12; DEL, deletion at the indicated position.
Positions −42, −18, and −15 are involved in the alternate promoter; positions −32 and −11 are part of the wild-type promoter; positions −13.1 and −13.2 are part of the spacer region of the wild-type promoter; nucleotides between positions +17 and +37 are located in the attenuator; and positions +63, +70, and +81 are located in the coding region.
Eighty-six percent (42 of 49) of the unique types did involve mutations that created a promoter that more closely resembled the E. coli consensus promoter (12). Mutations to the wild-type promoter occurred in 34 sequence types (45 strains) and included C→T at position −11, which changes the −10 box from TACAAT to TATAAT (the pertinent nucleotides are in boldface), and/or T→A at position −32, which changes the −35 box from TTGTCA to TTGACA, or an increase in the distance between the −35 and −10 boxes from 16 bp to either 17 or 18 bp by insertion of 1 or 2 bp between positions −13 and −14 (positions −13.1 and −13.2 in Table 3). Interestingly, the mutations in the −35 and −10 boxes and the insertions in the spacer region were mutually exclusive and were never found in the same promoter region.
Mutations that created an alternative displaced promoter also occurred in 11 sequence types (58 strains). Changes included G→A at position −18, which changed TGTCGT to TATCGT to create a −10 box, and/or C→T at position −42, which changed CTGACA to TTGACA to create a −35 box. The alternative −35 and −10 sequences were separated by 17 bp. Sequence type 15 (one strain) had, in addition to the G→A change at position −18, a G→A change at position −15, which created an alternate −10 box with the sequence TATCAT. Sequence type 48 (one strain) had the change at position −18 and the T→A change at position −32.
Changes at two other positions were found in the spacer region: a C→T change at position −29 in a single sequence type (type 28; one strain) and a G→A change at position −28 in 11 sequence types (35 strains). The sequence type 28 strain also had the change at position −32 and changes in the attenuator region and ampC-coding region. The change at position −28 was found to be associated with changes in the promoter region (positions −32, −13.1 and −13.2, and −11), the attenuator region, or other changes but never with the changes at position −42 and/or −18 that created the alternative −35 or −10 boxes. Due to their location in the spacer region, it is possible that nucleotides at these positions may interact with RNA polymerase. However, whether this has any affect on ampC expression awaits further evidence.
Mutations in the attenuator region are assumed to destabilize the stem-loop structure and allow increased levels of ampC transcription. Direct proof of this, by use of site-directed mutagenesis, for example, has not been obtained because of the difficulty of introducing mutations in the region due to the stem-loop structure (5). Mutations in the attenuator region (positions +17 to + 37) were found in 27 sequence types (63 strains). Most commonly, four changes were found together: C→T at position +22, T→G at position +26, A→T at position +27, and G→A at position +32. These changes occurred in nine sequence types (34 strains). The changes at positions +26 and +27 always occurred together and with the changes at positions +22 and +32, although the reverse was not true. Three sequence types (four strains) had single-base-pair deletions in the attenuator region, and in two types these occurred with the four most common changes to the attenuator region mentioned above. All 17 other variants with changes in the attenuator region (29 strains) had single-base-pair mutations. We note that changes in the attenuator region were often found in conjunction with changes in the promoter region, with this occurring in 25 sequence types.
Changes at three positions were found outside of the promoter and attenuator regions: at positions −1, +6, and +58. Two of these were the two most commonly found changes; the C→T change at position +58 was found in 19 sequence types (85 strains), and the C→T change at position −1 was found in 11 sequence types (59 strains). These two changes were most often found together (nine sequence types; 57 strains) and may represent strain-specific polymorphisms.
As mentioned above, changes were found at three positions in the ampC-coding region, and all led to changes in the amino acid sequence. A T→C change at position +63 causes a Phe→Leu change, a C→T change at position +70 causes a Thr→Met change, and a G→A change at position +81 causes a Ala→Thr change. In fact, changes at these three positions were very common, being found in 84% (42 of 49) of the sequence types (107 strains). Interestingly, the change at position +70 was never found with changes at position +63 or +81. The change at position +63 was always found with the change at position +81, although the reverse was not the case. The contribution, if any, of these three amino acid changes to increased AmpC activity is unknown, as all occurred within the first 8 residues of the AmpC enzyme in the leader peptide region. It could be that changes here lead to the more efficient transfer of the enzyme precursor into the periplasmic space. Previous evidence (27) indicated that in the SHV-7 beta-lactamase an Ile8Phe change could lead to a more mature enzyme in the periplasmic space and hence increased MICs of some beta-lactams.
In the collection of strains that we analyzed, the most common sequence type was 03, and 36 strains were of this type. Type 03 contained the strong alternate promoter caused by the changes at positions −42 and −18 and also included the changes at positions −1 and +58. Interestingly, the third most common sequence type, type 18 (n = 19 strains), differed from type 03 only in that it did not contain the mutation at position −42. The second most common sequence type, type 04, contained the four changes most often found in the attenuator region, as well as the change at position +70 in the ampC-coding region. As mentioned above, the effect of changes in the attenuator on transcription efficiency is not directly known, but we note that type 04 contains no changes in the promoter region.
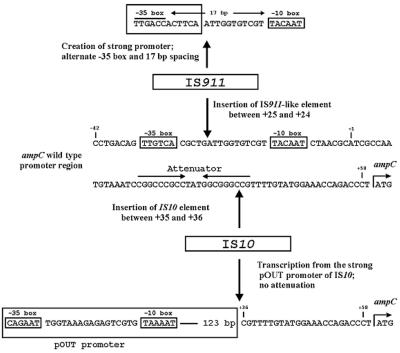
PCR promoter amplification from three isolates yielded amplicons of approximately 1.5 kb, which was significantly larger than the expected 191-bp amplicon. Sequence analysis of these amplicons revealed the presence of IS elements in the ampC promoter regions in these strains (Fig. 3). In one strain an IS911 variant was inserted 11 bp upstream of the wild-type −10 box, which, as a result, was located 17 bp downstream of the IS911-specific sequence TTGACC, which is expected to act as a −35 box. Thus, in this strain the resulting hybrid promoter most likely drives the expression of ampC. IS911 is flanked by the trimer ATT; thus, insertion appears to have caused duplication at positions −22 to −24. In two strains IS10 was inserted between the ampC attenuator region and the start codon. It is likely that the strong pOUT promoter of IS10 drives the expression of ampC in these strains (28). As well, the effect of the attenuator would be negated by the location of IS10. In both strains IS10 is flanked by the 9-bp direct repeat CGTTTTGTA; thus, insertion appears to have caused a duplication of positions +38 to +46. Interestingly, the two strains with the IS10 insertion had very different macrorestriction profiles (more than seven band differences).
FIG. 3.
Schematic diagram showing the insertion of the IS10 (two strains) and IS911-like (one strain) elements in the ampC promoter region, as found in this study.
Acquired ampC genes.
All cefoxitin-resistant isolates were subjected to PCR and sequence analysis to identify any Ambler class C genes that may have been acquired. Primer sets that identify CMY-2-, FOX-, or ACT-1/MIR-1-related genes were used. Of the 25 (13.5%) strains that produced an amplicon, all were identified by sequence analysis to contain the CMY-2 gene. The strains were identified from 7 of the 12 surveillance sites, 5 from the east (18 strains) and 2 from the west (7 strains). The relationship between the promoter type observed and the CMY-2 gene is presented in Table 4. Among the 14 strains with the wild-type ampC promoter, 7 were found to contain CMY-2, thus explaining the resistance to cefoxitin in these strains. The presence of CMY-2 also explains the cefoxitin resistance in four strains with changes in positions not known to cause AmpC overproduction: two strains with sequence type 11, one strain with sequence type 17, and one strain with sequence type 52.
TABLE 4.
Promoter sequence types of strains containing CMY-2 gene
| Promoter sequence type | No. of strains | No. of strains with CMY-2 |
|---|---|---|
| Wild type | 14 | 7 |
| 03 | 36 | 1 |
| 04 | 19 | 1 |
| 07 | 8 | 5 |
| 11 | 10 | 2 |
| 17 | 1 | 1 |
| 18 | 12 | 5 |
| 22 | 1 | 1 |
| 48 | 1 | 1 |
| 52 | 1 | 1 |
The mechanism of cefoxitin resistance remains to be elucidated in a number of other strains. When we discounted strains with changes that create a strengthened ampC promoter (changes at positions −42, −32, −18, −15, and −11 and insertions between positions −13 and −14), strains with changes at positions −29 and −28 in the spacer region, strains with changes in the attenuator region (positions +17 to +37), the 3 strains containing insertion elements, and strains containing CMY-2, we found that 17 strains (9.7%) had an unknown mechanism of cefoxitin resistance. Ten strains were of sequence type 11 (change at position +70), one strain was of sequence type 12 (changes at positions +58, +63, and +81), one strain was of sequence type 45 (change at position +81), and seven strains had the wild-type promoter. Possible explanations for the cefoxitin resistance in these strains include the presence of other acquired Ambler class C genes, porin or permeability mutations, or other changes outside of the region analyzed here that affect the expression of the ampC gene or that produce changes in the native AmpC enzyme that increase its activity enough to produce cefoxitin resistance, despite its constitutive low-level production.
DISCUSSION
The AmpC resistance phenotype in E. coli can be a result of overexpression of the chromosomal ampC gene, acquisition of a plasmidic ampC, alterations in the permeability of the cell to cefoxitin, or a combination of these factors. The fact that a standardized phenotypic method for screening and detection of this type of resistance does not exist makes the surveillance and characterization of strains problematic. Data regarding AmpC-type resistance in human isolates of E. coli is almost nonexistent in Canada. Forward et al. (10) provided the first detailed study examining 30 cefoxitin-resistant E. coli isolates identified in nine Toronto-area hospitals. They identified only two strains that did not contain mutations in the promoter or the attenuator region, one of which was identified as harboring a plasmidic AmpC.
This report describes the first national study of cefoxitin-resistant strains in Canadian hospitals. The number of cefoxitin-resistant E. coli strains did not dramatically increase over the study, which is in contrast to the findings for the classical Ambler class A ESBLs, a steady increase in the numbers of which occurred during the same study period in Canada (16). We note that these strains were a subgroup (cefoxitin resistant) of a collection of E. coli isolates selected as having reduced susceptibilities to extended-spectrum cephalosporins (NCCLS ESBL screen test) and so produced data for only a minimum number of cefoxitin-resistant E. coli strains in Canadian hospitals. Interestingly, although the strains were potential ESBL producers by screen tests, only a low percentage of the AmpC producers were clinically resistant to extended-spectrum cephalosporins (Table 2). As well, although cefotetan is a cephamycin, no strain was resistant to this drug, and hence, it is inadequate to use MICs of cefotetan as an indicator of possible AmpC activity. Although intrahospital outbreaks occurred at almost half of the sites, the sizes of the outbreaks remained small (2 to 4 strains), with the exception of one large outbreak (17 strains), which occurred at a site in western Canada. The lack of clonal strain dissemination, the high numbers of mutations in the promoter and attenuator regions, and the small number of acquired AmpC resistance genes suggests that antimicrobial selective pressure may be playing a large role in the potential emergence of cefoxitin resistance in a patient.
An analysis of 168 promoter regions in E. coli clearly showed the presence of two conserved regions, the −35 box and the −10 box, also called the Pribnow box (12). For most promoters the degree of homology to the −35 box consensus sequence, TTGACA, and the −10 box consensus sequence, TATAAT, is directly related to promoter strength. Furthermore, the spacing between these two regions plays a role in promoter strength, with the optimal distance being 17 bp. Numerous studies have reported that variations in the promoter and attenuator regions of ampC are a mechanism of hyperproduction of the AmpC protein that results in resistance to cefoxitin (3, 4, 5, 7, 10, 14, 20, 21, 22, 29). While some of the mutations described in this report may be polymorphisms that do not alter ampC expression, some of the changes observed have been shown to increase gene expression in previous studies. Changes at position −42 (C→T) in the ampC promoter were shown to increase transcription by 43-fold (5) or 18-fold (20), depending on the type of transcriptional reporter system used. The C→T transition at position −42 results in a consensus TTGACA box upstream of the native −35 sequence and in modification of the transcriptional initiation site up to 5 bp upstream from the original site (20). Interestingly, almost all of the promoters harboring this change (five of six) also had a corresponding T→A transversion at position −18. This change resulted in a new −10 box separated by 17 bp from the new −42 box, resulting in the formation of a strong promoter, as has been described in other clinical cefoxitin-resistant E. coli strains (5, 10, 20). These mutations (those at both positions −42 and −18 or position −18 only) were the most common promoter-region variations found in this study (32%; 59 of 183 isolates), suggesting that these changes may be favored in situ over other promoter-region mutations which may effect ampC expression.
Changes in the −35 and −10 boxes or in the spacer region that result in ampC promoter sequences more closely resembling the E. coli consensus promoter sequence have been described previously (4, 7, 10, 13, 29). A T→A transversion at position −32 resulted in the formation of a −35 consensus box and increased the level of ampC expression in two independent studies (4, 13). This mutation was the second most common promoter alteration observed in the present study, with 19% (33 of 183) of promoters sequenced containing this change. By use of a reporter system, only a single strain that had the C→T transition at the −11 position has been characterized, and the results showed a sixfold increase in promoter strength (7). In the present study only two strains were observed to have this mutation. Insertions of nucleotides between the −10 and −35 regions of the ampC promoter which produce 17 or 18 bp of separation between these regions have been reported previously and are thought to result in a more efficient promoter (10, 13, 29). In addition to the previously reported changes in the spacer region, this report describes two promoter variants with a doublet of T-A inserted between nucleotides −14 and −13, creating a novel 18-bp spacer region.
Jaurin et al. (13) first reported on a transcriptional attenuator region in the E. coli ampC promoter region. It was hypothesized that mutations in this region contribute to AmpC overproduction by causing destabilization of the hairpin structure, resulting in increased transcription. Of the 27 attenuator-region variants observed in the present study, 9 involved changes at 4 positions, and 2 of these also had 1-bp deletions. All of the 19 remaining variants had single-base-pair changes or a 1-bp deletion, which was detected in a single variant. All the changes resulted in a reduction of the thermodynamic strength of formation of the stem-loop structure to some extent (data not shown), providing further evidence that these mutations may be associated with increased transcription. We note, however, that 85% (23 of 27) of sequence types with attenuator mutations also had changes that create a stronger promoter (Table 3). Nonetheless, the only changes that we detected in type 14 strains (two strains) were the mutations at positions +22, +26, +27, and +32; hence, these strains may prove useful for quantification of the contributions of these attenuator mutations to cefoxitin resistance.
The insertion of IS elements in promoter regions that cause the up-regulation of gene expression by producing an alternate promoter has been described. A previous report (15) has described a laboratory mutant of E. coli that overproduced AmpC by the insertion of an IS2 element. Additionally, a recent study identified a possible IS element inserted upstream of the ampC gene in Acinetobacter baumannii, although the nucleotide sequence of the element was never elucidated (8). To our knowledge, the present report is the first to describe clinical isolates of E. coli with the AmpC phenotype as a result of the insertion of mobile elements. We provide sequence-based evidence that the insertion of IS elements creates new promoters which may be responsible for the up-regulation of ampC gene expression (Fig. 3). Insertion of IS911 in the promoter spacer region resulted in the possible generation of a stronger −35 sequence and a 17-bp spacer region, thereby creating a hybrid promoter sequence that more closely resembles the E. coli consensus promoter sequence. Insertion of the IS10 element between the attenuator and the start codon presumably allows ampC to be transcribed via the strong pOUT promoter of IS10 and also negates any effect from the attenuator region (28). Interestingly, the two AmpC strains identified as harboring identical IS10 sequences had unique DNA fingerprints, suggesting that the insertion site for IS10 may not be a random event.
In the present study, 13.5% of the isolates harbored a gene that correlated with acquired AmpC-type resistance, and in all strains the gene was identified as CMY-2. Although this gene was identified in 25 strains, only 7 of these strains were found to contain the wild-type ampC promoter sequence (Table 4). In two strains with sequence types containing only two changes, the change at position −28 and the change at either position +70 (type 17) or position +81 (type 52), CMY-2 is likely the major factor contributing to cefoxitin resistance. All other CMY-2-containing strains have sequence types with changes in the promoter and/or attenuator region. CMY-2-mediated resistance has been documented in Canadian isolates of Salmonella serovars (1, 26). In addition, a dramatic increase in a multidrug-resistant S. enterica serovar Newport containing the CMY-2 gene on a plasmid has been observed in the United States (6, 9, 32, 34). Furthermore, it has been shown that plasmidic CMY-2 can be transferred between Salmonella and E. coli strains isolated from food animals and humans (33). It will be important to examine the plasmids carrying the CMY-2 genes in the E. coli strains identified in this study and compare them to the CMY-2-harboring Salmonella strains to determine if similar genetic vectors are responsible for the resistance observed. That work is under way.
To our knowledge, this report describes the largest study to date on the contribution of ampC promoter mutations to cefoxitin resistance in E. coli, and the report also provides evidence that supports previous work involving mutations which affect ampC expression. However, as described above, this report describes additional mutations that affect promoter strength and that possibly result in increased gene expression. To substantiate these claims, further work is under way to examine gene expression in the strains containing the altered promoters.
As has been observed in previous studies related to nosocomial AmpC-type resistance, most strains remain genetically distinct, have resistance phenotypes that coincide with mutations in the promoter and the attenuator regions, and do not harbor acquired AmpC-type resistance (10, 20). This study supports this general observation for nosocomial E. coli isolates. Taken together, these findings suggest that E. coli AmpC-type resistance in the hospital setting is not primarily due to the dissemination of clonal strains or the spread of resistant plasmids but is due to the emergence of resistant strains in patients, possibly through antimicrobial selective pressure. Further studies focusing on antimicrobial use and resistance in patient populations will be required to substantiate this claim.
Acknowledgments
We thank Romeo Hizon for contributions to strain identification and antimicrobial susceptibility testing and Jennifer Campbell for molecular typing. We also acknowledge the contributions of Rebekah Murphy and Darrell Johnstone to work involving plasmid profiling and Shaun Tyler and the staff of the DNA Core Facility at NML for generating the sequence information and synthesizing oligonucleotides.
This project was partially funded by Astra Zeneca Canada.
The following individuals are members of CNISP: E. Bryce, Vancouver General Hospital, Vancouver, British Columbia; J. Conly, University of Calgary, Calgary, Alberta; J. Embil, Health Sciences Centre, Winnipeg, Manitoba; J. Embree, Health Sciences Centre, Winnipeg, Manitoba; M. Gardam, University Health Network, Toronto, Ontario; M. Gourdeau, Hôpital de l'Enfant-Jésus, Quebec City, Quebec; K. Green, Community and Hospital Infection Control Association-Canada; M. John, St. Joseph's Health Centre, London, Ontario; B. A. Henderson, Peter Lougheed Centre, Calgary, Alberta; J. Hutchinson, Health Sciences Centre, St. John's, Newfoundland; M. Ishak, Centre Hospitalier Angrignon, Verdun, Quebec; V. Roth, The Ottawa Hospital, Ottawa, Ontario; L. Johnston, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia; J. Langley, I.W.K. Grace Health Science Centre, Halifax, Nova Scotia; M. Loeb, Hamilton Health Sciences Corp., Hamilton, Ontario; A. Matlow, Hospital for Sick Children, Toronto, Ontario; A. McGeer, Mount Sinai Hospital, Toronto, Ontario; M. Miller, Jewish General Hospital, Montreal, Quebec; D. Moore, Montreal Children's Hospital, Montreal, Quebec; M. Mulvey, Canadian Science Centre for Human and Animal Health, Health Canada; M. Ofner-Agostini, Centre for Infectious Disease Prevention and Control, Health Canada; S. Paton, Centre for Infectious Disease Prevention and Control, Health Canada; A. Simor, Sunnybrook and Women's College Health Sciences Centre, Toronto, Ontario; G. Taylor, University of Alberta, Edmonton, Alberta; A. Khan, The Moncton Hospital, Moncton, New Brunswick; M. Vearncombe, Sunnybrook and Women's College Health Sciences Centre, Toronto, Ontario; K. Weiss, Hôpital Maisonneuve-Rosemont, Montreal, Quebec; A. Wong, Royal University Hospital, Saskatoon, Saskatchewan; and D. Zoutman, Kingston General Hospital, Kingston, Ontario.
The members of the Canadian Hospital Epidemiology Committee (CHEC) are Elizabeth Bryce, Vancouver General Hospital, Vancouver, British Columbia; John Conly, Foothills Medical Centre, Calgary, Alberta; John Embil, Health Sciences Centre, Winnipeg, Manitoba; Joanne Embree, Health Sciences Centre, Winnipeg; Michael Gardam, University Health Network, Toronto, Ontario; Gordon Dow, The Moncton Hospital, Moncton, New Brunswick; Elizabeth Henderson, Peter Lougheed Centre, Calgary; James Hutchinson, Health Sciences Centre, St. John’s, Newfoundland; Michael John, London Health Sciences Centre, London, Ontario; Lynn Johnston, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia; Pamela Kibsey, Victoria General Hospital, Victoria, British Columbia; Joanne Langley, I. W. K. Grace Health Science Centre, Halifax; Mark Loeb, Hamilton Health Sciences Corporation, Hamilton, Ontario; Anne Matlow, Hospital for Sick Children, Toronto; Allison McGeer, Mount Sinai Hospital, Toronto; Sophie Michaud, CHUS-Hôpital Fleurimont, Sherbrooke, Quebec; Mark Miller, SMBD-Jewish General Hospital, Montreal, Quebec; Dorothy Moore, Montreal Children’s Hospital, Montreal; Virginia Roth, The Ottawa Hospital, Ottawa, Ontario; Andrew Simor, Sunnybrook and Women’s College Health Sciences Centre, Toronto; Geoffrey Taylor, University of Alberta Hospital, Edmonton, Alberta; Mary Vearncombe, Sunnybrook and Women’s College Health Sciences Centre, Toronto; Alice Wong, Royal University Hospital, Saskatoon, Saskatchewan; and Dick Zoutman, Kingston General Hospital, Kingston, Ontario.
REFERENCES
- 1.Allen, K. J., and C. Poppe. 2002. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by β-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can. J. Vet. Res. 66:137-144. [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, M., J. H. Tran, N. Chow, and G. A. Jacoby. 2004. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 48:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briñas, L., M. Zarazaga, Y. Sáenz, F. Ruiz-Laerra, and C. Torres. 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46:3156-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caroff, N., E. Espaze, I. Berard, H. Richet, and A. Reynaud. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiol. Lett. 173:459-465. [DOI] [PubMed] [Google Scholar]
- 5.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January-April 2002. Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 7.Corvec, S., N. Caroff, E. Espaze, J. Marraillac, and A. Reynaud. 2002. −11 mutation in the ampC promoter increasing resistance to β-lactams in a clinical Escherichia coli strain. Antimicrob. Agents Chemother. 46:3265-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corvec, S., N. Caroff, E. Espaze, C. Giraudeau, H. Drugeon, and A. Reynaud. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629-635. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with ampC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 10.Forward, K. R., B. M. Willey, D. E. Low, A. McGeer, M. A. Kapala, M. M. Kapala, and L. L. Burrows. 2001. Molecular mechanisms of cefoxitin resistance in Escherichia coli from Toronto area hospitals. Diagn. Microbiol. Infect. Dis. 41:57-63. [DOI] [PubMed] [Google Scholar]
- 11.Hanson, N. D. 2003. AmpC beta-lactamases: what do we need to know for the future? J. Antimicrob. Chemother. 52:2-4. [DOI] [PubMed] [Google Scholar]
- 12.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaurin, B., T. Grundström, T. Edlund, and S. Normark. 1981. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221-225. [DOI] [PubMed] [Google Scholar]
- 14.Jaurin, B., T. Grundström, and S. Normark. 1982. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaurin, B., and S. Normark. 1983. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell 32:809-816. [DOI] [PubMed] [Google Scholar]
- 16.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, S. Paton, and The Canadian Hospital Epidemiology Committee of the Canadian Nosocomial Infection Surveillance Program, Health Canada. 2004. Ambler class A extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for disk susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.National Committee for Clinical Laboratory Standards. 2003. Performance standards for disk susceptibility tests, 8th ed. Approved standard M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsson, O., S. Bergström, and S. Normark. 1982. Identification of a novel ampC β-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1:1411-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson, O., S. Bergström, F. P. Lindberg, and S. Normark. 1983. ampC β-lactamase hyperproduction in Escherichia coli: natural ampicillin resistance generated by horizontal chromosomal DNA transfer from Shigella. Proc. Natl. Acad. Sci. USA 80:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papanicolaou, G. A., A. A. Medeiros, and G. A. Jacoby. 1990. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 34:2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitout, J. D. D., M. D. Reisbig, M. Mulvey, L. Chui, M. Louie, L. Crowe, D. L. Church, S. Elsayedd, D. Gregson, R. Ahmed, P. Tilley, and N. D. Hanson. 2003. Association between handling of pet treats and infection with Salmonella enterica serotype Newport expressing the AmpC β-lactamase, CMY-2. J. Clin. Microbiol. 41:4578-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randegger, C. C., A. Keller, M. Irla, A. Wada, and H. Hächler. 2000. Contribution of natural amino acid substitutions in SHV extended-spectrum β-lactamases to resistance against various β-lactams. Antimicrob. Agents Chemother. 44:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons, R. W., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1983. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell 34:673-682. [DOI] [PubMed] [Google Scholar]
- 29.Siu, L. K., P.-L. Lu, J.-Y. Chen, F. M. Lin, and S.-C. Chang. 2003. High level expression of AmpC β-lactamase due to insertion of nucleotides between −10 and −35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum cephalosporin treatment. Antimicrob. Agents Chemother. 47:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swaminathan, B., T. Barrett, S. B. Hunter, and R. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne disease surveillance in the United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson, K. S. 2001. Controversies about extended spectrum and AmpC beta-lactamases. Emerg. Infect. Dis. 7:333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winokur, P. L., A. Bruggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, S., S. Qaiyumi, S. Friedman, R. Singh, S. L. Foley, D. G. White, P. F. McDermott, T. Donkar, C. Bolin, S. Munro, E. J. Baron, and R. D. Walker. 2003. Characterization of Salmonella enterica serotype Newport isolated from humans and food animals. J. Clin. Microbiol. 41:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]